Abstract
The function of orf4 in the sigB cluster in Bacillus cereus ATCC 14579 remains to be explored. Amino-acid sequence analysis has revealed that Orf4 is homologous with bacterioferritins and Dps. In this study, we generated an orf4-null mutant and produced recombinant protein rOrf4 to establish the role of orf4. In vitro, the purified rOrf4 was found to exist in two distinct forms, a dimeric form and a polymer form, through size exclusion analysis. The latter form exhibited a unique filament structure, in contrast to the typical spherical tetracosamer structure of bacterioferritins; the former can be induced to form rOrf4 polymers immediately after the addition of FeCl2. Catalysis of the oxidation of ferrous irons by ferroxidase activity was detected with rOrf4, and the mineralized irons were subsequently sequestered only in the rOrf4 polymer. Moreover, rOrf4 exerted DNA-protective activity against oxidative damage via DNA binding in a nonspecific manner, as is seen with Dps. In vivo, deletion of orf4 had no effect on activation of the alternative sigma factor σB, and therefore, orf4 is not associated with σB regulation; however, orf4 can be significantly upregulated upon environmental stress but not H2O2 treatment. B. cereus strains with constitutive Orf4 expression exhibited a viability higher than that of the orf4-null mutant, under specific oxidative stress or heat shock. Taken together, these results suggest that Orf4 functions as a Dps-like bacterioferritin in response to environmental stress and can provide cell protection from oxidative damage through iron sequestration and DNA binding.
The Bacillus cereus group is composed of six Bacillus species, including B. cereus, B. anthracis, B. thuringiensis, B. mycoides, B. pseudomycoides, and B. weihenstephanensis (28). Although these bacterial species are very similar in terms of genetic organization, they exhibit divergent gene regulation activities and distinct physiologies. Bacillus anthracis causes anthrax, and Bacillus thuringiensis is used widely as an insecticide. Bacillus cereus produces a large number of potential virulence factors, including cereulide and the tripartite hemolysin BL, as emetic and diarrheal toxins, which frequently give rise to two types of food poisoning, with mild symptoms of vomiting and diarrhea (42). The characteristics of endospore formation enable B. cereus to overcome extreme conditions (49), and therefore, B. cereus research has focused on the issues of food safety and medical instrument hygiene.
It has been reported that the alternative sigma factor σB is upregulated to transcribe a set of genes in B. subtilis as cells encounter general stress during the exponential phase or entry into the stationary phase (39). σB activity is regulated by eight proteins encoded in the sigB operon through protein-protein interactions in B. subtilis (15, 30). Similarly, σB activation has also been observed to increase cell survival in B. cereus under the above-described conditions (45). In B. cereus, σB activity is controlled by the gene products encoded by the sigB cluster, which includes rsbV, rsbW, sigB, orf4, and rsbY. The roles of these genes have all been described, with the exception of the function of the fourth putative open reading frame, orf4 (GenBank accession number GI:29894703) (17, 46). As orf4 is situated in the sigB cluster, it would be interesting to know whether orf4 is involved in σB regulation. The orf4 gene is conserved across members of the B. cereus group, showing a very high (95 to 96%) amino acid sequence similarity (47). Analysis of the orf4 mRNA transcript by Northern blotting after 42°C heat stress has shown orf4 downstream to be a σB-dependent promoter (45). This result implies that orf4 can be induced to respond to environmental stress. On the other hand, based on sequence analysis, Orf4 is homologous with bacterioferritins and Dps (DNA protection proteins produced during starvation); however, experimental evidence of the hypothesized functions of orf4 relevant to bacterioferritins and Dps is still lacking (21, 45).
Iron participates in redox reactions and has a wide range of potential through the two interchangeable stable oxidation states (II and III) (4); it is also required for cell growth and development. Living cells retain iron at the effective concentration in the range of 10−3 to 10−5 M. Most organisms have evolved ferritin-like proteins with the function of iron storage, particularly under the condition of iron depletion (38). Moreover, ferritin-like proteins are also involved in a protective function against oxidative damage, a common environmental stress for bacteria (20, 50, 54). Oxidative damage is attributed to reactive oxygen species (ROS) attacking biomacromolecules, especially nucleic acids, lipids, and proteins (37). For example, the phagolysosome in the cell produces ROS, such as dioxygen, superoxide, and hydrogen peroxide, during biosynthesis and metabolism (9, 38). In an aerobic environment, superoxide and hydrogen peroxide can be converted to the hydroxyl radical, a more toxic ROS, via the Fenton reaction, catalyzed by Fe2+ (37, 43). When iron concentrations are very high, the level of oxidants increases accordingly by Fenton chemistry. Therefore, ferritins perform an alternate antioxidating function by sequestering iron inside the cavity, away from ROS (11, 44).
