Abstract
Acinetobacter baumannii isolate KAR was uncommonly more resistant to cefepime and cefpirome than to ceftazidime and cefotaxime. Cloning and expression of the β-lactamase gene content of this isolate into Escherichia coli TOP10 identified ß-lactamase RTG-4 (or CARB-10), which corresponds to the first reported extended-spectrum CARB-type enzyme. RTG-4 is a plasmid-encoded Ambler class A β-lactamase whose sequence differs by 4 amino acid substitutions from the narrow-spectrum β-lactamase RTG-3. RTG-4 hydrolyzes cefepime and cefpirome and weakly hydrolyzes ceftazidime due to the single Ser-to-Thr substitution at Ambler position 69. RTG-4 is less susceptible to inhibition by tazobactam and sulbactam than RTG-3. Expression of β-lactamase RTG-4 in a wild-type A. baumannii reference strain showed that it conferred resistance to cefepime and cefpirome. The genetic environment of the blaRTG-4 gene was made of a peculiar transposon located on a ca. 50-kb plasmid. ISAba9, located upstream of blaRTG-4, may be responsible for its acquisition by recognizing a secondary right inverted repeat sequence, thus acting by a one-ended transposition process.
Acinetobacter baumannii is an opportunistic pathogen that is an important source of nosocomial infections such as pneumonia, septicemia, urinary tract infections, and wound infections (2). Treatment of infections due to this microorganism is becoming a serious clinical concern since A. baumannii is frequently resistant to multiple classes of antibiotics (23, 37). The main mechanism of resistance to β-lactam molecules in A. baumannii is the production of β-lactamases. Resistance to carbapenems is mostly related to the production of metallo-β-lactamases or carbapenem-hydrolyzing oxacillinases (33), whereas resistance to expanded-spectrum cephalosporins mostly results from the overexpression of the natural AmpC-type enzyme (3) or from the acquisition of extended-spectrum β-lactamases (ESBLs). Those ESBLs may correspond to TEM or SHV derivatives but mostly correspond to β-lactamases of the VEB or PER type in A. baumannii (21). Carbenicillin-hydrolyzing β-lactamases (also named CARB enzymes) are narrow-spectrum class A penicillinases that share less than 50% amino acid identity with SHV and TEM β-lactamases (7). The 10 ß-lactamase variants of this family show similar hydrolytic properties but are divided into two subgroups, named the CARB and RTG subgroups, according to their amino acid sequences. The CARB subgroup includes CARB-1 (formerly PSE-4) (18), CARB-2 (formerly PSE-1) (18), CARB-3 (14), CARB-4 (26), Proteus mirabilis N29 β-lactamase (11), CARB-6 (6), CARB-7 (20), and CARB-9 (25). The RTG subgroup includes RTG-1 (P. mirabilis GN79 enzyme) (34), RTG-2 (CARB-5; described in Acinetobacter calcoaceticus var. anitratus [24]), and RTG-3 (CARB-8; identified from an Oligella urethralis clinical isolate [17]). Although those three RTG-type ß-lactamases possess low levels of amino acid identity with members of the CARB family (44%), they might be considered the ancestors of that enzymatic group, as proposed by Choury et al. (7).
The carbenicillin-hydrolyzing enzymes belonging to the CARB group were first identified from Pseudomonas aeruginosa clinical isolates (38). The blaCARB genes are now widespread among distantly related bacteria, including Vibrio spp. (19), because of their location on mobile genetic structures. Indeed, blaCARB genes such as blaCARB-1 (22), blaCARB-2 (28), and blaCARB-4 (36) were found as a form of gene cassettes of class 1 integrons. In contrast, little is known about the genetic environment of the blaRTG genes.
Analysis of the β-lactamase content of an A. baumannii clinical isolate exhibiting an atypical higher level of resistance to cefepime and cefpirome than to ceftazidime and cefotaxime allowed us to identify a novel plasmid-encoded ESBL-encoding gene belonging to the blaRTG group.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The identification of A. baumannii KAR was performed by using the API 32GN system (bioMérieux, Marcy l'Etoile, France) and was confirmed by sequencing of the 16S rRNA gene, as described previously (8). O. urethralis COH-1 expressing the CARB-8 (RTG-3) β-lactamase was used as the control strain and has been characterized previously (17). Escherichia coli TOP10 was used for site-directed mutagenesis. Reference strains E. coli TOP10 and A. baumannii CIP70.10 were used as the hosts for cloning. Kanamycin resistance-conferring plasmid pBK-CMV (Stratagene, Amsterdam, The Netherlands) and rifampin (rifampicin) resistance-conferring broad-host-range plasmid pAT801-RA (10) were used as cloning vectors in E. coli and A. baumannii, respectively. Reference strains E. coli TOP10 and A. baumannii CIP70.10 were also used as the recipients for mating-out and electroporation assays.
Antimicrobial agents and MIC determinations.
The antimicrobial agents and their sources have been described elsewhere (27). Susceptibility testing was performed by the disk diffusion assay (Sanofi-Diagnostic Pasteur, Marnes-la-Coquette, France), as described previously (27). The MICs were determined by Etest (AB Biodisk, Solna, Sweden) on Mueller-Hinton agar plates at 37°C (28). The production of an ESBL was assessed by a synergy test with disks containing cefepime and ticarcillin-clavulanic acid.
