Abstract
The gp41-encoding sequence of the env gene contains in two separate regions the Rev-responsive elements (RRE) and the alternative open reading frame of the second exon of the regulatory protein Rev. The binding of Rev to the RRE allows the transport of unspliced/singly spliced viral mRNAs out of the nucleus, an essential step in the life cycle of human immunodeficiency virus type 1 (HIV-1). In this study, we have investigated whether the fusion-inhibitor enfuvirtide (ENF) can induce mutations in Rev and if these mutations correlate with the classical ENF resistance gp41 mutations and with viremia and CD4 cell count. Specific Rev mutations were positively associated with ENF treatment and significantly correlated with classical ENF resistance gp41 mutations. In particular, a cluster was observed for the Rev mutations E57A (E57Arev) and N86Srev with the ENF resistance gp41 mutations Q40H (Q40Hgp41) and L45Mgp41. In addition, the presence at week 48 of the E57Arev correlates with a significant viremia increase from baseline to week 48 and with a CD4 cell count loss from baseline to week 48. By modeling the RRE structure, we found that the Q40gp41 and L45gp41 codons form complementary base pairs in a region of the RRE involved in Rev binding. The conformation of this Rev-binding site is disrupted when Q40Hgp41 and L45Mgp41 occur alone while it is restored when both mutations are present. In conclusion, our study shows that ENF pressure may also affect both Rev and RRE structures and can provide an excellent example of compensatory evolution. This highlights the multiple roles of ENF (and perhaps other entry inhibitors) in modulating the correct interplay between the different HIV-1 genes and proteins during the HIV-1 life cycle.
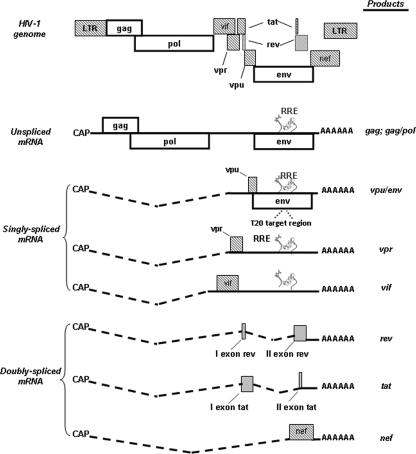
Retroviruses, such as human immunodeficiency virus (HIV), employ a variety of different overlapping reading frames and splicing events to express a large array of messenger RNAs (mRNAs) (over 30) and proteins (at least 15) from a single primary transcript (5). The HIV type 1 (HIV-1) RNA sequence contains at least four different 5′ splice sites and eight different 3′ splice sites that are used alternatively (28, 32, 34). During HIV-1 replication cycle, three groups of viral mRNAs are produced (Fig. 1). One group includes the doubly spliced 2-kb transcripts that encode the regulatory proteins Tat, Rev, and Nef (32). Another group includes the singly spliced mRNAs of approximately 4 kb that serve for the production of Vif, Vpr, Vpu, and Env proteins (gp120 and gp41). The last group includes the 9-kb unspliced mRNAs that encode the Gag and Gag-Pol polyprotein products and serve as genomic RNA for packaging into virions (32, 34). The doubly spliced mRNAs are produced early and are transported out of the nucleus into the cytoplasm by the ordinary cell machinery. In contrast, the export of the singly spliced and unspliced viral mRNAs from the nucleus to the cytoplasm is mediated by the interaction between the regulatory protein Rev and a nucleotide RNA sequence named Rev-responsive element (RRE), located in the Env gene and present in singly spliced and unspliced RNAs (Fig. 1) (21, 40, 31).
FIG. 1.
Schematic view of HIV-1 genome and transcripts produced during HIV-1 replication cycle (modified from reference 5 with permission). The ENF target region encompassing the amino acids 36 to 45 in gp41 is also shown. During the HIV-1 replication cycle, three classes of viral RNAs are produced: (i) the 9-kb unspliced mRNAs that are packaged into progeny virions as genomic RNA and can also serve for the expression of Gag/Pol genes; (ii) the singly spliced mRNAs encoding Vif, Vpr, Vpu, and env; and (iii) doubly spliced 2-kb transcripts encoding Tat, Rev, and Nef (29). The doubly spliced mRNAs encoding Tat, Rev, and Nef are the first produced and are transported into the cytoplasm by the ordinary cell machinery. When regulatory protein Rev is produced, it returns into the nucleus and binds the RRE, thus allowing the shuttling of unspliced and singly spliced mRNAs from the nucleus to the cytoplasm of HIV-1-infected cells.
Rev is an 18-kDa protein composed of 116 amino acids and is encoded by two exons spanning 26 and 90 amino acids, respectively (Fig. 1). The second exon is encoded by an alternative open reading frame overlapping the gp41-encoding sequence of the env gene. The Rev protein is composed of several domains harboring distinct functions. In particular, the amino-terminal basic domain of Rev contains the nuclear localization signal (NLS) (amino acids [aa] 35 to 50) that is an arginine-rich motif mediating both Rev nuclear/nucleolar import and RRE binding (8, 19, 30, 40, 41). The NLS is flanked by regions implicated in the oligomerization of Rev along the RRE. It has been demonstrated that multiple Rev molecules bind to the RRE and that this oligomerization of Rev along the RRE is necessary for the efficient shuttle of mRNAs from the nucleus to the cytoplasm (4, 7, 12, 14, 21-23, 38, 41). Finally, the carboxy-terminal domain contains the leucine-rich nuclear export signal (NES) (aa 73 to 84) that acts as an NES and also contains another NLS (3, 11, 21, 29).
