Abstract
β-Lactam resistance in methicillin (meticillin)-resistant Staphylococcus aureus (MRSA) is caused by the production of an additional low-affinity penicillin-binding protein 2a, which is encoded by the mecA gene. The disruption of mecA may inhibit mecA expression and thereafter lead to the restoration of MRSA susceptibility to β-lactams. In this study, we developed a novel anionic liposome for encapsulating and delivering the complexes of a specific anti-mecA phosphorothioate oligodeoxynucleotide (PS-ODN833) and polycation polyethylenimine (PEI). The efficiencies of liposome encapsulation of the complexes were around 79.7% ± 2.7%. The liposomes showed sustained release of PS-ODN833 at 37°C but very low levels of release at 4°C and room temperature. The addition of the encapsulated anti-mecA PS-ODN833-PEI complex to cultures of MRSA strains caused 45, 76, 82, and 93% reductions in mecA expression, accompanied by the inhibition of MRSA growth on Mueller-Hinton agar containing oxacillin (6 μg/ml) in a concentration-dependent manner. The encapsulated-PS-ODN833 treatment also reduced the MICs of five of the most commonly used antibiotics for MRSA clinical isolates to values within the sensitivity range and rescued mice from MRSA-caused septic death by downregulating mecA. The survival rates of septic mice increased from 0% for the control group to 53% for the PS-ODN833-treated group. The results were associated with reductions of bacterial titers in the blood of surviving mice. The findings of the present study indicate that an antisense oligodeoxynucleotide targeted to mecA can significantly restore the susceptibility of MRSA to existing β-lactam antibiotics, providing an apparently novel strategy for treating MRSA infections.
Methicillin (meticillin)-resistant Staphylococcus aureus (MRSA) is a major human pathogen and the leading cause of hospital-associated infections. The hospital outbreaks due to MRSA have revealed the potential of MRSA to produce invasive infections, particularly in vulnerable patients, and its resistance to almost all antibiotics, which limits the therapeutic options available (23, 25, 28).
According to a report of the National Nosocomial Infections Surveillance System, the rates of methicillin resistance in S. aureus isolates slowly increased from 20 to 25% in the early 1990s and then rapidly increased to about 60% in 2006 (36). Furthermore, the mortality from severe MRSA infections is reported to be as high as 20 to 50% (18, 39). The seriousness of the situation has recently been addressed by an article in the Journal of the American Medical Association in which the authors estimated that more patients died because of invasive MRSA infections than because of human immunodeficiency virus infection/AIDS in the United States in 2005 (20).
Vancomycin has become the drug of choice for treating MRSA infections (2). However, treatment failure, adverse side effects, and the emergence of vancomycin-resistant MRSA strains are leading to urgent requirements for alternative strategies to combat MRSA with mechanisms radically different from existing strategies.
The β-lactam resistance of MRSA is caused most commonly by the production of penicillin-binding protein 2a (PBP2a), a novel PBP which, unlike the intrinsic set of PBPs of S. aureus, has remarkably reduced binding affinities for β-lactam antibiotics. Despite the presence of otherwise inhibitory concentrations of β-lactam antibiotics, MRSA can continue cell wall synthesis by depending solely upon the uninhibited activity of PBP2a (9). PBP2a is encoded by a mecA gene located on the chromosome of MRSA, and the transcription of mecA of MRSA is inducible and regulated by the sensor-transducer proteins MecR1 and MecI, the genes for which are located immediately upstream from the mecA promoter on SCCmec elements and are transcribed in the opposite direction from mecA (1, 30, 43).
Previous results have demonstrated that the blockage of mecR1 mRNA expression by phosphorothioate oligodeoxynucleotides (PS-ODNs) or deoxyribozymes leads to significant reductions of both mecR1 and PBP2a mRNAs. Consequently, the susceptibility of MRSA to oxacillin is restored to a significant level (15, 16, 26). However, in previous studies, antisense oligonucleotides targeting antibiotic resistance genes were delivered into MRSA strains by electroporation and the inhibition of antibiotic resistance gene expression was limited by low delivery efficiency.
Effective antisense inhibition in cells requires efficient cell entry, and the delivery of antisense agents across stringent bacterial cell walls can be a particular problem for relatively large, hydrophilic molecules such as PS-ODNs (14, 40). Numerous chemical methods have been developed for nonviral DNA transfer, but the development of cationic liposome-DNA complexes has progressed the furthest. Cationic liposomes are utilized mainly for gene therapy in eukaryotic cells (32). It has been reported previously that anionic liposomes may efficiently deliver antisense oligonucleotides into bacteria and inhibit gene expression to eventually eliminate the resistance of bacterial strains to conventional antibiotics (8, 34).
To our present knowledge, the persistence of anion liposome-encapsulated antibiotic antisense oligonucleotides targeting the mecA gene in MRSA has not been investigated yet. The present study was undertaken, firstly, to prepare a novel anionic liposome for the encapsulation of PS-ODN-polyethylenimine (PEI) complexes; secondly, to explore the use of the entrapped PS-ODN-PEI complex targeting mecA transcripts as a potential therapeutic agent in inhibiting the expression of mecA and thereafter leading to the reduction of PBP2a expression; and finally, to investigate the restoration of MRSA susceptibility to β-lactam antibiotics in vitro and in vivo.
MATERIALS AND METHODS
Chemicals.
Oxacillin, floxacillin (flucloxacillin), cephalothin (cefalotin), cefoxitin, and cefoperazone were purchased from the National Institute for the Control of Pharmaceutical and Biological Products (Beijing, China). Egg yolk phosphatidylcholine (EPC) was obtained from Xi'an Libang Pharmaceutical Co., Ltd. (Xi'an, China). N-(Carbonyl-methoxy-polyethylene glycol 2000)-1,2-distearoyl-sn-glycero-3-phosphoethanolamine (PEG2000-DSPE) was purchased from HeliCan Biotechnology Ltd. (Vancouver, Canada). Dimyristoyl phosphatidylglycerol (DMPG) was purchased from Lipoid (Ludwigshafen, Germany). PEI was purchased from Sigma-Aldrich (St. Louis, MO). Sephadex G-200 was purchased from Pharmacia Fine Chemicals (Uppsala, Sweden). All other chemicals and solvents were of analytical grade.
Organisms.
The MRSA strain WHO-2 used in this study was obtained from the Chinese National Center for Surveillance of Antimicrobial Resistance, and four clinical isolates, MRSA 071001, MRSA 071002, MRSA 071003, and MRSA 071004, were isolated from cultures of sputum or catheter samples from patients in Xijing Hospital (Xi'an, China). All strains expressed mecA, which was confirmed by PCR detection (data not shown).
Animals.
Specific-pathogen-free male or female BALB/c mice, 5 to 6 weeks of age, with body weights between 18 and 20 g were used in this study. Animals were maintained in the Animal Housing Unit of the Fourth Military Medical University in an environment with controlled temperatures (21 to 24°C) and lighting (12-h/12-h light-darkness cycle). Standard laboratory chow and drinking water were provided ad libitum. A period of 3 days was allowed for the animals to acclimatize before any experimental manipulations were undertaken. The experimental and animal care procedures were approved by the animal care and use committees at the university.
Antisense oligonucleotides.
The sequence of the most active PS-ODN used in this study, PS-ODN833, was as follows: CGAGTCCCTTTTTACCAA. This sequence is complementary to nucleotides 833 to 850 in the coding region of mecA mRNA in MRSA. The control mismatched sequence corresponding to this antisense PS-ODN, which was randomly aligned with a sequence of the same number of bases, was AAACTCTTCTCCTACGTG. The PS-ODNs were synthesized by Aoke Biotechnology Co., Ltd. (Beijing, China), and were fully phosphorothioated.
