Abstract
European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoints classify Candida strains with a fluconazole MIC ≤ 2 mg/liter as susceptible, those with a fluconazole MIC of 4 mg/liter as representing intermediate susceptibility, and those with a fluconazole MIC > 4 mg/liter as resistant. Machine learning models are supported by complex statistical analyses assessing whether the results have statistical relevance. The aim of this work was to use supervised classification algorithms to analyze the clinical data used to produce EUCAST fluconazole breakpoints. Five supervised classifiers (J48, Correlation and Regression Trees [CART], OneR, Naïve Bayes, and Simple Logistic) were used to analyze two cohorts of patients with oropharyngeal candidosis and candidemia. The target variable was the outcome of the infections, and the predictor variables consisted of values for the MIC or the proportion between the dose administered and the MIC of the isolate (dose/MIC). Statistical power was assessed by determining values for sensitivity and specificity, the false-positive rate, the area under the receiver operating characteristic (ROC) curve, and the Matthews correlation coefficient (MCC). CART obtained the best statistical power for a MIC > 4 mg/liter for detecting failures (sensitivity, 87%; false-positive rate, 8%; area under the ROC curve, 0.89; MCC index, 0.80). For dose/MIC determinations, the target was >75, with a sensitivity of 91%, a false-positive rate of 10%, an area under the ROC curve of 0.90, and an MCC index of 0.80. Other classifiers gave similar breakpoints with lower statistical power. EUCAST fluconazole breakpoints have been validated by means of machine learning methods. These computer tools must be incorporated in the process for developing breakpoints to avoid researcher bias, thus enhancing the statistical power of the model.
The Antifungal Susceptibility Testing Subcommittee of the European Committee on Antimicrobial Susceptibility Testing (EUCAST) has recently established the breakpoints for fluconazole and Candida spp. (13). EUCAST considers a strain of a Candida sp. with a fluconazole MIC ≤ 2 mg/liter to be susceptible, a strain with a fluconazole MIC of 4 mg/liter to be intermediate in susceptibility, and a strain with a fluconazole MIC > 4 mg/liter to be resistant. These breakpoints do not apply to Candida glabrata and C. krusei (13).
The process for establishing breakpoints designed by EUCAST takes into account pharmacokinetic-pharmacodynamic data and other factors, such as dosing regimens, toxicology, resistance mechanisms, wild-type MIC distributions, and clinical outcome data.
The clinical data used in the process of setting breakpoints for fluconazole were reported in a previous study (12). That study analyzed, in the classical way, the correlation of MICs and the proportion between the dose administered and the MIC of the isolate (dose/MIC) in relationship to the clinical outcome seen with patients with candidemia and oropharyngeal candidosis (OPC) who had been treated with fluconazole. The results showed that 93.7% (136 of 145 episodes) of infections due to isolates with fluconazole MICs ≤ 2 mg/liter responded to fluconazole treatment. A response of 66% (8 of 12 episodes) was observed when the infections were caused by isolates with a fluconazole MIC of 4 mg/liter and a response of 11.8% (12 of 101 episodes) when the infection was caused by isolates with a fluconazole MIC > 4 mg/liter. Clinical outcome data used for setting breakpoints have usually been analyzed following the 90-60 rule (10). This rule states that infections due to susceptible isolates respond to therapy approximately 90% of the time, whereas infections due to resistant isolates respond approximately 60% of the time. However, data mining tools have been developed that allow better analysis and interpretation of the data. An extraordinary development in contemporary computer science is the introduction and application of methods of machine learning. These enable a computer program to analyze automatically a large body of data and decide what information is most relevant. This information can then be used to make decisions faster and more accurately. Machine learning tools for data mining tasks contain implementations of most of the algorithms for supervised classification such as decision trees, rule sets, Bayesian classifiers, support vector machines, logistic and linear regression, multilayer perceptrons, and nearest-neighbor methods, as well as metalearner methods such as bagging, boosting, and stacking. Machine learning models are supported by complex statistical analyses that include such performance measures as sensitivity, specificity, false-positive rate, and area under the receiver operating characteristic (ROC) curve, which enable researchers to assess whether the results have statistical relevance.
The aim of this work was to employ supervised classification algorithms to analyze the clinical data used to produce EUCAST fluconazole breakpoints to determine whether or not the breakpoints chosen are similar to those produced by these tools.