Cloning experiments, recombinant plasmid analysis, and DNA sequencing.
Whole-cell DNA of A. baumannii KAR was extracted as described previously (27). HindIII- and XbaI-restricted DNA was ligated into the corresponding sites of plasmid pBK-CMV and then introduced into E. coli TOP10 by electroporation, as described previously (17). Recombinant plasmids were selected on Mueller-Hinton agar plates containing amoxicillin (amoxicilline; 50 μg/ml) and kanamycin (30 μg/ml). Recombinant plasmid pHindIII-RTG-4, which possessed a 2-kb HindIII insert, was retained for further biochemical analysis. Recombinant plasmid pXbaI-RTG-4, which possessed an 11-kb XbaI insert, was used to determine the blaRTG-4 genetic context. HindIII-restricted DNAs from A. baumannii KAR (expressing RTG-4) and O. urethralis COH-1 (expressing RTG-3) were ligated into the broad-host-range plasmid pAT801-RA, which gave rise to recombinant plasmids pAT-RTG-4 and pAT-RTG-3, respectively. The recombinant plasmids were transformed into electrocompetent A. baumannii CIP70.10. A. baumannii CIP70.10 harboring recombinant plasmids was selected on ticarcillin (50 μg/ml) and rifampin (30 μg/ml). Both strands of the cloned DNA inserts of the recombinant plasmids were sequenced by using an Applied Biosystems sequencer (ABI 377). The nucleotide and deduced protein sequences were analyzed with software available from the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/BLAST/).
Determination of transcription initiation sites by 5′-RACE.
Total RNA was isolated from A. baumannii KAR isolate with an RNeasy Midi kit (Qiagen, Courtaboeuf, France) by using the recommendations of the manufacturer. The 5′ rapid amplification of cDNA ends (5′-RACE) reactions were performed with 500 ng of total RNA of A. baumannii KAR and a 5′-RACE system kit (version 2.0; Invitrogen), following the recommendations of the manufacturer and by using specific primers Gsp1, Gsp2, and Gsp3 (Table 1).
TABLE 1.
Primers used in this study
| Primer | Experiment | Sequence (5′ to 3′) | Gene |
|---|---|---|---|
| RTG-A | PCR | CTCACGCTATCATTAAATGC | blaRTG |
| RTG-B | PCR | TCAAACGAGGCGTCTGTCTCTG | blaRTG |
| RTG-4-T69S-A | Site-directed mutagenesis | CGTTTTCCTCTAAGTAGTACCTTTAAAACACTTGCC | |
| RTG-4-T69S-B | Site-directed mutagenesis | GGCAAGTGTTTTAAAGGTACTACTTAGAGGAAAACG | |
| RTG-4-A49V-A | Site-directed mutagenesis | GAATTGGTCTAGCTGTGCATGATTTGGAAACG | |
| RTG-4-A49V-B | Site-directed mutagenesis | CGTTTCCAAATCATGCACAGCTAGACCAATTC | |
| P13B | Gene rrn (16S rRNA) | CGGGATCCCAGGCCCGGGAAC | rRNA |
| UNI14 | Gene rrn (16S rRNA) | GTGCCAGCAGCCGCGGTAAT | rRNA |
| Gsp1 | 5′-RACE | TACAGGTGAGTATGTAACGAGATTGC | |
| Gsp2 | 5′-RACE | TTCCCGTTTCCAAATCATGCGCAGC | |
| Gsp3 | 5′-RACE | TTCAGCATTGGTAACCGCTTC |
IEF analysis.
The β-lactamase extracts from cultures of A. baumannii KAR and E. coli TOP10 harboring the plasmid pHindIII-RTG-4 were subjected to analytical isoelectric focusing (IEF) analysis, as described previously (17).
β-Lactamase purification.
Cultures of E. coli TOP10 harboring recombinant plasmid pHindIII-RTG-4 were grown overnight at 37°C in 2 liters of tryptic soy broth containing amoxicillin (50 μg/ml) and kanamycin (30 μg/ml). β-Lactamase RTG-4 was purified by ion-exchange chromatography. Briefly, the bacterial suspension was pelleted; resuspended in 40 ml of 100 mM sodium phosphate buffer (pH 7.0); and then sonicated, cleared by ultracentrifugation, and treated with DNase. The extract was then dialyzed against 20 mM Tris-HCl buffer (pH 7.5) and loaded onto a preequilibrated Q-Sepharose column in 20 mM Tris-HCl buffer (pH 7.5). The β-lactamase-containing fractions were eluted with a linear NaCl gradient (0 to 500 mM). The same procedure was repeated with a 20 mM bis-Tris buffer (pH 6.3). Finally, fractions containing the highest β-lactamase activities were pooled and subsequently dialyzed overnight against 100 mM phosphate buffer (pH 7.0). The β-lactamase activity was determined qualitatively by nitrocefin hydrolysis (Oxoid, Dardilly, France). The protein content was measured by the Bio-Rad DC protein assay.
Kinetic studies.
Kinetic measurements (kcat and Km) of purified β-lactamase RTG-4 were performed as described previously (30). The 50% inhibitory concentrations (IC50) were determined for RTG-3 and RTG-4 as the concentrations of clavulanate, tazobactam, or sulbactam that reduced the rate of hydrolysis of 100 μM benzylpenicillin by 50% under conditions in which RTG-3 and RTG-4 were preincubated with various concentrations of inhibitor for 10 min at 30°C before addition of the substrate.