Determination of PS-ODN833 stability in Mueller-Hinton broth.
PS-ODN833 at a concentration of 330 μg/ml was mixed with Mueller-Hinton broth in the absence or presence of cations (Ca2+ at 20 mg/liter, Mg2+ at 12.5 mg/liter, and Na+ at 20 g/liter), and then the mixtures were incubated at 35°C with agitation at 120 rpm for 12 h. At different time points, aliquots of 10 μl (1% of the total mixture volume) were removed from each mixture and loaded directly onto 3.0% agarose gels for electrophoresis.
Preparation of anionic liposome-encapsulated nanosized PS-ODN833-PEI complexes.
PS-ODN833-PEI complexes were prepared according to the method of Chen et al., with slight modifications (6). Briefly, PS-ODN833-PEI complexes were formed at a molar ratio of PEI nitrogen to PS-ODN phosphate (N/P ratio) of 8 and an optimum charge (+/−) ratio of 8.0 (PEI/PS-ODN ratio, 1.1:1 [wt/wt]). The PEI solution (adjusted to pH 7.4) and the PS-ODN833 solution were freshly diluted in distilled water to obtain the desired concentrations. Complexes were then prepared by subjecting equal volumes of PEI and PS-ODN solutions to a vortex for 2 min. Then the complexes were allowed to form for at least 15 min at room temperature. The final PS-ODN concentration was 0.462 mg/ml.
Anionic liposomes composed of EPC, DMPG, and PEG2000-DSPE in a molar ratio of 14:0.9:1 were prepared by the film dispersion method, with minor modifications (7). Briefly, appropriate amounts of a lipid mixture were dissolved in chloroform and dried by a rotary evaporator (Buchi Rotavapor-KRvr 65/45) at 50°C under a vacuum controlled by a Buchi 168-vacuum/distillation controller to obtain a lipid film. This film was kept under high-vacuum conditions for at least 1 h to remove the traces of chloroform. The dry lipid film was hydrated with an aqueous solution of PS-ODN-PEI complexes by subjection to a vigorous vortex for 10 min to obtain a lipid suspension. After hydration, the liposomes were extruded one by one through 0.4-, 0.2-, and 0.1-mm-pore-size polycarbonate membranes (Nuclepore Corp., Pleasanton, CA) in an extruder (Northern Lipids Inc., Vancouver, Canada). Control liposomes were prepared similarly, but phosphate-buffered saline (PBS) was used instead of PS-ODN-PEI complexes. The mean sizes of PS-ODN-PEI particles and liposomes were examined by a laser light-scattering particle size analyzer (submicron particle sizer model no. 370; Nicomp, Santa Barbara, CA). A 500-μl sample of liposome suspensions was applied to Sephadex G-200 columns (30 by 1.0 cm) and eluted with 0.05 M phosphate buffer (pH 7.4) at a flow rate of 15 ml/h for removing nonencapsulated PS-ODNs.
To determine the efficiency of PS-ODN encapsulation in liposomes, 0.5 ml of a liposome suspension was added to 4.5 ml of methanol for complete lysis of the liposomes and then the concentration of the PS-ODN in the lysis solution was determined using a UV spectrophotometer (model no. DU800; Beckman Coulter Inc.) with absorbance at 260 nm. The encapsulation efficiency was determined as the percentage of the PS-ODN incorporated into the liposomes relative to the initial total amount of the PS-ODN, as follows: % entrapment efficiency = [amount of entrapped PS-ODN/(amount of free PS-ODN + amount of entrapped PS-ODN)] × 100.
The storage stabilities of liposome preparations in phosphate buffer (pH 7.4) were tested after storage at 4°C, room temperature (20 to 25°C), and 37°C for an extended period of time. Samples were taken at certain time points, liposomes were separated from the free PS-ODN, and the amount of the PS-ODN was determined spectrophotometrically.
RNA extraction and real-time RT-PCR.
The total RNAs from the bacterial cultures were extracted with Trizol reagent according to the instructions of the manufacturer (Invitrogen, Carlsbad, CA). Briefly, 1 μg of total mRNA from each sample was mixed with 0.1 μg of a random primer and a 4-μl solution of 2.5 mM deoxynucleoside triphosphate, and the mixture was heated to 65°C for 5 min and then cooled on ice for at least 1 min. Subsequently, a mixture of 1 μl of 0.1 M dithiothreitol, 4 μl of 5× reverse transcriptase (RT) buffer, 0.5 μl of 40-U/μl RNase inhibitor, and 0.5 μl of 200-U/μl SuperScript III RT (Invitrogen, Carlsbad, CA) was added to obtain a final volume of 20 μl. The samples were then incubated at 25°C for 5 min, at 50°C for 45 min, and at 70°C for 15 min.
The resulting cDNA was amplified by real-time PCR with a thermal cycler and a Dice real-time PCR system (TaKaRa, Japan) using the following gene-specific oligonucleotide primers: mecA primers corresponding to nucleotides 310 to 333 (5′-AACTACGGTAACATTGATCGCAAC-3′) and nucleotides 423 to 402 (5′-GCTTTGGTCTTTCTGCATTCCT-3′), yielding a 114-bp product, and primers for 16S rRNA, an internal housekeeping gene in MRSA, corresponding to nucleotides 1155 to 1174 (5′-GTGACAAACCGGAGGAAGGT-3′) and nucleotides 1298 to 1274 (5′-ATCCGAACTGAGAACAACTTTATGG-3′), yielding a 144-bp product. The PCR was run using Sybr green I. The PCR reagents consisted of 10 μl of 2× Sybr premix Ex Taq (TaKaRa, China), 1 μl of each primer (5 μM), and 2 μl of sample cDNA in a final volume of 20 μl. The thermal cycling conditions were an initial denaturation step at 95°C for 2 min and 40 cycles at 95°C for 30 s, 58°C for 30 s, and 72°C for 30 s. The melting curves for the PCR products were acquired by the stepwise increase of the temperature from 60 to 94°C.
The cDNA from the control sample, which was not treated with a PS-ODN or PEI, was diluted in a fivefold dilution series, and the following analysis was performed. For each dilution, the difference in the cycle threshold (ΔCT) values for mecA and 16S rRNA was calculated by taking the mean cycle threshold (CT; the cycle number at which the fluorescence intensity reaches the threshold) for duplicate mecA sample tubes and subtracting the mean CT for duplicate 16S rRNA sample tubes, with the mean CT values measured using aliquots from the same RT reaction mixtures, as follows: ΔCT = (CT for mecA) − (CT for 16S rRNA).
The relative expression of mecA mRNA was calculated using the comparative CT method. The 16S rRNA gene, which is expressed in bacteria at nearly the same level throughout the developmental cycle, was used as the control to normalize the quantity of a cDNA target in order to determine the differences in the amounts of total cDNAs in a reaction mixture. The ΔCT for the treated sample was subtracted from the ΔCT for the untreated, control sample to generate a ΔΔCT value, as follows: ΔΔCT = ΔCT (treated sample) − ΔCT (control). The mean of the ΔΔCT measurements was then used to calculate the expression of mecA (2−ΔΔCT) relative to that of the 16S rRNA gene, and the result was normalized according to the level in the untreated control, as follows: relative mecA expression = 2−ΔΔCT. Analyses of gene expression data using the 2−ΔΔCT method have appeared in the literature (35, 41). The 2−ΔΔCT value indicates the level of change (n-fold) in gene expression relative to that in the untreated control.
