Abstract
The bacterial killing efficiency of a dynamic nanoplatform (DNP) was evaluated. The polyacrylamide (PAA) hydrogel matrix of the DNP was loaded with methylene blue (MB) and was previously applied successfully to killing rat C6 glioma tumor cells in culture. This series of experiments is aimed at determining the suitability of this nanoplatform for elimination of bacterial infections. Suspended cultures of Staphylococcus aureus, Pseudomonas aeruginosa, Escherichia coli, and Acinetobacter sp. were exposed to activated (∼650-nm laser light) MB-PAA-DNPs. The killing efficiency of nanoparticle mass concentration, light irradiance and fluence, and dark incubation time was determined on each of the bacterial species. Moreover, the ability of activated MB-PAA-DNPs to inhibit biofilm growth and eradicate and disperse preformed biofilms, preformed on glass and polystyrene surfaces, was demonstrated. The data revealed that activated MB-PAA-DNPs eradicated all species of bacteria examined. Also, encapsulation of MB into the PAA-DNP matrix significantly diminished the observed dark toxicity of free dye. The photobactericidal efficacy of MB-PAA-DNP was found to be higher for gram-positive bacteria than for gram-negative bacteria. In addition, activated MB-PAA-DNP can inhibit biofilm growth and eradicate almost all of the early-age biofilms that are formed by all of the bacteria examined.
Biofilms are a group of microorganisms growing collectively in an adhesive matrix, consisting mainly of extracellular polysaccharide, on biological or nonbiological surfaces. The collective growth provides enormous protection to microorganisms against various environmental stresses, including antibiotic treatment (4, 13, 29). Treatment of biofilm-related infections requires high doses and long-term use of antibiotics and the resultant inactivation is not always satisfactory. Furthermore, the widening spread of antibiotic resistance among bacterial pathogens makes the remediation of environmental media and clinically relevant infections rather difficult. Both situations have gained public attention and concern is on the rise (17). Novel and innovative approaches are needed for the treatment of biofilm-related infectious diseases (3). Photodynamic therapy (PDT) may emerge as one of the promising alternative treatments for infectious diseases (14, 44). PDT is a process that combines localized application of a nontoxic dye, termed a photosensitizer (PS), in a diseased tissue or cell, and subsequent illumination with low-intensity visible light to produce cytotoxic species (singlet oxygen and other consequent free radicals) that are able to kill target cells via oxidative stress to cell membranes and other cellular components (10). PDT has been previously shown to kill a variety of microorganisms, including viruses, bacteria, fungi, yeast, and parasites (14). Gram-positive species were found to be more susceptible than gram-negative species to PDT with many commonly used PSs, which may be due to the difference in their cell wall structures (41, 42). Previous studies have also shown that multi-antibiotic-resistant strains are as susceptible to PDT as their native counterparts (45). DNA damage and damage to the cytoplasmic membrane are thought to be two primary mechanisms for the lethal damage caused to bacteria by PDT. It is worthwhile to mention that PDT will not only be effective in killing pathogenic bacteria but also in destroying certain secreted virulence factors (21). There have been several reports on the use of PDT to treat infections in selected animal models (15, 16, 24, 39) and on some clinical trials for treatment of viral and bacterial infections (20, 25, 33, 35, 36).
Several PSs with favorable PDT properties have been developed for optimizing PDT (2). One of the traditional PSs that has been used for a variety of applications, including PDT, is methylene blue (MB) (11, 34). This dye has been approved for use by the United States Food and Drug Administration (FDA) for treatment of methemoglobinemia in a nonphotodynamic mode. However, when activated, MB produces a high quantum yield of 1O2 (ΦΔ ∼ 0.5) (32) in the therapeutic window (ca. 600 to 900 nm). When coupled with its inherently low toxicity, MB is a promising candidate for use as a PDT agent (9). Despite its promise as a clinically relevant PDT agent, use of MB has been limited by its lack of activity when used in vivo. The weak pharmaceutical effect of MB (26, 43, 47) results in part from poor penetration of MB into the cellular compartment of tumors and its inactivation via reduction of the cation to the neutral leukomethylene blue (LMB). The latter results in almost complete loss of photodynamic activity (28, 46, 47).
The encapsulation of PDT agents in nanoparticles as a means of addressing the aforementioned systemic problems with photodynamic dyes has been the subject of much recent research (22). A dynamic nanoplatform (DNP) containing MB that has been developed for use in cancer study (37) has demonstrated utility in vitro (27, 37, 51) and in vivo (23, 31). By encapsulating the singlet-oxygen-producing dyes within a targeted sub-200-nm nanoparticle, 1O2 still can be generated under the requisite excitation wavelengths and delivered outside the DNP to reach adjacent cell membranes. Also, encapsulation prevents direct contact between MB and cellular or plasma components that may reduce the dye to its inactive form. The DNPs, therefore, are not designed to deliver drugs or photodynamic molecules directly into the cell; rather, they deliver massive localized doses of 1O2 that represent several orders of magnitude greater molar concentrations of the dye delivered to a given site. The protective effect of the DNPs was demonstrated by our recent study (38).
The main objective of the present study is to evaluate the potential application of MB containing PAA-DNP for bactericidal, targeted, and localized control of biofilm-related infections. This DNP design offers several distinctive advantages compared to conventional PDT approaches: (i) targeted application in the infected sites; (ii) reduction of phototoxic side effect from the free PS's posttreatment accumulation; (iii) high degree of loading of PS molecules to reach critical mass of PS for effective therapy without introduction of an unacceptable level of side effect; and (iv) prevention of PS from being transformed to nonfunctional derivative by host biological systems. The results discussed here show that these MB-loaded PAA-DNPs would have great potential in bacterial killing and eradication of bacterial biofilms.
MATERIALS AND METHODS
PSs and MB-PAA-DNPs.