The RRE is a 353-nucleotide RNA segment spanning the junction between the gp120- and gp41-encoding sequences of the env gene and is present exclusively in unspliced and singly spliced mRNAs (21, 31, 40) (Fig. 1) (Los Alamos HIV Sequence Database, www.hiv.lanl.gov). To date, on the RRE structure, five putative Rev-binding sites have been identified. These sites consist of a 6-bp helical segment and three adjacent guanosines, and they involve the RRE stem-loops I, IIA, IIB, IIC, and III (18). The correct conformation of these Rev binding sites has been demonstrated to be essential for the correct Rev-RRE interaction (27).
Enfuvirtide (ENF; Fuzeon) is the first peptide fusion inhibitor approved for clinical use. The peptide sequence of ENF is derived from the HIV-1 gp41 C terminus heptad repeat (HR) sequence, which corresponds to a linear region of 36 aa; ENF inhibits fusion by binding to the N-terminal HR of gp41 and preventing six-helix bundle formation (33, 14). To date, 18 mutations at eight positions within the ENF target region encompassing aa 36 to 45 of HR1 in gp41 have been associated with ENF resistance (6, 15, 24, 26, 33). All the codons encoding the gp41 residues associated with ENF resistance are localized within the RRE, and some of them have been demonstrated to impair the ability of RRE to bind Rev, in turn reducing viral replication capacity (27). In this light, it is conceivable that during ENF pressure, mutations in Rev can appear in order to restore the correct binding between Rev and the RRE and, consequently, HIV-1 replication. To date, no study has shed light on this point.
Thus, the goal of our study was (i) to investigate whether mutations in Rev could be associated with ENF treatment, (ii) to correlate Rev mutations with the classical ENF resistance mutations in gp41 and with viremia and CD4 cell count, and (iii) to investigate the impact of ENF resistance mutations in the secondary structure of the RRE.
MATERIALS AND METHODS
Study population.
The study included 88 highly drug-experienced, multiply failing, patients infected with HIV-1 subtype B. ENF was administered with one to three nucleoside reverse transcriptase inhibitors (NRTIs) plus at least one protease inhibitors (PI) in 60 (68.2%) patients and with one to three NRTIs plus 1 non-NRTI in 16 (18.2%) patients. In seven patients ENF was administered with only two to three NRTIs and in the remaining patients with only two PIs. For each patient, viremia and CD4 cell count were accurately monitored every month during ENF treatment. A total of 193 plasma samples were collected from 88 patients at virological failure and at different time points later during ENF treatment. Baseline samples from 44/88 patients (50.0%) were also available for the gp41 sequencing. As a control, another 39 HIV-1-infected patients, naïve to ENF, were included in the analysis of gp41 and Rev mutations.
gp41 and Rev sequencing.
The sequencing of the entire gp41 was performed on plasma samples as previously described (1, 36). Briefly, RNA was extracted, retrotranscribed, and amplified by using two different sequence-specific primers. gp41-amplified products were sequenced full-length in sense and antisense orientations by using eight different overlapping sequence-specific primers for an automated sequencer (ABI 3100). Due to the overlapping reading frames between gp41 and Rev, the obtained sequences were analyzed for both the full coding sequence of gp41 and the second exon of Rev, which encodes the Rev region from aa 26 to 116. We focused our attention on the second exon of Rev, which contains all functional residues and domains important for the activity of the protein. Sequences having a mixture of wild-type and mutant residues at single positions were considered to have the mutant(s) at that position. Subtypes were assessed by the construction of phylogenetic trees generated with Kimura's two-parameter model. The statistical robustness within each phylogenetic tree was confirmed with a bootstrap analysis using 1,000 replicates. All calculations were performed with PAUP, version 4.0, software (37).
Statistical analysis.
(i) Mutation prevalence. To identify mutations in Rev associated with ENF treatment, we calculated the frequency of all mutations in the 90 residues of the second exon of Rev in isolates from 83 ENF-naïve patients and in isolates from 88 patients with virological failure to ENF (virological failure was defined as viremia of >50 copies/ml at two consecutive tests). Chi-square tests of independence were used to verify whether the differences in frequencies between the two groups of patients were statistically significant. The chi-square statistic was based on a two-by-two contingency table containing the numbers of isolates from patients either untreated or treated with ENF and the numbers of isolates with and without mutations. The chi-square test was performed to assess whether the prevalence of mutations in ENF-naïve and ENF-treated patients differed significantly from what would be expected under an independence assumption. In our analysis, the Cochran rule, which is a conventional criterion for the chi-square test to be valid, was fully satisfied. In fact, in each contingency table performed with our data set, 80% of the expected frequencies exceed 5, and all the expected frequencies exceed 1. For patients with more than one Rev sequence available during ENF treatment, the latest sequence obtained while receiving treatment was analyzed. ENF-associated mutations were defined as mutations that were significantly more common in ENF-treated than in ENF-naïve persons after adjusting for multiple comparisons.
To assess the association of specific ENF-associated mutations with a change in viremia and CD4 cell count, we compared the mean change in viremia and CD4 cell count from baseline to week 48 and to time of virological failure between the subset of patients harboring viral isolates with a specific ENF-associated mutation and the subset of patients harboring viral isolates without such mutation. Mann-Whitney tests were used to assess statistically significant differences.
For all statistical tests, we used the method of Benjamini and Hochberg to identify results that were statistically significant in the presence of multiple-hypothesis testing (2). In particular, we used a false discovery rate of 0.05 to have an expected number of false positives of 5%. In this method, the P values were ranked in ascending order. Each P value (with the exception of the largest) was then multiplied by the total number of hypotheses tested, divided by its rank. All the new resulting P values that were less than 0.05 were considered to be statistically significant.
(ii) Pairwise correlation.