Site-directed mutagenesis.
The RTG-4 β-lactamase that was identified contained a threonine residue at Ambler position 69; this residue replaced a serine residue identified in RTG-3. In order to analyze the role of this substitution, a site-directed mutagenesis protocol was used as described by the manufacturer (QuikChange site-directed mutagenesis kit; Stratagene). Recombinant plasmid pHindIII-RTG-4 was used as the template in the PCR amplification with primers RTG-4-T69S-A and RTG-4-T69S-B (Table 1) to generate recombinant plasmid pRTG4-S69. Similarly, the Ala residue at position 49 in RTG-4 was replaced by a Val residue by using primers RTG-4-A49V-A and RTG-4-A49V-B (Table 1). Recombinant plasmids pHindIII-RTG-4 and pRTG4-S69 were used as the templates, and recombinant plasmids pRTG4-V49 and pRTG4-S69-V49, respectively, were obtained.
Plasmid content, hybridizations, and mating-out assays.
Plasmid DNA of A. baumannii KAR was extracted by using the method of Kieser (13). E. coli NCTC50192 harboring four plasmids of 154, 66, 48, and 7 kb was used as the size marker for plasmids. Plasmid DNAs were analyzed by agarose gel electrophoresis, as described previously (29). In order to search for a possible chromosomal location of the blaRTG-4 gene, restriction with endonuclease I-CeuI (Ozyme; New England Biolabs, Saint-Quentin-en Yvelines, France) was performed as described previously (15). Both gels containing plasmid DNAs and I-CeuI-restricted DNA fragments were transferred to a nylon membrane (Hybond N+; GE Healthcare, Orsay, France) by the Southern technique (35). The DNAs were then UV cross-linked (Stratalinker; Stratagene). A 758-bp PCR-generated probe (primers RTG-A and RTG-B [Table 1]) was used for hybridization of the genes encoding the RTG group of ß-lactamases. The I-CeuI-restricted fragments were also hybridized with a PCR-generated probe for rRNA genes (Table 1).
The direct transfer of the ticarcillin resistance marker into A. baumannii CIP70.10 was attempted by liquid mating-out assays at 37°C and by electroporation of a plasmid DNA suspension extracted from A. baumannii KAR into A. baumannii CIP70.10 and into E. coli TOP10. Selection was performed on agar plates supplemented with ticarcillin (50 μg/ml).
Nucleotide sequence accession number.
The nucleotide sequence data reported in this work has been deposited in the GenBank nucleotide database under accession no. EU850412.
RESULTS AND DISCUSSION
Susceptibility testing.
A. baumannii KAR was isolated from a wound of a 31-year-old patient previously hospitalized in Morocco and then transferred to the surgery unit at the Grenoble University Hospital in September 2007. A. baumannii KAR showed resistance to penicillins and β-lactamase inhibitor (clavulanic acid and tazobactam)-penicillin combinations and decreased susceptibility to cefepime, cefpirome, cefotaxime, and ceftazidime (Table 2). Clear synergy between cefepime and clavulanic acid and atypical higher MICs of cefepime and cefpirome compared to those of ceftazidime and cefotaxime were observed, suggesting the production of a peculiar ESBL.
TABLE 2.
MICs of β-lactams for A. baumannii KAR, A. baumannii CIP70.10(pAT-RTG-4), A. baumannii CIP70.10(pAT-RTG-3), and reference strain A. baumannii CIP70.10
| β-Lactama | MIC (μg/ml)
|
|||
|---|---|---|---|---|
| A. baumannii KAR | A. baumannii CIP70.10(pAT-RTG-4) | A. baumannii CIP70.10(pAT-RTG-3) | A. baumannii CIP70.10 | |
| Amoxicillin | >256 | >256 | >256 | 32 |
| Amoxicillin + CLA | 32 | 32 | 32 | 8 |
| Ticarcillin | >256 | >256 | >256 | 8 |
| Ticarcillin + CLA | >256 | >256 | >256 | 8 |
| Piperacillin | >256 | >256 | >256 | 16 |
| Piperacillin + TZB | >256 | >256 | 128 | 4 |
| Ceftazidime | 8 | 16 | 4 | 2 |
| Cefotaxime | 16 | 16 | 8 | 8 |
| Cefepime | 32 | 64 | 4 | 1 |
| Cefpirome | 32 | 64 | 2 | 2 |
| Imipenem | 0.25 | 0.25 | 0.25 | 0.25 |
CLA, clavulanic acid (4 μg/ml); TZB, tazobactam (4 μg/ml).
Cloning and sequencing of β-lactamase gene.
Cloning of the β-lactam resistance markers from A. baumannii resulted in E. coli TOP10 harboring recombinant plasmid pHindIII-RTG-4. This recombinant E. coli strain was resistant to penicillins and cephalothin (cefalotin) and showed decreased susceptibility to ceftazidime, cefepime, and cefpirome (Table 3). Clavulanic acid was not able to restore the activities of ticarcillin and amoxicillin, whereas synergy between clavulanate- and cefpirome-containing disks was observed. DNA sequence analysis of the 2-kb insert of pHindIII-RTG-4 revealed an open reading frame (ORF) of 897 bp encoding a 298-amino-acid protein that had 4 amino acid substitutions compared to the amino acid sequence of the Ambler class A β-lactamase RTG-3 from Oligella urethralis COH-1 (Fig. 1). This β-lactamase was designated RTG-4.
