Abstract
A deficiency of the Escherichia coli Lon protease blocked paradoxical survival occurring at very high nalidixic acid concentrations. The absence of Lon also blocked a parallel increase in cell lysate viscosity likely to reflect DNA size. Thus, Lon may participate in repairing quinolone-mediated DNA lesions formed at high drug concentrations.
The quinolones are lethal antibacterials that form drug-gyrase/topoisomerase IV-DNA complexes in which DNA is broken. Release of DNA ends from protein-mediated constraint allows chromosome fragmentation (1, 9). An unexplained aspect of quinolone action is a paradoxical increase in survival occurring at very high drug concentrations (2). To explore protease involvement in paradoxical survival, we treated Escherichia coli with nalidixic acid and examined effects of lon mutations on survival and on the presence of chromosomal DNA lesions, the latter assayed by an empirical measure of cell lysate viscosity. The Lon protease is known to degrade damaged proteins and proteins produced in excess (15); it also serves as a chaperone (6, 15). However, a role in chromosome maintenance has not been reported. In the present work a deficiency of Lon protease activity lowered survival at high concentrations of nalidixic acid with little effect at maximal bactericidal concentration.
E. coli K-12 strains, listed in Table 1, were grown in Luria-Bertani (LB) liquid medium and on LB agar plates (10) at 30°C; nalidixic acid was obtained from Sigma Chemical (St. Louis, MO). The effect of nalidixic acid was monitored by measuring lethal action and cell lysate viscosity (9). Lethal action was assayed by incubating exponentially growing cultures at 30°C for 180 min with nalidixic acid followed by drug removal using brief centrifugation, dilution, and enumeration of CFU by incubation on drug-free agar. Data were expressed relative to an untreated control taken at the time of drug addition. For viscosity, cell lysates were prepared as for isolation of bacterial nucleoids (cells were harvested by centrifugation, treated with lysozyme, and incubated for several minutes at 20°C with nonionic detergents [9]), and then sodium dodecyl sulfate was added to 0.8% to release DNA breaks from constraint by protein. Viscosity was estimated by measuring the time required to fill a 25-μl glass capillary (9) at several DNA concentrations to estimate specific viscosity.
TABLE 1.
Bacterial strains
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| DM4100 | Wild type | 13 |
| KD2140 | DM4100 gyrA67 (A67S) gyrB225 | This work by transduction (17) from strain KD1911 (9) and gyrB225 from strain SD104-20 (3); large-colony varianta |
| KD2372 | DM4100 lon-100 | This work by transduction (17) from strain SG20252 (14) |
| KD2377 | KD2372 (lon-100) sulA3 | This work by transduction (17) from strain KL789 (11) (CGSC7699b) |
| KD2510 | DM4100 sulA3 | This work by transduction (17) from strain KL789 (11) (CGSC7699b) |
| KD3037 | KD2377 pBAD24 | 16c |
| KD3039 | KD2377 pBAD24lon S679A | 16c |
| KD3041 | KD2377 pBAD24lon K362Q | 16c |
| KD3045 | KD2377 pBAD24lon+ | 16c |
Chloramphenicol blocks lethal action of nalidixic acid with wild-type cells but not with gyrA67 mutants; large-colony variants show a strong paradoxical effect.
E. coli Stock Center collection.
Plasmids were introduced by bacterial transformation with selection for ampicillin resistance. In these plasmids lon was under the control of the pBAD arabinose promoter. The Lon S679A variant was deficient in protease activity; the K362Q variant was deficient in ATPase activity (16).
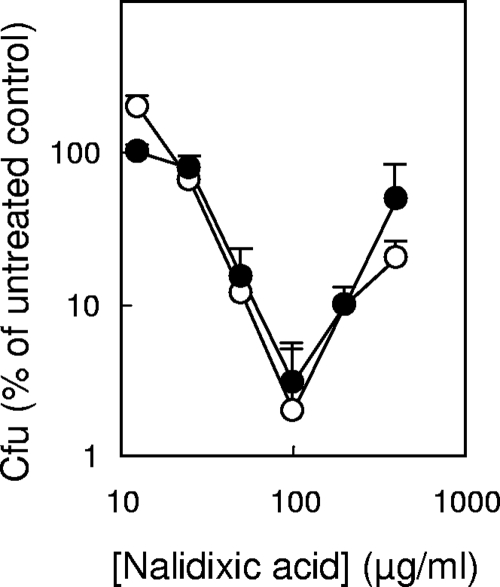
We began by confirming general features of paradoxical survival: it was observed with a variety of quinolones (nalidixic acid, norfloxacin, and ciprofloxacin), several incubation temperatures (30, 37, and 42°C), and two growth media (LB and nutrient broth [not shown]). We also found that paradoxical survival associated with nalidixic acid treatment was unaffected by a noninducible lexA3 mutation (not shown) or by concurrent chloramphenicol treatment of a gyrA67 gyrB225 mutant (Fig. 1) (the two gyrase mutations separately interfere with the ability of chloramphenicol to block quinolone-mediated killing [7, 9]; together they show strong paradoxical survival). Thus, neither the SOS response (8) nor protein synthesis is required for observation of paradoxical survival, a phenomenon that is common to several different quinolones and growth conditions.