Bacterial growth assay and susceptibility testing.
The bacterial strain WHO-2 was cultured for 12 h (to an optical density [OD] of 0.5 to 0.6), and then cells were diluted to 1.5 × 108 CFU/ml. A 160-μl sample of diluted bacterial broth medium was mixed with 40 μl of free PBS, liposome-encapsulated PBS, liposome-encapsulated mismatched PS-ODN (18 μM), free PEI (0.2 μM), free PS-ODN833 (18 μM), or liposome-encapsulated PS-ODN833 (0.7, 2, 6, or 18 μM). The OD values for each well at different time points were determined.
The cultures were incubated at 35°C with moderate agitation for 6 h. Samples of 50 μl of diluted cells were spread onto plates of Mueller-Hinton agar that contained 6 μg/ml oxacillin, and the plates were incubated for 24 h at 35°C. The colonies were counted, and the total number of CFU per sample was determined by correcting the colony count for the dilution factor.
To determine the growth curve for WHO-2 in the broth medium, cells were diluted and mixed with liposome-encapsulated PBS, liposome-encapsulated mismatched PS-ODN (with PS-ODN at a concentration of 18 μM), free PEI (0.2 μM), free PS-ODN833 (18 μM), and liposome-encapsulated PS-ODN833 (with PS-ODN833 at a concentration of 0.7, 2, 6, or 18 μM). Aliquots of 100 μl of a mixture containing 6 μg/ml oxacillin were added to the wells of a 96-well microtiter plate, and the plate was incubated at 35°C with agitation at 120 rpm. The growth rate of the cells was monitored by measuring the OD values at 630 nm with a microplate reader (Bio-Rad Laboratories, Tokyo, Japan) at different time points.
The MICs of oxacillin, floxacillin, cephalothin, cefoxitin, and cefoperazone for MRSA strains were determined by the standard broth dilution method according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI). Briefly, serial dilutions of antibiotics in Mueller-Hinton broth were prepared, and bacterial cultures were mixed with liposome-encapsulated PS-ODN833, liposome-encapsulated PBS, liposome-encapsulated mismatched PS-ODN, free PEI, or free PS-ODN833. A 50-μl sample of a bacterial suspension was then added to each tube to achieve a final inoculum of 5 × 105 CFU/ml. Following incubation at 35°C for 16 h, the cultures were examined for evidence of bacterial growth in the form of turbidity. The lowest concentration of an antimicrobial which prevents the visible growth of bacteria is considered to be the MIC.
Infection of mice with MRSA and therapeutic effects of PS-ODN833 on infected animals.
MRSA strain WHO-2 was grown for 6 h at 35°C in 2 ml of Mueller-Hinton medium, and the culture was diluted 1:100 in 100 ml of Mueller-Hinton broth and incubated under the conditions described above for 16 to 20 h. The culture was concentrated by centrifugation at 12,000 × g for 5 min at 26°C. The bacterial pellet was suspended in normal saline, pH 7.2, to a final concentration of 5 × 105 CFU/ml. Fifteen mice were used in each group, and the bacterial suspension in a volume of 25 ml/kg of body weight was injected intraperitoneally into each mouse. The total number of dead mice was tallied every 12 h for 7 days, and the cumulative percent survival was determined.
Immediately after bacterial challenge, the mice were randomly chosen to receive normal saline (control group), 100 mg/kg oxacillin (twice daily for 7 days), empty liposomes (once daily for 3 days) and 100 mg/kg oxacillin (twice daily for 7 days), 10 mg/kg free PS-ODN833 (once daily for 3 days) and 100 mg/kg oxacillin (twice daily for 7 days), or 2.5, 5, or 10 mg/kg liposome-encapsulated PS-ODN833 (once daily for 3 days) and 100 mg/kg oxacillin (twice daily for 7 days). The injections were given intravenously via the tail vein. The blood samples for culture were obtained from the tail vein by aseptic percutaneous puncture 24 h after bacterial challenge. CFU in blood samples were enumerated on Mueller-Hinton agar plates.
Statistical analysis.
The results were expressed as means ± standard deviations (SD). The resultant data were analyzed using a one-way analysis of variance, followed by the Student-Newman-Keuls test, and the animal survival curves associated with different treatments were tested by Kaplan-Meier analyses. A value of P of <0.05 was considered statistically significant.
RESULTS
Stability of PS-ODN833 in Mueller-Hinton broth.
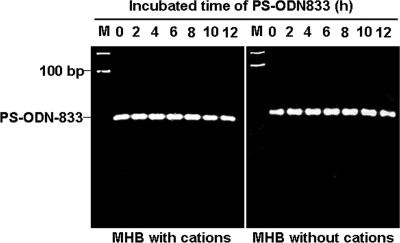
The results showed that PS-ODN833 remained stable in broth culture medium, and no degradation in any sample was observed during the experimental period (Fig. 1).
FIG. 1.
Stability of PS-ODN833 in Mueller-Hinton broth (MHB). The mixtures of PS-ODN833 and MHB in the absence or presence of cations were incubated at 35°C with agitation at 120 rpm for 12 h. Ten-microliter aliquots of the mixtures at different time points were analyzed by electrophoresis on 3.0% agarose gels. Lane M, molecular size markers.
Characterizations of anionic liposome-encapsulated nanosized PS-ODN-PEI complexes.
In order to enhance encapsulation efficiency, anti-mecA PS-ODN833 was condensed with PEI. The hydrodynamic mean diameter of the PS-ODN-PEI complex particles was 82.0 ± 21.0 nm (n = 3), and the polydispersity index was lower than 0.2. To determine the percentage of PS-ODN833 associated with PEI, the amounts of PS-ODN833 in the pellet and in the supernatant after centrifugation were assessed. It was found that 97.3% ± 1.1% of PS-ODN833 was associated with PEI. PS-ODN833-PEI complexes were encapsulated within EPC-DMPG-PEG2000-DSPE liposomes with average particle sizes of about 217 ± 78 nm. The encapsulation efficiency was found to be 79.7% ± 2.7%, and the amount of encapsulated PS-ODN833-PEI within the anionic liposomes, measured by a fluorospectrophotometer, was 16.4 μg/mg of liposomal lipid.
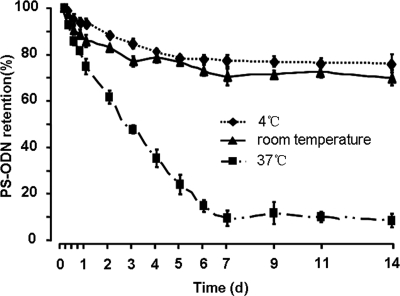
To examine the stability of PS-ODN833-PEI complex-containing liposomes, a long-term release study was carried out. The results showed that the liposomes retained 76.3 and 71.5% of the encapsulated drug until day 14 at 4°C and room temperature, respectively (Fig. 2). When the liposomes were maintained at 37°C, the sustained rapid release of PS-ODN833 was observed, and the samples collected at 18 h from liposome preparations showed that the amount of PS-ODN833 released was comparable to the amounts released from liposomes maintained at 4°C and room temperature over the 14-day study. At the end of study, 92.1% of the whole PS-ODN833 of the encapsulated liposomes was released, which were maintained at 37°C for 14 days (Fig. 2).
FIG. 2.