TABLE 3.
MICs of β-lactams for E. coli TOP10(pHindIII-RTG-4), E. coli TOP10(pRTG4-S69), E. coli TOP10(pRTG4-V49), E. coli TOP10(pRTG4-S69-V49), E. coli TOP10(pRTG-3), and E. coli TOP10 reference strain
| β-Lactam | MIC (μg/ml)
|
|||||
|---|---|---|---|---|---|---|
| E. coli TOP10(pHindIII-RTG-4) | E. coli TOP10(pRTG4-S69) | E. coli TOP10(pRTG4-V49) | E. coli TOP10(pRTG4-S69-V49) | E. coli TOP10(pRTG-3) | E. coli TOP10 | |
| Amoxicillin | >256 | >256 | >256 | >256 | 512 | 2 |
| Amoxicillin + CLAa | 32 | 32 | 32 | 32 | 4 | 2 |
| Ticarcillin | >256 | >256 | >256 | >256 | >512 | 2 |
| Ticarcillin + CLA | >256 | >256 | >256 | >256 | 16 | 2 |
| Piperacillin | >256 | >256 | >256 | >256 | 32 | 1 |
| Cephalothin | 64 | 8 | 64 | 8 | 4 | 4 |
| Cefuroxime | 4 | 2 | 2 | 2 | 2 | 2 |
| Cefotaxime | 0.25 | 0.25 | 0.25 | 0.25 | 0.06 | 0.06 |
| Ceftazidime | 1 | 0.5 | 1 | 0.5 | 0.12 | 0.12 |
| Cefepime | 4 | 0.5 | 4 | 0.5 | 0.06 | 0.06 |
| Cefpirome | 1 | 0.25 | 1 | 0.25 | 0.06 | 0.06 |
| Imipenem | 0.12 | 0.12 | 0.12 | 0.12 | 0.12 | 0.12 |
CLA, clavulanic acid (4 μg/ml).
FIG. 1.
Alignment of the amino acid sequences of the RTG-4 and RTG-3 β-lactamases. Dashes represent identical amino acids. Amino acids with gray shading represent conserved residues across multiple β-lactamases, according to Joris et al. (12).
Sequence analysis of the RTG-4 β-lactamase.
Analysis of the deduced amino acid sequence showed that RTG-4 contained the conserved motif of serine β-lactamases, S-T-F-K, at positions 70 to 73, according to the standard numbering scheme of Ambler (1), and consensual boxes I to VI described by Joris et al. were conserved (12) (Fig. 1). Amino acids of the consensus sequence of box VII of the Ambler class A β-lactamases at positions 234 to 236 consisted of an RTG triad. The RTG-4 β-lactamase is another representative of a group of CARB-type β-lactamases known as the RTG group. The RTG group includes RTG-1 from P. mirabilis GN79 (34), RTG-2 (CARB-5) from A. calcoaceticus var. anitratus (24), RTG-3 (CARB-8) (17), and now RTG-4 (which should be also defined as CARB-10). In the members of the RTG group, the typical KTG motif of class A, C, and D β-lactamases is replaced by an RTG motif. Pairwise alignment of the RTG-4 and RTG-3 amino acid sequences revealed 4 amino acid substitutions in the coding region (Lys to Gln at position 5, Val to Ala at position 49, Ser to Thr at position 69, and Ile to Val at position 290, according to the nomenclature of Ambler et al. [1]) (Fig. 1).
Biochemical properties of RTG-4 and RTG-3.
IEF analysis of a β-lactamase extract of A. baumannii KAR and E. coli TOP10(pHindIII-RTG-4) revealed a pI value of 6.4 (data not shown), as has been already described for RTG-3 (17). The values of the kinetic parameters of purified RTG-4 β-lactamase indicated that its hydrolytic profile includes benzylpenicillin, amoxicillin, ticarcillin, piperacillin, cephalothin, cefepime, and cefpirome (Table 4). Surprisingly, cefepime and cefpirome were hydrolyzed, even at low levels, whereas no hydrolysis of cefotaxime or aztreonam was detected. No hydrolysis of imipenem was detected (Table 4). Interestingly, RTG-4 represents the first CARB-type β-lactamase for which ESBL properties have been shown.
TABLE 4.
Kinetic parameters for β-lactamase RTG-4a
| Substrate | kcat (s−1) | Relative Vm (%)b | Km (μM) | kcat/Km (μM−1 s−1) | Relative Vm/Km (%)b |
|---|---|---|---|---|---|
| Benzypenicillin | 20 | 100 | 10 | 2 | 100 |
| Amoxicillin | 30 | 175 | 15 | 2 | 120 |
| Ticarcillin | 15 | 94 | 10 | 1.5 | 100 |
| Piperacillin | 5 | 31 | 10 | 0.5 | 35 |
| Cephalothin | 0.3 | 1.6 | 50 | 0.006 | 0.04 |
| Cefuroxime | <0.01 | <0.01 | NHc | ||
| Cefotaxime | <0.01 | <0.01 | NH | ||
| Ceftazidime | 0.1 | 0.6 | NDd | ||
| Cefepime | >1 | >5 | >1,000 | 0.001 | 0.06 |
| Cefpirome | 0.15 | 0.9 | 300 | 0.0005 | 0.03 |
| Aztreonam | <0.01 | <0.01 | NH | ||
| Imipenem | <0.01 | <0.01 | NH |
Data are the means of three independent experiments. Standard deviations were within 10% of the means.
