Abstract
The extent to which clonal spread contributes to emerging antimicrobial resistance in Escherichia coli is incompletely defined. To address this question within a recent, nationally representative strain collection, three established drug-resistant E. coli clonal groups (i.e., clonal group A, E. coli O15:K52:H1, and sequence type 131 [ST131]) were sought among 199 E. coli urine isolates recovered from across Canada from 2002 to 2004, with stratification by resistance to trimethoprim-sulfamethoxazole (TS) and fluoroquinolones (FQs). The isolates' clonal backgrounds, virulence genotypes, and macrorestriction profiles were assessed. The three clonal groups were found to account for 37.2% of isolates overall, but accounted for 0% of TS-susceptible (TS-S) and FQ-susceptible (FQ-S) isolates, 20% of TS-resistant (TS-R) and FQ-S isolates, 60% of TS-S and FQ-R isolates, and 68% of TS-R and FQ-R isolates (P < 0.001). E. coli ST131, the most prevalent clonal group, accounted for 23.1% of isolates overall and for 44% of the FQ-R isolates. Nearly all ST131 isolates were FQ-R (96%) but, notably, cephalosporin susceptible (98%). Although the distinctive virulence profiles of the FQ-R clonal group isolates were less extensive than those of the susceptible isolates, they were significantly more extensive than those of the other FQ-R isolates. These findings indicate that among the E. coli urine isolates studied, resistance to TS and FQs has a prominent clonal component, with the O15:K52:H1 clonal group and especially E. coli ST131 being the major contributors. These clonal groups appear to be more virulent than comparably resistant isolates, possibly contributing to their success as emerging multi-drug-resistant pathogens.
Resistance to first-line antimicrobial agents has become increasingly prevalent in Escherichia coli. This leads to a greater reliance on newer, more expensive agents and delays the institution of appropriate antimicrobial therapy for extraintestinal E. coli infections, which increases the associated rates of morbidity and mortality and costs (9, 10, 28, 30, 33). Better understandings of the basis for this phenomenon are needed to guide the rational development of preventive measures.
Emerging resistance in E. coli is likely due to a combination of (i) the conversion of susceptible strains to resistance and (ii) the expansion and dissemination of already resistant clones. Examples of the latter phenomenon include the outbreak in South London, United Kingdom, involving trimethoprim-sulfamethoxazole (TS)-resistant (TS-R) strains of E. coli O15:K52:H1 (1986 to 1987) (25, 26); disseminated urinary tract infections (UTIs) in the United States due to TS-R strains of E. coli clonal group A (CGA) (1990s) (2, 12, 22); and widespread infections in Canada, Europe, and Asia due to strains of E. coli sequence type 131 (ST131; O25:H4) that exhibit extended-spectrum cephalosporin (ESC) resistance through the production of CTX-M-15, an extended-spectrum beta-lactamase (2000s) (6, 7, 24). Determining the importance of clonal spread has immediate practical implications, since a finding that clonal spread is a major contributor to emerging resistance would suggest a need to define and interrupt transmission pathways, in addition to reducing selection pressure by limiting antimicrobial use.
Several recent studies from Canada have suggested a clonal component to emerging antimicrobial resistance in E. coli. Relevant findings include the prominence of two anonymous clonal groups among fluoroquinolone (FQ)-resistant (FQ-R) urine isolates (2002 to 2004) (19); a high prevalence of CGA and ST131 among TS-R and FQ-R E. coli urine isolates, respectively, from university students (2008) (23); and regionwide extended-spectrum beta-lactamase-positive E. coli infections due to ST131 and other clonal groups (2004 to 2005) (24, 27). To further address this question with a broadly representative sample, we assessed the clonal backgrounds of TS-R and FQ-R E. coli isolates from patients with UTIs from across Canada. These isolates were collected systematically from 2002 to 2004 through the North American Urinary Tract Infection Collaborative Alliance (NAUTICA) project. We screened 199 randomly selected NAUTICA urine isolates (33, 34), stratified by TS and FQ resistance phenotype, for CGA, E. coli O15:K52:H1, and ST131 and then compared clonal group members with other isolates for phylogenetic group, virulence genotype, and pulsed-field gel electrophoresis (PFGE) profile.
MATERIALS AND METHODS
Subjects and isolates.
NAUTICA is a UTI surveillance study involving 40 medical centers (30 from the United States, 10 from Canada). From 2002 through 2004, each center submitted up to 50 consecutive midstream urine isolates. The isolates were identified to the species level by each laboratory's existing protocol. It was not known whether the urine samples submitted came from patients with symptomatic UTIs or from patients with asymptomatic bacteriuria. The isolates (one per patient) and accompanying basic demographic data were submitted to the coordinating laboratory (Health Sciences Centre, Winnipeg, Manitoba, Canada), where broth microdilution susceptibility testing was done, including testing for susceptibility to TS, ciprofloxacin (an FQ), and cefdinir, by using the methods, control strains, and interpretive criteria specified by the CLSI (formerly the NCCLS). For the present study, four subsets of approximately 50 isolates each were randomly selected from the total Canadian NAUTICA isolate collection according to the combined TS and FQ (ciprofloxacin) phenotype, i.e., TS susceptible (TS-S) and FQ susceptible (FQ-S), TS-R and FQ-S, TS-S and FQ-R, and TS-R and FQ-R.
Molecular analysis.
XbaI PFGE profiles were generated according to the standard protocol and were used to construct a Dice coefficient-based similarity dendrogram within the BioNumerics program (Bio-Rad), as described elsewhere (19). Major E. coli phylogenetic groups (group A, B1, B2, or D) and extended virulence genotypes for 58 extraintestinal pathogenic E. coli-associated virulence genes and variants thereof were determined in duplicate by using established PCR-based methods in which boiled lysates were used as the template DNA (4, 11, 16, 18). A virulence profile dendrogram was inferred by the unweighted pair group method with averaging on the basis of the pairwise similarity relationship according to the presence or the absence of all virulence markers. The virulence score was the number of unique virulence markers detected.