MB, the commercially available photosensitizing agent (certified 97% dye content), was acquired from Aldrich (Milwaukee, WI) without further purification. MB-loaded polyacrylamide nanoparticles (MB-PAA-DNP) were prepared as previously described (37). The free MB and MB-PAA-DNP stock solutions were made by weight percent and diluted to the desired concentrations using 0.45% saline. The MB-PAA-DNP suspensions were sonicated for 60 min before use to prevent aggregation of particles. The concentration of MB in a 10-mg/ml MB-PAA-DNP suspension is equivalent to 3 μM free MB.
Bacterial strains and growth.
The following gram-positive and gram-negative organisms were examined: Staphylococcus aureus ATCC 25923, Escherichia coli K-12, Pseudomonas aeruginosa PAO1, and Acinetobacter sp. strain AC811. The initial bacterial concentration was estimated to be ∼106 CFU per ml by measuring the suspension turbidity with a spectrophotometer and was verified by using CFU/ml count on LB agar. All of these organisms were aerobically grown in LB media at 37°C overnight.
Irradiation of planktonic bacteria.
The red excitation light used in these experiments was generated by the combination of a mercury-xenon lamp (X-Cite 120 fluorescence illumination system; EXFO Corp., Mississauga, Ontario, Canada) and a broadband interference filter (650 ± 15 nm; NT46-160; Edmund Optics, Inc., Barrington, NJ). During the measurements, aliquots of 150 μl of the bacterial-0.45% saline suspension, both with and without PSs (MB-PAA DNP or MB alone), were added to the flat-bottom wells (1.6-cm diameter) of 24-well microplates and irradiated from the bottom at a 1.5-cm distance from the filter to ensure that the excitation light beam would cover the whole well surface. The thickness of the solution was ∼0.07 cm.
Determining the effect of various MB-PAA-DNP mass concentrations, light irradiances, and fluences on bacterial killing.
Suspensions containing the same concentration of microorganisms and different concentrations of MB-PAA-DNP were prepared as follows. A suitable volume of the stock solution of the MB-PAA-DNP at a concentration of 100 mg/ml was added to an equal volume of stock bacterial suspension at a concentration of 1 × 106 to 5 × 106 CFU placed in tubes so that the final MB-PAA-DNP mass concentration was 5 to 20 mg/ml, equivalent to 1.5 to 6 μM free MB. The appropriate volume of 0.45% saline was added to each tube for a total volume of 1 ml. Four types of experiments were performed for each mass concentration of MB-PAA-DNP: (i) L-N-, 150 μl of the prepared bacterial suspension was incubated in the dark without MB-PAA-DNP; (ii) L+N-, 150 μl of the suspension was exposed to red light without MB-PAA-DNP; (iii) L-N+, 150 μl of the suspension was incubated in the dark in the presence of MB-PAA-DNP; and (iv) L+N+, 150 μl of the suspension was exposed to red light in the presence of MB-PAA-DNP.
Determining the effect of MB-PAA-DNP on inhibition of biofilm growth and killing of preformed biofilms.
To visualize the killing of biofilms, P. aeruginosa biofilms were grown in a MicroSlide flowcell (Aline, Inc., Redondo Beach, CA) with a polystyrene membrane at the bottom following a protocol similar to what was described previously (6, 18, 50). At 3 h after the inoculation of overnight culture of P. aeruginosa into the flow cell, 1/10 tryptic soy agar liquid media were replaced with 0.45% saline solution containing 10 mM HEPES pH buffer, 0.1 mM SYTO-9 green, 10 mM propidium iodide (PI) (4), and 3 mg of MB-PAA nanoparticles/ml. Cell images were acquired with a Perkin-Elmer UltraView confocal microscope system equipped with an Argon-Krypton laser. Preexposure images were taken with an oil immersion 100× objective lens in two channels (488-nm excitation for SYTO-9 and 568-nm excitation for PI). The cells were then exposed for five minutes at 647 nm. After the illumination, images were taken every 2 min, for up to 2 h, to monitor cell death. During the time intervals, the cells were continuously exposed to the 647-nm light. Finally, a 60× objective was used to broaden the field of view and allow concurrent comparison of the cells exposed to light, along with those that had not been exposed.
To quantify the efficiency of killing biofilms, biofilms of S. aureus and Acinetobacter sp. strain AC811 were developed in a 96-well microtiter plate for 24 h following the protocol as described above with or without the presence of 5 mg of MB-PAA-DNP/ml. Planktonic cells were separated from biofilm biomass without the presence of MB-PAA-DNP by aspiration. Biofilm cells were dispersed by using a homogenizer (OMNI TH, Marietta, GA) to obtain more than 99% single cells without damaging the cells. Both planktonic cells and dispersed biofilm cells were adjusted to ∼106 cells per ml in 0.45% saline solution with 5 mg of MB-PAA particles/ml. The mixtures were exposed to red light with a light irradiance of 38 mW/cm2 and a light fluence of 68.4 J/cm2 as described above. After illumination, serial 10-fold dilutions were plated on LB plates for colony counts. Biofilms developed with 5 mg of MB-PAA-DNP particles/ml were exposed to red light with a light irradiance of 38 mW/cm2 and a light fluence of 68.4 J/cm2 after the suspensions were removed. The biofilm cells were resuspended in 1 ml of 0.45% saline solution and dispersed using homogenization as described above, and serial 10-fold dilutions were plated on LB plates for colony counts.
The crystal violet (CV) staining method (30) was also used to assess the effect of MB-PAA-DNP and red light illumination on the inhibition of biofilm growth and the dispersal of preformed biofilms. A 1% concentration of overnight culture was inoculated into the wells of a 96-well microplate containing 100 μl of 1/10 LB liquid medium and 5 mg of MB-PAA-DNP/ml. After 10, 24, and 48 h of biofilm formation at room temperature, the wells of a 96-well microplate were exposed to red light at 650 nm as described above, resulting in a light fluence of 68.4 J/cm2 with a light irradiance of 20 mW/cm2. The wells without red light treatment were set as a control. After red light illumination, 15 μl of a 1% solution of CV was added to each well to stain the cells for 15 min. Then, the wells were rinsed thoroughly and repeatedly with water. Biofilm biomass was quantified by the addition of 100 μl of 95% ethanol to each CV-stained microplate well, and the absorbance was determined with a plate reader at 600 nm (series 700 microplate reader; Cambridge Technology).
