Abstract
Petite mutations have been described in Saccharomyces cerevisiae and pathogenic yeasts. However, previous studies of the phenotypic traits of these petite mutants reported that they express azole resistance. We describe a clinical isolate of Candida glabrata with a striking association between increased susceptibility to azoles and respiratory deficiency. This isolate was obtained from a urine sample together with a respiration-competent C. glabrata isolate which exhibited azole resistance. The respiratory status of the two isolates was confirmed by cultivation on glycerol-containing agar and oxygraphy. Flow cytometry revealed the normal incorporation of rhodamine 123, and mitochondrial sections with typical cristae were seen by transmission electron microscopy for both isolates. Together, these results suggested a nuclear origin for the reduced respiratory capacity of the hypersusceptible isolate. The sterol contents of these isolates were similar to the sterol content of a reference strain. Sequencing of the ERG11 and PDR1 genes revealed that the sequences were identical in the two isolates, demonstrating their close relatedness. In addition to silent mutations, they carried a nonsense mutation in PDR1 that led to the truncation of transcription factor Pdr1p. They also overexpressed both PDR1 and one of its targets, CDR1, providing a possible explanation for the azole resistance of the respiration-competent isolate. In conclusion, in addition to azole resistance, which is a common feature of C. glabrata mitochondrial petite mutants, the mutation of a nuclear gene affecting aerobic growth may lead to azole hypersusceptibility; however, the mechanisms underlying this phenotype remain to be determined.
Petite mutations in budding yeasts were first described by Ephrussi et al. in 1949 (13). These petite mutants produce small colonies on fermentative culture media but are unable to grow on nonfermentative media. Since then, mutations that result in the petite phenotype have been described in other yeast species, and these species are called petite positive; in other species, called petite negative, such mutations are lethal. Petite mutants share the particularity of being unable to respire. However, this phenomenon may have two distinct origins. Indeed, most petite mutants result from the partial loss of mitochondrial DNA (petite mutants [rho−]) or total loss of mitochondrial DNA (petite mutants [rho0]), although the petite phenotype can also result from mutations in the nuclear genome affecting mitochondrial biogenesis or assembly and thereby impairing respiratory activity.
Unlike Candida albicans, in which mutations that result in the petite phenotype occur under a very limited range of conditions (2, 8), Candida glabrata produces petite mutants with a high frequency in vitro (6, 19). As in Saccharomyces cerevisiae (15) and C. albicans (8), petite mutants of C. glabrata, which may also occur in vivo (4), exhibit decreased susceptibilities to azole antifungals (6, 19). This azole resistance is due to the overexpression of various ATP-binding cassette (ABC) efflux pumps (6, 19) linked to the upregulation of the transcription factor Pdr1p (24).
Azole drugs, one of the four classes of antifungals used in clinical practice, act by inhibition of a key enzyme of the ergosterol bisoynthesis pathway in fungi, the lanosterol 14-alpha-demethylase (7). These drugs are currently the “gold standard” for the treatment of fungal infections. Unfortunately, their extensive use for both prophylaxis and therapy has led to an increased occurrence of resistant isolates. Four main mechanisms of azole resistance have been described (18): (i) mutations in the lanosterol 14-alpha-demethylase gene, ERG11, that lead to a modification of the target, which decreases the affinity of azoles without diminishing the activity of the enzyme; (ii) increases in the copy number of the azole target resulting from gene amplification or an increase in the mRNA half-life; (iii) blockage of the ergosterol biosynthesis pathway, which allows the fungal cell to remain viable, despite the presence of the drug; and (iv) the overexpression of genes coding some ABC or major facilitator superfamily efflux pumps, leading to the increased efflux of azole drugs.
Although the overexpression of genes encoding the efflux pumps has been reported in several studies, the cause of the deregulation of their expression is poorly understood. In C. glabrata, as in S. cerevisiae and C. albicans, the ABC genes are controlled by a pleiotropic drug resistance (PDR) transcription factor, but the mechanisms leading to the increased activity of this transcription factor have not been clearly described (3, 11). In C. albicans, the expression of the PDR transcription factor Tac1p can be increased by duplication of the entire chromosome harboring the TAC1 gene (9). The increased activity of Tac1p may also be due to a gain-of-function mutation of the TAC1 gene (10). Brun et al. (6) suggested that given the close communication between the nuclear and the mitochondrial genomes (16, 23), deregulation of the expression of the gene encoding the ABC membrane transporter (Cdr1p) in C. glabrata petite mutants and, consequently, their azole resistance may be linked to the absence of mitochondrial DNA. In the Parasitology-Mycology Laboratory of Angers University Hospital in Angers, France, we recently recovered an isolate with decreased respiratory capacity and increased susceptibility to azoles. We report on an investigation of the molecular mechanisms involved in this unusual phenotypic association.
MATERIALS AND METHODS
Strains and culture conditions.
Two Candida glabrata isolates were used throughout this study. They were isolated in the Parasitology-Mycology Laboratory of Angers University Hospital in January 2008 from a urine sample collected from a 28-year-old woman with cystic fibrosis. The disease was diagnosed in 1992, and the patient had been treated with voriconazole (V-Fend; Pfizer) since April 1996 for an Aspergillus infection. The patient had been colonized with multiresistant Pseudomonas aeruginosa since 1994 and with Aspergillus fumigatus since 1997. She was hospitalized at the end of November 2007 for severe asthenia and a substantial deterioration of lung function due to repeated respiratory infections since August 2007. On 5 December 2007, the voriconazole regimen was increased from 200 mg twice a day to 400 mg twice a day, because of a very low plasma concentration of the drug (<0.1 mg/liter on 27 November 2007). The plasma voriconazole concentration was 1.6 mg/ml on 14 December 2007, 3.5 mg/ml on 28 December 2007, and 1 mg/liter on 14 January 2008. Mycological examination of a urine sample collected on 14 January 2008 revealed the growth of two types of colonies on CHROMagar Candida medium (Becton-Dickinson, Franklin Lakes, NJ); both were identified as C. glabrata by the use of ID32C test strips (bioMérieux, Marcy l'Etoile, France). In vitro susceptibility testing with ATB Fungus 3 strips (bioMérieux) indicated that the two isolates had distinct phenotypes: one of them was hypersusceptible to fluconazole and voriconazole, and the other was totally resistant to both these triazoles. Both isolates were deposited at the Institute of Hygiene and Epidemiology (IHEM), Mycology Section (Scientific Institute of Public Health, Brussels, Belgium) and are publicly available (accession number 22852 for the hypersusceptible isolate [referred to here as clinical isolate 22852] and accession number 22853 for the resistant isolate [referred to here as clinical isolate 22853]). Since no matched susceptible isolate was available, a wild-type clinical isolate, IHEM accession number 21231 (referred to here as clinical isolate 21231), already described by our group (6, 26), was used as a control.