For the detection of CGA, all group D isolates were screened by PCR for CGA-associated single-nucleotide polymorphisms (SNPs) in fumC (14) and gyrB (i.e., gyrB T162G, C174A, T411C, and T417C). The presence of all these SNPs identifies a strain as belonging to clonal complex 69 (CC69; which corresponds to CGA), to the exclusion of closely related CC394 (32). The primers used for the detection of gyrB SNPs were gyrB27f (5′-GGTGCGTTTCTGGCCA-3′) and gyrB27r (5′-GACGCCGATACCATC-3′). Amplification was done with 4.0 mM MgCl2 and a cycling protocol of 95°C for 10 min; then 32 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min; and then a final extension at 72°C for 10 min and a hold at 4°C. In validation experiments, these gyrB-specific primers, when they were multiplexed with the published fumC primers (14), yielded a 259-bp gyrB amplicon and a 175-bp fumC amplicon with all 143 CC69 control strains (sensitivity, 100%; 95% confidence interval, 97.5% to 100%) and no product with 119 E. coli strains representing either CC394 (n = 10) or 77 unique non-CC69 and non-CC394 STs (n = 109) (specificity, 100%; 95% confidence interval, 97% to 100%).
For the detection of the O15:K52:H1 clonal group, all group D isolates were screened by PCR for (i) the O15 rfb allele (5) and (ii) an SNP in fumC that is specific for CC31 (which corresponds to the O15:K52:H1 clonal group) (15). Isolates that were positive by these assays, which yielded 100% concordant results, underwent random amplified polymorphic DNA (RAPD) analysis, in which the isolates were compared with O15:K52:H1 reference strains (1, 17); this uniformly confirmed the isolates' O15:K52:H1 clonal group status.
For the detection of ST131, all group B2 isolates were screened by PCR for (i) the ST131-associated O25b rfb variant (Clermont) and (ii) ST131-associated SNPs in mdh (i.e., C288T and C525T) and gyrB (i.e., C621T, C729T, and T735C). The primers used for the detection of mdh SNPs were mdh36_forward (5′-GTTTAACGTTAACGCCGGT-3′) and mdh36_reverse (5′-GGTAACACCAGAGTGACCA-3′), and the primers used for the detection of gyrB SNPs were gyrB47_forward (5′-CGCGATAAGCGCGAC-3′) and gyrB47_reverse (5′-ACCGTCTTTTTCGGTGGAA-3′). Amplification was done with 4.0 mM MgCl2 and a cycling protocol of 95°C for 10 min; then 32 cycles of 94°C for 30 s, 65°C for 30 s, and 68°C for 2 min; and then a hold at 4°C. In validation experiments, these primers yielded 275-bp and 132-bp amplicons, respectively, with 34 diverse ST131 control strains (sensitivity, 100%; 95% confidence interval, 90% to 100%) and no product with 86 E. coli strains representing unique STs other than ST131 (specificity, 100%; 95% confidence interval, 96% to 100%).
Isolates that tested positive by both assays (which yielded 100% concordant results) were further assessed for ST131 status by RAPD analysis in which the isolates were compared with reference ST131 strains. They were also tested for blaCTX-M-15 by PCR with established primers and control strains (6, 21).
To confirm these presumptive clonal group assignments, selected isolates underwent confirmatory multilocus sequence typing based on the partial sequences of adk, fumC, gyrB, icd, mdh, purA, and recA (http://mlst.ucc.ie). Assignment to CC69, CC31, or ST131 by multilocus sequence typing, which occurred precisely as anticipated in each instance, was regarded as confirming membership in CGA, E. coli O15:K52:H1, and ST131, respectively.
Statistical methods.
Unpaired comparisons involving proportions or continuous variables were tested by using Fisher's exact test or the Kruskal-Wallis or the Mann-Whitney U test, respectively (all tests were two tailed). Paired comparisons of proportions were tested by McNemar's test. For prevalence assessments of the clonal groups (individually and combined) by resistance phenotype, the four individual susceptibility subgroups (TS-S and FQ-S, TS-R and FQ-S, TS-S and FQ-R, and TS-R and FQ-R) and various combinations thereof (e.g., all TS-S versus all TS-R or all FQ-S versus all FQ-R) were compared. For prevalence assessments of bacterial traits by clonal group, an initial four-way comparison was done (for the three clonal groups and all other isolates, which were combined as a reference group), and only if this yielded a P value of <0.05 were additional comparisons tested.
RESULTS
Description of population.
The 199 study isolates from Canadian ambulatory patients with UTIs collected from 2002 to 2004 were selected to give four susceptibility subgroups representing all four TS-FQ phenotypes, i.e., TS-S and FQ-S (n = 49), TS-R and FQ-S (n = 50), TS-S and FQ-R (n = 50), and TS-R and FQ-R (n = 50). The isolates were derived from eight southern Canadian provinces (British Columbia east through Quebec, plus New Brunswick and Nova Scotia), with each province except New Brunswick contributing from 21 to 37 isolates; New Brunswick contributed 13 isolates. The subjects were predominantly female (81%), with a median age of 50 years (age range, 1 to 93 years); 13% were <18 years old.
Prevalence and distribution of clonal groups.
According to molecular typing, CGA accounted for 9 (4.5%) of the 199 study isolates, E. coli O15:K52:H1 accounted for 19 (9.5%), and ST131 accounted for 46 (23.1%). Thus, the three clonal groups collectively accounted for 37.2% of the total population. ST131 was significantly more prevalent than either of the other two groups (P ≤ 0.001 for each comparison, McNemar's test), whereas the O15:K52:H1 clonal group was borderline significantly more prevalent than CGA (P = 0.087, McNemar's test).
The three clonal groups were widely distributed across the eight provinces, and there was no statistically significant geographical clustering. The three clonal groups were not associated with the age of the host, after adjustment for susceptibility status (data not shown). In contrast, they were significantly distributed according to susceptibility status, with all three clonal groups being confined to the three resistance-associated subgroups (Table 1). CGA was concentrated within the TS-R and FQ-S group and thus was positively associated with TS-R and was negatively associated with FQ-R. Conversely, the O15:K52:H1 and ST131 clonal groups were concentrated within the FQ-R subgroups (whether they were TS-S or TS-R) and thus were strongly associated with FQ-R but not TS-R. Collectively, the three clonal groups accounted for 44% of the TS-R isolates, 64% of the FQ-R isolates, and 68% of the dually resistant isolates. Notably, ST131 alone accounted for 44% of the FQ-R isolates.
TABLE 1.