RESULTS
Killing of bacterial cells in suspension.
In order to evaluate the efficiency with which MB-PAA-DNP kills bacterial cells in suspension, cultures of S. aureus, P. aeruginosa, E. coli, and Acinetobacter strains were incubated with MB-PAA-DNPs and exposed to red light at 650 nm for preset lengths of time. The number of survived cells was determined after treatment by using the plate counting method and the results are summarized in Table 1. These experiments demonstrated that various portions of all bacterial types were eradicated by red light in the presence of MB-PAA-DNP at a concentration of 10 mg/ml. However, the degree of light-induced killing was dependent on the light as well as the genus of bacteria (Table 1). When the light irradiance was constant, the photobactericidal efficiency of MB-PAA-DNP against planktonic bacteria increased with the increase of light fluence and would reach its maximum when the light fluence was ∼68.4 J/cm2. The maximum photodamage to the gram-positive bacterium (S. aureus strain) can reach more than 4 log10 CFU reduction in the viable counts. This photodamage effect is better than that to all gram-negative bacteria (E. coli, P. aeruginosa, and Acinetobacter strains) tested, which reaches ∼2.2 log10 CFU reduction in the viable counts. In the absence of light, MB-PAA-DNP exhibited no dark toxicities against all of the microorganisms (P < 0.001). The results from experiment 1 (L-N-) and experiment 2 (L+N-) showed that there was no damage to planktonic bacterial cells when they were exposed only to red light (data not shown).
TABLE 1.
Bactericidal efficacy of MB-PAA-DNP against gram-positive and gram-negative bacteria
| Strain | Treatmenta | Viable bacteria (mean log10 CFU) ± SE at a light fluence (J/cm2)b of:
|
|||||
|---|---|---|---|---|---|---|---|
| 0 | 11.4 | 22.8 | 45.6 | 68.4 | 136.8 | ||
| S. aureus ATCC 25923 | L-N+ | 6.16 ± 0.04 | 6.14 ± 0.03 | 6.15 ± 0.07 | 6.15 ± 0.14 | 6.14 ± 0.08 | 6.12 ± 0.11 |
| L+N+ | 6.16 ± 0.04 | 6.13 ± 0.04 | 4.58 ± 0.13 | 3.38 ± 0.09 | 2.00 ± 0.10 | 1.95 ± 0.08 | |
| E. coli K-12 | L-N+ | 6.22 ± 0.03 | 6.20 ± 0.04 | 6.18 ± 0.06 | 6.16 ± 0.03 | 6.17 ± 0.04 | 6.15 ± 0.04 |
| L+N+ | 6.22 ± 0.03 | 6.05 ± 0.05 | 5.88 ± 0.08 | 4.52 ± 0.19 | 3.66 ± 0.09 | 3.27 ± 0.17 | |
| P. aeruginosa PAO1 | L-N+ | 6.15 ± 0.05 | 6.14 ± 0.02 | 6.14 ± 0.07 | 6.13 ± 0.07 | 6.10 ± 0.07 | 6.09 ± 0.11 |
| L+N+ | 6.15 ± 0.05 | 6.03 ± 0.07 | 5.83 ± 0.12 | 4.93 ± 0.07 | 3.86 ± 0.14 | 3.77 ± 0.09 | |
| Acinetobacter sp. strain AC481 | L-N+ | 6.25 ± 0.01 | 6.23 ± 0.01 | 6.20 ± 0.05 | 6.17 ± 0.05 | 6.17 ± 0.04 | 6.17 ± 0.07 |
| L+N+ | 6.16 ± 0.01 | 5.77 ± 0.03 | 5.20 ± 0.16 | 4.64 ± 0.07 | 4.17 ± 0.13 | 3.96 ± 0.10 | |
| Acinetobacter sp. strain AC811 | L-N+ | 6.15 ± 0.03 | 6.13 ± 0.03 | 6.15 ± 0.07 | 6.13 ± 0.11 | 6.10 ± 0.09 | 6.09 ± 0.09 |
| L+N+ | 6.15 ± 0.03 | 6.03 ± 0.04 | 5.84 ± 0.05 | 4.92 ± 0.07 | 4.09 ± 0.06 | 3.92 ± 0.09 | |
As described in Materials and Methods, L-N+ represents the bacterial suspensions incubated in the dark in the presence of MB-PAA-DNP, and L+N+ represents the bacterial suspensions exposed to red light in the presence of MB-PAA-DNP. The concentration of MB-PAA-DNP used in these experiments was 10 mg/ml.
Light irradiance used was 38 mW/cm2. Values are means of three determinations.
Effect of various MB-PAA-DNP mass concentrations, light irradiances, and initial bacterial concentrations on bacteria killing.
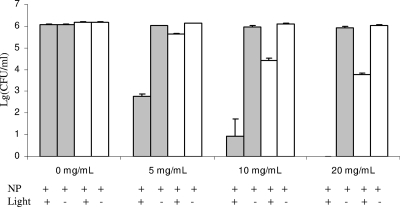
When the MB-PAA-DNP mass concentration was increased, the photodamage to both gram-positive and gram-negative bacteria increased (Fig. 1). As shown in Fig. 1, when the mass concentration of MB-PAA-DNP was increased up to 20 mg/ml, the complete photodestruction of the gram-positive bacterial strain (S. aureus) was achieved, illuminated with a light irradiance of 38 mW/cm2 and a light fluence of 68.4 J/cm2, while the photodamage to the gram-negative bacterial strain (Acinetobacter sp. strain AC811) also increased to ∼2 log10 reduction.
FIG. 1.
Effect of various MB-PAA-DNP concentrations on the killing of S. aureus (░⃞) and Acinetobacter sp. strain AC811 (□) in suspension. The light irradiance used was 38 mW/cm2, and the light fluence was 68.4 J/cm2. NP stands for MB-PAA-DNP. Error bars indicate the standard errors of three replicates.