All isolates were maintained by regular passage on yeast extract-peptone-glucose (YEPD) agar plates containing yeast extract, 5 g/liter; peptone, 10 g/liter; glucose, 20 g/liter; chloramphenicol, 0.5 g/liter; and agar, 20 g/liter. They were preserved by lyophilization and by freezing at −80°C in 20% (wt/vol) glycerol.
Susceptibility testing.
Susceptibility to polyene and azole drugs was determined by a disk diffusion method on Casitone agar plates (Bacto-Casitone, 9 g/liter; glucose, 20 g/liter; yeast extract, 5 g/liter; chloramphenicol, 0.5 g/liter; agar, 18 g/liter; pH 7.2) with Neosensitabs tablets (Rosco Diagnostica, Taastrup, Denmark), as described previously (25).
The MICs of amphotericin B, fluconazole, and voriconazole were determined by the Etest procedure (AB Biodisk, Solna, Sweden) on Casitone agar plates, according to the manufacturer's recommendations.
Characterization of respiratory capacity.
The respiratory status of clinical isolates 22852 and 22853 was investigated on yeast extract-peptone-glycerol (YEPG) agar medium, which contains glycerol, a nonfermentable carbohydrate, as the sole carbon source. Thus, cells unable to respire are negatively selected on this medium.
The respiratory status was then confirmed by oxygraphy with an Oxytherm oxygraph (Hansatech Instruments Ltd., Norfolk, England), as described previously (25). Respiration activities are expressed as the maximal rates of oxygen consumption (in nanomoles of oxygen consumed per ml and per minute), determined from the curves of the oxygen concentration in the oxygraph chamber plotted against the duration of incubation. Three independent experiments were performed. The data presented here were derived from one of these experiments, as less than 10% variation was seen in the maximal oxygen consumption rates between the three experiments.
The activity of the mitochondrial respiratory chain was investigated by flow cytometry with rhodamine 123. The incorporation of this fluorochrome into mitochondria is dependent on the potential difference between the intermembrane space and the mitochondrial lumen, and therefore, the level of fluorochrome incorporation allows the detection of a defect in the electron flow through the mitochondrial respiratory chain (20). Briefly, blastoconidia from YEPD agar plates were washed twice in sterile distilled water and resuspended in 0.15 M phosphate-buffered saline (PBS; pH 7.2). Cells (2 × 106) were incubated for 30 min at 37°C with 250 μl of a 10-μg/ml rhodamine 123 solution. To inhibit the respiratory chain, aliquots of the cell suspensions were first incubated for 2 h at 37°C with 1 mM sodium azide (which blocks the fourth complex of the respiratory chain, the cytochrome oxidase, and therefore the electron flow) before the addition of rhodamine 123. Other samples were incubated in the absence of the fluorescent dye to verify that there was no autofluorescence. After the cells were washed with cold PBS, the fluorescence of 10,000 cells was quantified with a FACS CantoII flow cytometer (Becton Dickinson) and the data were analyzed with FACSDiva software from Becton Dickinson.
Sterol analysis.
Total sterols were analyzed by high-performance liquid chromatography (HPLC). Aliquots of 50 mg of lyophilized yeast obtained from stationary-growth-phase cultures in 50 ml of YEPD broth were resuspended in 250 μl HPLC-grade methanol, the mixture was incubated for 40 s at 47°C with shaking, and 50-μl samples were injected into the HPLC system. The reverse-phase elution was conducted with 100% acetonitrile (1.2 ml/min) at 37°C with a Chromolith Performance RP-18 capped column (100 mm by 4.6 mm; Merck, Darmstadt, Germany). Solutions of known concentrations of squalene, lanosterol, and ergosterol were also injected into the column by the same procedure, to serve as references for the qualitative and the quantitative analysis of each sample. The data collected were the concentrations, in mg of sterol species per ml of sample, and each result corresponds to the results for two independent cultures analyzed in duplicate.
Transmission electron microscopy (TEM).
Samples for electron microscopy were processed by the method of Aoki and Ito-Kuwa (2), with slight modifications, as described previously (4). Cells grown on yeast-peptone-dextrose agar plates were washed in PBS, fixed with glutaraldehyde in cacodylate buffer, washed in the same buffer, fixed again with 1.5% KMnO4, and finally, postfixed in osmium tetroxide. The samples were dehydrated and embedded in Epon, and ultrathin sections were prepared, stained with uranyl acetate, and examined under a JEM-2010 transmission electron microscope (JEOL, Paris, France).
Nucleic acid extraction and purification.
DNA was recovered from 10 ml of YEPD broth cultures with a DNeasy plant minikit (Qiagen Inc., Valencia, CA), according to the manufacturer's recommendations. Total RNA was recovered from early-exponential-growth-phase cultures in 50 ml of YEPD broth with a phenol-chloroform protocol, as described previously (26).
ERG11 and PDR1 gene sequencing.
The sequences of the ERG11 gene of C. glabrata isolates 22852 and 22853 were determined as described previously (26). Briefly, the gene was amplified with the primers described in Table 1. The PCR products were purified with a High Pure PCR product purification kit (Roche Diagnostics GmbH, Mannheim, Germany) and then quantified by use of the Nanodrop technology (Nanodrop, Wilmington, DE) and used as the template for sequencing PCR with a Dye Terminator cycle sequencing quick-start kit (Beckman Coulter Inc., Fullerton, CA). The sequencing products were purified with an Ilustra AutoSeq G50 column (GE Healthcare, Little Chalfont, United Kingdom) and resolved on a CEQ8000 sequencer (Beckman Coulter). The sequences were compared by alignment with the sequence of the ERG11 gene of wild-type isolate 21231 (GenBank accession number DQ060157) by use of the ALIGNn program (http://bioinfo.hku.hk/services/analyseq/cgi-bin/alignn_in.pl).