Distribution of three clonal groups by resistance subgroup among 199 Escherichia coli urine isolates from across Canada, 2002 to 2004
| Clonal group | Total no. (n = 199) | Prevalence (no.) of clonal group
|
P value for 4-group comparison | Prevalence (no.) of clonal group
|
P value for TS-S vs TS-R | Prevalence (no.) of clonal group
|
P value for FQ-S vs FQ-R | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FQ-S
|
FQ-R
|
|||||||||||
| TS-S (n = 49) | TS-R (n = 50) | TS-S (n = 50) | TS-R (n = 50) | TS-S (n = 99) | TS-R (n = 100) | FQ-S (n = 99) | FQ-R (n = 100) | |||||
| CGA | 9 (4.5)b | 0 (0) | 8 (16) | 0 (0) | 1 (2) | <0.001 | 0 (0) | 9 (9) | 0.003 | 8 (8) | 1 (1) | (0.018)c |
| O15:K52:H1 | 19 (9.5) | 0 (0) | 0 (0) | 7 (14) | 12 (24) | <0.001 | 7 (7) | 12 (12) | 0 (0) | 19 (19) | <0.001 | |
| ST131 | 46 (23.1) | 0 (0) | 2 (4) | 23 (46) | 21 (42) | <0.001 | 23 (23) | 23 (23) | 2 (2) | 44 (44) | <0.001 | |
| Any | 74 (37.2) | 0 (0) | 10 (20) | 30 (60) | 34 (68) | <0.001 | 30 (30) | 44 (44) | 10 (10) | 64 (64) | <0.001 | |
a P values (by Fisher's exact test) are shown when P was <0.05.
Values in parentheses indicate the percentage on the basis of the total number of isolates for that column.
The P value in parentheses indicates a negative association with resistance.
Molecular characteristics.
Of the 58 virulence genes for whose presence the isolates were evaluated, 54 (93%) were detected within the population (Table 2). Comparisons of virulence gene prevalence values among the three clonal groups and the remaining isolates identified significant differences for 31 (57%) of the 54 virulence genes detected. Distinctive characteristics of CGA (from phylogenetic group D) included the presence of the pap operon (with the F16 papA allele and papG allele II), iha, sat, iutA, traT, and ompT, plus the absence of many group B2-associated traits. Distinctive characteristics of the O15:K52:H1 clonal group (also from phylogenetic group D) included all of the traits mentioned above except traT and ompT, plus fyuA and kpsM II. Distinctive characteristics of ST131 (from phylogenetic group B2) included the absence of many typical group B2-associated traits but the presence of the F10 papA allele, iha, sat, fyuA, iutA, kfiC (K5 capsule), usp, ompT, and malX. Additionally, all of the ST131 isolates except one (i.e., 98%) were susceptible to cefdinir and negative for blaCTX-M-15. Although the prevalence of fimH did not differ significantly among the three individual clonal groups and the nonclonal group strains (data not shown), the three clonal groups collectively exhibited a significantly higher prevalence of fimH than the remaining isolates (67/67 [100%] versus 120/132 [91%]; P = 0.009).
TABLE 2.
Distinctive virulence characteristics of three clonal groups among 199 Escherichia coli urine isolates from across Canada, 2002 to 2004
| Category | Specific traita | Total no. (n = 199) | Prevalence (no.) of trait by clonal group
|
P valueb
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| None (n = 125) | CGA (n = 9) | O15:K52:H1 (n = 19) | ST131 (n = 46) | Overall (four-group test) | Any clonal group vs none | CGA vs none | O15:K52:H1 vs none | ST131 vs none | |||
| Adhesin | papA | 73 (37)c | 48 (38) | 8 (89) | 17 (90) | 0 (0) | <0.001 | 0.004 | <0.001 | (<0.001) | |
| F10 allele | 77 (34) | 31 (25) | 1 (11) | 0 (0) | 45 (98) | <0.001 | (0.001) | (0.01) | <0.001 | ||
| F16 allele | 27 (14) | 2 (2) | 8 (89) | 17 (90) | 0 (0) | <0.001 | <0.001 | <0.001 | <0.001 | ||
| papG II | 58 (29) | 33 (26) | 8 (89) | 17 (90) | 0 (0) | <0.001 | 0.001 | <0.001 | (<0.001) | ||
| papG III | 20 (10) | 20 (16) | 0 (0) | 0 (0) | 0 (0) | 0.002 | (<0.001) | . | (0.002) | ||
| sfa/foc | 29 (15) | 29 (23) | 0 (0) | 0 (0) | 0 (0) | <0.001 | (<0.001) | (0.01) | (<0.001) | ||
| focG | 17 (9) | 17 (14) | 0 (0) | 0 (0) | 0 (0) | 0.01 | (0.001) | (0.007) | |||
| iha | 114 (57) | 43 (34) | 9 (100) | 18 (95) | 44 (96) | <0.001 | <0.001 | 0.001 | <0.001 | <0.001 | |
| hra | 47 (24) | 47 (36) | 0 (0) | 0 (0) | 0 (0) | <0.001 | (<0.001) | (0.03) | (0.001) | (<0.001) | |
| Toxin | hlyD | 38 (19) | 38 (30) | 0 (0) | 0 (0) | 0 (0) | <0.001 | (<0.001) | (0.004) | (<0.001) | |
| cnf1 | 29 (15) | 29 (23) | 0 (0) | 0 (0) | 0 (0) | <0.001 | (<0.001) | (0.01) | (<0.001) | ||
| sat | 111 (56) | 38 (30) | 9 (100) | 18 (95) | 46 (100) | <0.001 | <0.001 | 0.004 | <0.001 | <0.001 | |
| vat | 62 (31) | 62 (50) | 0 (0) | 0 (0) | 0 (0) | <0.001 | (<0.001) | (0.009) | (<0.001) | (<0.001) | |
| pic | 14 (7) | 14 (11) | 0 (0) | 0 (0) | 0 (0) | (0.03) | (0.001) | (0.02) | |||
| astA | 14 (7) | 14 (11) | 0 (0) | 0 (0) | 0 (0) | (0.03) | (0.001) | (0.02) | |||
| Siderophore | iroN | 40 (20) | 39 (31) | 0 (0) | 0 (0) | 1 (2) | <0.001 | (<0.001) | (0.002) | (<0.001) | |
| fyuA | 168 (84) | 95 (76) | 9 (100) | 19 (100) | 45 (98) | <0.001 | <0.001 | 0.01 | 0.001 | ||
| ireA | 26 (13) | 26 (21) | 0 (0) | 0 (0) | 0 (00) | <0.001 | (<0.001) | (<0.001) | |||