Values are relative to those for benzylpenicillin, which were set equal to 100.
NH, no detectable hydrolysis was observed with 1 μM of purified RTG-4 and up to 500 μM of substrate.
ND, not determinable due to a Km value that was too high.
Inhibition studies, carried out by measuring the IC50s, showed that the activities of RTG-4 and RTG-3 were similarly inhibited by clavulanic acid (Table 5). Therefore, the resistance to clavulanic acid-penicillin combinations by an RTG-4-producing strain may be due to the high level of expression of RTG-4 rather than to the reduced susceptibility of the enzyme to clavulanic acid. However, the IC50s of tazobactam and sulbactam were higher for RTG-4 than for RTG-3 (Table 5), suggesting that RTG-4 might be more resistant to those inhibitors than RTG-3. This is in accordance with what has been observed for inhibitor-resistant TEM-type β-lactamases, for which a single amino acid substitution at position 69 is responsible for resistance to inhibitors (5).
TABLE 5.
IC50s of clavulanic acid, tazobactam, and sulbactam for β-lactamases RTG-3 and RTG-4a
| Inhibitor | IC50 (μM)
|
|
|---|---|---|
| RTG-3 | RTG-4 | |
| Clavulanic acid | 0.1 | 0.07 |
| Tazobactam | 0.1 | 0.35 |
| Sulbactam | 0.2 | 1.5 |
Data are the means of three independent experiments. Standard deviations were within 10% of the means.
Site-directed mutagenesis and consequences for MICs.
Among the four amino acid substitutions of RTG-4 (Lys5Gln, Val49Ala, Ser69Thr, and Ile290Val), two, Val49Ala and Ser69Thr, could have been involved in the resistance profile conferred by this enzyme. Therefore, the role of the Thr residue at position 69, immediately adjacent to the active serine site, was first investigated by generating plasmid pRTG4-S69, which contained the exact same insert as pHindIII-RTG-4 except for a C-to-G change that led to the Thr-to-Ser change at amino acid position 69. Comparison of the β-lactam susceptibility profile of E. coli TOP10(pHindIII-RTG-4) and E. coli TOP10(pRTG4-S69) showed lower MICs of cephalothin, ceftazidime, cefepime, and cefpirome for E. coli TOP10(pRTG4-S69) (Table 3). This substitution was thus at least in part responsible for the extended hydrolysis spectrum of RTG-4. Then, to evaluate the possible additional involvement of the Ala residue at position 49, we performed an Ala-to-Val substitution at position 49. E. coli TOP10(pHindIII-RTG-4) and E. coli TOP10(pRTG4-S69) were used as templates. The MICs for recombinant strains E. coli TOP10(pRTG4-V49) and E. coli TOP10(pRTG4-S69-V49) were compared (Table 3). The MICs of β-lactams for E. coli TOP10(pHindIII-RTG-4) and E. coli TOP10(pRTG4-V49) were identical, indicating that the substitution at position 49 did not play a role in the RTG-4 hydrolysis profile. The MICs for E. coli TOP10(pRTG4-S69) and E. coli TOP10(pRTG4-S69-V49) were also identical, confirming the role of the Ser-to-Thr substitution at position 69. As has already been observed for TEM- and SHV-type β-lactamases, β-lactamases of the RTG group may extend their substrate profile toward expanded-spectrum cephalosporins by a single amino acid substitution.
Expression of the blaRTG-4 and blaRTG-3 genes in A. baumannii.
In order to compare the expression of RTG-4 and RTG-3 in A. baumannii, cloning of the corresponding genes was achieved by using a broad-host-range plasmid subsequently transformed into wild-type reference strain A. baumannii CIP70.10. The MICs for the recombinant strains were then compared (Table 2). Our results showed an increased level of resistance to expanded-spectrum cephalosporins in A. baumannii CIP70.10(pAT-RTG-4), especially to cefepime and cefpirome. In contrast, RTG-3 expression in A. baumannii CIP70.10 did not lead to decreased susceptibility to expanded-spectrum cephalosporins and mirrored the narrow-spectrum resistance profile of the RTG-3 ß-lactamase.
Genetic environment of the blaRTG-4 gene.