MATERIALS AND METHODS
Patients and diseases. (i) Candidemia.
A total of 126 candidemia patients treated with fluconazole were recruited from a population-based surveillance study performed in Barcelona, Spain, during 2002 and 2003 (1). Characteristics of the patients and analyses of the correlation of the MICs with patient outcomes have been published previously (12). Briefly, a case was defined as representing a Candida infection when recovery of any Candida species from blood cultures was determined. Candidemia that occurred >30 days after the initial case was considered to represent a new case. Cure was defined by eradication of candidemia and resolution of the associated signs and symptoms. Failure was defined as persistent candidemia despite 4 days of fluconazole treatment. The recommended dose of fluconazole for candidemia is 400 mg/day, but the dose was adjusted to 200 mg/day when the creatinine clearance was between 10 and 50 ml/min and to 100 mg/day when the creatinine clearance was <10 ml/min. Four episodes of candidemia were treated with 100 mg of fluconazole/day, 25 with 200 mg/day, 92 with 400 mg/day, 2 with 600 mg/day, and 3 with 800 mg/day (12).
(ii) Oropharyngeal candidiasis (OPC).
A total of 110 patients with human immunodeficiency virus infection in another study had been treated with fluconazole for a total of 132 episodes of oral thrush caused by C. albicans (6). Sixty-five episodes of OPC were treated with 100 mg of fluconazole/day, 44 with a dose of 200 mg/day, and 23 with 400 mg/day (12). Characteristics of the patients and analyses of the correlation of the MICs with patient outcomes were published previously (12). Briefly, clinical resolution was defined as the absence of lesions compatible with oral thrush after 10 days of therapy. Mycological cure was not evaluated. All episodes were used to evaluate clinical outcome irrespective of the dose of fluconazole given.
Hence, a total of 258 episodes of Candida infection were available for analysis.
Antifungal susceptibility testing.
Antifungal susceptibility testing was performed following the guidelines of the Antifungal Susceptibility Testing Subcommittee of EUCAST for fermentative yeasts (11). Briefly, fluconazole was distributed in RPMI medium supplemented with 2% glucose in flat-bottomed microtitration trays that were inoculated with 105 CFU of yeast/ml, incubated at 35°C, and then read after 24 h at 530 nm using a spectrophotometer. The endpoint was defined as the concentration that resulted in 50% inhibition compared with the growth seen with the control well. C. krusei ATCC 6258 and C parapsilosis ATCC 22019 were included for quality control.
Computational methods.
The following information for each patient was entered in an Excel database (Microsoft Iberica, Spain): the MIC of the isolate, the dose/MIC values, and the treatment outcome for the patient (following the definitions stated above as representative of success or failure). The database contains 156 successes (60.5%) and 102 failures (39.5%). When required, data were transformed to log2 values to approximate a normal distribution. To build up the models, the target variable was the outcome of the infections and the predictor variables were MIC or dose/MIC.
The models were built up using WEKA software (version 3.4.13) (15) and Correlation and Regression Trees (CART) software (version 6.0; Salford Systems, San Diego, CA).
Five classifiers were used to analyze the database: J48 and CART decision trees, OneR decision rule, Naïve Bayes, and Simple Logistic regression. These classifiers cover a wide spectrum of methodologies (trees, rules, and probabilistic classifiers) and were chosen because of their sound theoretical basis and their suitability for intuitive interpretation. The main characteristics of the classifiers are as follows.
J48 and CART decision trees.
A decision tree basically defines at its nodes a series of tests of predictor variables organized in a tree-like structure. Each terminal node (called a “leaf”) gives a classification that applies to all instances that reach the leaf, after being routed down the tree according to the values of the predictors tested in successive nodes. The tree is constructed by recursively splitting the data into smaller and smaller subsets so that after each split the new data subset is purer (i.e., represents less entropy) than the old data subset. The J48 algorithm is the Weka implementation of the C4.5 decision tree (9), which is an outgrowth of the basic original ID3 algorithm. It incorporates a method for postpruning the tree to avoid overfitting. A default of 25% for the confidence level for the error rate was kept. The CART system was proposed by Breiman et al. (2). This system is characterized by binary-split searches, automatic self-validation procedures, and surrogate splitters. The CART method used to build up the model was Gini together with the minimum-cost tree, regardless of tree size.