TABLE 1.
Oligonucleotides used for gene sequencing and evaluation of gene expression
| Gene name (gene product) | GenBank accession no. | Primer | Nucleotide sequence (5′ to 3′) | Nucleotide coordinatesa |
|---|---|---|---|---|
| ERG11 (lanosterol 14-α-demethylase) | L40389 | ERG11-1F | CTACAATCGAGTGAGCTTG | 17 to 35 |
| ERG11-1R | GTAGAACACAAGTGGTGG | 729 to 746 | ||
| ERG11-2F | GGTCGTTGAACTATTGGAG | 584 to 602 | ||
| ERG11-2R | GGACCCAAGTAGACAGTC | 863 to 880 | ||
| ERG11-3Fb | CCATCACATGGCAATTGC | 688 to 705 | ||
| ERG11-3Rb | GGTCATCTTAGTACCATCC | 1445 to 1463 | ||
| ERG11-4F | CGTGAGAAGAACGATATCC | 1380 to 1398 | ||
| ERG11-4R | CACCTTCAGTTGGGTAAC | 2047 to 2064 | ||
| ERG11-5F | CGCTTACTGTCAATTGGG | 1991 to 2008 | ||
| ERG11-5R | GTCATATGCTTGCACTGC | 2397 to 2414 | ||
| PDR1 (transcription factor involved | AY700584 | PDR1-Fb | TGGAAAAGCTTGTGATAGCTG | 81 to 101 |
| in pleiotropic drug resistance) | PDR1-Rb | TCTGATGACTGTAGTAGCCGA | 498 to 518 | |
| NC005967 | PDR1-SF | CGTTATTGAGAGAATATGCAA | 47542 to 47562 | |
| PDR1-SR | AGGCTATGCACACTGTCTAA | 50891 to 50910 | ||
| CDR1 (ABC transporter) | AF109723 | CDR1-Fb | GAAGTCTATGGAAGGTGC | 1084 to 1101 |
| CDR1-Rb | GTCTAGCGTAAGTCTCTC | 1383 to 1400 | ||
| CDR2 (ABC transporter) | AF251023 | CDR2-Fb | GTTGAGTACTGGCACAAC | 362 to 379 |
| CDR2-Rb | GATGGCAAAGAACATGGC | 695 to 712 | ||
| HSP12 (heat shock protein) | NC006033 | HSP12-Fb | ACTTGGAGACGTATTCGACGG | 391031 to 391051 |
| HSP12-Rb | TGTCTGACGCTGGTAGAAAGA | 391299 to 391308 | ||
| ACT (β-actin) | AF069746 | ACT-Fb | TATTGACAACGGTTCCGG | 949 to 966 |
| ACT-Rb | TAGAAAGTGTGATGCCAG | 1177 to 1194 |
Nucleotide coordinates refer to those for the corresponding gene sequence in the GenBank database.
Primers used for evaluation of gene expression.
The sequences of the PDR1 gene of C. glabrata isolates 22852 and 22853 were resolved by Qiagen by the use of PCR products covering the entire PDR1 open reading frame. These PCR products were obtained with primers designed with the WebPrimer program (http://seq.yeastgenome.org/cgi-bin/web-primer) from the PDR1 sequence of C. glabrata strain CBS138, available in the GenBank database under accession number AY700584.
Gene expression study.
The levels of expression of the genes coding proteins potentially involved in the azole susceptibilities of C. glabrata clinical isolates 22852 and 22853 were evaluated by real-time reverse transcription-PCR (RT-PCR), and the corresponding level of expression of the genes in wild-type isolate 21231 was used as the reference. The genes whose expression was studied were ERG11, which encodes the target enzyme of azoles; PDR1, which encodes a transcription factor mediating the azole response in C. glabrata; and three of the known targets of PDR1 in C. glabrata, CDR1 and CDR2 (which encode two ABC efflux pumps) and HSP12 (which encodes a heat shock protein central to the stress response [17]). The RT-PCR experiments were performed as described previously (26) with the primers specified in Table 1. Four independent experiments were performed, and the mean values (± standard deviations) of the differences in gene expression were determined.
Nucleotide sequence accession numbers.
The nucleotide sequences of the PDR1 genes of C. glabrata isolates 22852 and 22853 and the ERG11 genes of C. glabrata isolates 22852 and 22853 are available in the GenBank database under accession numbers FJ167406, FJ167407, FJ167408, and FJ167409, respectively.
RESULTS
Susceptibility to azole and polyene drugs.
Clinical isolate 22852 showed increased susceptibility to all the azole drugs tested, as assessed by the disk diffusion method on Casitone agar plates (Table 2). The diameters of the growth inhibition zones were, on average, double those for the control isolate (wild-type isolate 21231). Likewise, clinical isolate 22852 presented a higher level of susceptibility to amphotericin B: the diameter of the growth inhibition zone was 40 mm for isolate 22852, whereas it was 27 mm for the control. Conversely, isolate 22853 exhibited decreased susceptibility to azoles and even total resistance to fluconazole and miconazole, whereas its susceptibility to amphotericin B was similar to that of the control isolate (Table 2). These observations were confirmed by determination of the MICs of fluconazole, voriconazole, and amphotericin B (Table 3). The MICs of fluconazole and voriconazole for clinical isolate 22852 were 6- and 16-fold lower than those for the control, respectively, confirming its hypersusceptibility to azole drugs. Likewise, the decreased susceptibility to azoles of clinical isolate 22853 was confirmed by the Etest procedure, which revealed total resistance to fluconazole (MIC > 256 μg/ml) and a voriconazole MIC 100-fold higher than that for the control isolate.
TABLE 2.
Susceptibilities of Candida glabrata isolates 21231, 22852, and 22853 to antifungalsa
| Antifungal | Diameter (mm) of growth inhibition zones for isolate:
|
||
|---|---|---|---|
| 21231 | 22852 | 22853 | |
| Amphotericin B | 27 | 40 | 31 |
| Fluconazole | 21 (M) | 50 | TR |
| Clotrimazole | 25 (M) | 50 | 12 |
| Miconazole | 21 (M) | 35 | TR |
| Ketoconazole | 33 (M) | 50 | 16 |
| Econazole | 25 (M) | 50 | 13 |
In vitro susceptibility testing was performed by the disk diffusion method on Casitone agar plates with Neosensitab tablets containing 10 μg of drug for amphotericin B, econazole, miconazole, and clotrimazole and 15 μg of drug for fluconazole and ketoconazole. The values reported are the diameters of the growth inhibition zones after 48 h of incubation at 37°C. TR, total resistance (no inhibition zone); (M), presence of resistant colonies randomly distributed in the growth inhibition zone.