We used the binomial correlation coefficient (phi) to assess covariation among gp41 and Rev mutations in the set of 88 ENF-treated patients. For a given pair of mutations X and Y, the phi coefficient is calculated as follows: (N × NXY − NX × NY)/{square root of [NX × NY × (N − NX) × (N − NY)]}, where NXY represents the number of sequences containing X and Y, NX represents the number of sequences containing X, and NY represents the number of sequences containing Y. The binomial correlation coefficient was calculated for any pair of mutations as a measure of association. The matrix of pairwise phi values contains values between −1 and 1, with values close to 1 indicating strongly positive association and values close to −1 indicating strongly negative association. Samples having a mixture of two or more mutations at a given pair of positions were ignored in calculating the covariation since it is not possible to identify whether these mutations are indeed located in the same viral genome. A Fisher exact test was performed to assess whether cooccurrence of mutations X and Y differed significantly from what would be expected under an independence assumption. Again, the Benjamini-Hochberg method (2) was used to correct for multiple testing at an false discovery rate of 0.05.
(iii) Cluster analysis.
To analyze the covariation structure of mutations in more detail, we performed average linkage hierarchical agglomerative clustering, as described elsewhere (35).
Hierarchical clustering methods, which under different names are also widely used in phylogenetic tree building, rely on a matrix of pairwise dissimilarities between entities, based on which groups are associated into hierarchical clusters of increasingly less strong association. As such, it is in the firsthand an explorative and not a predictive tool. Briefly, in average linkage clustering, clusters of increasing size are formed starting from one-element groups by iteratively joining two clusters with minimum average intercluster distances between pairs of mutations. The distance between a pair of mutations was derived from the phi correlation coefficient, which is a measure of the association between two binary random variables, with 1 and −1 representing maximal positive and negative association, respectively. This similarity measure was transformed into a distance by mapping phi = 1 to distance 0 and phi = −1 to distance 1, with linear interpolation in between. The distance between different mutations at a single position was left undefined as such pairs never cooccur in a single sequence (except from mixtures) and would lead to distorted dendrograms owing to their high distance values. The resulting partial distance matrix was then used as input for the clustering algorithm, ignoring undefined distances in computing averages. To assess the stability of the resulting dendrogram, confidence values for all subtrees in the dendrogram were computed by 100 replications of the clustering procedure on sequence sets bootstrapped from the original 88 sequences (35, 36). For instance, a bootstrap value of 1 simply means that out of 100 runs, all 100 had these two mutations (or groups of mutations) linked closest together.
RRE structural model analysis.
The RRE secondary structure was modeled by using the Vienna RNA Fold computer program (10) and the complete sequence (175 nucleotides) of the HIV-1 B subtype RRE. By using this program, we obtained an RRE secondary structure that was superimposable on one confirmed by previous in vitro experiments (10). Then, we introduced specific mutations, either alone or in combination, in the RRE secondary structure and repeated the predictions in order to evaluate the ability of these mutations to induce conformational changes in the RRE.
Nucleotide sequence accession numbers.
Nucleotide sequences obtained in this were submitted to GenBank under accession numbers EU251192 to EU251378 and EU281662 to EU281733.
RESULTS
Patient characteristics.
Table 1 summarizes the demographic and clinical characteristics of the 88 ENF-treated patients. All patients were heavily treatment experienced, with resistance to multiple NRTIs, non-NRTIs, and PIs. They were experiencing virological failure in response to their last antiretroviral regimen, with high and stable viremia levels (median HIV RNA, 5 log10 copies/ml; interquartile range [IQR], 4.9 to 5.1 log10 copies/ml) and CD4 cell counts in progressive decline during the last 12 weeks prior to ENF therapy (87 cells/μl at baseline; IQR, 36 to 149 cells/μl). After 8 weeks of ENF treatment, a significant decline in median viremia (3.1 log10 copies/ml; IQR, 2.2 to 4.8 log10 copies/ml) (P < 0.001) and a significant increase in the median CD4 cell count from 87 cells/μl at baseline to 129 cells/μl (IQR, 73 to 196 cells/μl) (P < 0.001) were noted. Eight (9.1%) patients achieved at week 8 viremia levels of <50 copies/ml. Median viremia rebounded to 4.4 log10 copies/ml (IQR, 2.9 to 5.5 log10 copies/ml) at week 24 and then was maintained stable at this value up to week 48, while the CD4 cell count continued to increase up to a median value of 183 cells/μl (IQR, 69 to 294 cells/μl) at week 48. The increase in CD4 cell count up to week 48 was observed in 71 (80.7%) out of 88 ENF-treated patients.
TABLE 1.
Patient characteristics at baseline
| Characteristic | Value |
|---|---|
| No. of ENF-treated patients | 88 |
| No. of male patients (%) | 57 (64.8) |
| Median age (yr [IQR]) | 43 (38.5-48.5) |
| No. (%) of risk factors | |
| Sexual behavior | 36 (40.9) |
| Drug addiction | 20 (22.7) |
| Unknown | 31 |
| No. (%) of patients at CDC stage C | 42 (47.7) |
| Median viremia at baseline (log HIV RNA | |
| copies/ml [IQR]) | 5.0 (4.9-5.1) |
| Median CD4 cell count at baseline (cells/μl [IQR]) | 87 (36-149) |
| Median time since diagnosis (yr [IRQ]) | 12 (8.5-16.3) |
| Median no. of ENF-coadministered drugs (IQR) | 3 (2-3) |
| Median no. of resistance mutations (IQR)a | |
| NRTI | 5 (4-6) |
| Non-NRTI | 1 (1-2) |
| PI | 8 (6-12) |
The drug resistance mutations considered in this study were those listed by the International AIDS Society.
Novel mutations in Rev associated with ENF treatment.