Ferritins constitute a broad superfamily of iron storage proteins, including eukaryotic maxi-ferritins, bacterial maxi-ferritins, and miniferritin Dps proteins, which are widespread in aerobic and anaerobic organisms (32, 33, 34). Ferritin isolated from bacteria contains at least 12 heme groups and is named bacterioferritin (Bfr). The function of these heme groups is still not clear, but they may be involved in the regulation of iron ion release (5). Both ferritin and bacterioferritin have the same architecture, being assembled from 24 identical subunits to form a hollow, roughly spherical construction with a diameter of ∼120 Å (11). In addition, a common scheme in ferritins and bacterioferritins is ferroxidase activity, which controls the reversible phase transition between hydrated Fe(II) in solution and the solid mineral (III) core inside its cavity for the accommodation of up to 4,500 iron ions (44).
Dps was first identified as stress-induced protein from Escherichia coli (3); it is found in a wide range of bacteria (13, 27, 33, 43). Dps is under σB control in B. subtilis, induced by stress and starvation (7). The secondary and tertiary structures of Dps molecules are largely homologous with those of ferritin and bacterioferritin (22). Differing from ferritin, which has a tetracosamer structure, Dps exhibits a dodecameric structure with a central cavity (∼45 Å in diameter), which accommodates about 500 iron ions (16, 33). Not all Dps proteins possess the capability of binding DNA and sequestering iron simultaneously; some display either DNA binding or iron storage capability; for instance, Agrobacterium tumefaciens Dps does not bind DNA in vitro (13). Regarding DNA binding capability, Dps and DNA form large condensed Dps-DNA complexes in a nonspecific associative manner (36, 38) and thereby provide a physical shield for DNA (14). The extension of the C terminus and the positive charge at the N terminus are able to stabilize DNA binding in Dps (13, 38, 40). Dps plays an important role in protecting cells from oxidative damage by both iron sequestration and DNA binding. In summary, sequential multiple steps are employed to protect against oxidative damage. First, the conserved ferroxidase center in Dps binds ferrous irons rapidly to diminish the Fenton reaction; second, oxidation of iron in the ferroxidase center is accomplished by the reduction of hydrogen peroxide, which decreases the toxicity of the hydrogen peroxide; and third, the formation of mineralized ferric irons gives rise to a microcrystalline core in the central cavity of Dps (26). In addition to protection from oxidative damage, certain genes are regulated by Dps in order to enhance the bacterial survival rate during the stationary phase and environmental stress in the exponential phase (37).
In the present study, we generated an orf4 deletion mutant, produced an Orf4 recombinant protein in order to investigate the role of orf4 in cell protection in vivo, and discovered biochemical evidence of the function of Orf4 in iron sequestration and DNA binding in vitro. Our data showed that Orf4 exhibited not only a DNA binding property like that of Dps but also an iron sequestration capacity like that of bacterioferritin. Furthermore, the effect of orf4 on cell protection against oxidative stress was also investigated by t-BOOH (tert-butylhydroperoxide) treatment and 50°C heat stress using the orf4 deletion mutant or preloaded Orf4 under the control of a constitutive promoter, the results of which suggested that orf4 can confer to B. cereus relatively effective protection under 50°C heat stress compared to protection against the specific oxidative damage of t-BOOH treatment.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The genotypes and sources of the bacterial strains and plasmids used in this study are listed in Table 1. B. cereus and E. coli were grown in Luria-Bertani (LB) broth or LB agar at 37°C, with vigorous shaking (41). The bacterial growth rate was monitored by measuring the absorbance of the medium with a GeneQuant Pro spectrophotometer (GE Healthcare) at 600 nm.
TABLE 1.
Bacteria strains and plasmids used in this study
| Strain or plasmid | Genotype and/or description | Source or reference |
|---|---|---|
| B. cereus strains | ||
| ATCC 14579 | Wild type | ATCC Biological Resource Center |
| WT708 | Δorf4 mutant | This work |
| WT709 | Δorf4-pRF304 orf4 complementary strain | This work |
| WT710 | Wild-type-pRF305 orf4 overexpression strain | This work |
| WT711 | Δorf4-pRF305 orf4 overexpression strain | This work |
| E. coli strains | ||
| DH5α | General-purpose cloning | Invitrogen |
| BL21(DE3) | For protein expression | Novagen |
| Plasmids | ||
| pMAD | ermC bgaB | 8 |
| pMAD-B-S-Y | sigB anti-spc rsbY | This work |
| pHY300PLK | Tcr | Takara |
| pRF304 | pHY300PLK-orf4, Tcr, orf4 original promoter | This work |
| pRF305 | pHY300PLK-orf4, Tcr, orf4 tufA promoter | This work |
| pDG1728 | Spr | Bacillus Genetic Stock Center |
| pET11-a | E. coli overexpression vector, Apr | Novagen |
| pET14-b | E. coli overexpression vector, Apr | Novagen |
| pET14-b-orf4 | orf4 | This work |
| pGEX-6p-3 | For DNA assays | GE Healthcare Biosciences |
As required, ampicillin (50 μg/μl), tetracycline (20 μg/μl), erythromycin (3 μg/μl), and spectinomycin (300 μg/μl) were added to the cultures for cloning and mutant screening.