TABLE 3.
MICs of amphotericin B, fluconazole, and voriconazole for Candida glabrata isolates 21231, 22852, and 22853a
| Antifungal | MIC (μg/ml) for isolate:
|
||
|---|---|---|---|
| 21231 | 22852 | 22853 | |
| Amphotericin B | 0.047 | 0.032 | 0.047 |
| Fluconazole | 6 | 0.94 | >256b |
| Voriconazole | 0.047 | 0.003 | 4 |
Results were obtained with Etest antifungal strips on Casitone agar plates.
>256, no inhibition zone.
Respiratory capabilities.
Both clinical isolates and the control were able to grow on YEPG agar plates. However, azole-susceptible clinical isolate 22852 produced very small colonies and did so only after 48 h of incubation, whereas the two other isolates produced standard-sized colonies after 24 h of incubation (data not shown). This suggests a diminished respiratory capacity for clinical isolate 22852.
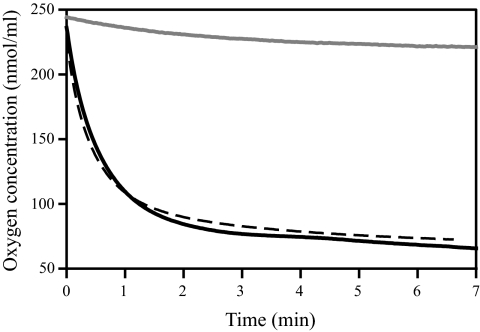
The diminished respiratory capacity was confirmed by oxygraphy experiments. The oxygen concentration in the oxygraph chamber remained almost unchanged even after 7 min of incubation with 108 cells of isolate 22852, whereas cells of isolate 22853 or control isolate 21231 consumed almost all the oxygen in less than 3 min (Fig. 1). The maximal oxygen consumption rates determined from the curves representing the oxygen concentration according to the duration of incubation were 75.8, 6.6, and 68.9 nmol of O2 consumed per ml and per min for wild-type isolate 21231 and clinical isolates 22852 and 22853, respectively.
FIG. 1.
Oxygen consumption by cells of C. glabrata isolates 21231 (continuous black line), 22852 (continuous gray line), and 22853 (dashed black line). Almost all the oxygen in the oxygraph chamber was consumed after 3 min of incubation by the wild-type isolate and azole-resistant isolate 22853, whereas the oxygen concentration remained almost unchanged after 7 min for hypersusceptible isolate 22852. Three independent experiments were performed. The data presented were derived from one of these experiments, but less than 10% variation in the maximal oxygen consumption rates was seen between the three experiments.
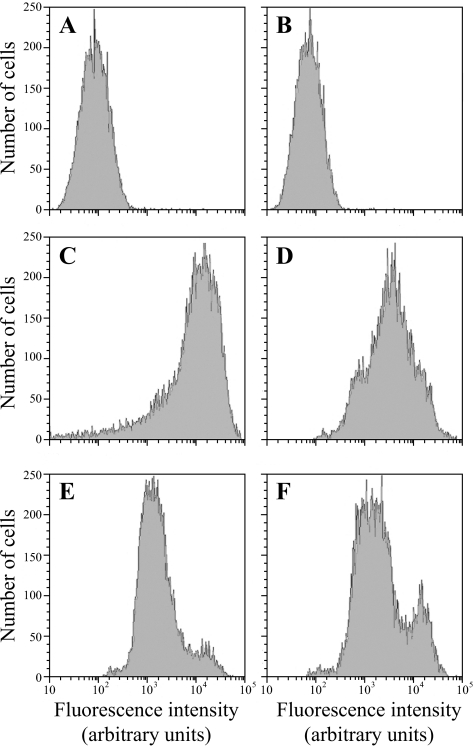
Flow cytometry experiments with rhodamine 123 showed greater incorporation of the fluorochrome by isolate 22852 than isolate 22853 (mean fluorescence values, 33,671 and 24,980 arbitrary units, respectively; Fig. 2). However, the intercell variability was higher for isolate 22852 (standard deviations, 22,281 arbitrary units for isolate 22853 and 30,430 arbitrary units for isolate 22852). Following incubation with sodium azide, the level of rhodamine 123 incorporation was similar for the two isolates (mean fluorescence values, 10,529 and 10,207 arbitrary units). Similar results were obtained in two independent experiments, and incubation of the cells without rhodamine 123 confirmed the absence of autofluorescence.
FIG. 2.
Flow cytometry analysis of rhodamine 123 staining of cells of C. glabrata isolates 22852 (A, C, and E) and 22853 (B, D, and F). The cells were preincubated (E and F) or not preincubated (C and D) for 2 h at 37°C in the presence of 1 mM sodium azide before the addition of rhodamine 123. The fluorescence of cells incubated without the fluorescent probe (A and B) is presented as a control.
Sterol content.
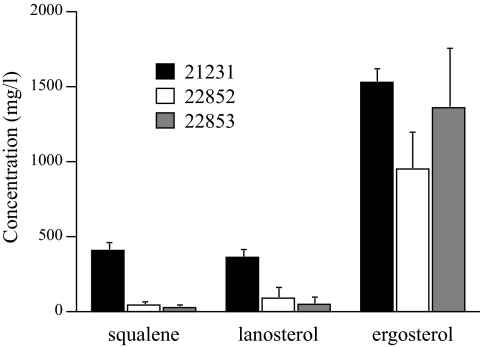
The sterol contents of C. glabrata isolates 22852 and 22853 and control isolate 21231 were studied qualitatively and quantitatively. The peak profiles obtained by HPLC were identical for the three isolates (data not shown). The use of standards permitted the quantification of ergosterol and sterol intermediates (squalene and lanosterol) in the three isolates (Fig. 3). No significant difference in the ergosterol contents was observed: 1,528, 1,359, and 951 mg per ml of sample for isolates 21231, 22852, and 22853, respectively. However, the amounts of squalene and lanosterol were greatly diminished (4- to 10-fold) in isolates 22852 and 22853 compared to the amount in the wild-type isolate.