In vitro stability of anionic liposomes. Liposomes encapsulating PS-ODN833-PEI nanosized particles were incubated in PBS at 4°C, room temperature, and 37°C. At the times indicated, samples were collected and eluted on a Sephadex G-200 column, and then the concentrations of free and encapsulated PS-ODN833 were determined by UV spectrophotometry (A260). Data are shown as the means ± SD of results from three independent experiments. d, day.
Real-time quantitation assays for mecA expression.
To ascertain whether anti-mecA PS-ODN833 inhibits the target gene, the encapsulated anti-mecA PS-ODN833-PEI complexes were applied to MRSA strain WHO-2, and the expression of mecA mRNA in WHO-2 was detected by real-time PCR. The values of the two standard curves were used to determine the absolute value of the slope of the log [cDNA] versus the ΔCT (the difference in the CT values obtained in two PCR systems with the same cDNA dilution) for the respective dilution. This validation experiment involved pairwise comparisons between mecA and 16S rRNA. The slope was 0.0645, demonstrating approximately equal PCR amplification efficiencies for mecA and 16S rRNA. The analysis of the melting curve for the PCR product from each sample of each gene showed a single-peak graph for all amplicons, indicating that a single PCR product was formed. This finding was confirmed by running 5 μl of each product on a 1% agarose gel. The negative controls did not show any amplification product (data not shown).
The addition of the encapsulated anti-mecA PS-ODN833 (at 0.7, 2, 6, and 18 μM) to WHO-2 cultures with 6 h of incubation caused significant reductions of mecA expression, to 55, 24, 18, and 7% of control values, respectively, in a concentration-dependent manner. Free PS-ODN833 also showed certain downregulation of mecA expression, but 18 μM free PS-ODN833 reduced mecA expression only by 30%. Moreover, the expression of target mecA mRNA was not altered by treatment with empty liposomes, liposome-encapsulated mismatched PS-ODN, and free PEI. These results demonstrate that the encapsulated antisense oligonucleotides not only enter the bacterial cells, but also interact with the target mRNA (Table 1).
TABLE 1.
Relative mRNA expression of mecA of MRSA strain WHO-2 as detected by real-time PCRa
| Treatment | Concn (μM) | CT (mecA) | CT (16S rRNA) | ΔCT | ΔΔCT | 2−ΔΔCT |
|---|---|---|---|---|---|---|
| Control | 0 | 20.82 ± 0.13 | 16.01 ± 0.09 | 4.81 ± 0.08 | 0 | 1 |
| L-PBS | 0 | 20.81 ± 0.17 | 15.93 ± 0.06 | 4.89 ± 0.11 | 0.07 ± 0.03 | 0.95 ± 0.02 |
| F-PEI | 0.2 | 20.41 ± 0.09 | 15.73 ± 0.10 | 4.68 ± 0.05 | −0.14 ± 0.03 | 1.10 ± 0.02 |
| L-M-ODN | 18 | 20.10 ± 0.08 | 15.35 ± 0.07 | 4.75 ± 0.09 | −0.06 ± 0.02 | 1.05 ± 0.02 |
| F-ODN833 | 18 | 23.44 ± 0.12 | 18.11 ± 0.12 | 5.32 ± 0.10 | 0.51 ± 0.03 | 0.70 ± 0.01 |
| L-ODN833 | 0.7 | 24.04 ± 0.10 | 18.36 ± 0.07 | 5.68 ± 0.04 | 0.87 ± 0.04 | 0.55 ± 0.02 |
| 2 | 22.79 ± 0.09 | 15.93 ± 0.11 | 6.86 ± 0.20 | 2.04 ± 0.25 | 0.24 ± 0.05 | |
| 6 | 22.42 ± 0.10 | 15.15 ± 0.10 | 7.27 ± 0.20 | 2.46 ± 0.19 | 0.18 ± 0.02 | |
| 18 | 26.84 ± 0.31 | 18.22 ± 0.18 | 8.62 ± 0.29 | 3.80 ± 0.35 | 0.07 ± 0.02 |
Bacteria were treated with free PBS (control), liposome-encapsulated PBS (L-PBS), free PEI (F-PEI; 0.2 μM), liposome-encapsulated mismatched PS-ODN (L-M-ODN; 18 μM), free PS-ODN833 (F-ODN833; 18 μM), or liposome-encapsulated PS-ODN833 (L-ODN833) at different concentrations. ΔCT = (CT.for mecA) − (CT.for 16S rRNA); DΔCT = (ΔCT for treated sample) − (ΔCT for control sample). Results are expressed as means ± SD (n = 3).
Reversal of antibiotic resistance of MRSA strain WHO-2 by encapsulated PS-ODN833.
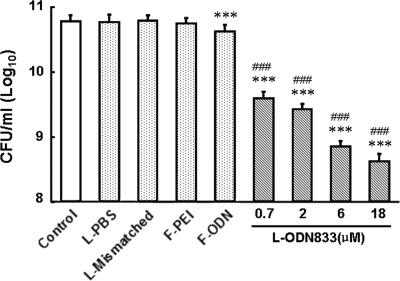
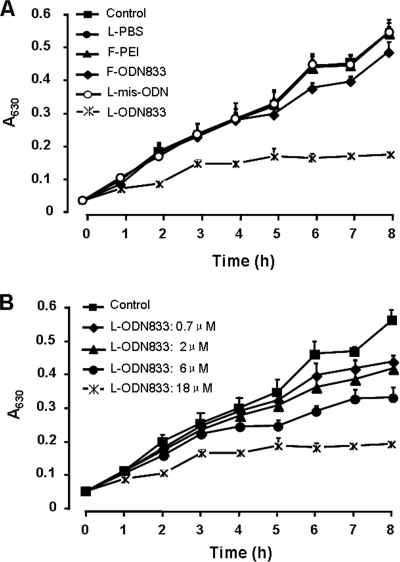
We found that the downregulation of mecA by the addition of anti-mecA PS-ODN833 correlated with the restoration of MRSA strain WHO-2 susceptibility to β-lactam antibiotics. The numbers of WHO-2 colonies in all encapsulated-PS-ODN833-treated cultures on Mueller-Hinton agar containing oxacillin (6 μg/ml) strikingly decreased to 6.5, 4.9, 1.2, and 0.7% of the control value in a concentration-dependent manner. In liquid medium containing oxacillin (6 μg/ml), the growth of encapsulated-PS-ODN833-treated WHO-2 cells was also inhibited in a concentration-dependent manner. However, the growth of WHO-2 was not influenced by treatment with empty liposomes, liposome-encapsulated mismatched PS-ODN, and free PEI and was only slightly inhibited by treatment with free PS-ODN833 (Fig. 3 and 4).
FIG. 3.
Effects of anti-mecA encapsulated PS-ODN833 on the growth of WHO-2 colonies. Liposome-encapsulated PS-ODN833 (L-ODN833) was added to cell cultures containing 1.5 × 108 CFU/ml WHO-2 to a final concentration of 0.7, 2, 6, or 18 μM. Additional cell cultures were treated with free PBS in a volume equal to that of the encapsulated-PS-ODN833 preparation as a control, liposome-encapsulated PBS (L-PBS), 18 μM liposome-encapsulated mismatched PS-ODN (L-Mismatched), 0.2 μM free PEI (F-PEI), or 18 μM free PS-ODN833 (F-ODN). Aliquots of each culture were collected at 6 h, diluted, and inoculated onto solid agar containing 6 μg/ml of oxacillin. The number of CFU was calculated from the number of colonies growing on plates. The data are shown as means ± SD for 10 samples. ***, P < 0.01 for comparison to control values; ###, P < 0.01 for comparison to results for free PS-ODN833.