FIG. 1.
Paradoxical survival in the presence of chloramphenicol. An exponentially growing culture of strain KD2140 (gyrA67 gyrB225) was treated with 20 μg/ml chloramphenicol for 10 min prior to addition of the indicated concentrations of nalidixic acid for an additional 180 min, at which time samples were diluted, applied to drug-free agar, and incubated to determine bacterial survival. Symbols: filled circles, cultures with chloramphenicol; open circles, cultures without chloramphenicol. Similar results were obtained in a replicate experiment.
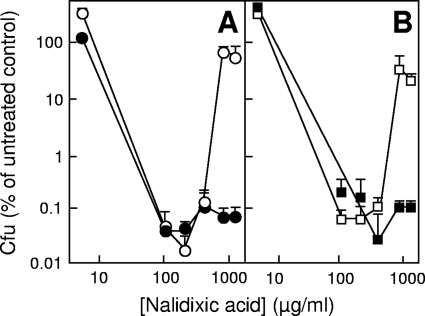
To examine the involvement of Lon protease, we treated a lon-deficient strain (KD2372) with nalidixic acid at a variety of concentrations. At maximal bactericidal concentrations, survival was similar to that of wild-type cells, but at very high concentrations, wild-type survival was about 3 orders of magnitude greater (Fig. 2A). Since accumulation of SulA (SfiA) causes lon-deficient mutants to be sensitive to DNA-damaging agents (4, 5), we also examined lethal action in a lon sulA double mutant. Such a mutant behaved similarly to a Lon-deficient strain, and a sulA mutant exhibited paradoxical survival similar to the wild-type strain (Fig. 2B). Thus, paradoxical survival requires Lon activity but not through its effect on SulA stability.
FIG. 2.
Effect of lon-100 and sulA3 mutations on survival during treatment with nalidixic acid. Exponentially growing cultures were treated with the indicated concentrations of nalidixic acid for 180 min. The cells were then concentrated by centrifugation, resuspended in medium, diluted, and plated. (A) Lon deficiency. Open circles, wild-type strain DM4100; filled circles, lon-100 strain KD2372. (B) SulA deficiency. Open squares, sulA3 strain KD2510; filled squares, lon-100 sulA3 strain KD2377. Similar results were observed in three replicate experiments.
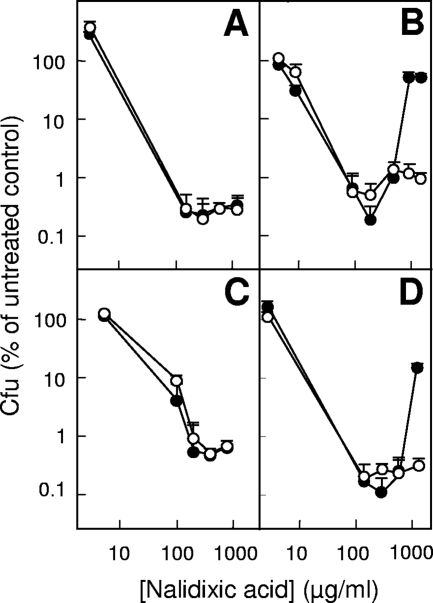
Lon protease has multiple domains, one functioning as an ATPase and another as a protease (16). We asked whether both or only one Lon activity is required for paradoxical survival by using mutant lon genes expressed from plasmids. A lon-deficient strain (KD3037) containing the pBAD24 vector-only control showed little paradoxical survival in the presence or absence of 0.1% arabinose, the inducer used for this expression vector (Fig. 3A). These data demonstrated that no plasmid component contributed to survival at high quinolone concentration. When the plasmid contained wild-type lon (strain KD3045), paradoxical survival was observed when arabinose was present but not when arabinose was absent (Fig. 3B). A strain (KD3039) containing pBAD24lon S679A that lacked protease activity failed to show the paradoxical effect (Fig. 3C), while a strain (KD3041) having pBAD24lon K362Q deficient in ATPase activity did (Fig. 3D). Neither strain showed paradoxical survival in the absence of arabinose (Fig. 3C and D). These data confirmed that Lon is necessary for paradoxical survival and indicated that activity of its ATPase is dispensable. A comparable experiment using complementation of UV sensitivity and capsule overproduction as assays led to the conclusion that protease activity rather than ATPase activity is dispensable (16). The reason for this difference is currently not understood.