FIG. 3.
HPLC analysis of the squalene, lanosterol, and ergosterol contents of C. glabrata isolates 21231 (black bars), 22852 (white bars), and 22853 (gray bars). The ergosterol content did not differ significantly between the three isolates. Conversely, very small amounts of sterol intermediates were found in both clinical isolates compared with the amount in the wild-type isolate.
Cell ultrastructure.
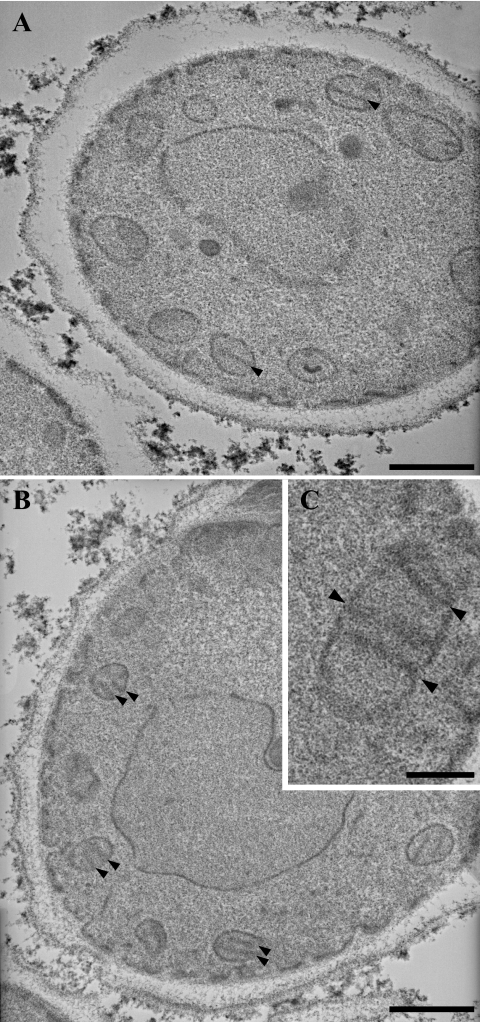
TEM revealed that the three C. glabrata isolates studied had a standard morphology, with each isolate presenting as solitary, ovoid blastoconidia of 1 to 5 μm in diameter, some of which were budding (Fig. 4). Moreover, functional mitochondria with typical cristae were seen in all three isolates, including respiration-deficient isolate 22852.
FIG. 4.
Transmission electron micrographs of cells of C. glabrata isolates 22852 (A) and 22853 (B and C). Numerous mitochondrial sections with typical cristae (arrowheads) can be seen in both isolates, consistent with a nuclear origin for the decreased respiratory capacity of hypersusceptible isolate 22852. Bars, 0.5 μm (A and B) and 0.2 μm (C).
Sequence analysis of ERG11 and PDR1 genes.
All nucleotide differences in the ERG11 and PDR1 sequences relative to the sequences of the same genes in strain CBS138 were shared by the two clinical isolates (Table 4). In addition to silent mutations in the nucleotide sequences, both isolates carried a nonsense mutation in PDR1 that introduced a stop codon at position 728 in the protein sequence of the transcription factor (as determined by use of the Multiple Translation tool [http://bioinfo.hku.hk/services/analyseq/cgi-bin/traduc_in.pl]).
TABLE 4.
Point mutations in ERG11 and PDR1 genes from C. glabrata isolates 22852 and 22853
| Gene | Point mutation in gene of isolatea:
|
|
|---|---|---|
| 22852 | 22853 | |
| ERG11 | T834C | T834C |
| T846C | T846C | |
| T1275C | T1275C | |
| PDR1 | G307A | G307A |
| T329C | T329C | |
| G763A | G763A | |
| G1699A | G1699A | |
| C1892T | C1892T | |
| G2219A | G2219A | |
Mutations are described as follows: the first letter indicates the nucleotide in the GenBank database sequence for the corresponding gene (GenBank accession numbers AY700584 for PDR1 and L40389 for ERG11), the number gives the position relative to the start of the open reading frame, and the second letter indicates the nucleotide found in the gene sequence of isolate 22852 or 22853. The nonsense mutation found in the PDR1 gene sequence is underlined. Other mutations are silent. The GenBank accession numbers of the sequences are indicated in Materials and Methods.
Expression of genes potentially involved in resistance or hypersusceptibility to azoles.
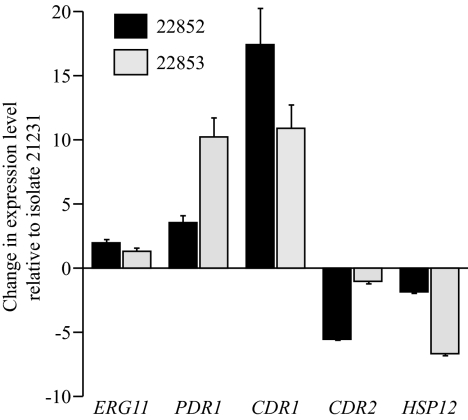
The RT-PCR experiments revealed that both clinical isolates had higher levels of expression of the PDR1 gene than the wild type, with induction factors of 3.5 for isolate 22852 and 10.2 for isolate 22853 (Fig. 5). Accordingly, the CDR1 gene was also overexpressed by 10.9-fold in isolate 22853, but curiously, overexpression of this gene was greater in isolate 22852 (17.4-fold; Fig. 5). In contrast, the level of expression of CDR2 gene in azole-resistant isolate 22853 was not significantly different from that in the wild type, whereas this gene was underexpressed fivefold in the hypersusceptible isolate relative to the level of expression by the wild-type isolate. Likewise, the HSP12 gene was underexpressed in azole-resistant isolate 22853, but its expression remained unchanged in the hypersusceptible isolate, with the relative expression values being 0.54 for isolate 22852 and 0.15 for 22853 (Fig. 5).
FIG. 5.