FIG. 4.
Effects of anti-mecA encapsulated PS-ODN833 on the growth of MRSA strain WHO-2 in liquid culture medium. The cells were cultured in liquid medium containing 6 μg/ml of oxacillin. The growth of different groups of WHO-2 cells was monitored by using OD measurements. (A) Bacteria were treated with free PBS (control), liposome-encapsulated PBS (L-PBS), free PEI (F-PEI; 0.2 μM), free PS-ODN833 (F-ODN833; 18 μM), liposome-encapsulated mismatched PS-ODN (L-mis-ODN; 18 μM), or liposome-encapsulated PS-ODN833 (L-ODN833; 18 μM). (B) Bacteria were treated with a final concentration of encapsulated PS-ODN833 of 0.7, 2, 6, or 18 μM or with an equal volume of free PBS as a control. The data are shown as means ± SD for 10 samples.
The results presented in Table 2 revealed that encapsulated anti-mecA PS-ODN833 lowered the MICs of oxacillin for WHO-2 in a concentration-dependent manner. At the concentrations of 6 and 18 μM, the encapsulated PS-ODN833 reduced the MIC of oxacillin from 1,024 μg/ml to 2 and 1 μg/ml, respectively, values which are within the oxacillin sensitivity range for the MRSA strain on the basis of the interpretive criteria recommended by the CLSI, representing the restoration of WHO-2 susceptibility to oxacillin. Free PS-ODN833 decreased the MIC of oxacillin merely from 1,024 μg/ml for the control sample to 256 μg/ml for the treated sample, which suggested that free PS-ODN833 could not efficiently penetrate WHO-2 cells. The wrapped mismatched PS-ODN (18 μM), free PEI (0.2 μM), or empty liposomes did not alter the MIC of oxacillin for WHO-2 (Table 2). These results indicate that the bactericidal effect of oxacillin was restored through the inhibition of mecA mRNA expression by PS-ODN833 and that the antibacterial efficiency of oxacillin was enhanced by increasing the concentration of encapsulated PS-ODN833.
TABLE 2.
MICs of oxacillin for oxacillin-sensitive strain ATCC 29213 and MRSA strain WHO-2 in the presence and absence of anti-mecA PS-ODN833a
| Cultureb | Concn (μM) of PS-ODN833 | MIC (μg/ml) of oxacillin |
|---|---|---|
| ATCC 29213 culture | 0 | 0.5 |
| Untreated WHO-2 control culture | ||
| WHO-2 cultures treated with: | 0 | 1,024 |
| L-PBS | 0 | 1,024 |
| F-PEI | 0.2 | 1,024 |
| L-M-ODN | 18 | 1,024 |
| F-ODN833 | 18 | 256 |
| L-ODN833 | 0.7 | 16 |
| 2 | 8 | |
| 6 | 2 | |
| 18 | 1 |
Classification of S. aureus strains by MIC of oxacillin: sensitive, MIC ≤ 2 μg/ml; resistant, MIC ≥ 4 μg/ml.
L-PBS, liposome-encapsulated PBS; F-PEI, free PEI; L-M-ODN, liposome-encapsulated mismatched PS-ODN; F-ODN833, free PS-ODN833; L-ODN833, liposome-encapsulated PS-ODN833.
Restoration of susceptibility of MRSA clinical isolates to β-lactam antibiotics by encapsulated PS-ODN833.
The results reported above prompted us to examine whether these observations could be extended to strains of present MRSA clinical isolates. Four MRSA clinical isolates obtained from a hospital affiliated with the Fourth Military Medical University showed strong resistance to five of the most commonly used β-lactam antibiotics, oxacillin, floxacillin, cephalothin, cefoxitin, and cefoperazone. The treatment with encapsulated PS-ODN833 (18 μM) could reduce the MICs of all five antibiotics from a range of 32 to 1,024 μg/ml to a range of 1 to 8 μg/ml, within the margin defining the sensitivity of MRSA strains to these antibiotics on the basis of the interpretive criteria recommended by the CLSI (Table 3).
TABLE 3.
MICs of β-lactam antibiotics for MRSA clinical isolatesa
| Isolate | MIC (μg/ml) of:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Oxacillin
|
Floxacillin
|
Cephalothin
|
Cefoxitin
|
Cefoperazone
|
||||||
| Control sample | ODN833-treated sample | Control sample | ODN833-treated sample | Control sample | ODN833-treated sample | Control sample | ODN833-treated sample | Control sample | ODN833-treated sample | |
| 071001 | 128 | 2 | 1,024 | 2 | 512 | 2 | 32 | 2 | 128 | 8 |
| 071002 | >1,024 | 2 | >1,024 | 2 | 512 | 8 | 512 | 1 | 1,024 | 2 |
| 071003 | >1,024 | 2 | >1,024 | 2 | 512 | 2 | 512 | 1 | 1,024 | 4 |
| 071004 | >1,024 | 1 | >1,024 | 2 | 512 | 8 | 512 | 4 | 1,024 | 4 |
MICs of β-lactam antibiotics in the presence and absence of anti-mecA PS-ODN833 encapsulated in anionic liposomes for four clinically isolated MRSA strains in broth cultures were determined. The concentration of liposome-encapsulated anti-mecA PS-ODN833 was 18 μM. The classification of S. aureus strains by MICs is as follows. Oxacillin MIC: ≤2 μg/ml, sensitive; ≥4 μg/ml, resistant. Floxacillin MIC: ≤2 μg/ml, sensitive; ≥4 μg/ml, resistant. Cephalothin MIC: ≤8 μg/ml, sensitive; 16 μg/ml, intermediate; ≥32 μg/ml, resistant. Cefoxitin MIC: ≤8 μg/ml, sensitive; 16 μg/ml, intermediate; ≥32 μg/ml, resistant. Cefoperazone MIC: ≤16 μg/ml, sensitive; 32 μg/ml, intermediate; ≥64 μg/ml, resistant.
The encapsulated PS-ODN833 rescued mice from MRSA-caused lethal sepsis by the downregulation of mecA.
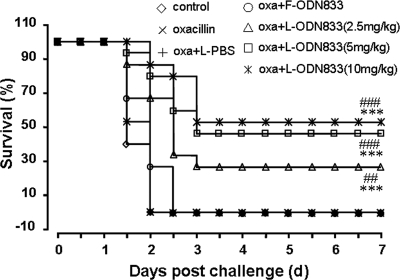
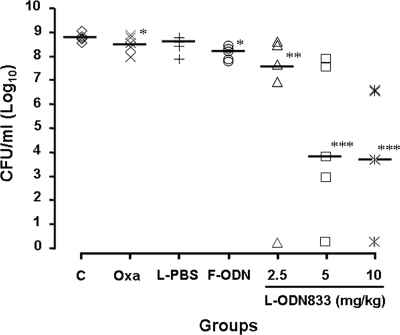
In the light of the capacity of encapsulated PS-ODN833 to downregulate mecA expression and restore the susceptibility of MRSA strains to β-lactams, we explored its efficacy in an animal model of MRSA-caused lethal sepsis. We firstly observed that the administration of one of three doses of encapsulated PS-ODN833 (2.5, 5, and 10 mg/kg) once daily for 3 days, along with 100 mg/kg oxacillin twice daily for 7 days, significantly improved the animal survival rate (from 0% for the control group to 26.7, 46.7, and 53.3% for encapsulated-PS-ODN833-treated groups) in a dose-dependent manner (Fig. 5). Furthermore, the rescues were associated with reductions of the bacterial titers in the blood of mice inoculated with MRSA strain WHO-2, from 5.2 × 108 CFU/ml to 4.1 × 107, 4.6 × 102, and 2.7 × 102 CFU/ml, respectively (Fig. 6). Treatments with oxacillin alone, oxacillin combined with empty liposomes, or free PS-ODN833 did not show any protective effects by decreasing the numbers of deaths of WHO-2-infected mice or altering CFU counts in blood samples from the septic mice. Since the rise of the colony counts in blood samples was directly related to an increase in the probability of death, the present results suggested that PS-ODN833 protected animals against lethal endotoxemia mainly through the restoration of the susceptibility of WHO-2 to oxacillin by inhibiting mecA expression.