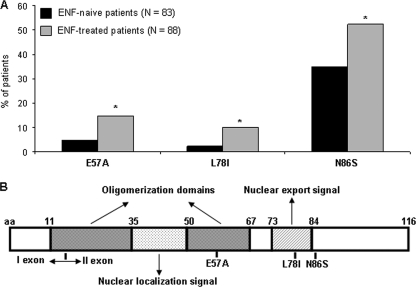
By evaluating Rev sequences derived from 83 ENF-naïve patients and 88 ENF-treated patients, we identified three Rev mutations significantly associated with ENF treatment (Fig. 2A). In particular, Rev mutation E57A (E57Arev) and L78Irev occurred at a frequency of 4.8% and 2.4% in isolates from ENF-naïve patients and increased to 14.8% and 10.2% in isolates from ENF-treated patients (P = 0.03 and P = 0.02, respectively). N86Srev was already present in isolates from ENF-naïve patients at a frequency of 35% and increased after ENF treatment to a frequency of 52.3% (P = 0.02). The E57Arev and L78Irev mutations are localized in the oligomerization domain II (close to the RRE binding site) and in the NES of Rev, respectively, while the N86Srev mutation is localized close to the NES (Fig. 2B).
FIG. 2.
Prevalence and localization of Rev mutations associated with ENF treatment. (A) The frequency of mutations was calculated in isolates from 83 ENF-näive patients and 88 patients who experienced virological failure to ENF. Statistically significant differences were assessed by chi-square tests of independence (based on a 2-by-2 contingency table). (B) Localization of Rev mutations in the schematic structure of HIV-1 Rev protein. The HIV-1 Rev is composed of several domains harboring distinct functions: (i) an NLS that mediates Rev import in the nucleus and the RRE binding; (ii) two oligomerization domains, flanking the NLS, that are implicated in the oligomerization of Rev along the RRE; (iii) an NES that acts as an NES and contains also another NLS.
Associations among Rev mutations and known gp41 mutations.
To identify significant patterns of pairwise associations between Rev and gp41 mutations observed in 88 ENF-treated patients, we calculated the phi coefficient and its statistical significance for each pair of mutations (Table 2). A positive and statistically significant correlation between mutations at two specific positions (0 < phi < 1; P < 0.05) indicates that under drug pressure these two positions mutate in a correlated manner in order to confer an advantage in terms of viral fitness, indicating that the cooccurrence of mutations is not due to chance. The novel mutation E57Arev was positively correlated with the classical ENF resistance mutations Q40H and L45M in gp41 (Q40Hgp41 L45Mgp41; phi = 0.24; P = 0.04). In addition, E57Arev also showed a positive correlation with L54Mgp41 (phi = 0.24; P = 0.03) and with N86Srev (phi = 0.23; P = 0.03). In addition, we also found that the presence of N86Srev at baseline significantly correlated with the on-treatment development of Q40Hgp41 and L45Mgp41 (P = 0.018). No other correlations between Rev and gp41 mutations were observed.
TABLE 2.
Significantly correlated mutations of gp41 and Rev with the novel mutation E57Arev
| E57Arev-correlated mutation(s) | Frequency (no. of positive patients [%])a | Covariation frequency (no. [%])b | Phi | P |
|---|---|---|---|---|
| L54Mgp41 | 38 (43.2) | 10 (76.9) | 0.24 | 0.03 |
| Q40Hgp41 L54Mgp41 | 7 (7.9) | 3 (23.0) | 0.24 | 0.04 |
| N86Srev | 46 (52.3) | 10 (76.9) | 0.23 | 0.03 |
The frequency was determined in isolates from 88 ENF-treated patients. The E57Arev mutation was present in 13 patients (14.8%).
Percentages were calculated in patients positive for the E57Arev mutation.
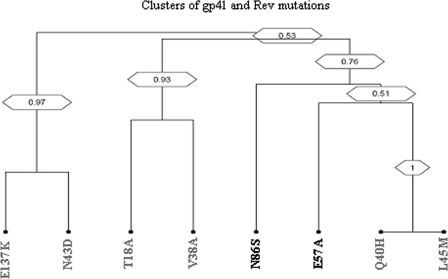
Clusters of correlated mutations.
The topology of the dendrogram suggests the existence of distinct clusters of mutations involved in ENF resistance. In particular, Q40Hgp41 L45Mgp41 mutations formed a strong cluster that was linked to E57Arev and N86Srev as well (Fig. 3). None of the other ENF resistance mutations in gp41 showed any correlation with Rev mutations (Fig. 3).
FIG. 3.
Covariation analysis among gp41 and Rev mutations. The dendrogram was obtained from average linkage hierarchical agglomerative clustering, showing significant clusters among Rev and gp41 mutations. The length of branches reflects distances between mutations in the original distance matrix. Bootstrap values, indicating the significance of clusters, are reported in the boxes.
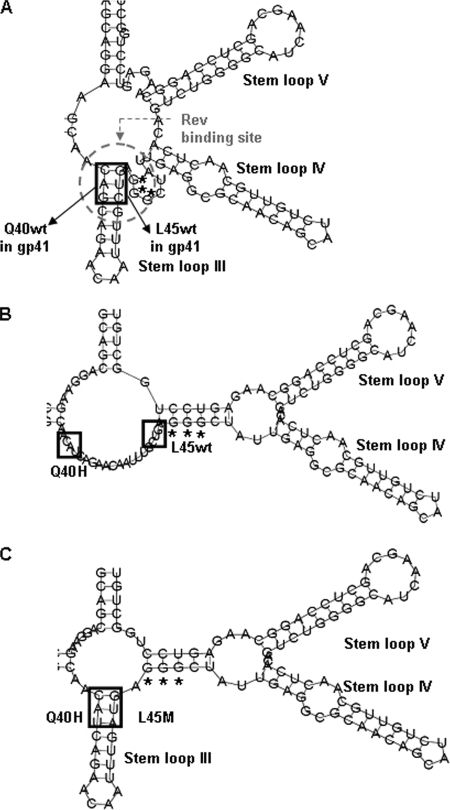
Effect of the gp41 mutations on the secondary structure of the RRE.