FIG. 3.
Effect of expression of Lon variants on survival during treatment with nalidixic acid. Exponentially growing cells deficient in chromosomal lon and containing pBAD24 vector (A) (strain KD3037), pBAD24lon+ (B) (strain KD3045), pBAD24lon S679A (C) (strain KD3039), or pBAD24lon K362Q (D) (strain KD3041) were cultured with 0.1% arabinose (filled circles) or without arabinose (open circles) and treated with nalidixic acid as described in the legend to Fig. 1.
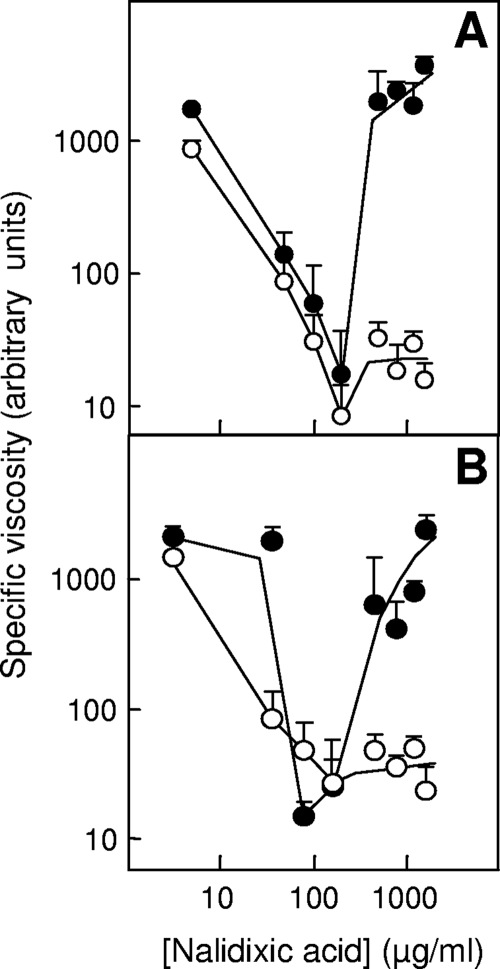
An empirical viscometric assay was used to examine the effect of nalidixic acid concentration on chromosomal quinolone-gyrase-DNA complexes. Cells were exposed to various concentrations of nalidixic acid, gently lysed, and treated with sodium dodecyl sulfate to unfold chromosomes and release broken DNA from cleaved complexes. Viscosity of lysates from cells exposed to various nalidixic acid concentrations for 180 min exhibited a response similar to that seen when survival was measured with both wild-type and lon-deficient cells (Fig. 4A). These data are consistent with chromosomal DNA breaks being more prevalent when cells are treated with bactericidal concentrations of nalidixic acid than with much higher concentrations and with Lon playing a role in reducing the number of DNA breaks.
FIG. 4.
Viscosity of nucleoids from wild type and lon mutant exposed to nalidixic acid. Wild-type (filled circles) and lon mutant (KD2372, open circles) cultures were incubated with nalidixic acid for 180 min (A) or 15 min (B). Cells were lysed gently, and sodium dodecyl sulfate was added to denature proteins. Similar results were obtained with five replicate experiments.
Since quinolone-gyrase-DNA complexes form quickly after addition of quinolone (12), while cell death is a slower process (9), we also measured lysate viscosity after a brief, 15-min treatment with nalidixic acid. Wild-type bacterial lysates exhibited a minimum in viscosity that occurred at the same nalidixic acid concentration as that with drug treatment for 180 min; when drug concentration was very high, lysates from a lon-deficient mutant were less viscous than those from wild-type cells (Fig. 4B). Thus, cell death is not required for observation of lon-dependent changes in lysate viscosity.
In eukaryotic systems proteosome activity facilitates processing of DNA lesions generated by trapping of DNA topoisomerase II (18). Lon may serve a similar role for an undefined repair process with bacterial gyrase trapped on DNA by quinolones. In the present case, we observed the Lon effect only at very high concentrations of nalidixic acid, suggesting that complexes formed at high drug concentrations differ from those formed at optimal bactericidal concentrations. An understanding of the structural differences between complexes formed at optimal and at very high quinolone concentrations should help solve the paradox of increased survival at high quinolone concentrations. We are now examining Lon action biochemically to determine whether it acts directly on the complexes.