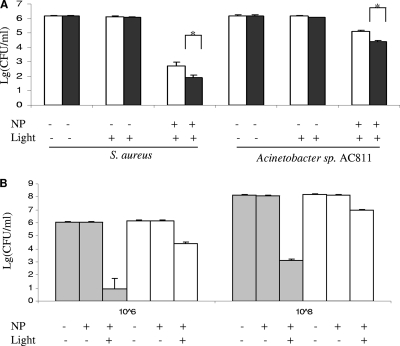
The light irradiance tested also had a significant effect on bacterial killing (Fig. 2). When MB-PAA-DNP was 10 mg/ml, and the light fluence was kept constantly at 68.4 J/cm2, the photodamage to both gram-positive and gram-negative bacterial strains increased with the increase of light irradiance (Fig. 2A). This may be due to a higher concentration of 1O2 generated by a higher light irradiance than by a lower light irradiance, which may cause greater damage that bacterial cells could not repair. When initial bacterial concentrations were increased up to 108 CFU/ml, almost the same log10 reduction was obtained when the light irradiance, light fluence, and nanoparticle concentration were kept constant (10 mg of MB-PAA-DNP/ml, 38-mW/cm2 light irradiance, and 68.4-J/cm2 light fluence) (Fig. 2B).
FIG. 2.
Effect of various light irradiances (A) and starting cell concentrations (B) on the killing of S. aureus and Acinetobacter sp. strain AC811 in the presence of MB-PAA-DNP in suspension. In panel A, light irradiances of 20 mW/cm2 (□) and 38 mW/cm2 (▪) and the same light fluence (68.4 J/cm2) were used. *, significant difference (P < 0.05) between two tests. A statistical analysis was done by using Student t tests. In panel B, a light irradiance of 38 mW/cm2 and a light fluence of 68.4 J/cm2 were used. Suspensions of S. aureus (░⃞) and Acinetobacter (□) strains with starting concentrations of 106 CFU/ml (left columns) and 108 CFU/ml (right columns) were used. NP, MB-PAA nanoparticle. Error bars indicate the standard errors of three replicates.
Effect of activated MB-PAA-DNP on killing bacterial cells in biofilms and on dispersing preformed biofilms and inhibiting biofilm growth.
The above-mentioned results have demonstrated effective kill of gram-positive and gram-negative strains under planktonic conditions using the MB-PAA-DNP. With the intention to test the potential of these nanoparticles in eradicating and dispersing preformed biofilms, three sets of experiments were performed.
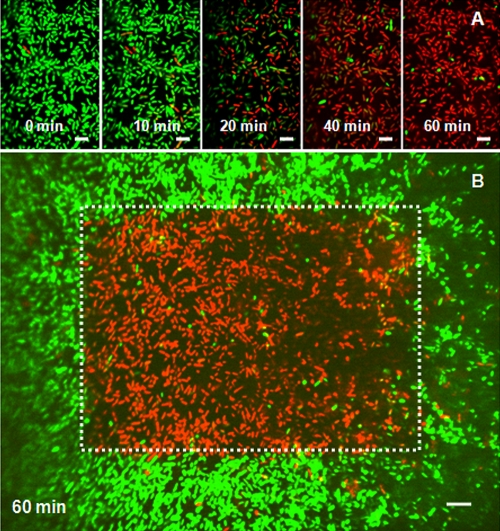
Confocal microscopy was used to obtain a direct visual observation of 3-h P. aeruginosa biofilms treated with the MB-PAA-DNP under red light irradiation. The treated samples also contained SYTO-9 and PI to monitor live and dead cells based on the integrity of membrane, respectively. Figure 3 shows typical confocal images (green SYTO-9 and red PI channels overlaid) of cells treated with 3 mg of MB-PAA-DNP/ml, which is equivalent to 0.9 μM free MB. Before illumination, all of the cells in the first image were stained by SYTO-9 showing green fluorescence, which verified the cell membrane integrity. Subsequent images (Fig. 3) indicate that the cytoplasm of the cultured cells lost its green fluorescence in a time-dependent manner. Concomitant with the loss of green fluorescence, DNA of the compromised bacterial cells were observed to increase their incorporation of PI, indicated by the red fluorescence. Internal controls were obtained by capturing images sequentially, using a 60× objective lens with the irradiated area in the center (Fig. 3B). Local illumination induced death of only those cells located in the beam of light, while other cells, in contact with the MB-PAA-DNP but not illuminated by the laser, remained intact (represented by green fluorescence only). These observations suggest that the exposure of cells to the combination of MB-PAA-DNP and light resulted in the production of sufficient quantities of 1O2 to damage and kill P. aeruginosa biofilms at its very early age.
FIG. 3.
Eradication of 3-h P. aeruginosa biofilms using MB-PAA-DNP. (A) Series of confocal images (SYTO 9 [green] and PI [red] channels overlaid) of P. aeruginosa biofilms at very early age (3 h after inoculation) treated with 3 mg of MB-PAA-DNP/ml and continuously exposed to the red light. The times indicated in each image are times after light illumination when images were taken with a 100× objective. (B) Confocal image (SYTO-9 [green] and PI [red] channels overlaid) of P. aeruginosa biofilms taken at 1 h after illumination with a 60× objective lens. The red area in the white frame indicating dead cells was the area illuminated when a 100× objective lens was used. Scale bars, 5 μm.