Expression of ERG11, PDR12, CDR1, and CDR2 genes in C. glabrata clinical isolates 22852 (black bars) and 22853 (gray bars) relative to that in wild-type isolate 21231. Expression was determined by RT-PCR. Relative to the level of expression by the control, PDR1 and CDR1 were overexpressed in the two clinical isolates, CDR2 was underexpressed in hypersusceptible isolate 22852, and HSP12 was underexpressed in azole-resistant isolate 22853. The levels of expression of ERG11 were similar in the clinical isolates and the control. The data presented are mean values (± standard deviations) of the changes from four independent experiments.
DISCUSSION
In all yeast species that have been studied, i.e., S. cerevisiae, C. albicans, and C. glabrata, mutations that result in the petite phenotype have been found to be associated with azole resistance. However, we report here on the isolation of a C. glabrata isolate with a striking association between decreased respiratory capacity and increased susceptibility to azole drugs.
This isolate was recovered together with an azole-resistant C. glabrata isolate from a 28-year-old woman undergoing antifungal therapy with voriconazole. The selection of this susceptible isolate despite the antifungal treatment may appear to be surprising. The plasma voriconazole concentration at the time of urine sampling was 1 mg/liter, which corresponds to the lower limit of the therapeutic range (1 to 6 mg/liter), as recently proposed by Brüggemann et al. (5). However, the sample used for determination of the plasma concentration of the azole drug was collected at the time of the peak concentration and not just before the morning dose.
The antifungal susceptibility patterns of these isolates were determined by a disk diffusion method on Casitone agar and were confirmed by determination of the MICs of amphotericin B, fluconazole, and voriconazole with Etest strips. Compared with a wild-type isolate, one of the clinical isolates exhibited a markedly lower level of susceptibility to azole drugs and even complete resistance to fluconazole and miconazole, whereas its susceptibility to polyenes was not modified. In contrast, the other clinical isolate exhibited higher levels of susceptibility to azoles and polyene drugs.
As azole resistance can be the result of mutations that result in the petite phenotype, we studied the respiratory status of the two isolates. Surprisingly, the azole-resistant isolate displayed a wild-type respiratory status, whereas the growth of the hypersusceptible isolate was markedly reduced on a medium containing 2% glycerol as the sole carbon source. Consistent with this result, oxygraphy showed that the cells of the azole-susceptible isolate were almost unable to consume oxygen. Therefore, this isolate was first considered to be a petite mutant. However, in contrast to previous observations of C. glabrata petite mutants (6, 19), this isolate accumulated rhodamine 123 (and to a greater extent than the respiration-competent azole-resistant isolate did), and TEM revealed numerous mitochondrial sections in blastoconidia. Also in contrast to previous observations, this isolate was able to grow on glycerol-containing agar, although it produced very small colonies which appeared only after 48 h of incubation (whereas the wild-type isolate and respiration-competent azole-resistant isolate 22853 gave larger colonies on this culture medium within 24 h). All petite mutants previously studied in our laboratory exhibited cross-resistance to all azole drugs tested in association with a complete lack of mitochondrial sections as assessed by TEM; all were unable to grow on glycerol-containing agar and were not stained by rhodamine 123 (4, 6). The respiratory deficiency of these mitochondrial petite mutants originated from the total or partial loss of mitochondrial DNA. Along with mitochondrial mutations that result in the petite phenotype, the petite phenotype may also have a nuclear origin because of the dysfunction of a nuclear gene necessary for respiratory activity. Therefore, the decreased respiratory capacity of the hypersusceptible isolate that we report on here seems to have a nuclear cause. Indeed, the normal accumulation of rhodamine 123 indicated that the electron flow through the mitochondrial respiratory chain, which is possible only if complex V and complexes III and/or IV are functional, was maintained (1). Moreover, oxygraphy revealed that the isolate was unable to consume oxygen, which is the final acceptor of electrons in complex IV. The most probable hypothesis is that a nuclear gene coding one of the subunits of complex IV of the respiratory chain is mutated. About 40 genes encode enzymes of the mitochondrial respiratory chain, but only 7 of them are mitochondrial (1). Denaturating HPLC experiments performed with the PCR products of the nuclear genes encoding subunits of the mitochondrial respiratory chain will be performed to confirm the nuclear origin of this decreased respiratory capacity by the identification of the mutated gene. Nevertheless, numerous other nuclear genes are involved in the targeting of the various subunits of the respiratory chain to the mitochondrial membrane and in their assembly. Steinmetz et al. (21) reported that deletion of any of 201 of 353 genes known to encode proteins involved in mitochondrial function or biogenesis resulted in a decrease in growth capacity on nonfermentative culture media and therefore that these 201 genes are required for normal respiratory function. Deletion of most of the nuclear genes encoding proteins of the respiratory chain also resulted in reduced growth on nonfermentative culture media (21). Using a similar experimental procedure, Dimmer et al. (12) identified 64 genes whose deletion severely impaired growth on nonfermentative culture media but did not abolish respiration. However, as far as we know, the azole susceptibilities of such nuclear mutants with diminished respiratory capacity have not previously been investigated.
Additionally, the distribution of fluorescence of the cells after rhodamine 123 staining was wider for the hypersusceptible isolate than for a wild-type isolate or the azole-resistant isolate. It is unlikely that petite mutants with mitochondrial defects arise from isolate 22852, and the reasons for the variability of cell fluorescence after rhodamine staining remain to be elucidated. Although antifungal susceptibility testing by the disk diffusion method was repeated several times for both isolates, we never observed randomly distributed, standard-sized colonies within the growth inhibition zone for isolate 22852 on fluconazole, whereas such colonies were regularly observed on fluconazole for wild-type isolates of C. glabrata.
Susceptibility to azole and polyene drugs is associated with the ergosterol biosynthesis pathway, so we analyzed the sterol contents of the two isolates. The amounts of sterol intermediates (squalene and lanosterol) in the two clinical isolates were lower than the amount in the wild-type control, suggesting increased activity of the ergosterol biosynthesis pathway. However, there were no significant differences regarding the ergosterol content. RT-PCR experiments did not reveal any difference in ERG11 gene expression. These findings indicate that the azole susceptibility patterns of these isolates were not due to changes in the amounts of the azole target. In addition, they may appear to be in conflict with the increased susceptibility to polyenes observed for one of these isolates. We must therefore consider another mechanism, like decreased tolerance or fitness.