The Q40Hgp41 and L45Mgp41 mutations derive from the nucleotide substitutions (in boldface) CAG to CAU and CUG to AUG, respectively. Due to the overlapping between the gp41 nucleotide sequence and the RRE, we examined the impact of these nucleotide substitutions in the secondary structure of the RRE. By using the Vienna RNA Fold software, we modeled a secondary structure of the RRE that was superimposable onto one confirmed by previous in vitro experiments (9). The codons CAG and CUG form complementary base pairs in stem-loop III, which has been proposed to form a well-defined Rev-binding site in the RRE (Fig. 4A) (18). The nucleotide substitution (in boldface) from CAG to CAU (corresponding to Q40Hgp41) completely abrogates stem-loop III and induces new base pairing among nucleotides (for instance, with the three G residues important in Rev binding) and consequently abolishes the conformation of this Rev-binding site in the RRE (Fig. 4B). A similar result was observed for the nucleotide substitution (in boldface) from CUG to AUG (corresponding to L45Mgp41). The conformation of this Rev-binding site in the RRE is restored (even if not completely) only in the copresence of double mutations CAU AUG (corresponding to Q40Hgp41 L45Mgp41) (Fig. 4C). Therefore, these results suggest that the highly frequent cooccurrence of Q40Hgp41 and L45Mgp41 is mostly related to the need to maintain a proper structure in the RRE (i.e., codons CAU and AUG) rather than to compensate modifications in the tertiary structure of gp41; this further confirms the relevance of mutations occurring at sites of the HIV genome encoding different proteins and functions.
FIG. 4.
Putative secondary structure of the HIV-1 RRE. HIV-1 RRE wild-type stem-loops III, IV, and V, together with the single strand of three guanosines important for the binding with Rev, are shown in panel A. The HIV-1 RRE with a nucleotide substitution at 40 position is shown in panel B and with nucleotide substitutions at positions 40 and 45 is shown in panel C. The putative secondary structure of the HIV-1 RRE with a nucleotide substitution only at position 45 is not shown since it is superimposable on the structure shown in panel B.
Association of ENF resistance mutations with viremia and CD4 cell count.
By investigating the correlation of Rev mutations with viremia and CD4 cell count, we found that the E57Arev mutation was associated, during ENF-based treatment, with a mean change in viremia from baseline to week 48 significantly higher than that observed in the absence of this mutation (+0.93 log copies/ml versus −0.84 log copies/ml; P = 0.006) (Table 3). At the same time, E57Arev was associated with a mean change in CD4 cell count from baseline to week 48 of ENF treatment that was significantly lower than that observed in the absence of this mutation (−42 cells/μl versus +56 cells/μl; P = 0.04) (Table 3). Additionally, we found that the copresence of E57Arev with Q40Hgp41 L45Mgp41 was associated with an increase in viremia from baseline to week 48 and with a decrease in CD4 cell count from baseline to week 48 compared to Q40Hgp41 L45Mgp41 alone (mean change in viremia, +0.40 log copies/ml versus −0.94 log copies/ml; mean change in CD4 cell count, −48 cells/μl versus +43 cells/μl). These results suggest that this mutation may play a compensatory role in order to restore the correct binding between the Rev molecules and the RRE since this RNA structure could be impaired by the classical ENF resistance mutations.
TABLE 3.
Mean change in viremia and in CD4 cell counts from baseline to week 48 in patients treated with ENF according to the presence/absence of mutation E57Arev
| Amino acid at position 57 | Frequency (no. of positive patients [%])a | Viremia data
|
CD4 cell count data
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| No. of RNA copies/ml at:
|
Mean change | P valueb | No. of cells/μl at:
|
Mean change | P valueb | ||||
| Baseline | Wk 48 | Baseline | Wk 48 | ||||||
| E57 wild type | 75 (85.2) | 4.9 | 4.4 | −0.84 | 0.006 | 146 | 192 | +56 | 0.04 |
| E57A | 13 (14.8) | 4.9 | 5.4 | +0.93 | 176 | 104 | −42 | ||
The frequency was determined in 88 ENF-treated patients.
Significance was determined for the mean change from baseline to week 48 in the presence versus the absence of the mutation (Mann-Whitney test).
For the other two Rev mutations (L78Irev and N86Srev), we did not observe any statistically significant correlations with viremia or CD4 cell count.
DISCUSSION
This study, which involved one of the largest cohorts of ENF-treated patients assembled so far, shows that HIV-1 mutations arising as a result of ENF treatment may affect both the regulatory protein Rev and the secondary structure of the RRE.