OneR (4) is a simple classification rule. It is a one-level decision tree expressed as a set of rules testing only one particular predictor variable. The classification given to each branch is the class label that occurs most often in the data. The predictor variable chosen is the one that produces rules with the smallest number of errors, which are computed as the number of instances that do not represent the majority class.
The Naïve Bayes classifier (3) is a simple probabilistic classifier based on applying Bayes's theorem with strong (naïve) independence assumptions. A Naïve Bayes classifier assumes that given the class variable, the predictor variables are conditionally independent.
The Simple Logistic method builds logistic regression models, i.e., a linear model based on a transformed target variable determined using the logit transformation (5). The weights in the linear combination of the predictor variables are calculated from the training data and are used to maximize the likelihood function.
Every classifier develops a model when it searches for the MIC or the dose/MIC that best splits the populations of successes and failures. Tenfold cross-validation was the method used to estimate the performance of each classifier. This was assessed by determining values for (i) sensitivity, (ii) specificity, (iii) the false-positive rate or 1-specificity, (iv) the area under the ROC curve, and (v) the Matthews correlation coefficient (MCC). This coefficient is a measure of the quality of two-class classifications. It takes into account true and false positives and negatives and is generally regarded as a balanced measure which can be used even when classes are of very different sizes. Analysis using the MCC returns a value between −1 and +1. A coefficient of +1 represents a perfect prediction, 0 an average random prediction, and −1 an inverse prediction.
RESULTS
As stated above, the cohort with oral thrush and candidemia was previously described in detail (6, 12). Therefore, the results of the present study represent only the performance of the classifiers and their statistical power.
Table 1 shows the values for the MICs predicting failure for each classifier and the sensitivity, specificity, false-positive rate, area under the ROC curve, and MCC index for the cohorts of patients with OPC and candidemia.
TABLE 1.
Sensitivity, specificity, false-positive rate, area under the ROC curve, and MCC for MIC values as a predictor of failure
| Disease(s) | Classifier | MIC for failures (mg/liter) | Sensitivity (%) | Specificity (%) | False-positive rate (%) | Area under ROC curve | MCC |
|---|---|---|---|---|---|---|---|
| OPC | J48 | >2 | 97 | 72 | 28 | 0.81 | 0.74 |
| CART | >4 | 96 | 83 | 17 | 0.89 | 0.81 | |
| OneR | >4 | 97 | 72 | 28 | 0.84 | 0.74 | |
| Naïve Bayes | >2 | 99 | 74 | 26 | 0.94 | 0.79 | |
| Simple Logistic | >4 | 95 | 77 | 23 | 0.95 | 0.75 | |
| Candidemia | J48 | NCa | |||||
| CART | >4 | 31 | 96 | 4 | 0.63 | 0.31 | |
| OneR | NC | ||||||
| Naïve Bayes | >2 | 31 | 96 | 4 | 0.48 | 0.31 | |
| Simple Logistic | NC | ||||||
| All | J48 | >4 | 87 | 92 | 8 | 0.86 | 0.80 |
| CART | >4 | 87 | 92 | 8 | 0.89 | 0.80 | |
| OneR | >4 | 87 | 92 | 8 | 0.89 | 0.80 | |
| Naïve Bayes | >2 | 91 | 87 | 13 | 0.91 | 0.77 | |
| Simple Logistic | >2 | 91 | 87 | 13 | 0.91 | 0.77 |
NC, not calculated (the classifier did not find a value for splitting the populations of successes and failures).
All classifiers yielded good results, as the OPC data set MCC values show. J48 and Naïve Bayes analyses selected a MIC of >2 mg/liter and CART, OneR, and Simple Logistic a MIC of >4 mg/liter for predicting failures. The best values for the area under the ROC curve were those obtained with Naïve Bayes (0.94) and Simple Logistic (0.95), but those methods had a higher false-positive rate than CART (Table 1). However, for candidemia analyses, the results were poor. Three of the classifiers, J48, OneR, and Simple Logistic, were unable to determine a MIC that could split the populations of successes and failures (Table 1). The CART and Naïve Bayes methods found values of >4 mg/liter and >2 mg/liter, respectively, to be best for predicting failures, although the statistical power was limited. The area under the ROC curve for the Naïve Bayes method was 0.48 and for CART was 0.63 (Table 1). When the whole set was analyzed, MICs similar to those found with the OPC predictive model were obtained. CART and Naïve Bayes were the classifiers showing the same results for both OPC and candidemia. The areas under the ROC curves were 0.89 and 0.91, respectively, but the false-positive rate was slightly lower for CART (8% versus 13%).