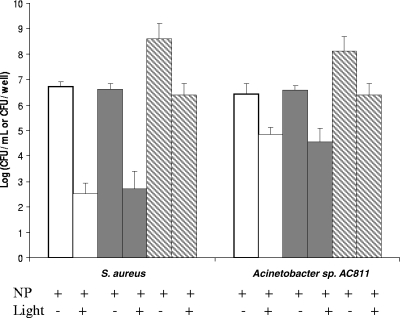
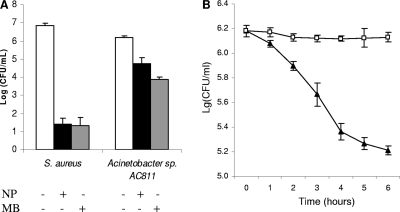
To quantify the efficiency of MB-PAA-DNP on killing bacterial cells in preformed biofilms, biofilms of S. aureus and Acinetobacter sp. strain AC811 were developed in a 96-well microtiter plate for 24 h at room temperature. Planktonic cells and dispersed biofilm cells were incubated with 5 mg/ml of MB-PAA-DNP solutions and exposed to red light at 650 nm with a light fluence of 68.4 J/cm2. After illumination, the remaining viable cells were quantified on LB plates. It was found that there was no observable difference in killing efficacy on planktonic cells and dispersed biofilm cells (∼4 log10 reduction of viable cells for S. aureus cells and 2 log10 reduction for Acinetobacter sp. strain AC811 cells) (Fig. 4). However, when the biofilm biomass was not dispersed, there was only an ∼2-log10 reduction for S. aureus cells and a 1.7-log10 reduction for Acinetobacter sp. strain AC811 cells (Fig. 4), indicating reduced killing efficacy on cells in intact biofilms.
FIG. 4.
Biofilm killing assay with MB-PAA-DNP and red light illumination. Biofilms of S. aureus and Acinetobacter sp. strain AC811 were developed in a 96-well microtiter plate for 24 h. Planktonic cells (□) and dispersed biofilm cells (░⃞) were adjusted to ∼106 cells per ml in 0.45% saline solution with 5 mg of MB-PAA-DNP/ml. The mixtures were exposed to red light with a light irradiance of 38 mW/cm2 and a light fluence of 68.4 J/cm2. After eradication, serial 10-fold dilutions were plated on LB plates for colony counts. The same biofilms developed with 5 mg of MB-PAA-DNP/ml (▧) were exposed to red light with a light irradiance of 38 mW/cm2 and a light fluence of 68.4 J/cm2 after the suspension was removed. The biofilm cells were resuspended in 1 ml of 0.45% saline solution and dispersed by using homogenization, and serial 10-fold dilutions were plated onto LB plates for colony counts. Error bars indicate the standard errors of three replicates.
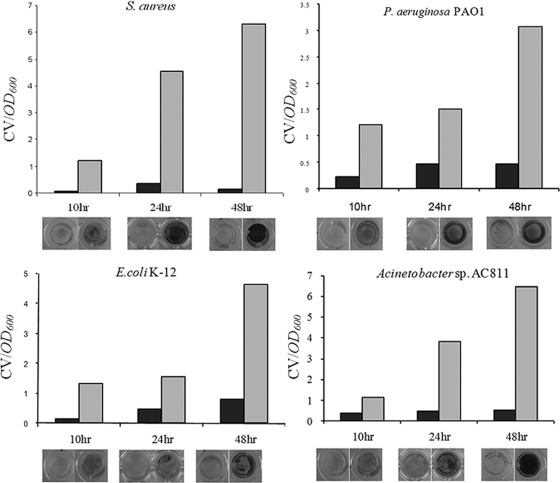
In addition, in order to quantify the photodynamic efficacy of MB-PAA-DNP on the dispersion of preformed biofilms and inhibition of biofilm growth, the CV staining method (30) was used to quantify the remaining biomass of biofilms at up to 2 days after exposure to red light in the presence of MB-PAA-DNP. Bacterial cells were cultured in the wells of a 96-well microplate together with 100 μl of 5-mg/ml MB-PAA-DNP solutions to form biofilms on the well surface. After 10, 24, and 48 h of incubation at room temperature, wells of 96-well microplate were exposed to red light at 650 nm with a light fluence of 68.4 J/cm2; meanwhile, the wells without red light irradiation were set as control. After red light treatment, CV staining was used to measure the remaining biofilm biomass attached to the wall surface.
After 10 h of biofilm formation at room temperature, all of the bacteria strains (S. aureus, P. aeruginosa, E. coli, and Acinetobacter sp.) formed thin biofilms at the bottoms and walls of the wells; however, the biofilm biomass dramatically decreased after being exposed to red light at 650 nm with a light fluence of 68.4 J/cm2 (Fig. 4). With increased time of biofilm formation, thicker biofilms (indicated by CV staining at 24 and 48 h of biofilm formation) were formed by all four tested strains (Fig. 4); however, similar to the 10-h biofilms, only limited biofilm biomass was left in the wells exposed to the red light at 650 nm. These data clearly indicate that the application of the MB-PAA-DNP and light illumination dispersed preformed biofilms and inhibited biofilm growth. The data also suggest that the MB-PAA-DNP has no observable effect on the biofilm formation of these tested strains. In addition, red light illumination alone has no observable effect on dispersing preformed biofilms and inhibiting biofilm growth (data not shown).
DISCUSSION
Photodynamic production of singlet oxygen species is a promising treatment for killing bacteria (20, 45) and for the removal of biofilms (48, 49). As mentioned earlier, the DNP offers more advantages than traditional photodynamic methods and has been successfully used to kill tumor cells in vitro (37) and in vivo (23, 31). Encapsulated within PAA nanoparticles, the PS, MB, still retained its absorption spectrum and the ability to generate singlet oxygen (37). In our study, the ability of the MB-PAA-DNP to kill bacteria in suspension, to kill and disperse preformed biofilms, and to inhibit biofilm formation was confirmed in vitro.
Upon illumination, the encapsulated MB is able to generate sufficient 1O2 that can diffuse out of the matrix, perforate adjacent cell membranes and kill cells. The results presented here revealed that all tested bacteria could be killed by the 1O2 generated from the activated MB-PAA-DNP while the killing efficiency for gram-positive strains (i.e., S. aureus strain) was much higher than that for gram-negative strains (E. coli, P. aeruginosa, and Acinetobacter sp.). This indicated that 1O2 was more effective in damaging the cell membranes, and walls of gram-positive bacteria than those of gram-negative bacteria and/or could more easily diffuse into the cytoplasm of gram-positive bacteria to damage cellular components.