FIG. 5.
Effects of encapsulated PS-ODN833 in a mouse model of MRSA-induced sepsis. BALB/c mice were intraperitoneally inoculated with MRSA strain WHO-2, and the survival rates of the mice were recorded every 12 h for 7 days. The mice were randomly chosen to receive, intravenously via the tail vein, isotonic sodium chloride solution (control group), 100 mg/kg oxacillin (twice daily for 7 days; oxacillin group), liposome-encapsulated PBS (once daily for 3 days) and 100 mg/kg oxacillin (twice daily for 7 days) (oxa+L-PBS), 10 mg/kg free PS-ODN833 (once daily for 3 days) and 100 mg/kg oxacillin (twice daily for 7 days) (oxa+F-ODN833), or 2.5, 5, or 10 mg/kg liposome-encapsulated PS-ODN833 (once daily for 3 days) and 100 mg/kg oxacillin (twice daily for 7 days) (oxa+L-ODN833). n = 15; ***, P < 0.01 for comparison to control results; ##, P < 0.05, and ###, P < 0.01 for comparison to results with free PS-ODN833.
FIG. 6.
Reductions of bacterial titers in the blood samples of mice inoculated with MRSA strain WHO-2 by anti-mecA encapsulated PS-ODN833. Mice were randomly chosen to receive, intravenously via the tail vein, isotonic sodium chloride solution (control group; C), 100 mg/kg oxacillin (Oxa; twice daily for 7 days), liposome-encapsulated PBS (L-PBS; once daily for 3 days) and 100 mg/kg oxacillin (twice daily for 7 days), 10 mg/kg free PS-ODN833 (F-ODN; once daily for 3 days) and 100 mg/kg oxacillin (twice daily for 7 days), or 2.5, 5, or 10 mg/kg liposome-encapsulated PS-ODN833 (L-ODN833; once daily for 3 days) and 100 mg/kg oxacillin (twice daily for 7 days). Horizontal lines represent the average CFU value in each group. *, P > 0.05; **, P < 0.05; and ***, P < 0.01 for comparison to control results.
DISCUSSION
MRSA has become a global problem in hospitals and in communities (23, 25, 27, 28, 31). This organism is resistant to all commercially available β-lactam antibiotics and also represents a therapeutic challenge because effective therapeutic options are becoming limited. The loss of effectiveness of commonly used antibacterial antibiotics such as β-lactam drugs further adds to the dilemma, calling for the immediate need to develop a new strategy and agents that can specifically combat MRSA infection. The concept of using antisense antibiotics is innovative, and antisense antibiotics have been proposed as a new hope for bacterial infection therapy through the targeting of specific genes in bacteria (10, 29).
The antisense oligonucleotide-based downregulation of murA1 gene expression in Bacillus anthracis hypersensitizes cells to the MurA-specific antibiotic fosfomycin (19). An antisense phosphorodiamidate morpholino oligomer targeted to acpP significantly inhibits the viability of a strain of Escherichia coli with a normal, intact outer membrane both in pure culture and in infected mice (11, 38). Treatment with antisense PS-ODNs against the mRNA for inositol-1-phosphate synthase in Mycobacterium tuberculosis reduces the level of mycothiol, enhances susceptibility to antibiotics, and thereafter inhibits the proliferation of M. tuberculosis (22). In our previous study, the knockdown of mRNA for the sensor-transducer protein gene mecR1 by phosphorothioate-modified antisense oligomers or deoxyribozymes inhibited mecR1 expression and was followed by the repression of the downstream PBP2a gene mecA, thereafter leading to the restoration of MRSA susceptibility to β-lactam antibiotics (15, 16, 26). Although antisense agents have been shown to function promisingly in bacteria, the lack of an efficient delivery system has been a major problem that hinders their usefulness against bacteria in vivo.
In this study, we designed anionic liposomes for delivering the nanosized complexes obtained by the spontaneous binding of a mecA antisense PS-ODN and PEI. The cationic polymer PEI has been widely used for nonviral gene delivery in vitro and in vivo and has an advantage over other polycations in that it combines a strong DNA compaction capacity with an intrinsic endosomolytic activity (24). It was reported previously that circulating proteins can bind to and inactivate PEI-DNA complexes (12). There are two ways to minimize the effects of protein binding to PEI-DNA, the manipulation of the PEI amine/DNA phosphate ratio and surface modification (13). Therefore, we prepared PS-ODN833-PEI complexes with the optimum charge (+/−) ratio of 8:1, which resulted in an efficiency of PS-ODN binding to PEI of 97.3% ± 1.1%, and then wrapped the PS-ODN833-PEI complex in anionic liposomes composed of EPC-DMPG-PEG2000-DSPE with efficiencies of up to 80%. PEG-DSPE is essential for increasing the circulation times of anionic liposomes (5) and facilitates uptake by MRSA strains (4). The results of the long-term release study indicated that PS-ODN833-PEI complex-containing liposomes are relatively stable at 4°C and room temperature. Under these conditions, 70 to 75% of PS-ODN833-PEI was retained in the liposomes at the end of the 14-day period (Fig. 2). However, at 37°C, the liposome showed instability and relatively rapid release of PS-ODN833-PEI, perhaps because this temperature is close to the phase transition temperature for liposomes (36°C). The fluctuation of the negatively charged liposomes at 37°C might have caused transitions from the solid phase to liquid-crystalline phases (3, 17, 34). Therefore, it is possible that, due to the fluidity or plasticity of the EPC-DMPG-PEG2000-DSPE liposomal formulation, the liposomes may fuse with the bacterial cells. Such a fusion process would improve the penetration of PS-ODN833 into bacterial cell walls. This process is in accordance with the fact that the liposome-wrapped PS-ODN833-PEI complex showed significant downregulation of mecA gene expression in the MRSA strains. Consequently, the phenotypes of MRSA clinical isolates reverted to β-lactam sensitivity when bacteria were treated with encapsulated PS-ODN833. MICs of five β-lactam antibiotics commonly used in clinical practice, oxacillin, floxacillin, cefoxitin, cephalothin, and cefoperazone, for MRSA strains were significantly reduced to values within the range defining sensitivity (Table 3). More importantly, we demonstrated that the administration of the encapsulated PS-ODN833 and oxacillin rescued mice challenged with a MRSA strain from lethal sepsis. The rescue was associated with significant decreases in the bacterial titers in blood samples. The results obtained in these experiments are encouraging, since our findings support the development of an antisense agent strategy as an alternative therapy to confer superior protection of animals against MRSA infection in the near future.