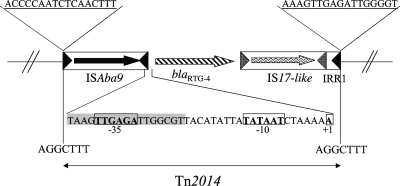
Sequencing of the entire XbaI insert of recombinant plasmids revealed several ORFs. Analysis of the sequences located upstream of the blaRTG-4 gene identified an insertion sequence (IS) named ISAba9 in the same orientation. ISAba9 is an IS that belongs to the IS982 family (16), which has recently been characterized to be associated with the blaOXA-51 gene in A. baumannii (9). ISAba9 possesses two imperfect inverted repeat (IRs) of 17 bp with 2 mismatches. Downstream of the blaRTG-4 gene, an IS17-like IS was identified. Its transposase shared 98% amino acid identity with that of IS17 that was identified in Acinetobacter haemolyticus and that belongs to the IS5 family. Downstream of this IS17 isoform, a 31-bp sequence was identified that matched the left IR (IRL) of ISAba9 and that was consequently named right IR 1 (IRR1). In fact, 17 of those 31 bp corresponded to the IRL of ISAba9 (Fig. 2), with the other 14 bp constituting a perfect inverted copy of the 14-bp sequence immediately close to the IRL of ISAba9. The identification of a target site duplication at the left-hand extremity of ISAba9 and at the right-hand extremity of IRR1 was likely the signature of a transposition event. It led us to identify a new transposon named Tn2014, which comprised ISAba9, blaRTG-4, an IS17-like element, and IRR1. The direct repeat (DR) sequences generated by transposon insertion were 7 bp (Fig. 2). As the ISAba9 IRL perfectly matched IRR1, we hypothesized that ISAba9 could be responsible for the mobilization of the overall structure that included blaRTG-4 by a transposition process involving the IRL and IRR1 sequences. IRR1 could be recognized more easily by the transposase of ISAba9 since IRR1 and the IRL of ISAba9 are perfect IRs, whereas the IRL and the IRR of ISAba9 are imperfect ones.
FIG. 2.
Schematic map of transposon Tn2014 harboring the blaRTG-4 gene. The nucleotide sequences of regions identified upstream of the blaRTG-4 gene in A. baumannii KAR are indicated. The genes and their corresponding transcriptional orientations are indicated by horizontal arrows. The +1 transcription start site and the −35 and −10 sequences of the promoter are indicated in boldface and are boxed. The IRR of ISAba9 is shaded in gray.
By using the 5′-RACE technique, the sites of the initiation of transcription of the blaRTG-4 gene were mapped (Fig. 2). The +1 transcription start site was located 29 bp upstream of the start codon of blaRTG-4 gene. Upstream of this transcriptional start site, a −35 sequence (TTGAGA) separated by 16 bp from a −10 sequence (TATAAT) constituted a promoter. The −35 sequence belonged to the ISAba9 sequence, and the −10 sequence might correspond to that of its original promoter. Thus, ISAba9 contributes to the expression of the blaRTG-4 gene in A. baumannii clinical isolate KAR by bringing a −35 sequence and thus creating a hybrid promoter.
We describe here a new type of transposon made of a single antibiotic resistance-conferring gene, two different ISs, and an additional sequence used as an IR. Our findings suggest that only a single transposase (likely that of ISAba9, which is already known to generate 7-bp target site duplications) was at the origin of the mobility of that transposon. That kind of transposition involving only a single IS element and a second IRR-like sequence has already been well documented with ISEcp1 at the origin of mobilization of blaCTX-M genes but also of the qnrB19 gene (4, 31, 32). Sequencing of the upstream and downstream regions surrounding the Tn2014 sequence identified several ORFs encoding different proteins that share high degrees of identity with several ORFs of A. baumannii, which might suggest a chromosomal location for the blaRTG-4 gene.
Genetic support of blaRTG-4 gene.
Plasmid DNA analysis showed that A. baumannii KAR harbored a 50-kb plasmid (data not shown). Conjugation experiments failed to transfer any plasmid encoding the RTG-4 β-lactamase from A. baumannii KAR to the A. baumannii CIP70.10 recipient strain. Attempts to transfer a blaRTG-4-carrying plasmid into A. baumannii or E. coli TOP10 recipient strains by electroporation also remained unsuccessful.
In order to determine the genetic location of blaRTG-4, we also used the endonuclease I-CeuI technique. Three fragments were generated from A. baumannii KAR. The DNA probe for rRNA genes hybridized with the I-CeuI-generated fragments, whereas the blaRTG-4-specific probe did not hybridize with any of the DNA fragments (data not shown). In addition, the plasmid extracted from A. baumannii KAR hybridized with the blaRTG-4-specific probe, likely indicating that the blaRTG-4 gene is located on that 50-kb plasmid (data not shown). This result does not fit with the analysis of the sequences surrounding Tn2014, suggesting a chromosomal location for this transposon.
Conclusion.
This study characterized a new type of ESBL from an A. baumannii clinical isolate. RTG-4 exhibits strong amino acid identity with the RTG group of enzymes, notably, RTG-3. Interestingly, a single amino acid change located just near the serine residue of the catalytic site confers hydrolytic activity against cefepime and cefpirome. This is the very first description of a β-lactamase with ESBL properties among the CARB group. However, further work may evaluate if any of the previously identified CARB-type enzymes might not in fact have similar ESBL activity. Indeed, in many cases, hydrolytic activities toward cefepime and cefpirome, which were not available at that time, have not been evaluated. However, none of the previously reported RTG-type enzymes possessed a similar threonine residue at position 69.