Table 2 shows the dose/MIC values predicting treatment success for each classifier and the values for sensitivity, specificity, false-positive rate, area under the ROC curve, and MCC index for the cohort of patients with OPC and candidemia.
TABLE 2.
Sensitivity, specificity, false-positive rate, area under the ROC curve, and MCC for dose/MIC values as a predictor of treatment success
| Disease(s) | Classifier | Dose/MIC for success | Sensitivity (%) | Specificity (%) | False-positive rate (%) | Area under ROC curve | MCC |
|---|---|---|---|---|---|---|---|
| OPC | J48 | >50.0a | 93 | 72 | 28 | 0.95 | 0.68 |
| CART | >75.0a | 84 | 100 | 0 | 0.98 | 0.80 | |
| OneR | >37.5 | 97 | 81 | 19 | 0.89 | 0.81 | |
| Naïve Bayes | >41.7 | 97 | 81 | 19 | 0.85 | 0.81 | |
| Simple Logistic | >38.8 | 98 | 72 | 28 | 0.97 | 0.76 | |
| Candidemia | J48 | NCb | |||||
| CART | >75.0 | 31 | 96 | 4 | 0.63 | 0.34 | |
| OneR | NC | ||||||
| Naïve Bayes | >1,578.0 | 0 | 100 | 0 | 0.56 | 0.0 | |
| Simple Logistic | NC | ||||||
| All | J48 | >50.0 | 87 | 90 | 10 | 0.87 | 0.77 |
| CART | >75.0 | 91 | 90 | 10 | 0.90 | 0.80 | |
| OneR | >37.5 | 86 | 93 | 7 | 0.90 | 0.80 | |
| Naïve Bayes | >674.0 | 91 | 59 | 41 | 0.86 | 0.50 | |
| Simple Logistic | NC |
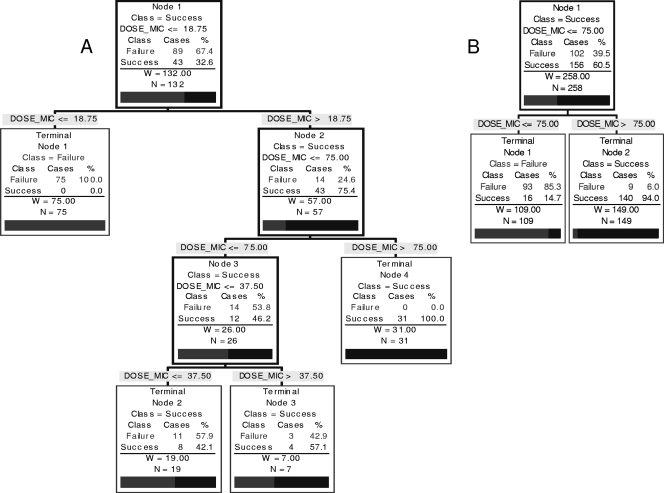
The J48 and CART classifiers produced complex trees for dose/MIC versus outcome (Fig. 1). The initial values splitting the tree for successes and failures were 12.5 and 18.75, respectively. Therefore, the table shows the dose/MIC values predicting the absence of failures.
NC, not calculated (the classifier did not find a value for splitting the populations of successes and failures).
All classifiers yielded a dose/MIC value that predicted treatment success for the OPC data set. In all cases, the value for dose/MIC was >37.5 and that for the area under the ROC curve was ≥0.85 (Table 2).