Singlet oxygen produced by MB-PAA-DNP not only killed bacteria in suspension but also killed and dispersed biofilms formed on surfaces (Fig. 3, 4, and 5). There was no observable difference in killing efficiency on cells in dispersed biofilms compared to cells in suspension (Fig. 4); however, the killing efficacy was reduced on cells in intact biofilms. Biofilm matrix may neutralize or reduce the diffusion of singlet oxygen produced, resulting in reduced killing efficacy. Different from the killing efficacy of the MB-PAA-DNP, the dispersal efficacy seemed to be about the same for both gram-positive and gram-negative bacteria, although the biofilm dispersing mechanism by MB-PAA-DNP is not yet known. One possibility might be that the singlet oxygen produced by MB-PAA-DNP would cause damage to cell membranes (lipid), pili (proteins), extracellular DNA, and other biological molecules, all of which play important roles in biofilm formation (1, 5, 7, 12, 19). These hypothetical targets warrant further studies to elucidate the dispersion mechanisms.
FIG. 5.
Biofilm dispersal and inhibition assay with MB-PAA-DNP and red light illumination. Biofilm biomass was determined by using the CV staining method. Photo images of CV-stained biofilms are shown below each treatment. Biofilms incubated with MB-PAA-DNP and with red light illumination are indicated by gray columns and biofilm incubated with MB-PAA-DNP but without red light illumination are indicated by black columns.
It is known that free MB can diffuse into bacterial cytoplasm and bind to DNA and other cellular components (40), resulting in damage of these cellular components without light illumination, which is described as dark toxicity (41). The dark toxicity of MB embedded in PAA nanoparticles was also determined and compared to that of free MB by using S. aureus strain as the target, since gram-positive bacteria are more sensitive to free MB (41). The result revealed that free MB had a significant dark toxicity to S. aureus cells, whereas MB-embedded PAA nanoparticles had no observable dark toxicity to S. aureus cells (Fig. 6B) and also retained the similar killing efficacy compared to free MB (Fig. 6A). It is likely, therefore, that the encapsulation of MB in the PAA nanoparticles prevents its direct contact with bacterial cells and host tissues, and thus MB-PAA-DNP together with localized illumination provides greater control and may be useful for the eradication of biofilms formed on specific locations without affecting the host tissues or commensal organisms outside of the treated area. Examples of potential applications of MB-PAA-DNP could be treatment of wound infections and dental plagues by topical spraying MB-PAA-DNP onto the infection site, followed by red light illumination, which have been demonstrated by other studies (16, 24, 49, 52). Animal model studies and clinical trial of such application are warranted.
FIG. 6.
Comparison of killing efficiency (A) and dark toxicity (B) of free MB and MB-PAA-DNP. (A) Cultures of S. aureus or Acinetobacter sp. strain AC811 were incubated with free MB (3 μM, ░⃞) and MB-PAA-DNP (10 mg/ml, equivalent to 3 μM free MB, ▪) and exposed to red light with a light irradiance of 38 mW/cm2 and a light fluence of 68.4 J/cm2. Viable cells were determined after light illumination. (B) The culture of S. aureus was incubated with MB (3 μM, ▴) and MB-PAA nanoparticles (10 mg/ml, equivalent to 3 μM free MB, □) without light illumination, and viable cells were determined at each incubation time point. Error bars indicate the standard errors of three replicates.
It should be noted that, in our study, the bacterial killing efficacy could be further improved by an increase in light irradiance, light fluence, and the concentration of the MB-PAA-DNP. The optimal relationships between parameters such as mass concentration of the MB-PAA-DNP, intensity of illumination, duration of illumination, and required O2 concentrations, for achieving the most efficient cell kill still remain to be determined. For example, in the present study, initial cell density tested had no effect on killing efficiency. However, one previous study showed that initial microbial cell density did affect killing efficacy (8). Our different observation may be due to the ratio of mass concentration of nanoparticles to cell density, different property of PSs, and other conditions used in these studies.
Encapsulation of the dye within the body of the polyacrylamide matrix obviates the need for heroic measures to limit the effects of ambient (endogenous) reductive enzymes that diminish the effectiveness of MB. Moreover, the tendency of MB to dimerize to an inactive form is virtually eliminated by encapsulation in the body of the nanoparticles (42). The red-shifted excitation wavelength of MB makes it an ideal candidate for use in biological systems and allows for greater penetration of the activating light with minimal diffusion.
In summary, these data demonstrate the usefulness of encapsulating photosensitizing dyes such as MB in the matrix of hydrogel nanoparticles for the reduction or control of microbial growth and biofilm-related infections. Additional optimization is required to maximize the efficiency of these nanoparticles and to target them to specific microbial subpopulations of nuisance environmental species and/or pathogenic strains.
Acknowledgments
Funding for this study came from internal discretionary funds at the University of Michigan School of Public Health (C.X.) and also partially from the National Science Foundation (NSF-DMR 0455330 [R.K.]).
Footnotes
Published ahead of print on 4 May 2009.