| iutA | 147 (74) | 74 (59) | 9 (100) | 18 (95) | 46 (100) | <0.001 | <0.001 | 0.01 | 0.002 | <0.001 | |
| Capsule | kpsM II | 135 (68) | 82 (66) | 9 (100) | 19 (100) | 25 (54) | <0.001 | 0.001 | |||
| K1 kpsM | 37 (19) | 37 (30) | 0 (0) | 0 (0) | 0 (0) | <0.001 | (<0.001) | (0.004) | (<0.001) | ||
| K5 kfiC | 35 (18) | 12 (10) | 1 (11) | 1 (5) | 21 (46) | <0.001 | 0.001 | <0.001 | |||
| Miscellaneous | usp | 113 (57) | 66 (53) | 0 (0) | 1 (5) | 46 (100) | <0.001 | (0.003) | (<0.001) | <0.001 | |
| traT | 120 (60) | 79 (63) | 9 (100) | 3 (16) | 29 (63) | <0.001 | 0.03 | (<0.001) | |||
| ompT | 141 (71) | 76 (61) | 9 (100) | 10 (53) | 46 (100) | <0.001 | <0.001 | 0.03 | <0.001 | ||
| H7 fliC | 16 (8) | 16 (13) | 0 (0) | 0 (0) | 0 (0) | 0.01 | (0.002) | (0.007) | |||
| malX | 121 (10) | 74 (59) | 0 (0) | 1 (5) | 46 (100) | <0.001 | (<0.001) | (<0.001) | <0.001 | ||
| ibeA | 20 (10) | 19 (13) | 0 (0) | 0 (0) | 1 (2) | 0.02 | (0.001) | (0.02) | |||
The 31 traits shown yielded P values of <0.05 in four-group comparisons across the three clonal groups and all other isolates combined (none). Definitions: astA, enteroaggregative E. coli toxin; cnf1, cytotoxic necrotizing factor 1; focG, F1C fimbria; fyuA, yersiniabactin receptor; H7 fliC, flagellin variant; hra, heat-resistant agglutinin; hlyD, alpha-hemolysin; ibeA, invasion of brain endothelium; iha, adhesin-siderophore; ireA, siderophore receptor; iroN, salmochelin receptor; iutA, aerobactin receptor; kpsM II, group 2 capsule; K1, group 2 capsule variant; K5 kfiC, K5 group 2 capsule variant; malX, pathogenicity island marker; ompT, outer membrane protease; papA, P fimbria structural subunit, with its F10 and F16 alleles (note that the results for papC [assembly], papEF [tip pilins], and papG [adhesin molecule] were similar to those shown for papA); papG alleles II and III, P adhesin variants; pic, autotransporter protease; sat, secreted autotransporter toxin; sfa/foc, S and F1C fimbriae; traT, serum resistance associated; vat, vacuolating autotransporter toxin; and usp, uropathogenic specific protein. Twenty three traits detected in one or more isolate each but not yielding a P value of <0.05 in the four-group comparison (numbers in parentheses are the numbers of isolates positive): afa/dra, Dr-binding adhesins (n = 14); afaE8, variant afimbrial adhesin (n = 5); bmaE, M fimbria (n = 4); cdtB, cytolethal distending toxin (n = 5); cvaC, microcin (colicin) V (n = 9); fimH, type 1 fimbria (n = 188); gafD, G fimbria (n = 3); hlyF, variant hemolysin (n = 12); iss, increased serum survival (n = 12); K2, group 2 capsule variant (n = 7); K15, group 2 capsule variant (n = 1); kpsM III, group 3 capsule (n = 9); papA alleles F7-1 (n = 3), F7-2 (n = 3), F8 (n = 3), F9 (n = 5), F11 (n = 11), F12 (n = 4), F13 (n = 10), F14 (n = 3), F15 (n = 1); rfc, O4 lipopolysaccharide (n = 8); and sfaS, S fimbria adhesin (n = 10). The following were not detected: clpG, mannose-resistant adhesin; F17, mannose-resistant adhesin; F536, papA allele from strain 536; and papG allele I.
P values (by Fisher's exact test) are shown when P was <0.05. P values in parentheses indicate a negative association with the indicated clonal group(s).
Values in parentheses indicate the percentage on the basis of the total number of isolates for that column.
Virulence scores.
The aggregate virulence scores for the total population ranged from 0 to 20 (median, 9.0) and varied significantly by susceptibility subgroup, decreasing along a gradient from the TS-S and FQ-S isolates (median score, 13; range, 0 to 20) through the TS-R and FQ-S isolates (median score, 9.5; range, 0 to 19.75) to the FQ-R isolates, irrespective of TS susceptibility status (median score, 8.0; range, 0 to 15) (P < 0.001). The virulence scores differed only slightly among the three clonal groups and all other isolates combined, i.e., median scores of 8.0 (O15:K52H1), 9.0 (CGA), 9.0 (ST131), and 9.0 (combined nonclonal group isolates) (P = 0.03, four-group Kruskal-Wallis test) (Table 3). Consistent with their genetic homogeneity, the three clonal groups, whether individually or combined, exhibited a narrower range of virulence scores than the nonclonal group isolates (Table 3).
TABLE 3.
Virulence scores according to clonal group and resistance status among 199 Escherichia coli urine isolates from across Canada, 2002 to 2004
| Clonal groupa status and subset | No. of isolates | Median (range) virulence scoreb |
|---|---|---|
| Member | ||
| Total | 74 | 9.0 (3-12) |
| CGA | 9 | 9.0 (8.25-9.0) |
| O15:K52:H1 | 19 | 8.0 (3-9) |
| ST131 | 46 | 9.0 (8-12) |
| Nonmemberc | ||
| Total | 125 | 9.75 (0-20) |
| TS-S, FQ-S | 49 | 13.0 (0-20) |
| TS-R and/or FQ-R | 76 | 7.125 (0-19.75) |
| TS-R, FQ-S | 40 | 12.0 (1-19.75) |
| TS-S, FQ-R | 20 | 7.0 (1-10.75) |
| TS-R, FQ-R | 16 | 5.0 (0-9) |
Clonal group refers to one of the three clonal groups of interest, i.e., CGA, the O15:K52:H1 clonal group, and ST131.