Our covariation analysis showed that the classical ENF resistance gp41 mutations Q40Hgp41 and L45Mgp41 are strongly correlated with each other and are the only gp41 mutations significantly correlated with mutations in Rev, in particular, with E57Arev and N86Srev. By modeling the gp41 structure, we observed that Q40Hgp41 and L45Mgp41 do not establish a direct interaction in the secondary structure of gp41 (data not shown); thus, we explored the reason(s) of the coevolution of these two mutations by analyzing the gp41 nucleotide sequence. The Q40Hgp41 and L45Mgp41 derive from the nucleotide substitutions (in boldface) CAG to CAU and CUG to AUG, respectively. While the methionine (M) is the only amino acid encoded by a single codon (AUG), the histidine (H) can be encoded by either CAU or CAC. Despite this, in our cohort of patients, we always observed the codon CAU at the gp41 residue 40. By modeling the RRE structure, we observed that the Q40Hgp41- and L45Mgp41-corresponding nucleotides are base paired in stem-loop III of the RRE, known to form a well-defined Rev-binding site in the RRE (10, 18). The conformation of stem-loop III is completely abrogated when CAU (Q40Hgp41) or AUG (L45Mgp41) mutations occur alone while it is restored when CAU and AUG (Q40Hgp41 L45Mgp41) mutations are copresent. This result can explain why these two ENF resistance mutations are found together and at the same time can provide an excellent example of compensatory evolution. Indeed, compensatory evolution has been shown to play a key role in the evolution of the stem regions in the RNA secondary structure since it allows the maintenance of the correct base pairing among nucleotides in the stem (16, 17, 39). In addition, we observed that L45M-corresponding nucleotides fail to restore the conformation of stem-loop III when the H at the gp41 position 40 is encoded by the codon CAC (another possible codon for H). This may explain why this codon is completely absent in our cohort of patients. Our result is also consistent with two previous studies showing that the ENF resistance gp41 mutations determine nucleotide changes in the RRE, influencing the stability of this RNA element (27, 36). We recently showed that gp41 residues T18 and V38 localize as complementary nucleotides with each other in stem-loop IIA of the RRE (36), which is involved in Rev interaction. The copresence of T18A (GCU) with V38A (GCG) was associated with a release of free energy even higher than that observed with the wild-type base pair (a ΔG of −3.4 kcal/mol versus −2.1 kcal/mol) (36). Similarly, it has been suggested that the gp41 mutations A30V and G36D may appear in the gp41 protein to readjust the secondary structure of the RRE, thus rescuing RRE stability (27). Thus, overall findings support the idea that viral evolution under ENF pressure may be constrained by the secondary structure of the RRE.
The modeling of the RRE also showed that the single strand of three guanosines that has been demonstrated to bind Rev (18) is completely lost (even when Q40Hgp41 and L45Mgp41 are copresent), thus suggesting an impairment in the binding between the RRE and Rev. Consistent with these findings, the presence of Q40Hgp41 L45Mgp41 is associated with a −0.70 log copies/ml decrease in viremia from baseline to week 48 of ENF treatment. It is conceivable to hypothesize that mutations in Rev, such as E57Arev, can appear in order to rescue the correct binding between Rev and the RRE, thus increasing viral replication capacity. The E57Arev mutation corresponds to the D239Hgp41 mutation that is localized in the cytoplasmic tail of gp41, outside any important functional domain of gp41. In contrast, E57Arev is localized in oligomerization domain II very close to the RRE binding site and is strongly associated with the resistance gp41 mutations Q40Hgp41 L45Mgp41. In addition, E57Arev is significantly correlated with an increase in viremia from baseline to week 48 as well as with a significant decrease in CD4 cell count, when in the presence of Q40Hgp41 L45Mgp41.
The E57Arev mutation results in the loss of a negative charge and the acquisition of a hydrophobic residue. Previous studies demonstrated that hydrophobic residues are necessary for a correct oligomerization of Rev along the RRE and consequently for an efficient shuttle of mRNAs from the nucleus to the cytoplasm (4, 14). Thus, we hypothesize that E57Arev may act as a compensatory mutation able to restore the correct shuttling of mRNAs impaired by the classical ENF resistance mutations. We emphasize that this is a working hypothesis that needs to be confirmed by analyses of enlarged databases (that include genotypic and clinical data) and in in vitro experiments.
Another mutation that should be discussed is L78Irev. This mutation, associated with ENF treatment, does not correspond to any change in gp41, is localized in the activation domain of Rev, and has been shown to reduce the export of Rev from the nucleus to the cytoplasm (13). Consistent with the negative impact of this mutation on viral fitness (13), we observed that the presence of L78Irev is associated with a 1-log decrease in viremia from baseline to week 48 of ENF treatment (even if not statistically significant) (data not shown). Further studies are necessary to investigate the impact of this mutation on disease progression.
As a final point, it should be stressed that some peptidomimetics of the Rev protein have been recently demonstrated to efficiently suppress HIV-1 replication by blocking the export of mRNAs into the cytoplasm (25, 20). The ability of ENF to affect the interaction between Rev and the RRE, shown in this paper, should be taken into account in the design of these compounds and in the possible cooperation between these two drug classes.
In conclusion, our study suggests that ENF pressure could also affect Rev-RRE interactions. This highlights the importance of the correct interplay between the different HIV-1 genes and proteins during the HIV-1 life cycle and extends the knowledge about the evolutionary ability of HIV to select for mutations able to restore viral functions and replication capacity.
Acknowledgments
We thank Caterina Gori, Fabio Continenza, Daniele Pizzi, Andrea Biddittu, and Amalia Mastrofrancesco for sequencing and data management.
This work was financially supported by grants from the Italian National Institute of Health, the Ministry of University and Scientific Research, Current and Finalized Research of the Italian Ministry of Health, and the European Community (QLK2-CT-2000-00291 and the Descartes Prize HPAW-90001).
Footnotes
Published ahead of print on 5 January 2009.