The classification trees generated by CART and J48 for the OPC data set were rather complex (Fig. 1 shows the CART); the values for the area under the ROC curve were 0.98 and 0.95, with false-positive rates of 0 and 28%, respectively (Table 2). Figure 1 shows the trees produced by CART for the OPC data set and for the OPC and candidemia datasets combined. In the OPC cohort, none of the patients responded with a dose/MIC ≤ 18.75. With a dose/MIC > 18.75, there were 14 (24.6%) failures and 43 (75.4%) successes. All patients with a dose/MIC > 75 were successes. The percentage of failures seen with a dose/MIC between 18.75 and 37.5 was 57.9% (11 patients) and with a dose/MIC between 37.5 and 75 was 42.9% (3 patients) (Fig. 1A). The CART dose/MIC results for the candidemia cohort also showed a value of >75 as the splitter. In this case, it was a two-leaf tree with a 4% false-positive rate but a limited sensitivity of 31%. The area under the ROC curve was 0.63.
FIG. 1.
(A) CART showing values for dose/MIC versus outcome for patients with OPC; (B) CART for both datasets (OPC and candidemia).
Similar values were obtained with the J48 tree for the OPC data set. No response was obtained when the dose/MIC was ≤12.5, and no failures were detected when the dose/MIC was >50.
For the whole data set, only CART results were considered because the breakpoints for the OPC and candidemia cohorts were identical. The tree is displayed in Fig. 1B. Only 6% of failures were detected with a dose/MIC > 75, whereas 85.3% of failures were observed when the dose/MIC was ≤75. The area under the ROC curve was 0.90, and the false-positive rate was 10% (Table 2).
DISCUSSION
Machine learning methods are being used in genomics, proteomics, microarrays, system biology evolution, and text mining (7). However, this is the first time that they have been applied to validation of antifungal breakpoints. The 90-60 rule has defined the accuracy of antimicrobial susceptibility testing for predicting the outcome of bacterial infections. This rule was adopted by mycologists because the clinical correlation studies analyzed showed similar predictive patterns (10). However, the correlation of MIC values or pharmacodynamic parameters with patient outcome should include statistical evaluations similar to those used with any diagnostic tool. Machine learning methods represent an opportunity to use statistical theory for building a model using a data set. However, it is crucial to find the optimal solution; to achieve such a solution, several classifiers must be employed for comparisons of the models obtained with each one. In this work, five classifiers (J48, CART, OneR, Naïve Bayes, and Simple Logistic) were compared to identify which values for MIC or dose/MIC split the populations of successes and failures. The statistical power of each model has been evaluated by means of analyses of the sensitivity, specificity, false-positive rate, area under the ROC curve, and MCC index.
The classifiers choose the MIC that best split the populations of successes and failures. This value is presented as ≤ω mg/liter for successes and >x mg/liter for failures. However, breakpoints usually have three categories, namely, susceptible, intermediate (with susceptibility dependent on dose level), and resistant, presented as ≤ω mg/liter for susceptible isolates, >x and <y mg/liter for intermediate isolates, and >y mg/liter for resistant isolates. The definition for the intermediate category implies that an infection due to the isolate may be appropriately treated in body sites where the drugs are physically concentrated or when a high dosage of drug can be used. It also indicates a buffer zone that should prevent small, uncontrolled, technical factors from causing major discrepancies in interpretations. The main target of any susceptibility testing is to identify resistant strains or, in other words, to identify the drugs that are less likely to eradicate the infection (14). Thus, a very major error is considered to have occurred when a resistant strain is characterized as susceptible in cases in which a patient has been treated with a drug lacking activity against the microorganism causing the infection. On the other hand, a major error is defined as having occurred when a susceptible strain is classified as resistant. In summary, the aim is to minimize very major errors; as a consequence, the rules for any analysis trying to predict the outcome of patients should be based on failures. The crude mortality rate for Candida bloodstream infections is 40% (8); thus, inappropriate treatments must be avoided by all possible means.
All classifiers were able to yield a predictive model for the OPC data set. The CART method gave the best statistical power for a MIC > 4 mg/liter for detecting failures, although the rest of classifiers also exhibited high statistical power (Table 1). However, for candidemia the scenario was completely different. CART and Naïve Bayes methods were able to discover a MIC value that split the populations, but the statistical power was limited (although it was slightly better for CART).
Regarding dose/MIC targets, results were similar for the OPC data set. All classifiers were able to determine a dose/MIC value that split the populations of successes and failures, with areas under the ROC curves above 0.85. However, for candidemia, only CART produced the same dose/MIC value as that determined for the OPC data set, but the CART determination had little statistical power (Table 2).