P values (by Mann-Whitney U test) were as follows: for any clonal group versus nonclonal group, P > 0.10; for any clonal group versus nonclonal group (TS-S, FQ-S), P < 0.001; for any clonal group versus nonclonal group (TS-R and/or FQ-R), P = 0.04; for nonclonal group (TS-S, FQ-S) versus nonclonal group (TS-R and/or FQ-R), P < 0.001.
Nonmember indicates isolates other than those in one of the three clonal groups of interest.
The aggregate virulence scores of the 74 combined clonal group isolates did not differ significantly from those of the 125 combined nonclonal group isolates (median scores, 9.0 for each group) (Table 3). However, the nonclonal group isolates' virulence scores varied significantly with susceptibility subgroup, being higher for the 49 TS-S and FQ-S isolates (median, 13.0) than for the remaining 76 (resistance-associated) isolates (median score, 7.125; P < 0.001). Thus, although the virulence scores of the combined clonal group isolates were significantly lower than those of the 49 TS-S and FQ-S nonclonal group isolates (P < 0.001), they were significantly higher than those of the 76 TS-R and/or FQ-R nonclonal group isolates (P = 0.03) (Table 3).
When the virulence scores of the clonal group and the nonclonal group isolates within each resistance subgroup were compared, for the TS-R and FQ-S isolates the virulence scores of the clonal group members (mostly CGA) did not differ significantly from those of the nonclonal group members (data not shown). In contrast, within both the TS-S and FQ-R and the TS-R and FQ-R subgroups, the virulence scores of the clonal group members were significantly greater than those of the comparably resistant nonclonal group members (P = 0.002 and P < 0.001, respectively). The prevalence of clonal group members within a given TS-FQ subgroup exhibited a strong positive relationship with the difference in the median virulence scores between the subgroup's clonal group members and nonclonal group members (data not shown).
Extended virulence profiles.
In a dendrogram based on extended virulence profiles, which extended to the 70% similarity level overall, each of the three clonal groups was significantly concentrated within a group-specific cluster (data not shown), indicating considerable homogeneity of the virulence profiles within the clonal group and clonal group-specific virulence profiles. That is, CGA constituted 80% (8/10 isolates) of its specific cluster (100% identical profiles), whereas it constituted only 0.5% (1/189) of the other isolates (P < 0.001; 98% classification accuracy). Likewise, the O15:K52:H1 clonal group constituted 68% (17/25 isolates) of its specific cluster (97% similar profiles), whereas it constituted only 1.5% (2/174) of other isolates (P < 0.001; 95% classification accuracy). Finally, ST131 constituted 90% (46/51 isolates) of its specific cluster (93% similar profiles), whereas it constituted only 0% (0/148) of the other isolates (P < 0.001; 97% classification accuracy).
Pulsed-field analysis.
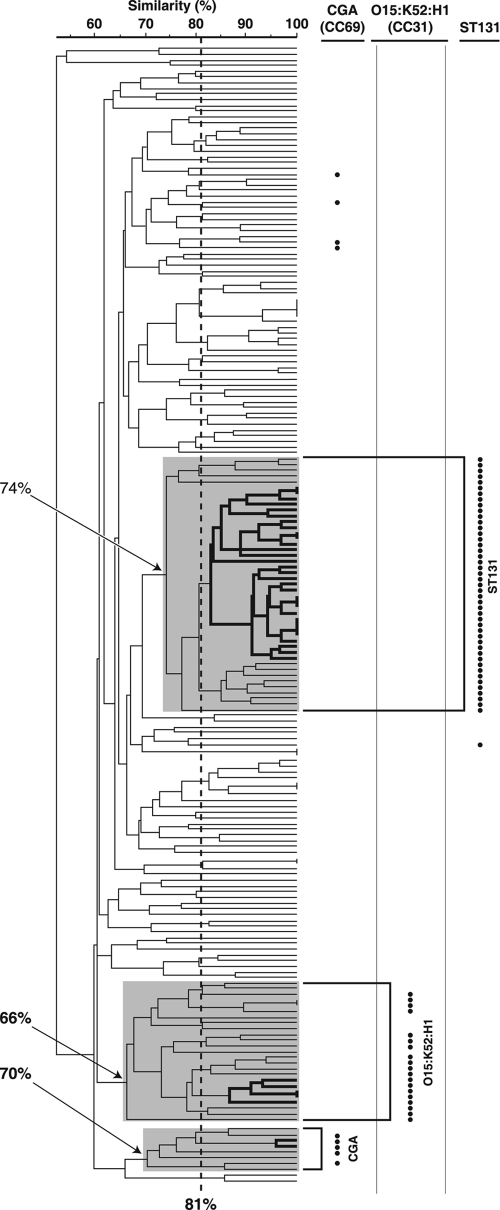
In an XbaI PFGE profile-based dendrogram, which extended to the 52% similarity level overall, each of the three clonal groups was significantly concentrated within a specific cluster (Fig. 1). CGA constituted 63% (5/8 isolates) of its specific cluster (70% similarity level), whereas it constituted only 2% (4/191) of the other isolates (P < 0.001; 96% classification accuracy). The O15:K52:H1 clonal group constituted 76% (19/25 isolates) of its specific cluster (66% similarity level), whereas it constituted only 0% (0/174) of the other isolates (P < 0.001; 97% classification accuracy). ST131 constituted 100% (45/45 isolates) of its main cluster (74% similarity level), whereas it constituted only 0.6% (1/154) of the other isolates (P < 0.001; 98% classification accuracy). However, at the 81% similarity level, the maximum numbers of clonal group members included within a single cluster was only 2 (22% of the group) for CGA, only 5 (26% of group) for E. coli O15:K52:H1, and only 31 (67% of the group) for ST131 (Fig. 1). A separate PFGE tree that included reference isolates from an earlier study of Canadian NAUTICA isolates confirmed that the two FQ-R-associated clonal clusters from that study (19) corresponded to ST131 and the O15:K52:H1 clonal group, respectively (data not shown).
FIG. 1.