REFERENCES
- 1.Aquaro, S., R. D'Arrigo, V. Svicher, G. D. Perri, S. L. Caputo, U. Visco-Comandini, M. Santoro, A. Bertoli, F. Mazzotta, S. Bonora, V. Tozzi, R. Bellagamba, M. Zaccarelli, P. Narciso, A. Antinori, and C. F. Perno. 2006. Specific mutations in HIV-1 gp41 are associated with immunological success in HIV-1-infected patients receiving enfuvirtide treatment. J. Antimicrob. Chemother. 58:714-722. [DOI] [PubMed] [Google Scholar]
- 2.Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57:289-300. [Google Scholar]
- 3.Demart, S., F. Ceccherini-Silberstein, S. Schlicht, S. Walcher, H. Wolff, M. Neumann, V. Erfle, and R. Brack-Werner. 2003. Analysis of nuclear targeting activities of transport signals in the human immunodeficiency virus Rev protein. Exp. Cell. Res. 291:484-501. [DOI] [PubMed] [Google Scholar]
- 4.Edgcomb, S. P., A. Aschrafi, E. Kompfner, J. R. Williamson, L. Gerace, and M. Hennig. 2008. Protein structure and oligomerization are important for the formation of export-competent HIV-1 Rev-RRE complexes. Protein Sci. 17:420-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freed, E. O., and M. A. Martin. 2001. HIVs and their replication, p. 1971-2041. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. II. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 6.Greenberg, M., N. Cammack, M. Salgo, and L. Smiley. 2004. HIV fusion and its inhibition in antiretroviral therapy. Rev. Med. Virol. 14:321-337. [DOI] [PubMed] [Google Scholar]
- 7.Heaphy, S., C. Dingwall, I. Ernberg, M. J. Gait, S. M. Green, J. Karn, A. D. Lowe, M. Singh, and M. A. Skinner. 1990. HIV-1 regulator of virion expression (Rev) protein binds to an RNA stem-loop structure located within the Rev response element region. Cell 60:685-693. [DOI] [PubMed] [Google Scholar]
- 8.Henderson, B. R., and P. Percipalle. 1997. Interactions between HIV Rev. and nuclear import and export factors: the Rev nuclear localisation signal mediates specific binding to human importin-beta. J. Mol. Biol. 274:693-707. [DOI] [PubMed] [Google Scholar]
- 9.Hofacker I. L. 2003. Vienna RNA secondary structure server. Nucleic Acids Res. 13:3429-3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holland, S. M., N. Ahmad, R. K. Maitra, P. Wingfield, and S. Venkatesan. 1990. Human immunodeficiency virus rev protein recognizes a target sequence in rev-responsive element RNA within the context of RNA secondary structure. J. Virol. 64:5966-5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hope, T. J., B. L. Bond, D. McDonald, N. P. Klein, and T. G. Parslow. 1991. Effector domains of human immunodeficiency virus type 1 Rev and human T-cell leukemia virus type I Rex are functionally interchangeable and share an essential peptide motif. J. Virol. 65:6001-6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang, X. J., T. J. Hope, B. L. Bond, D. McDonald, K. Grahl, and T. G Parslow. 1991. Minimal Rev-response element for type 1 human immunodeficiency virus. J. Virol. 65:2131-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iversen, A. K., E. G. Shpaer, A. G. Rodrigo, M. S. Hirsch, B. D. Walker, H. W. Sheppard, T. C. Merigan, and J. I. Mullins. 1995. Persistence of attenuated rev genes in a human immunodeficiency virus type 1-infected asymptomatic individual. J. Virol. 69:5743-5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jain, C., and J. G. Belasco. 2001. Structural model for the cooperative assembly of HIV-1 Rev multimers on the RRE as deduced from analysis of assembly-defective mutants. Mol. Cell 7:603-614. [DOI] [PubMed] [Google Scholar]
- 15.Johnson, V. A., F. Brun-Vezinet, B. Clotet, H. F. Gunthard, D. R. Kuritzkes, D. Pillay, J. M. Schapiro, and D. D. Richman. 2008. Update of the drug resistance mutations in HIV-1: spring 2008. Top. HIV Med. 16:62-68. [DOI] [PubMed] [Google Scholar]
- 16.Kern, A. D., and F. A. Kondrashov. 2004. Mechanisms and convergence of compensatory evolution in mammalian mitochondrial tRNAs. Nat. Gen. 36:1207-1212. [DOI] [PubMed] [Google Scholar]
- 17.Kirby, D. A., S. V. Muse, and W. Stephan. 1995. Maintenance of pre-messenger-RNA secondary structure by epistatic selection. Proc. Nat. Acad. Sci. USA 92:9047-9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kjems, J., M. Brown, D. D. Chang, and P. A. Sharp. 1991. Structural analysis of the interaction between the human immunodeficiency virus Rev protein and the Rev response element. Proc. Natl. Acad. Sci. USA 88:683-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kubota, S., H. Siomi, T. Satoh, S. Endo, M. Maki, and M. Hatanaka. 1989. Functional similarity of HIV-I Rev and HTLV-I Rex proteins: identification of a new nucleolar-targeting signal in Rev protein. Biochem. Biophys. Res. Commun. 162:963-970. [DOI] [PubMed] [Google Scholar]
- 20.Legiewicz, M., C. S. Badorrek, K. B. Turner, D. Fabris, T. E. Hamm, D. Rekosh, M. L. Hammarskjöld, and S. F. Le Grice. 2008. Resistance to RevM10 inhibition reflects a conformational switch in the HIV-1 Rev response element. Proc. Natl. Acad. Sci. USA 105:14365-14370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malim, M. H., J. Hauber, S. Y. Le, J. V. Maizel, and B. R. Cullen. 1989. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 338:254-257. [DOI] [PubMed] [Google Scholar]