REFERENCES
- 1.Allesen-Holm, M., K. B. Barken, L. Yang, M. Klausen, J. S. Webb, S. Kjelleberg, S. Molin, M. Givskov, and T. Tolker-Nielsen. 2006. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 59:1114-1128. [DOI] [PubMed] [Google Scholar]
- 2.Bonnett, R. 1995. Photosensitizers of the porphyrin and phthalocyanine series for photodynamic therapy. Chem. Soc. Rev. 24:19-33. [Google Scholar]
- 3.Cassell, G. H., and J. Mekalanos. 2001. Development of antimicrobial agents in the era of new and reemerging infectious diseases and increasing antibiotic resistance. JAMA 285:601-605. [DOI] [PubMed] [Google Scholar]
- 4.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 5.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 6.Davies, D. G., and G. G. Geesey. 1995. Regulation of the alginate biosynthesis gene algC in Pseudomonas aeruginosa during biofilm development in continuous culture. Appl. Environ. Microbiol. 61:860-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Bentzmann, S., M. Aurouze, G. Ball, and A. Filloux. 2006. FppA, a novel Pseudomonas aeruginosa prepilin peptidase involved in assembly of type IVb pili. J. Bacteriol. 188:4851-4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Demidova, T. N., and M. R. Hamblin. 2005. Effect of cell-photosensitizer binding and cell density on microbial photoinactivation. Antimicrob. Agents Chemother. 49:2329-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Derosa, M. C., and R. J. Crutchley. 2002. Photosensitized singlet oxygen and its applications. Coord. Chem. Rev. 233-234:351-371. [Google Scholar]
- 10.Dougherty, T. J., C. J. Gomer, B. W. Henderson, G. Jori, D. Kessel, M. Korbelik, J. Moan, and Q. Peng. 1998. Photodynamic therapy. J. Natl. Cancer Inst. 90:889-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gabrielli, D., E. Belisle, D. Severino, A. J. Kowaltowski, and M. S. Baptista. 2004. Binding, aggregation, and photochemical properties of methylene blue in mitochondrial suspensions. Photochem. Photobiol. 79:227-232. [DOI] [PubMed] [Google Scholar]
- 12.Giltner, C. L., E. J. van Schaik, G. F. Audette, D. Kao, R. S. Hodges, D. J. Hassett, and R. T. Irvin. 2006. The Pseudomonas aeruginosa type IV pilin receptor binding domain functions as an adhesin for both biotic and abiotic surfaces. Mol. Microbiol. 59:1083-1096. [DOI] [PubMed] [Google Scholar]
- 13.Hall-Stoodley, L., J. W. Costerton, and P. Stoodley. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2:95-108. [DOI] [PubMed] [Google Scholar]
- 14.Hamblin, M. R., and T. Hasan. 2004. Photodynamic therapy: a new antimicrobial approach to infectious disease? Photochem. Photobiol. Sci. 3:436-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamblin, M. R., D. A. O'Donnell, N. Murthy, C. H. Contag, and T. Hasan. 2002. Rapid control of wound infections by targeted photodynamic therapy monitored by in vivo bioluminescence imaging. Photochem. Photobiol. 75:51-57. [DOI] [PubMed] [Google Scholar]
- 16.Hamblin, M. R., T. Zahra, C. H. Contag, A. T. McManus, and T. Hasan. 2003. Optical monitoring and treatment of potentially lethal wound infections in vivo. J. Infect. Dis. 187:1717-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hinman, A. R. 1998. Global progress in infectious disease control. Vaccine 16:1116-1121. [DOI] [PubMed] [Google Scholar]
- 18.Hunter, R. C., and T. J. Beveridge. 2005. Application of a pH-sensitive fluoroprobe (C-SNARF-4) for pH microenvironment analysis in Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 71:2501-2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenkins, A. T., A. Buckling, M. McGhee, and R. H. ffrench-Constant. 2005. Surface plasmon resonance shows that type IV pili are important in surface attachment by Pseudomonas aeruginosa. J. R. Soc. Interface 2:255-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karrer, S., C. Abels, M. B. Wimmershoff, M. Landthaler, and R. M. Szeimies. 2002. Successful treatment of cutaneous sarcoidosis using topical photodynamic therapy. Arch. Dermatol. 138:581-584. [DOI] [PubMed] [Google Scholar]
- 21.Komerik, N., M. Wilson, and S. Poole. 2000. The effect of photodynamic action on two virulence factors of gram-negative bacteria. Photochem. Photobiol. 72:676-680. [DOI] [PubMed] [Google Scholar]
- 22.Konan, Y. N., R. Gurny, and E. Allemann. 2002. State of the art in the delivery of photosensitizers for photodynamic therapy. J. Photochem. Photobiol. B 66:89-106. [DOI] [PubMed] [Google Scholar]
- 23.Koo, Y. E., G. R. Reddy, M. Bhojani, R. Schneider, M. A. Philbert, A. Rehemtulla, B. D. Ross, and R. Kopelman. 2006. Brain cancer diagnosis and therapy with nanoplatforms. Adv. Drug Deliv. Rev. 58:1556-1577. [DOI] [PubMed] [Google Scholar]
- 24.Lambrechts, S. A., T. N. Demidova, M. C. Aalders, T. Hasan, and M. R. Hamblin. 2005. Photodynamic therapy for Staphylococcus aureus-infected burn wounds in mice. Photochem. Photobiol. Sci. 4:503-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lombard, G. F., S. Tealdi, and M. M. Lanotte. 1985. The treatment of neurosurgical infections by lasers and porphyrins, p. 363-366. In G. Jori and C. A. Perria (ed.), Photodynamic therapy of tumors and other disease. Edizione Libreria Progetto, Padua, Italy.