Acknowledgments
We thank the following for critical comments on the manuscript: Marila Gennaro, Susan Gottesman, Richard Pine, and Xilin Zhao. We also thank Susan Gottesman for generously providing plasmids and bacterial strains.
The work was supported by NIH grants AI35257, AI63431, and AI73491.
Footnotes
Published ahead of print on 4 May 2009.
REFERENCES
- 1.Chen, C.-R., M. Malik, M. Snyder, and K. Drlica. 1996. DNA gyrase and topoisomerase IV on the bacterial chromosome: quinolone-induced DNA cleavage. J. Mol. Biol. 258:627-637. [DOI] [PubMed] [Google Scholar]
- 2.Crumplin, G. C., and J. T. Smith. 1975. Nalidixic acid: an antibacterial paradox. Antimicrob. Agents Chemother. 8:251-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DiNardo, S., K. Voelkel, R. Sternglanz, A. Reynolds, and A. Wright. 1982. Escherichia coli DNA topoisomerase I mutants have compensatory mutations in DNA gyrase genes. Cell 31:43-51. [DOI] [PubMed] [Google Scholar]
- 4.Gayda, R., L. Yamamoto, and A. Markovitz. 1976. Second-site mutations in capR (lon) strains of Escherichia coli K-12 that prevent radiation sensitivity and allow bacteriophage lambda to lysogenize. J. Bacteriol. 127:1208-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.George, J., M. Castellazzi, and G. Buttin. 1975. Prophage induction and cell division in E. coli. III. Mutations sfiA and sfiB restore division in tif and lon strains and permit the expression of mutator properties of tif. Mol. Gen. Genet. 140:309-332. [PubMed] [Google Scholar]
- 6.Gottesman, S. 1996. Proteases and their targets in Escherichia coli. Annu. Rev. Genet. 30:465-506. [DOI] [PubMed] [Google Scholar]
- 7.Heddle, J., T. Lu, X. Zhao, K. Drlica, and A. Maxwell. 2001. gyrB-225, a mutation of DNA gyrase that compensates for topoisomerase I deficiency: investigation of its low activity and quinolone hypersensitivity. J. Mol. Biol. 309:1219-1231. [DOI] [PubMed] [Google Scholar]
- 8.Kelley, W. L. 2006. Lex marks the spot: the virulent side of SOS and a closer look at the LexA regulon. Mol. Microbiol. 62:1228-1238. [DOI] [PubMed] [Google Scholar]
- 9.Malik, M., X. Zhao, and K. Drlica. 2006. Lethal fragmentation of bacterial chromosomes mediated by DNA gyrase and quinolones. Mol. Microbiol. 61:810-825. [DOI] [PubMed] [Google Scholar]
- 10.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 11.Miller, J. H., and K. B. Low. 1984. Specificity of mutagenesis resulting from the induction of the SOS system in the absence of mutagenic treatment. Cell 37:675-682. [DOI] [PubMed] [Google Scholar]
- 12.Snyder, M., and K. Drlica. 1979. DNA gyrase on the bacterial chromosome: DNA cleavage induced by oxolinic acid. J. Mol. Biol. 131:287-302. [DOI] [PubMed] [Google Scholar]
- 13.Sternglanz, R., S. DiNardo, K. A. Voelkel, Y. Nishimura, Y. Hirota, A. K. Becherer, L. Zumstein, and J. C. Wang. 1981. Mutations in the gene coding for Escherichia coli DNA topoisomerase I affecting transcription and transposition. Proc. Natl. Acad. Sci. USA 78:2747-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trisler, P., and S. Gottesman. 1984. lon transcriptional regulation of genes necessary for capsular polysaccharide synthesis in Escherichia coli K-12. J. Bacteriol. 160:184-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsilibaris, V., G. Maenhaut-Michel, and L. VanMelderen. 2006. Biological roles of the Lon ATP-dependent protease. Res. Microbiol. 157:701-713. [DOI] [PubMed] [Google Scholar]
- 16.Van Melderen, L., and S. Gottesman. 1999. Substrate sequestration by a proteolytically inactive Lon mutant. Proc. Natl. Acad. Sci. USA 96:6064-6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wall, J. D., and P. D. Harriman. 1974. Phage P1 mutants with altered transducing abilities for Escherichia coli. Virology 59:532-544. [DOI] [PubMed] [Google Scholar]
- 18.Zhang, A., Y. Lyu, C.-P. Lin, N. Zhou, A. Azarova, L. Wood, and L. F. Liu. 2006. A protease pathway for the repair of topoisomerase II-DNA covalent complexes. J. Biol. Chem. 281:35997-36003. [DOI] [PubMed] [Google Scholar]