It has been demonstrated recently that the MRSA phenotype reverts to that of β-lactam-sensitive S. aureus when bacteria are grown in broth at pH 5.0. The mechanisms of this reversion may be related to the observations (i) that β-lactams interact with PBP2a more avidly at pH 5 than at higher pHs and (ii) that in the presence of β-lactams, PBP2a undergoes a conformational change consistent with the opening of the active site from the closed conformation occurring under low-pH conditions (21). These results further prove that PBP2a function is pivotal for MRSA survival. Although the results are promising, the global effects of the change of pH at the organism level may cause life-threatening acidosis and manifest themselves at sites other than PBP2a. Therefore, the therapeutic potential of this strategy is limited.
RNA interference is a process by which gene expression can be silenced in a sequence-specific manner, making it a powerful tool that is being pursued for many therapeutic applications. This silencing mechanism functions only in eukaryotes, because in prokaryotes RNase III, a very potent and fast double-strand-specific RNase, degrades double-stranded RNA substrates as short as 12 bp. There are two reports that parallel cRNA can function to inhibit gene expression in E. coli or S. aureus (37, 42). However, the efficacy of an RNA silencing mechanism for prokaryotes has not been confirmed (33).
In conclusion, the results of the present study indicate that the anionic liposomes could efficiently deliver anti-mecA PS-ODN833-PEI complexes inside MRSA cells and inhibit gene expression to eventually eliminate MRSA resistance to conventional antibiotics. The targeting of mecA with an antisense approach appears to be a novel strategy for treating MRSA infections.
Acknowledgments
This research was supported by grants from the National Natural Science Foundation of China (no. 30271556) and the Natural Science Foundation of Shaanxi Province (no. 2002C2-04).
Footnotes
Published ahead of print on 11 May 2009.
REFERENCES
- 1.Anstead, G. M., G. Quinones-Nazario, and J. S. Lewis II. 2007. Treatment of infections caused by resistant Staphylococcus aureus. Methods Mol. Biol. 391:227-258. [DOI] [PubMed] [Google Scholar]
- 2.Archer, G. L., and J. M. Bosilevac. 2001. Signaling antibiotic resistance in Staphylococci. Science 291:1915-1916. [DOI] [PubMed] [Google Scholar]
- 3.Beaulac, C., S. Clément-Major, J. Hawari, and J. Lagacé. 1996. Eradication of mucoid Pseudomonas aeruginosa with fluid liposome-encapsulated tobramycin in an animal model of chronic pulmonary infection. Antimicrob. Agents Chemother. 40:665-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beaulac, C., S. Sachetelli, and J. Lagacé. 1998. In-vitro bactericidal efficacy of sub-MIC concentration of liposome-encapsulated antibiotic against gram-negative and gram-positive bacteria. J. Antimicrob. Chemother. 41:35-41. [DOI] [PubMed] [Google Scholar]
- 5.Bucke, W. E., S. Leitzke, J. E. Diederichs, K. Borner, H. Hahn, S. Ehlers, and R. H. Müller. 1998. Surface-modified amikacin-liposomes: organ distribution and interaction with plasma proteins. J. Drug Target. 5:99-108. [DOI] [PubMed] [Google Scholar]
- 6.Chen, T., Z. Wang, R. Wang, T. Lu, and W. Wang. 2007. Polyethylenimine DNA solid particles for gene delivery. J. Drug Target. 15:714-720. [DOI] [PubMed] [Google Scholar]
- 7.Elhissi, A. M., M. A. O'Neill, S. A. Roberts, and K. M. Taylor. 2006. A calorimetric study of dimyristoylphosphatidylcholine phase transitions and steroid-liposome interactions for liposomes prepared by thin film and proliposome methods. Int. J. Pharm. 320:124-130. [DOI] [PubMed] [Google Scholar]
- 8.Fillion, P., A. Desjardines, K. Sayasith, and J. Lagacé. 2001. Encapsulation of DNA in negatively charged liposomes and inhibition of bacterial gene expression with fluid liposome-encapsulated antisense oligonucleotides. Biochim. Biophys. Acta 1515:44-54. [DOI] [PubMed] [Google Scholar]
- 9.Fuda, C., M. Suvorov, S. B. Vakulenko, and S. Mobashe. 2004. The basis for resistance to β-lactam antibiotics by penicillin binding protein 2a of methicillin resistant Staphylococcus aureus. J. Biol. Chem. 270:40802-40806. [DOI] [PubMed] [Google Scholar]
- 10.Geller, B. L. 2005. Antibacterial antisense. Curr. Opin. Mol. Ther. 7:109-113. [PubMed] [Google Scholar]
- 11.Geller, B. L., J. Deere, L. Tilley, and P. L. Iversen. 2005. Antisense phosphorodiamidate morpholino oligomer inhibits viability of Escherichia coli in pure culture and in mouse peritonitis. J. Antimicrob. Chemother. 55:983-988. [DOI] [PubMed] [Google Scholar]
- 12.Godbey, W. T., K. K. Wu, G. J. Hirasaki, and A. G. Mikos. 1999. Improved packing of poly(ethylenimine)/DNA complexes increases transfection efficiency. Gene Ther. 6:1380-1388. [DOI] [PubMed] [Google Scholar]
- 13.Godbey, W. T., K. K. Wu, and A. G. Mikos. 1999. Poly(ethylenimine) and its role in gene delivery. J. Control. Release 60:149-160. [DOI] [PubMed] [Google Scholar]
- 14.Harth, G., P. C. Zamecnik, J. Y. Tang, D. Tabatadze, and M. A. Horwitz. 2000. Treatment of Mycobacterium tuberculosis with antisense oligonucleotides to glutamine synthetase mRNA inhibits glutamine synthetase activity, formation of the poly-l-glutamate/glutamine cell wall structure, and bacterial replication. Proc. Natl. Acad. Sci. USA 97:418-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hou, Z., J. R. Meng, C. Niu, H. F. Wang, J. Liu, B. Q. Hu, M. Jia, and X. X. Luo. 2007. Restoration of antibiotic susceptibility in methicillin-resistant Staphylococcus aureus by targeting mecR1 with a phosphorothioate deoxyribozyme. Clin. Exp. Pharmacol. Physiol. 34:1160-1164. [DOI] [PubMed] [Google Scholar]
- 16.Hou, Z., J. R. Meng, J. R. Zhao, B. Q. Hu, J. Liu, X. J. Yan, M. Jia, and X. X. Luo. 2007. Inhibition of β-lactamase-mediated oxacillin resistance in Staphylococcus aureus by a deoxyribozyme. Acta Pharmacol. Sin. 28:1775-1782. [DOI] [PubMed] [Google Scholar]
- 17.Jones, M. N. 2005. Use of liposomes to deliver bactericides to bacterial biofilms. Methods Enzymol. 391:211-228. [DOI] [PubMed] [Google Scholar]
- 18.Kaye, K. S., D. J. Anderson, Y. Choi, K. Link, P. Thacker, and D. J. Sexton. 2008. The deadly toll of invasive methicillin-resistant Staphylococcus aureus infection in community hospitals. Clin. Infect. Dis. 46:1568-1577. [DOI] [PubMed] [Google Scholar]