- 26.Mellish, K. J., R. D. Cox, D. I. Vernon, J. Griffiths, and S. B. Brown. 2002. In vitro photodynamic activity of a series of methylene blue analogues. Photochem. Photobiol. 75:392-397. [DOI] [PubMed] [Google Scholar]
- 27.Moreno, M. J., E. Monson, R. G. Reddy, A. Rehemtulla, B. D. Ross, M. A. Philbert, R. J. Schneider, and R. Kopelman. 2003. Production of singlet oxygen by Ru(dpp(SO3)2)3 incorporated in polyacrylamide PEBBLES. Sens. Actuators B Chem. 90:82-89. [Google Scholar]
- 28.Orth, K., G. Beck, F. Genze, and A. Ruck. 2000. Methylene blue mediated photodynamic therapy in experimental colorectal tumors in mice. J. Photochem. Photobiol. B 57:186-192. [DOI] [PubMed] [Google Scholar]
- 29.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 30.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 31.Reddy, G. R., M. S. Bhojani, P. McConville, J. Moody, B. A. Moffat, D. E. Hall, G. Kim, Y. E. Koo, M. J. Woolliscroft, J. V. Sugai, T. D. Johnson, M. A. Philbert, R. Kopelman, A. Rehemtulla, and B. D. Ross. 2006. Vascular targeted nanoparticles for imaging and treatment of brain tumors. Clin. Cancer Res. 12:6677-6686. [DOI] [PubMed] [Google Scholar]
- 32.Redmond, R. W., and J. N. Gamlin. 1999. A compilation of singlet oxygen yields from biologically relevant molecules. Photochem. Photobiol. 70:391-475. [PubMed] [Google Scholar]
- 33.Roome, A. P., A. E. Tinkler, A. L. Hilton, D. G. Montefiore, and D. Waller. 1975. Neutral red with photoinactivation in the treatment of herpes genitalis. Br. J. Vener. Dis. 51:130-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharman, W. M., C. M. Allen, and J. E. van Lier. 1999. Photodynamic therapeutics: basic principles and clinical applications. Drug Discov. Today 4:507-517. [DOI] [PubMed] [Google Scholar]
- 35.Shikowitz, M. J., A. L. Abramson, K. Freeman, B. M. Steinberg, and M. Nouri. 1998. Efficacy of DHE photodynamic therapy for respiratory papillomatosis: immediate and long-term results. Laryngoscope 108:962-967. [DOI] [PubMed] [Google Scholar]
- 36.Shikowitz, M. J., A. L. Abramson, B. M. Steinberg, J. DeVoti, V. R. Bonagura, V. Mullooly, M. Nouri, A. M. Ronn, A. Inglis, J. McClay, and K. Freeman. 2005. Clinical trial of photodynamic therapy with meso-tetra (hydroxyphenyl) chlorin for respiratory papillomatosis. Arch. Otolaryngol. Head Neck Surg. 131:99-105. [DOI] [PubMed] [Google Scholar]
- 37.Tang, W., H. Xu, R. Kopelman, and M. A. Philbert. 2005. Photodynamic characterization and in vitro application of methylene blue-containing nanoparticle platforms. Photochem. Photobiol. 81:242-249. [DOI] [PubMed] [Google Scholar]
- 38.Tang, W., H. Xu, E. J. Park, M. A. Philbert, and R. Kopelman. 2008. Encapsulation of methylene blue in polyacrylamide nanoparticle platforms protects its photodynamic effectiveness. Biochem. Biophys. Res. Commun. 369:579-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teichert, M. C., J. W. Jones, M. N. Usacheva, and M. A. Biel. 2002. Treatment of oral candidiasis with methylene blue-mediated photodynamic therapy in an immunodeficient murine model. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 93:155-160. [DOI] [PubMed] [Google Scholar]
- 40.Tuite, E. M., and J. M. Kelly. 1993. Photochemical interactions of methylene blue and analogues with DNA and other biological substrates. J. Photochem. Photobiol. B 21:103-124. [DOI] [PubMed] [Google Scholar]
- 41.Usacheva, M. N., M. C. Teichert, and M. A. Biel. 2001. Comparison of the methylene blue and toluidine blue photobactericidal efficacy against gram-positive and gram-negative microorganisms. Lasers Surg. Med. 29:165-173. [DOI] [PubMed] [Google Scholar]
- 42.Usacheva, M. N., M. C. Teichert, and M. A. Biel. 2003. The role of the methylene blue and toluidine blue monomers and dimers in the photoinactivation of bacteria. J. Photochem. Photobiol. B 71:87-98. [DOI] [PubMed] [Google Scholar]
- 43.Wainwright, M. 1996. Non-porphyrin photosensitizers in biomedicine. Chem. Soc. Rev. 25:351-359. [Google Scholar]
- 44.Wainwright, M. 1998. Photodynamic antimicrobial chemotherapy (PACT). J. Antimicrob. Chemother. 42:13-28. [DOI] [PubMed] [Google Scholar]
- 45.Wainwright, M., D. A. Phoenix, S. L. Laycock, D. R. Wareing, and P. A. Wright. 1998. Photobactericidal activity of phenothiazinium dyes against methicillin-resistant strains of Staphylococcus aureus. FEMS Microbiol. Lett. 160:177-181. [DOI] [PubMed] [Google Scholar]
- 46.Wainwright, M., D. A. Phoenix, L. Rice, S. M. Burrow, and J. Waring. 1997. Increased cytotoxicity and phototoxicity in the methylene blue series via chromophore methylation. J. Photochem. Photobiol. B 40:233-239. [DOI] [PubMed] [Google Scholar]
- 47.Williams, J. L., J. Stamp, R. Devonshire, and G. J. Fowler. 1989. Methylene blue and the photodynamic therapy of superficial bladder cancer. J. Photochem. Photobiol. B 4:229-232. [DOI] [PubMed] [Google Scholar]
- 48.Wood, S., D. Metcalf, D. Devine, and C. Robinson. 2006. Erythrosine is a potential photosensitizer for the photodynamic therapy of oral plaque biofilms. J. Antimicrob. Chemother. 57:680-684. [DOI] [PubMed] [Google Scholar]
- 49.Wood, S., B. Nattress, J. Kirkham, R. Shore, S. Brookes, J. Griffiths, and C. Robinson. 1999. An in vitro study of the use of photodynamic therapy for the treatment of natural oral plaque biofilms formed in vivo. J. Photochem. Photobiol. B 50:1-7. [DOI] [PubMed] [Google Scholar]
- 50.Xi, C., D. Marks, S. Schlachter, W. Luo, and S. A. Boppart. 2006. High-resolution three-dimensional imaging of biofilm development using optical coherence tomography. J. Biomed. Opt. 11:34001. [DOI] [PubMed] [Google Scholar]
- 51.Xu, H., S. M. Buck, R. Kopelman, M. A. Philbert, M. Brasuel, B. D. Ross, and A. Rehemtulla. 2004. Photoexcitation-based nano-explorers: chemical analysis inside live cells and photodynamic therapy. Israel J. Chem. 44:317-337. [Google Scholar]
- 52.Zolfaghari, P. S., S. Packer, M. Singer, S. P. Nair, J. Bennett, C. Street, and M. Wilson. 2009. In vivo killing of Staphylococcus aureus using a light-activated antimicrobial agent. BMC Microbiol. 9:27. [DOI] [PMC free article] [PubMed] [Google Scholar]