In contrast to other RTG β-lactamases, which are encoded by the chromosome, the plasmidic location of blaRTG-4 may facilitate its spread. Analysis of the surrounding sequences of the blaRTG-4 gene showed that it was part of a peculiar transposon containing two different ISs, ISAba9 and an IS17-like IS. We therefore hypothesized that ISAba9 was responsible for the mobilization of that transposon. However, further in vitro experiments will be required to establish the conditions and the frequency of those transposition events, as well as the putative role of the IS17-like element. Interestingly, we have identified an IS that is located just downstream of the blaRTG-4 gene and that possesses significant identity with the IS17 element described for A. haemolyticus. Together with the fact that ISAba9 has been identified in Acinetobacter genomospecies 9 and that the GC content of blaRTG-4 (42%) corresponds to that of the genus Acinetobacter, this would suggest that blaRTG-4 originates from an Acinetobacter-like species.
Acknowledgments
This work was funded by a grant from the Ministère de l'Education Nationale et de la Recherche (grant UPRES-EA3539), Université Paris XI, Paris, France; by a grant from the European Community (6th PCRD, LSHM-CT-2005-018705); and mostly by the INSERM, France.
Footnotes
Published ahead of print on 20 April 2009.
REFERENCES
- 1.Ambler, R. P., A. F. W. Coulson, J.-M. Frère, J. M. Ghuysen, B. Joris, M. Forsman, R. C. Lévesque, G. Tirabi, and S. G. Waley. 1991. A standard numbering scheme for the class A β-lactamases. Biochem. J. 276:269-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergogne-Bérezin, E., and K. J. Towner. 1996. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin. Microbiol. Rev. 9:148-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonomo, R. A., and D. Szabo. 2006. Mechanisms of multidrug resistance in Acinetobacter species and Pseudomonas aeruginosa. Clin. Infect. Dis. 43(Suppl. 2):S49-S56. [DOI] [PubMed] [Google Scholar]
- 4.Cattoir, V., P. Nordmann, J. Silva-Sanchez, P. Espinal, and L. Poirel. 2008. ISEcp1-mediated transposition of qnrB-like gene in Escherichia coli. Antimicrob. Agents Chemother. 52:2929-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaïbi, E. B., D. Sirot, G. Paul, and R. Labia. 1999. Inhibitor-resistant TEM ß-lactamases: phenotypic, genetic and biochemical characteristics. J. Antimicrob. Chemother. 43:447-458. [DOI] [PubMed] [Google Scholar]
- 6.Choury, D., G. Aubert, M.-F. Szajnert, K. Azibi, M. Delpech, and G. Paul. 1999. Characterization and nucleotide sequence of CARB-6, a new carbenicillin-hydrolyzing β-lactamase from Vibrio cholerae. Antimicrob. Agents Chemother. 43:297-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choury, D., M.-F. Szajnert, M.-L. Joly-Guillou, K. Azibi, M. Delpech, and G. Paul. 2000. Nucleotide sequence of the blaRTG-2 (CARB-5) gene and phylogeny of a new group of carbenicillinases. Antimicrob. Agents Chemother. 44:1070-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dortet, L., P. Legrand, C. J. Soussy, and V. Cattoir. 2006. Bacterial identification, clinical significance, and antimicrobial susceptibilities of Acinetobacter ursingii and Acinetobacter schindleri, two frequently misidentified opportunistic pathogens. J. Clin. Microbiol. 44:4471-4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Figueiredo, S., L. Poirel, A. Papa, V. Koulourida, and P. Nordmann. 2008. Nosocomial dissemination of carbapenem-resistant Acinetobacter spp. isolates in a general hospital in Thessaloniki, Greece. Clin. Microbiol. Infect. 14(Suppl. 7):s434-s435. [Google Scholar]
- 10.Héritier, C., L. Poirel, T. Lambert, and P. Nordmann. 2005. Contribution of acquired carbapenem-hydrolyzing oxacillinases in carbapenem resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:3198-3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ito, Y., and T. Hirano. 1997. Carbenicillin-hydrolysing penicillinase mediated by a plasmid of Proteus mirabilis and its relationship to the PSE-type enzymes of Pseudomonas aeruginosa. J. Appl. Microbiol. 83:175-180. [DOI] [PubMed] [Google Scholar]
- 12.Joris, B., J. Ghuysen, G. Dive, A. Renard, O. Dideberg, P. Charlier, J.-M. Frère, J. Kelly, J. Boyington, P. Moews, and J. Knox. 1988. The active-site-serine penicillin-recognizing enzymes as members of the Streptomyces R61 dd-peptidase family. Biochem. J. 250:313-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kieser, T. 1984. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid 12:19-36. [DOI] [PubMed] [Google Scholar]
- 14.Lachapelle, J., J. Dufresne, and R. C. Levesque. 1991. Characterization of the blaCARB-3 gene encoding the carbenicillinase-3 ß-lactamase of Pseudomonas aeruginosa. Gene 102:7-12. [DOI] [PubMed] [Google Scholar]
- 15.Liu, S. L., A. Hessel, and K. E. Sanderson. 1993. Genomic mapping with I-CeuI, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc. Natl. Acad. Sci. USA 90:6874-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mammeri, H., L. Poirel, N. Mangeney, and P. Nordmann. 2003. Chromosomal integration of a cephalosporinase gene from Acinetobacter baumannii into Oligella urethralis as a source of acquired resistance to β-lactams. Antimicrob. Agents Chemother. 47:1536-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matthew, M. 1979. Plasmid-mediated β-lactamases of gram-negative bacteria: properties and distribution. J. Antimicrob. Chemother. 5:349-358. [DOI] [PubMed] [Google Scholar]