Dendrogram of XbaI PFGE profiles for 199 Escherichia coli urine isolates collected from across Canada from 2002 to 2004. The dendrogram was inferred according to the unweighted pair group method with averaging based on Dice similarity coefficients. The bullets to the right of the dendrogram indicate membership in the three clonal groups studied, i.e., CGA (which corresponds to CC69), the O15:K52:H1 clonal group (which corresponds to CC31), and ST131, each as defined by multiple complementary molecular methods. The shaded areas within the dendrogram denote the three clonal group-specific clusters (which are labeled to the right of the dendrogram; arrows show the minimum percent similarity values for the clusters), as defined on the basis of the optimal separation of clonal group members from other isolates. The vertical dashed line indicates the 81% profile similarity threshold. Within each clonal group-specific cluster, heavy lines indicate the largest subcluster with ≥81% similar profiles. The CGA-specific cluster includes three such subclusters, each with two isolates, but only one of these is marked.
DISCUSSION
In this molecular epidemiological analysis of 199 E. coli urine isolates from Canadian patients with UTIs, collected from 2002 to 2004, we found that the three resistance-associated clonal groups of interest contributed substantially to the study population (37.2% of isolates), particularly the TS-R and FQ-R subsets (44% and 64% of isolates, respectively). Our findings provide novel insights into the epidemiology of each of the three clonal groups, suggest possible explanations for the observed trends, and imply that interruption of the dissemination and emergence of these clonal groups could slow or halt the rise in FQ resistance in E. coli.
The three clonal groups contributed to the resistant population in distinctive patterns. That is, whereas CGA was confined almost exclusively to the TS-R and FQ-S subset and accounted for 16% of the isolates in that subset, the O15:K52:H1 clonal group and ST131 were confined almost exclusively to the FQ-R subset and accounted for 19% and 44% of the isolates in that subset, respectively. These data suggest that CGA, which in some U.S. locales accounted for 34 to 50% of cases of TS-R uncomplicated E. coli cystitis and pyelonephritis in the late 1990s (2, 12, 22), may not have achieved a comparable prevalence level in Canada or, by the time of the NAUTICA project (2002 to 2004), had waned in prevalence. Alternatively, many of the study isolates may have derived from patients with asymptomatic bacteriuria or complicated UTIs, among whom CGA may be inherently less prevalent than it is in patients with uncomplicated cystitis and pyelonephritis. Additionally, CGA appears not to have become widely FQ-R, instead retaining its historical TS-R phenotype (2, 12, 13, 22). The CGA isolates exhibited VF scores somewhat lower than those for the nonclonal group TS-R and FQ-S isolates, possibly limiting CGA's relative fitness and contributing to its modest epidemiological success.
In contrast, the O15:K52:H1 clonal group, which has attracted little attention since its clinical debut during the striking 1986 to 1987 South London outbreak of TS-R UTIs and other extraintestinal infections (8, 17, 25, 26, 29), appears to have made a dramatic resurgence in Canada and is now an FQ-R pathogen. This apparent reemergence may have been driven both by the clonal group's relatively new FQ-R phenotype and its virulence profiles, which were significantly more extensive than those of nonclonal group FQ-R isolates. The unexpected prominence of this clonal group and its new appearance as an FQ-R pathogen, as was also recently noted in Europe (3), illustrate the dynamic nature of the microbial ecology of extraintestinal E. coli infections, underscoring the need for ongoing surveillance to capture new emerging trends.
The most striking clonal group was ST131, which was significantly more prevalent than the other two clonal groups and which accounted for nearly half of the FQ-R population. Although ST131 has gained notoriety internationally because of its central role in the emergence of CTX-M-15 (6, 7, 24), all but one of the ST131 isolates in the present study (i.e., 98%) were susceptible to ESCs and lacked blaCTX-M-15. This, together with similar recent reports from Italy (3), France (20), and Montreal, Quebec, Canada (23), suggests that although ST131 is currently best known for its association with CTX-M-15, it may be much more prevalent (and, hence, an equal or greater public health threat) as an FQ-R but ESC-susceptible pathogen. Moreover, the widespread abundance of ST131, which is clearly receptive to CTX-M-15-containing genetic elements, may provide the substrate for a future large-scale epidemic of CTX-M-15-mediated ESC resistance if these elements begin to propagate through the receptive ST131 population. Notably, for ST131, as for the other two clonal groups, a seeming relationship was evident between epidemiological success (i.e., prevalence) and virulence profiles, which for ST131 were significantly more extensive than those of comparably resistant nonclonal group isolates.
These findings emphasize the importance of clonal spread and expansion in the unfolding epidemic of FQ-R in E. coli, since the three clonal groups studied, particularly ST131 and the E. coli O15:K52:H1 clonal group, accounted for 64% of the FQ-R population. This is even higher than the 35% prevalence of ST131 and O15:K52:H1 noted by Cagnacci et al. among FQ-R urine isolates collected in eight European countries from 2003 to 2006 (3). As such, interventions directed against the spread of FQ-R clones in general and the ST131 and the O15:K52:H1 clonal groups in particular could help prevent further erosion of the clinical utility of FQs. Epidemiological and environmental studies are needed to clarify the relevant risk factors, transmission pathways, and reservoirs for these and other drug-resistant clonal groups. With respect to preventive measures, the distinctive virulence profiles of these three clonal groups, which, interestingly, have in common certain adhesins (iha, fimH), siderophores (iha, fyuA, iutA), toxins (sat, usp), and genes of unknown function (ompT, malX), suggest a range of potential targets for vaccines or other interventions, particularly if a role in virulence or dissemination can be demonstrated for these (or associated) traits. Finally, more prudent antimicrobial use in humans and agriculture can be expected to reduce the selection pressure for the further expansion of these virulent and drug-resistant clonal groups, regardless of their origins and modes of dissemination.
Methodologically, the relationship between PFGE profile similarity and clonal group assignment that was observed indicates that profile similarity thresholds (e.g., 81%) (19) that approximately correspond to a specific number of band differences (e.g., less than or equal to six), which in turn may correspond to a specific number of genetic events (e.g., less than or equal to two) (31), however suitable for the identification of localized point-source outbreaks involving a single clone, are insensitive to broader phylogenetic relationships. Consequently, a previous analysis of NAUTICA FQ-R isolates likely appreciably underestimated the prevalence of the two prominent clonal groups that were observed (19), which we show correspond to the ST131 and the O15:K52:H1 clonal groups. This is consistent with previous findings involving international CTX-M-15-positive ST131 isolates, some of which exhibited PFGE profile similarity values as low as 68% (24) or 62% (20).