To investigate the molecular basis of the antifungal susceptibility phenotype of the two isolates, we studied the expression of genes encoding some ABC efflux pumps, Cdr1p and Cdr2p, and the transcription factor Pdr1p; we also sequenced the PDR1 gene. Those experiments demonstrated that the two isolates are very closely related, as their ERG11 and PDR1 gene sequences (more than 5,200 bp) were identical. Presumably, the hypersusceptible isolate was derived from the resistant one, although this cannot be proved because the two isolates were recovered simultaneously from the same clinical sample.
In addition to silent mutations in the ERG11 and the PDR1 genes, the two isolates also shared a nonsense mutation in PDR1. This mutation was a guanine-to-adenine substitution at position 2219, which led to a stop codon and, therefore, to a truncation of 380 amino acids from the C terminus of the encoded transcription factor. Recent studies with C. glabrata and S. cerevisiae demonstrate that the C terminus of Pdr1p contains a domain that can induce the expression of PDR genes in the presence of azoles (22), suggesting a decreased activity of the truncated transcription factor in the presence of azole drugs. RT-PCR experiments revealed for the azole-resistant isolate the overexpression of PDR1 and one of its targets, CDR1, which was thought to be responsible for the azole resistance. Surprisingly, another gene encoding an ABC efflux pump, CDR2, was not overexpressed in the resistant isolate, although this gene contains a pleiotropic drug-responsive element (PDRE) in its promoter and has been reported to be regulated by Pdr1p (27). Likewise, the expression of the HSP12 gene was decreased in the resistant isolate, although the expression of this gene is induced in a C. glabrata isolate with a constitutively hyperactive Pdr1p (27). One may speculate that the nonsense mutation in the PDR1 gene of the azole-resistant isolate disrupts the Pdr1p domains required to activate the expression of HSP12 and CDR2. However, in silico analysis confirmed the presence of two PDRE sequences in the promoter region of the CDR1 gene but only one in the promoter region of the CDR2 gene. Unlike the HSP12 genes in S. cerevisiae and C. albicans, no PDRE sequence was detected within 2,000 bp upstream from the start codon of the HSP12 gene in C. glabrata. Another hypothesis would be that the truncation of the C terminus of Pdr1p leads to the hyperactivity of the transcription factor, thus explaining the upregulation of CDR1 observed in our two clinical isolates. Indeed, some mutations in the TAC1 gene of C. albicans are associated with the upregulation of PDR genes and are called gain-of-function mutations (9). Moreover, recent work with C. glabrata described gain-of-function mutations in PDR1 and demonstrated that the nature of the mutation itself may have different consequences on the expression of Pdr1p target genes. For example, the Y584C mutation leads to the upregulation of CDR1 but does not affect CDR2 expression, as was observed in our clinical isolate (14).
Whereas the azole resistance of isolate 22853 may easily be explained by the increased expression of the PDR1 gene, the results obtained for the azole-susceptible isolate were quite surprising. Indeed, like the resistant isolate, the hypersusceptible isolate overexpressed PDR1, although to a lesser extent. However, it also overexpressed the CDR1 gene (17.4-fold relative to the level of expression by the wild-type isolate and 10.9-fold relative to the level of expression by the azole-resistant isolate). The CDR2 gene in the hypersusceptible isolate was underexpressed fivefold relative to the level of expression by the wild-type isolate, and the expression of HSP12 gene was not affected. The increased susceptibility to azoles of this clinical isolate, despite the overexpression of CDR1, is quite surprising, since the overexpression of ABC-coding genes is the most frequent mechanism of resistance to azole drugs found in clinical isolates of Candida yeasts (18). Further investigation is required to elucidate how a yeast overexpressing genes encoding ABC proteins can be susceptible to azole drugs. This apparent discrepancy may be explained by a posttranslational modification of Cdr1p, such as a mutation in the CDR1 gene that impairs the targeting of the transporter to the plasma membrane or by an epigenetic phenomenon like a decrease in the intracellular ATP level linked to the diminished aerobic growth capacity found for this isolate (given that ATP is the energy source for efflux through ABC proteins).
Previous observations of mitochondrial petite mutants of C. glabrata suggested a direct relationship between azole resistance and respiratory deficiency. The recovery of this isolate with an association between decreased respiratory capacity and azole hypersusceptibility complicates the understanding of the mechanisms governing the regulation of PDR genes.
Acknowledgments
This study was supported in part by a grant from the Institut de Parasitologie de l'Ouest, Rennes, France. We thank Angers University Hospital for financial support.
We thank the Service Commun d'Imagerie et d'Analyses Microscopiques of Angers University for help with TEM.
Footnotes
Published ahead of print on 20 April 2009.