- 19.Medeiros, A. A., R. W. Hedges, and G. A. Jacoby. 1982. Spread of a “Pseudomonas-specific” β-lactamase to plasmids of enterobacteria. J. Bacteriol. 149:700-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Melano, R., A. Petroni, A. Garutti, H. A. Saka, L. Mange, F. Pasteran, M. Rapoport, A. Rossi, and M. Galas. 2002. New carbenicillin-hydrolyzing β-lactamase (CARB-7) from Vibrio cholerae non-O1, non-O139 strains encoded by the VCR region of the V. cholerae genome. Antimicrob. Agents Chemother. 46:2162-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naas, T., L. Poirel, and P. Nordmann. 2008. Minor extended-spectrum ß-lactamases. Clin. Microbiol. Infect. 14:42-52. [DOI] [PubMed] [Google Scholar]
- 22.Partridge, S. R., H. J. Brown, and R. M. Hall. 2002. Characterization and movement of the class 1 integron known as Tn2521 and Tn1405. Antimicrob. Agents Chemother. 46:1288-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paterson, D. L. 2006. The epidemiological profile of infections with multidrug-resistant Pseudomonas aeruginosa and Acinetobacter species. Clin. Infect. Dis. 43:S43-S48. [DOI] [PubMed] [Google Scholar]
- 24.Paul, G., M. L. Joly-Guillou, E. Bergogne-Berezin, P. Névot, and A. Philippon. 1989. Novel carbenicillin-hydrolyzing β-lactamase (CARB-5) from Acinetobacter calcoaceticus var. anitratus. FEMS Microbiol. Lett. 59:45-50. [DOI] [PubMed] [Google Scholar]
- 25.Petroni, A., R. G. Melano, H. A. Saka, A. Garutti, L. Mange, F. Pasteran, M. Rapoport, M. Miranda, D. Faccone, A. Rossi, P. S. Hoffman, and M. Galas. 2004. CARB-9, a carbenicillinase encoded in the VCR region of Vibrio cholerae non-O1, non-O139 belongs to a family of cassette-encoded β-lactamases. Antimicrob. Agents Chemother. 48:4042-4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Philippon, A., G. Paul, A. Thabaut, and G. Jacoby. 1986. Properties of a novel carbenicillin-hydrolyzing β-lactamase (CARB-4) specified by an IncP-2 plasmid from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 29:519-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Philippon, L. N., T. Naas, A. T. Bouthors, V. Barakett, and P. Nordmann. 1997. OXA-18, a class D clavulanic acid-inhibited extended-spectrum β-lactamase from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 41:2188-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poirel, L., M. Guibert, S. Bellais, T. Naas, and P. Nordmann. 1999. Integron- and carbenicillinase-mediated reduced susceptibility to amoxicillin-clavulanic acid in isolates of multidrug-resistant Salmonella enterica serotype Typhimurium DT104 from French patients. Antimicrob. Agents Chemother. 43:1098-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poirel, L., T. Naas, M. Guibert, E. B. Chaibi, R. Labia, and P. Nordmann. 1999. Molecular and biochemical characterization of VEB-1, a novel class A extended-spectrum ß-lactamase encoded by an Escherichia coli integron gene. Antimicrob. Agents Chemother. 43:573-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poirel, L., D. Girlich, T. Naas, and P. Nordmann. 2001. OXA-28, an extended spectrum variant of OXA-10 β-lactamase from Pseudomonas aeruginosa and its plasmid- and integron-located gene. Antimicrob. Agents Chemother. 45:447-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poirel, L., J.-W. Decousser, and P. Nordmann. 2003. Insertion sequence ISEcp1B is involved in expression and mobilization of a blaCTX-M ß-lactamase gene. Antimicrob. Agents Chemother. 47:2938-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poirel, L., M. F. Lartigue, J.-W. Decousser, and P. Nordmann. 2005. ISEcp1B-mediated transposition of blaCTX-M in Escherichia coli. Antimicrob. Agents Chemother. 49:447-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poirel, L., and P. Nordmann. 2006. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin. Microbiol. Infect. 12:826-836. [DOI] [PubMed] [Google Scholar]
- 34.Sakurai, Y., K. Tsukamoto, and T. Sawai. 1991. Nucleotide sequence and characterization of a carbenicillin-hydrolyzing penicillinase gene from Proteus mirabilis. J. Bacteriol. 173:7038-7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 36.Sanschagrin, F., N. Bejaoui, and R. C. Levesque. 1998. Structure of CARB-4 and AER-1 carbenicillin-hydrolyzing β-lactamases. Antimicrob. Agents Chemother. 42:1966-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Looveren, M., H. Goossens, and the ARPAC Steering Group. 2004. Antimicrobial resistance in Acinetobacter spp. in Europe. Clin. Microbiol. Infect. 10:684-704. [DOI] [PubMed] [Google Scholar]
- 38.Williams, R. J., D. M. Livermore, M. A. Lindridge, A. A. Said, and J. D. Williams. 1984. Mechanisms of ß-lactam resistance in British isolates of Pseudomonas aeruginosa. J. Med. Microbiol. 17:283-293. [DOI] [PubMed] [Google Scholar]