The limitations of the study include the fact that the study period was in the somewhat remote past; the absence of clinical data regarding the specific UTI syndrome, illness severity, and outcomes; and the reliance on gene detection to make inferences regarding virulence. The strengths include the large sample size used, the broadly distributed and systematically collected strain set used, the extensive molecular typing performed, and the multiple complementary approaches used for statistical analysis.
In summary, our novel findings include the fact that the three E. coli clonal groups of interest (CGA, the O15:K52:H1 clonal group, and ST131) accounted for approximately half of the TS-R and/or FQ-R E. coli urine isolates from Canadian patients collected from 2002 to 2004 studied, evidence that the emergence of TS resistance and, particularly, FQ resistance in E. coli has important clonal components. CGA, a significant emerging pathogen in the 1990s, was overshadowed here by the O15:K52:H1 clonal group and especially ST131 for both prevalence and resistance phenotype, indicating that the last two groups may now represent more important public health threats, even without the ESC resistance for which ST131 is currently best known. The virulence profiles of the O15:K52:H1 and ST131 clonal groups were more extensive than those of their nonclonal group FQ-R counterparts, perhaps partly explaining these clonal groups' epidemiological success. Further study of these threatening FQ-R clonal groups is needed to clarify their broader role in drug-resistant E. coli infections and to identify risk factors, reservoirs, transmission pathways, and preventive measures.
Acknowledgments
This material is based upon work supported by the Office of Research and Development, Medical Research Service, U.S. Department of Veterans Affairs (to J.R.J.).
Dave Prentiss (Veterans Affairs Medical Center) prepared the figure. Sarah Arlein and Stefan Colinet-Adler assisted in the laboratory.
J.R.J. has received grant support and/or consultancies from Bayer, Wyeth-Ayerst, Ortho-McNeil, Merck, Procter and Gamble, and the Rochester Medical Group. G.G.Z. has received grant support from Abbott, Bayer, Ortho-McNeil, Pfizer, Procter and Gamble, and Wyeth. None of the other authors has conflicts to report with this work.
Footnotes
Published ahead of print on 27 April 2009.
REFERENCES
- 1.Berg, D. E., N. S. Akopyants, and D. Kersulyte. 1994. Fingerprinting microbial genomes using the RAPD or AP-PCR method. Methods Mol. Cell. Biol. 5:13-24. [Google Scholar]
- 2.Burman, W. J., P. E. Breese, B. E. Murray, K. V. Singh, H. A. Batal, T. D. MacKenzie, J. W. Ogle, M. L. Wilson, R. R. Revers, and P. S. Mehler. 2003. Conventional and molecular epidemiology of trimethoprim-sulfamethoxazole resistance among urinary Escherichia coli isolates. Am. J. Med. 115:358-364. [DOI] [PubMed] [Google Scholar]
- 3.Cagnacci, S., L. Gualco, E. Debbia, G. C. Schito, and A. Marchese. 2008. European emergence of ciprofloxacin-resistant Escherichia coli clonal groups O25:H4-ST 131 and O15:K52:H1 causing community-acquired uncomplicated cystitis. J. Clin. Microbiol. 46:2605-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clermont, O., J. R. Johnson, M. Menard, and E. Denamur. 2007. Determination of Escherichia coli O types by allele-specific polymerase chain reaction: application to the O types involved in human septicemia. Diagn. Microbiol. Infect. Dis. 57:129-136. [DOI] [PubMed] [Google Scholar]
- 6.Clermont, O., M. Lavollay, S. Vimont, C. Deschamps, C. Forestier, C. Branger, E. Denamur, and G. Arlet. 2008. The CTX-M-15-producing Escherichia coli diffusing clone belongs to a highly virulent B2 phylogenetic subgroup. J. Antimicrob. Chemother. 61:1024-1028. [DOI] [PubMed] [Google Scholar]
- 7.Coque, T. M., A. Novais, A. Carattoli, L. Poirel, J. Pitout, L. Peixe, F. Baquero, R. Canton, and P. Nordmann. 2008. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum beta-lactamase CTX-M-15. Emerg. Infect. Dis. 14:195-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalmau, D., F. Navarro, B. Mierlis, J. Blanco, J. Garau, and G. Prats. 1996. Escherichia coli bacteraemia. Serotype O15:K52:H1 as a urinary pathogen. J. Hosp. Infect. 34:233-234. (Letter.) [DOI] [PubMed] [Google Scholar]
- 9.Garau, J., M. Xercavins, M. Rodriguez-Carballeira, J. Gomez-Vera, I. Coll, D. Vidal, T. Llovet, and A. Ruiz-Bremon. 1999. Emergence and dissemination of quinolone-resistant Escherichia coli in the community. Antimicrob. Agents Chemother. 43:2736-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta, K., T. M. Hooton, and W. E. Stamm. 2001. Increasing antimicrobial resistance and the management of uncomplicated community-acquired urinary tract infections. Ann. Intern. Med. 135:41-50. [DOI] [PubMed] [Google Scholar]
- 11.Johnson, J. R., B. Johnston, M. A. Kuskowski, J. P. Nougayrede, and E. Oswald. 2008. Molecular epidemiology and phylogenetic distribution of the Escherichia coli pks genomic island. J. Clin. Microbiol. 46:3906-3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson, J. R., A. R. Manges, T. T. O'Bryan, and L. R. Riley. 2002. A disseminated multi-drug resistant clonal group of extraintestinal pathogenic Escherichia coli as a cause of pyelonephritis. Lancet 359:2249-2251. [DOI] [PubMed] [Google Scholar]