- 22.Malim, M. H., D. F. McCarn, L. S. Tiley, and B. R. Cullen. 1991. Mutational definition of the human immunodeficiency virus type 1 Rev activation domain. J. Virol. 65:4248-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mann, D. A., I. Mikaélian, R. W. Zemmel, S. M. Green, A. D. Lowe, T. Kimura, M. Singh, P. J. Butler, M. J. Gait, and J. Karn. 1994. A molecular rheostat. Co-operative rev binding to stem I of the Rev-response element modulates human immunodeficiency virus type-1 late gene expression. J. Mol. Biol. 241:193-207. [DOI] [PubMed] [Google Scholar]
- 24.Miller, M. D., and D. J. Hazuda. 2004. HIV resistance to the fusion inhibitor enfuvirtide: mechanisms and clinical implications. Drug Resist. Update 7:89-95. [DOI] [PubMed] [Google Scholar]
- 25.Mills, N. L., M. D. Daugherty, A. D. Frankel, and R. K. Guy. 2006. An alpha-helical peptidomimetic inhibitor of the HIV-1 Rev-RRE interaction. J. Am. Chem. Soc. 128:3496-3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mink, M., S. M. Mosier, S. Janumpalli, D. Davison, L. Jin, T. Melby, P. Sista, J. Erickson, D. Lambert, S. A. Stanfield-Oakley, M. Salgo, N. Cammack, T. Matthews, and M. L. Greenberg. 2005. Impact of human immunodeficiency virus type 1 gp41 amino acid substitutions selected during enfuvirtide treatment on gp41 binding and antiviral potency of enfuvirtide in vitro. J. Virol. 79:12447-12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nameki, D., E. Kodama, M. Ikeuchi, N. Mabuchi, A. Otaka, H. Tamamura, M. Ohno, N. Fujii, and M. Matsuoka. 2005. Mutations conferring resistance to human immunodeficiency virus type 1 fusion inhibitors are restricted by gp41 and Rev-responsive element functions. J. Virol. 79:764-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neumann, M., J. Harrison, M. Saltarelli, E. Hadziyannis, V. Erfle, B. K. Felber, and G. N. Pavlakis. 1994. Splicing variability in HIV type 1 revealed by quantitative RNA polymerase chain reaction. AIDS Res. Hum. Retrovir. 10:1531-1542. [DOI] [PubMed] [Google Scholar]
- 29.Neville, M., F. Stutz, L. Lee, L. I. Davis, and M. Rosbash. 1997. The importin beta family member Crm1p bridges the interaction between Rev. and the nuclear pore complex during nuclear export. Curr. Biol. 7:767-775. [DOI] [PubMed] [Google Scholar]
- 30.Perkins, A., A. W. Cochrane, S. M. Ruben, and C. A. Rosen. 1989. Structural and functional characterization of the human immunodeficiency virus Rev protein. J. Acquir. Immune Defic. Syndr. 2:256-263. [PubMed] [Google Scholar]
- 31.Pollard, V. W., and M. H. Malim. 1998. The HIV-1 Rev protein. Annu. Rev. Microbiol. 52:491-532. [DOI] [PubMed] [Google Scholar]
- 32.Purcell, D. F., and M. A. Martin. 1993. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J. Virol. 67:6365-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reeves, J. D., F. H. Lee, J. L. Miamidian, C. B. Jabara, M. M. Juntilla, and R. W. Doms. 2005. Enfuvirtide resistance mutations: impact on human immunodeficiency virus envelope function, entry inhibitor sensitivity, and virus neutralization. J. Virol. 79:4991-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwartz, S., B. K. Felber, D. M. Benko, E. M. Fenyo, and G. N. Pavlakis. 1990. Cloning and functional analysis of multiply spliced mRNA species of human immunodeficiency virus type 1. J. Virol. 64:2519-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Svicher, V., T. Sing, M. M. Santoro, F. Forbici, F. Rodríguez-Barrios, A. Bertoli, N. Beerenwinkel, M. C. Bellocchi, F. Gago, A. d'Arminio Monforte, A. Antinori, T. Lengauer, F. Ceccherini-Silberstein, and C. F. Perno. 2006. Involvement of novel human immunodeficiency virus type 1 reverse transcriptase mutations in the regulation of resistance to nucleoside inhibitors. J. Virol. 80:7186-7198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Svicher, V., S. Aquaro, R. D'Arrigo, A. Artese, S. Dimonte, S. Alcaro, M. M. Santoro, G. Di Perri, S. L. Caputo, R. Bellagamba, M. Zaccarelli, U. Visco-Comandini, A. Antinori, P. Narciso, F. Ceccherini-Silberstein, and C. F. Perno. 2008. Specific enfuvirtide-associated mutational pathways in HIV-1 Gp41 are significantly correlated with an increase in CD4+ cell count, despite virological failure. J. Infect. Dis. 197:1408-1418. [DOI] [PubMed] [Google Scholar]
- 37.Swafford, K. L. 1999. PAUP 4.0: phylogenetic analysis with parsimony (and other methods), version 4.0b2a. Sinauer Associates Inc, Sunderland, MS.
- 38.Thomas, S. L., M. Oft, H. Jaksche, G. Casari, P. Heger, M. Dobrovnik, D. Bevec, and J. Hauber. 1998. Functional analysis of the human immunodeficiency virus type-1 Rev protein oligomerization interface. J. Virol. 72:2935-2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilke, C., R. Lenski, and C. Adami. 2003. Compensatory mutations cause excess of antagonistic epistasis in RNA secondary structure folding. BMC Evol. Biol. 3:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zapp, M. L., and M. R. Green. 1989. Sequence-specific RNA binding by the HIV-1 Rev protein. Nature 342:714-716. [DOI] [PubMed] [Google Scholar]
- 41.Zapp, M. L., T. J. Hope, T. G. Parslow, and M. R. Green. 1991. Oligomerization and RNA binding domains of the type 1 human immunodeficiency virus Rev protein: a dual function for an arginine-rich binding motif. Proc. Natl. Acad. Sci. USA 88:7734-7738. [DOI] [PMC free article] [PubMed] [Google Scholar]