The determinations for the two cohorts had different values with respect to statistical power, and this merits an explanation. For the OPC cohort, 85 cases had a MIC > 4 mg/liter, but there were only 4 cases for candidemia that showed this value. The lack of strains with MIC > 4 mg/liter in the candidemia cohort explains the limited statistical power of the models. However, the models for the OPC and candidemia datasets gave the same values for MIC and for dose/MIC for at least one classifier. Therefore, the results obtained for the whole data set can be considered if the models produce the same MIC or dose/MIC when the datasets are analyzed separately, despite candidemia and OPC representing quite different clinical situations. If several models satisfy this circumstance, the statistical power of the analyses must be taken into account.
The CART model produced identical target values for the two datasets and the highest statistical power in satisfying both sets of circumstances. The CART model gave a MIC > 4 mg/liter as the breakpoint of resistance, with a sensitivity of 87%, a false-positive rate of 8%, an area under the ROC curve of 0.89, and an MCC index of 0.80 (Table 1). In addition, a dose/MIC > 75 is proposed as the target to achieve treatment success. This means that a fluconazole dose of 400 mg/day will cover all strains with a fluconazole MIC of 4 mg/liter or less. The sensitivity of this target is 91%, with a false-positive rate of 10%, an area under the ROC curve of 0.90, and an MCC index of 0.80 (Table 2).
In summary, the fluconazole breakpoints (susceptible, ≤2 mg/liter; intermediate, 4 mg/liter; resistant, >4 mg/liter) determined by the EUCAST Antifungal Susceptibility Testing Subcommittee have been validated by means of machine learning methods. This proves that these computer tools must be incorporated into the process for developing breakpoints because such an approach completely avoids researcher bias, thus enhancing the statistical power of the model. The use of machine learning tools allows the evaluation of antimicrobial susceptibility results in a manner similar to that used with other diagnostic tools.
Acknowledgments
This work was supported in part by Ministerio de Sanidad y Consumo, Instituto de Salud Carlos III, Spanish Network of Infection in Transplantation (RESITRA G03/075), and by the Spanish Network for the Research in Infectious Diseases (REIPI RD06/0008).
In the past 5 years, M.C.-E. has received grant support from Astellas Pharma, BioMerieux, Gilead Sciences, Merck Sharp and Dohme, Pfizer, Schering Plough, Soria Melguizo SA, the European Union, the ALBAN program, the Spanish Agency for International Cooperation, the Spanish Ministry of Culture and Education, The Spanish Health Research Fund, The Instituto de Salud Carlos III, The Ramon Areces Foundation, and The Mutua Madrileña Foundation. He has been an advisor/consultant to the Panamerican Health Organization, Gilead Sciences, Merck Sharp and Dohme, Pfizer, and Schering Plough. He has been paid for talks on behalf of Gilead Sciences, Merck Sharp and Dohme, Pfizer, and Schering Plough. In the past 5 years, J.L.R.-T. has received grant support from Astellas Pharma, Gilead Sciences, Merck Sharp and Dohme, Pfizer, Schering Plough, Soria Melguizo SA, the European Union, the Spanish Agency for International Cooperation, the Spanish Ministry of Culture and Education, The Spanish Health Research Fund, The Instituto de Salud Carlos III, The Ramon Areces Foundation, and The Mutua Madrileña Foundation. He has been an advisor/consultant to the Panamerican Health Organization, Gilead Sciences, Merck Sharp and Dohme, Myconostica, Pfizer, and Schering Plough. He has been paid for talks on behalf of Gilead Sciences, Merck Sharp and Dohme, Pfizer, and Schering Plough. In the past 5 years, B.A. has received grant support from Gilead Sciences, Pfizer, and the Instituto de Salud Carlos III. He has been paid for talks on behalf of Gilead Sciences, Merck Sharp and Dohme, Pfizer, and Novartis. The other authors have no potential conflicts of interest to declare.
Footnotes
Published ahead of print on 11 May 2009.