- 13.Johnson, J. R., A. C. Murray, M. A. Kuskowski, S. Schubert, M.-F. Prere, B. Picard, R. Colodner, R. Raz, and Trans-Global Initiative for Antimicrobial Resistance Analysis (TIARA) Investigators. 2005. Distribution and characteristics of Escherichia coli clonal group A. Emerg. Infect. Dis. 11:141-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson, J. R., K. Owens, A. R. Manges, and L. W. Riley. 2004. Rapid and specific detection of Escherichia coli clonal group A by gene-specific PCR. J. Clin. Microbiol. 42:2618-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson, J. R., K. Owens, T. T. O'Bryan, M. Sabate, and G. Prats. 2004. Rapid and specific detection of the O15:K52:H1 clonal group of Escherichia coli by gene-specific PCR. J. Clin. Microbiol. 42:3841-3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, J. R., and A. L. Stell. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181:261-272. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, J. R., A. L. Stell, T. T. O'Bryan, M. Kuskowski, B. Nowicki, C. Johnson, J. M. Maslow, A. Kaul, J. Kavle, and G. Prats. 2002. Global molecular epidemiology of the O15:K52:H1 extraintestinal pathogenic Escherichia coli clonal group: evidence of distribution beyond Europe. J. Clin. Microbiol. 40:1913-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson, J. R., A. L. Stell, F. Scheutz, T. T. O'Bryan, T. A. Russo, U. B. Carlino, C. C. Fasching, J. Kavle, L. van Dijk, and W. Gaastra. 2000. Analysis of F antigen-specific papA alleles of extraintestinal pathogenic Escherichia coli using a novel multiplex PCR-based assay. Infect. Immun. 68:1587-1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karlowsky, J. A., S. Kasloff, K. A. Nichol, D. J. Hoban, and G. G. Zhanel. 2007. Genetic relatedness of multidrug-resistant Escherichia coli cultured from geographically diverse outpatient, midstream urine specimens. Diagn. Microbiol. Infect. Dis. 58:283-287. [DOI] [PubMed] [Google Scholar]
- 20.Leflon-Guibout, V., J. Blanco, K. Amaqdouf, A. Mora, L. Guize, and M. H. Nicolas-Chanoine. 2008. Absence of CTX-M enzymes but high prevalence of clones, including clone ST131, among fecal Escherichia coli isolates from healthy subjects living in the area of Paris, France. J. Clin. Microbiol. 46:3900-3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leflon-Guibout, V., C. Jurand, S. Bonacorsi, F. Espinasse, M. C. Guelfi, F. Duportail, B. Heym, E. Bingen, and M. H. Nicolas-Chanoine. 2004. Emergence and spread of three clonally related virulent isolates of CTX-M-15-producing Escherichia coli with variable resistance to aminoglycosides and tetracycline in a French geriatric hospital. Antimicrob. Agents Chemother. 48:3736-3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manges, A. R., J. R. Johnson, B. Foxman, T. T. O'Bryan, K. E. Fullerton, and L. W. Riley. 2001. Widespread distribution of urinary tract infections caused by a multidrug-resistant Escherichia coli clonal group. N. Engl. J. Med. 345:1007-1013. [DOI] [PubMed] [Google Scholar]
- 23.Manges, A. R., H. Tabor, P. Tellis, C. Vincent, and P. P. Tellier. 2008. Endemic and epidemic lineages of Escherichia coli that cause urinary tract infections. Emerg. Infect. Dis. 14:1575-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicolas-Chanoine, M. H., J. Blanco, V. Leflon-Guibout, R. Demarty, M. P. Alonso, M. M. Canica, Y. J. Park, J. P. Lavigne, J. Pitout, and J. R. Johnson. 2008. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 61:273-281. [DOI] [PubMed] [Google Scholar]
- 25.O'Neill, P. M., C. A. Talboys, A. P. Roberts, and B. S. Azadian. 1990. The rise and fall of Escherichia coli O15 in a London teaching hospital. J. Med. Microbiol. 33:23-27. [DOI] [PubMed] [Google Scholar]
- 26.Phillips, I., S. Eykyn, A. King, W. R. Grandsden, B. Rowe, J. A. Frost, and R. J. Gross. 1988. Epidemic multiresistant Escherichia coli infection in West Lambeth health district. Lancet i:1038-1041. [DOI] [PubMed] [Google Scholar]
- 27.Pitout, J. D., D. L. Church, D. B. Gregson, B. L. Chow, M. McCracken, M. R. Mulvey, and K. B. Laupland. 2007. Molecular epidemiology of CTX-M-producing Escherichia coli in the Calgary Health Region: emergence of CTX-M-15-producing isolates. Antimicrob. Agents Chemother. 51:1281-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pitout, J. D., P. Nordmann, K. B. Laupland, and L. Poirel. 2005. Emergence of Enterobacteriaceae producing extended-spectrum beta-lactamases (ESBLs) in the community. J. Antimicrob. Chemother. 56:52-59. [DOI] [PubMed] [Google Scholar]
- 29.Prats, G., F. Navarro, B. Mirelis, D. Dalmau, N. Margall, P. Coll, A. Stell, and J. R. Johnson. 2000. Escherichia coli serotype O15:K52:H1 as a uropathogenic clone. J. Clin. Microbiol. 38:201-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sannes, M. R., M. San Roman, J. Moya, N. Mora, A. A. Eckhoff, D. N. Williams, J. R. Johnson, and P. K. Peterson. 2003. Antimicrobial resistance among Escherichia coli causing urinary tract infections in Costa Rica: a clinical dilemma. Int. J. Antimicrob. Agents 21:79-82. [DOI] [PubMed] [Google Scholar]
- 31.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Muray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wallace-Gadsden, F., J. R. Johnson, J. Wain, and I. N. Okeke. 2007. Enteroaggregative Escherichia coli related to uropathogenic clonal group A. Emerg. Infect. Dis. 13:757-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhanel, G. G., T. L. Hisanaga, N. M. Laing, M. R. DeCorby, K. A. Nichol, L. P. Palatnik, J. Johnson, A. Noreddin, G. K. Harding, L. E. Nicolle, and D. J. Hoban. 2005. Antibiotic resistance in outpatient urinary isolates: final results from the North American Urinary Tract Infection Collaborative Alliance (NAUTICA). Int. J. Antimicrob. Agents 26:380-388. [DOI] [PubMed] [Google Scholar]
- 34.Zhanel, G. G., T. L. Hisanaga, N. M. Laing, M. R. DeCorby, K. A. Nichol, B. Weshnoweski, J. Johnson, A. Noreddin, D. E. Low, J. A. Karlowsky, and D. J. Hoban. 2006. Antibiotic resistance in Escherichia coli outpatient urinary isolates: final results from the North American Urinary Tract Infection Collaborative Alliance (NAUTICA). Int. J. Antimicrob. Agents 27:468-475. [DOI] [PubMed] [Google Scholar]