- 19.Kedar, G. C., V. Brown-Driver, D. R. Reyes, M. T. Hilgers, M. A. Stidham, K. J. Shaw, J. Finn, and R. J. Haselbeck. 2008. Comparison of the essential cellular functions of the two murA genes of Bacillus anthracis. Antimicrob. Agents Chemother. 52:2009-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klevens, R. M., M. A. Morrison, J. Nadle, S. Petit, K. Gershman, S. Ray, L. H. Harrison, R. Lynfield, G. Dumyati, J. M. Townes, A. S. Craig, E. R. Zell, G. E. Fosheim, L. K. McDougal, R. B. Carey, and S. K. Fridkin. 2007. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298:1763-1771. [DOI] [PubMed] [Google Scholar]
- 21.Lemaire, S., C. Fuda, F. Van Bambeke, P. M. Tulkens, and S. Mobashery. 2008. Restoration of susceptibility of methicillin-resistant Staphylococcus aureus to beta-lactam antibiotics by acidic pH: role of penicillin-binding protein PBP 2a. J. Biol. Chem. 283:12769-12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, Y., Z. Chen, X. Li, H. Zhang, Q. Huang, Y. Zhang, and S. Xu. 2007. Inositol-1-phosphate synthetase mRNA as a new target for antisense inhibition of Mycobacterium tuberculosis. J. Biotechnol. 128:726-734. [DOI] [PubMed] [Google Scholar]
- 23.Lin, Y. C., T. L. Lauderdale, H. M. Lin, P. C. Chen, M. F. Cheng, K. S. Hsieh, and Y. C. Liu. 2007. An outbreak of methicillin-resistant Staphylococcus aureus infection in patients of a pediatric intensive care unit and high carriage rate among health care workers. J. Microbiol. Immunol. Infect. 40:325-334. [PubMed] [Google Scholar]
- 24.Lungwitz, U., M. Breunig, T. Blunk, and A. Göpferich. 2005. Polyethylenimine-based non-viral gene delivery systems. Eur. J. Pharm. Biopharm. 60:247-266. [DOI] [PubMed] [Google Scholar]
- 25.McDonald, J. R., C. M. Carriker, B. C. Pien, J. V. Trinh, J. J. Engemann, L. J. Harrell, M. A. Oden, D. T. Tanaka, R. N. Goldberg, D. J. Sexton, and K. S. Kaye. 2007. Methicillin-resistant Staphylococcus aureus outbreak in an intensive care nursery: potential for interinstitutional spread. Pediatr. Infect. Dis. J. 26:678-683. [DOI] [PubMed] [Google Scholar]
- 26.Meng, J. R., B. Q. Hu, J. Liu, Z. Hou, J. Meng, M. Jia, and X. X. Luo. 2006. Restoration of oxacillin susceptibility in methicillin-resistant Staphylococcus aureus by blocking the MecR1-mediated signaling pathway. J. Chemother. 18:360-365. [DOI] [PubMed] [Google Scholar]
- 27.Millar, B. C., B. D. Prendergast, and J. E. Moore. 2008. Community-associated MRSA (CA-MRSA): an emerging pathogen in infective endocarditis. J. Antimicrob. Chemother. 61:1-7. [DOI] [PubMed] [Google Scholar]
- 28.Murray, R. J., J. C. Pearson, G. W. Coombs, J. P. Flexman, C. L. Golledge, D. J. Speers, J. R. Dyer, D. G. McLellan, M. Reilly, J. M. Bell, S. F. Bowen, and K. J. Christiansen. 2008. Outbreak of invasive methicillin-resistant Staphylococcus aureus infection associated with acupuncture and joint injection. Infect. Control Hosp. Epidemiol. 29:859-865. [DOI] [PubMed] [Google Scholar]
- 29.Nekhotiaeva, N., S. K. Awasthi, P. E. Nielsen, and L. Good. 2004. Inhibition of Staphylococcus aureus gene expression and growth using antisense peptide nucleic acids. Mol. Ther. 10:652-659. [DOI] [PubMed] [Google Scholar]
- 30.Niemeyer, D. M., M. J. Pucci, J. A. Thanassi, V. K. Sharma, and G. L. Archer. 1996. Role of mecA transcriptional regulation in the phenotypic expression of methicillin resistance in Staphylococcus aureus. J. Bacteriol. 178:5464-5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Otter, J. A., and G. L. French. 2008. The emergence of community-associated methicillin-resistant Staphylococcus aureus at a London teaching hospital, 2000-2006. Clin. Microbiol. Infect. 14:670-676. [DOI] [PubMed] [Google Scholar]
- 32.Patil, S. D., G. David, D. G. Rhodes, and D. J. Burgess. 2005. DNA-based therapeutics and DNA delivery systems: a comprehensive review. AAPS J. 7:E61-E77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rasmussen, L. C., H. U. Sperling-Petersen, and K. K. Mortensen. 2007. Hitting bacteria at the heart of the central dogma: sequence-specific inhibition. Microb. Cell Fact. 6:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sachetelli, S., H. Khalil, T. Chen, C. Beaulac, S. Sénéchal, and J. Lagacé. 2000. Demonstration of a fusion mechanism between a fluid bactericidal liposomal formulation and bacterial cells. Biochim. Biophys. Acta 1463:254-266. [DOI] [PubMed] [Google Scholar]
- 35.Schmittgen, T. D., B. A. Zakrajsek, A. G. Mills, V. Gorn, M. J. Singer, and M. W. Reed. 2000. Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: comparison of endpoint and real-time methods. Anal. Biochem. 285:194-204. [DOI] [PubMed] [Google Scholar]
- 36.Siegel, J. D., E. Rhinehart, M. Jackson, and L. Chiarello. 2007. Management of multidrug-resistant organisms in health care settings, 2006. Am. J. Infect. Control 35:S165-S193. [DOI] [PubMed] [Google Scholar]
- 37.Tchurikov, N. A., L. G. Chistyakova, G. B. Zavigelsky, I. V. Manukhov, B. K. Chernov, and Y. B. Golova. 2000. Gene-specific silencing by expression of parallel complementary RNA in Escherichia coli. J. Biol. Chem. 275:26523-26529. [DOI] [PubMed] [Google Scholar]
- 38.Tilley, L. D., B. L. Mellbye, S. E. Puckett, P. L. Iversen, and B. L. Geller. 2007. Antisense peptide-phosphorodiamidate morpholino oligomer conjugate: dose-response in mice infected with Escherichia coli. J. Antimicrob. Chemother. 59:66-73. [DOI] [PubMed] [Google Scholar]
- 39.Wang, F. D., Y. Y. Chen, T. L. Chen, and C. Y. Liu. 2008. Risk factors and mortality in patients with nosocomial Staphylococcus aureus bacteremia. Am. J. Infect. Control 36:118-122. [DOI] [PubMed] [Google Scholar]
- 40.White, D. G., K. Maneewannakul, E. von Hofe, M. Zillman, W. Eisenberg, A. K. Field, and S. B. Levy. 1997. Inhibition of the multiple antibiotic resistance (mar) operon in Escherichia coli by antisense DNA analogs. Antimicrob. Agents Chemother. 41:2699-2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winer, J., C. K. Jung, I. Shackel, and P. M. Williams. 1999. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal. Biochem. 270:41-49. [DOI] [PubMed] [Google Scholar]
- 42.Yanagihara, K., M. Tashiro, Y. Fukuda, H. Ohno, Y. Higashiyama, Y. Miyazaki, Y. Hirakata, K. Tomono, Y. Mizuta, K. Tsukamoto, and S. Kohno. 2006. Effects of short interfering RNA against methicillin-resistant Staphylococcus aureus coagulase in vitro and in vivo. J. Antimicrob. Chemother. 57:122-126. [DOI] [PubMed] [Google Scholar]
- 43.Zhang, H. Z., C. J. Hackbarth, K. M. Chansky, and H. F. Chambers. 2001. A proteolytic transmembrane signaling pathway and resistance to beta-lactams in staphylococci. Science 291:1962-1965. [DOI] [PubMed] [Google Scholar]