REFERENCES
- 1.Almirante, B., D. Rodríguez, B. J. Park, M. Cuenca-Estrella, A. M. Planes, M. Almela, J. Mensa, F. Sanchez, J. Ayats, M. Gimenez, P. Saballs, S. K. Fridkin, J. Morgan, J. L. Rodriguez-Tudela, D. W. Warnock, and A. Pahissa. 2005. Epidemiology and predictors of mortality in cases of Candida bloodstream infection: results from population-based surveillance, Barcelona, Spain, from 2002 to 2003. J. Clin. Microbiol. 43:1829-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Breiman, L., J. H. Friedman, J. H. Olshen, and C. G. Stone. 1984. Classification and regression trees. Wadsworth International, Belmont, CA.
- 3.Cestnik, B., I. Kononenko, and I. Bratko. 1987. ASSISTANT-86: a knowledge elicitation tool for sophisticated users, p. 31-45. In: Progress in Machine Learning. Sigma Press, Ammanford, United Kingdom.
- 4.Holte, R. C. 1993. Very simple classification rules perform well on most commonly used datasets. Mach. Learn. 11:63-91. [Google Scholar]
- 5.Hosmer, D. W., and S. Lemeshow. 2000. Applied logistic regression. Wiley Interscience, New York, NY.
- 6.Laguna, F., J. L. Rodriguez-Tudela, J. V. Martinez-Suarez, R. Polo, E. Valencia, T. M. Diaz-Guerra, F. Dronda, and F. Pulido. 1997. Patterns of fluconazole susceptibility in isolates from human immunodeficiency virus-infected patients with oropharyngeal candidiasis due to Candida albicans. Clin. Infect. Dis. 24:124-130. [DOI] [PubMed] [Google Scholar]
- 7.Larrañaga, P., B. Calvo, R. Santana, C. Bielza, J. Galdiano, I. Inza, J. A. Lozano, R. Armananzas, G. Santafe, A. Perez, and V. Robles. 2006. Machine learning in bioinformatics. Brief. Bioinform. 7:86-112. [DOI] [PubMed] [Google Scholar]
- 8.Pfaller, M. A., and D. J. Diekema. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20:133-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quinlan, J. R. 1993. C4.5: programs for machine learning. Morgan Kaufmann Publishers Inc., San Francisco, CA.
- 10.Rex, J. H., and M. A. Pfaller. 2002. Has antifungal susceptibility testing come of age? Clin. Infect. Dis. 35:982-989. [DOI] [PubMed] [Google Scholar]
- 11.Rodriguez-Tudela, J. L., M. C. Arendrup, F. Barchiesi, J. Bille, E. Chryssanthou, M. Cuenca-Estrella, E. Dannaoui, D. W. Denning, J. P. Donnelly, F. Dromer, W. Fegeler, C. Lass-Florl, C. Moore, M. Richardson, P. Sandven, A. Velegraki, and P. Verweij. 2008. EUCAST definitive document EDef 7.1: method for the determination of broth dilution MICs of antifungal agents for fermentative yeasts. Clin. Microbiol. Infect. 14:398-405. [DOI] [PubMed] [Google Scholar]
- 12.Rodríguez-Tudela, J. L., B. Almirante, D. Rodriguez-Pardo, F. Laguna, J. P. Donnelly, J. W. Mouton, A. Pahissa, and M. Cuenca-Estrella. 2007. Correlation of the MIC and dose/MIC ratio of fluconazole to the therapeutic response of patients with mucosal candidiasis and candidemia. Antimicrob. Agents Chemother. 51:3599-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodríguez-Tudela, J. L., J. P. Donnelly, M. C. Arendrup, S. Arikan, F. Barchiesi, J. Bille, E. Chryssanthou, M. Cuenca-Estrella, E. Dannaoui, D. Denning, W. Fegeler, P. Gaustad, N. Klimko, C. Lass-Florl, C. Moore, M. Richardson, A. Schmalreck, J. Stenderup, A. Velegraki, and P. Verweij. 2008. EUCAST technical Note on fluconazole. Clin. Microbiol. Infect. 14:193-195. [DOI] [PubMed] [Google Scholar]
- 14.Sanders, C. C. 1991. ARTs versus ASTs—where are we going? J. Antimicrob. Chemother. 28:621-623. [DOI] [PubMed] [Google Scholar]
- 15.Witten, I. H., and E. Frank. 2005. Data mining: Practical machine learning tools and techniques. Elsevier, San Francisco, CA.