REFERENCES
- 1.Andreoli, C., H. Prokisch, K. Hortnagel, J. C. Mueller, M. Munsterkotter, C. Scharfe, and T. Meitinger. 2004. MitoP2, an integrated database on mitochondrial proteins in yeast and man. Nucleic Acids Res. 32:459-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aoki, S., and S. Ito-Kuwa. 1987. Induction of petite mutation with acriflavine and elevated temperature in Candida albicans. J. Med. Vet. Mycol. 25:269-277. [DOI] [PubMed] [Google Scholar]
- 3.Balzi, E., W. Chen, S. Ulaszewski, E. Capieaux, and A. Goffeau. 1987. The multidrug resistance gene PDR1 from Saccharomyces cerevisiae. J. Biol. Chem. 262:16871-16879. [PubMed] [Google Scholar]
- 4.Bouchara, J. P., R. Zouhair, S. Le Boudouil, G. Renier, R. Filmon, D. Chabasse, J. N. Hallet, and A. Defontaine. 2000. In-vivo selection of an azole-resistant petite mutant of Candida glabrata. J. Med. Microbiol. 49:977-984. [DOI] [PubMed] [Google Scholar]
- 5.Brüggemann, R. J., J. P. Donnelly, R. E. Aarnoutse, A. Warris, N. M. Blijlevens, J. W. Mouton, P. E. Verweij, and D. M. Burger. 2008. Therapeutic drug monitoring of voriconazole. Ther. Drug Monit. 30:403-411. [DOI] [PubMed] [Google Scholar]
- 6.Brun, S., T. Bergès, P. Poupard, C. Vauzelle-Moreau, G. Renier, D. Chabasse, and J. P. Bouchara. 2004. Mechanisms of azole resistance in petite mutants of Candida glabrata. Antimicrob. Agents Chemother. 48:1788-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrillo-Muñoz, A. J., G. Giusiano, P. A. Ezkurra, and G. Quindós. 2006. Antifungal agents: mode of action in yeast cells. Rev. Esp. Quimioter. 19:130-139. [PubMed] [Google Scholar]
- 8.Cheng, S., C. J. Clancy, K. T. Nguyen, W. Clapp, and M. H. Nguyen. 2007. A Candida albicans petite mutant strain with uncoupled oxidative phosphorylation overexpresses MDR1 and has diminished susceptibility to fluconazole and voriconazole. Antimicrob. Agents Chemother. 51:1855-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coste, A., A. Selmecki, A. Forche, D. Diogo, M. E. Bougnoux, C. d'Enfert, J. Berman, and D. Sanglard. 2007. Genotypic evolution of azole resistance mechanisms in sequential Candida albicans isolates. Eukaryot. Cell 6:1889-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coste, A., V. Turner, F. Ischer, J. Morschhäuser, A. Forche, A. Selmecki, J. Berman, J. Bille, and D. Sanglard. 2006. A mutation in Tac1p, a transcription factor regulating CDR1 and CDR2, is coupled with loss of heterozygosity at chromosome 5 to mediate antifungal resistance in Candida albicans. Genetics 172:2139-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coste, A. T., M. Karababa, F. Ischer, J. Bille, and D. Sanglard. 2004. TAC1, transcriptional activator of CDR genes, is a new transcription factor involved in the regulation of Candida albicans ABC transporters CDR1 and CDR2. Eukaryot. Cell 3:1639-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dimmer, K. S., S. Fritz, F. Fuchs, M. Messerschmitt, N. Weinbach, W. Neupert, and B. Westermann. 2002. Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Mol. Biol. Cell 13:847-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ephrussi, B., H. Hottinguer, and J. Tavlitzki. 1949. Action de l'acriflavine sur les levures. II. Etude génétique du mutant “petite colonie.” Ann. Inst. Pasteur 76:419-450. [Google Scholar]
- 14.Ferrari, S., F. Ischer, D. Calabrese, B. Posteraro, M. Sanguinetti, G. Fadda, B. Rohde, C. Bauser, O. Bader, and D. Sanglard. 2009. Gain of function mutations in CgPDR1 of Candida glabrata not only mediate antifungal resistance but also enhance virulence. PLoS Pathog. 5:e1000268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kontoyiannis, D. P. 2000. Modulation of fluconazole sensitivity by the interaction of mitochondria and erg3p in Saccharomyces cerevisiae. J. Antimicrob. Chemother. 46:191-197. [DOI] [PubMed] [Google Scholar]
- 16.Parikh, V. S., M. M. Morgan, R. Scott, L. S. Clements, and R. A. Butow. 1987. The mitochondrial genome can influence nuclear gene expression in yeast. Science 235:576-580. [DOI] [PubMed] [Google Scholar]
- 17.Praekelt, U. M., and P. A. Meacock. 1990. HSP12, a new small heat shock gene of Saccharomyces cerevisiae: analysis of structure, regulation and function. Mol. Gen. Genet. 223:97-106. [DOI] [PubMed] [Google Scholar]
- 18.Sanglard, D. 2002. Clinical relevance of mechanisms of antifungal drug resistance in yeasts. Enferm. Infec. Microbiol. Clin. 20:462-469. [DOI] [PubMed] [Google Scholar]
- 19.Sanglard, D., F. Ischer, and J. Bille. 2001. Role of ATP-binding-cassette transporter genes in high-frequency acquisition of resistance to azole antifungals in Candida glabrata. Antimicrob. Agents Chemother. 45:1174-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skowronek, P., G. Krummeck, O. Haferkamp, and G. Rödel. 1990. Flow cytometry as a tool to discriminate respiratory-competent and respiratory-deficient yeast cells. Curr. Genet. 18:265-267. [DOI] [PubMed] [Google Scholar]
- 21.Steinmetz, L. M., C. Scharfe, A. M. Deutschbauer, D. Mokranjac, Z. S. Herman, T. Jones, A. M. Chu, G. Giaever, H. Prokisch, P. J. Oefner, and R. W. Davis. 2002. Systematic screen for human disease genes in yeast. Nat. Genet. 31:400-404. [DOI] [PubMed] [Google Scholar]
- 22.Thakur, J. K., H. Arthanari, F. Yang, S. J. Pan, X. Fan, J. Breger, D. P. Frueh, K. Gulshan, D. K. Li, E. Mylonakis, K. Struhl, W. S. Moye-Rowley, B. P. Cormack, G. Wagner, and A. M. Näär. 2008. A nuclear receptor-like pathway regulating multidrug resistance in fungi. Nature 452:604-609. [DOI] [PubMed] [Google Scholar]
- 23.Traven, A., J. M. S. Wong, D. Xu, M. Sopta, and C. J. Ingles. 2001. Interorganellar communication: altered nuclear gene expression profiles in a yeast mitochondrial mutant. J. Biol. Chem. 276:4020-4027. [DOI] [PubMed] [Google Scholar]
- 24.Tsai, H. F., A. A. Krol, K. E. Sarti, and J. E. Bennett. 2006. Candida glabrata PDR1, a transcriptional regulator of a pleiotropic drug resistance network, mediates azole resistance in clinical isolates and petite mutants. Antimicrob. Agents Chemother. 50:1384-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vandeputte, P., G. Larcher, T. Bergès, G. Renier, D. Chabasse, and J. P. Bouchara. 2005. Mechanisms of azole resistance in a clinical isolate of Candida tropicalis. Antimicrob. Agents Chemother. 49:4608-4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vandeputte, P., G. Tronchin, T. Bergès, C. Hennequin, D. Chabasse, and J. P. Bouchara. 2007. Reduced susceptibility to polyenes associated with a missense mutation in the ERG6 gene in a clinical isolate of Candida glabrata with pseudohyphal growth. Antimicrob. Agents Chemother. 51:982-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vermitsky, J. P., K. D. Earhart, W. L. Smith, R. Homayouni, T. D. Edlind, and P. D. Rogers. 2006. Pdr1 regulates multidrug resistance in Candida glabrata: gene disruption and genome-wide expression studies. Mol. Microbiol. 61:704-722. [DOI] [PubMed] [Google Scholar]