Construction of orf4-null mutant.
All of the oligonucleotides used in this study are listed in Table 2. An 862-bp DNA fragment containing the sigB gene was amplified by PCR using the primer pair sigB-BamHI-fw and sigB-SalI-rw. Plasmid pMAD and the PCR product were double-digested with BamHI and SalI and then ligated into pMAD, resulting in pMAD-B. A 1.25-kb DNA fragment containing the anti-spectinomycin gene was amplified by PCR using the primer pair spc-SalI-fw and spc-NcoI-rw. Plasmid pMAD-B and the PCR product were double-digested with SalI and NcoI and ligated into pMAD-B, resulting in pMAD-B-S. A 1.19-kb DNA fragment containing the rsbY gene was amplified by PCR using the primer pair rsbY-NcoI-fw and rsbY-BglII-rw. Plasmid pMAD-B-S and the PCR product were double-digested with NcoI and BglII and ligated into pMAD-B-S, resulting in pMAD-Δorf4, which was electroporated into B. cereus ATCC 14579 as described previously (19), with slight modification. B. cereus was grown overnight in 10 ml of LB broth, and 3 ml of the overnight culture was inoculated into 300 ml LB broth. When the culture reached an optical density at 600 nm of 0.2 to 0.3, it was centrifuged at 6,000 × g for 6 min at 4°C, and the pellet was resuspended in 40 ml of cold electroporation buffer (0.5 mM MgCl2, 272 mM sucrose, 0.2 mM K2HPO4, 50 μM KH2PO4), filter sterilized, and stored at 4°C. The pellet was resuspended in 0.2 to 0.3 ml of cold electroporation buffer and placed on ice. Plasmid DNA (0.1 to 1.0 μg) was mixed with 60 μl cell suspension, incubated on ice for 5 min, and transferred to a chilled cuvette with a 0.1-cm interelectrode gap (Eppendrof). Cells were electroporated at 1.5 kV in an electroporator (Eppendorf), transferred to 1 ml of LB broth, incubated at 37°C, with shaking for 90 min, and then spread onto LB agar containing the appropriate antibiotics. Gene replacement was performed by the method of Arnaud et al. (8).
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′-3′)a | bp |
|---|---|---|
| Orf4 XhoI-F | CCGCTCGAGCGGATGAAAATGTCACACGATGTG | 33 |
| Orf4 BamHI-R | GCCGCGGATCCTTAATTTAACACCATTGC | 29 |
| SigB-BamHI-F | GCCGCGGATCCAATCTCAACCTACG | 25 |
| SigB-SalI-R | GCCCGCCGTCGACATTTTCATATATCTC | 28 |
| Spc-SalI-F | AATGTCGACAGTAGTTCACCACCTTTTCC | 29 |
| Spc-NcoI-R | CATGCCATGGTTTATTGTTTTCTAAAATC | 29 |
| RsbY-NcoI -F | CATGCCATGGGAAATAACTTGTGT | 24 |
| RsbY-BglII-R | TCGAGATCTGTTTGGATTTCATACG | 25 |
| Orf4-SalI-F | AATGTCGACATGAAAATGTCACACGATGTG | 30 |
| Orf4-BamHI-R | GCCGCGGATCCTTAATTTAACACCATTGCT | 30 |
| orf4-promoter-XbaI-F | CAGCTCTAGAAAAATTTAAAATAATGATGT | 30 |
| orf4-stop-SalI-R | AATGTCGACTTAATTTAACACCATTGCTTT | 30 |
| rsbV-probe-F | ATGATGAATTTGGCAATAAATATTTTGC | 28 |
| rsbV-probe-R | TCACCTTCTTTCTACTTTTTCAAAATCGGA | 30 |
| Tuf-XbaI-F | CAGCTCTAGATTGATTTTTATCGATTGTTC | 30 |
| Tuf-SalI-R | AATGTCGACTTCCTCCTTAGTTTATATAGG | 30 |
| qPCR-Orf4-F | ATGAAAATGTCACACGATGT | 20 |
| qPCR-Orf4-R | TACTTGTTTCACTGGTAAAG | 20 |
Introduced restriction sites are underlined.
Cloning, overexpression, and purification of Orf4 recombinant proteins.
The orf4 gene was amplified from B. cereus genomic DNA by PCR using the primer pair Orf4-XhoI-FW and Orf4-BamHI-BW. The PCR product was cloned into pET14-b plasmid (Novagen; Darmstadt, Germany), and the resulting vector pET14-b-orf4 was transformed into E. coli BL21. To produce Orf4, one liter of the bacterial culture was grown at 37°C with ampicillin. When the cells reached the mid-logarithmic phase (optical density at 600 nm of 0.5), isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM and was incubated for 3 h. Cell pellets were then harvested by centrifugation, washed three times with phosphate-buffered saline (PBS), resuspended in binding buffer (20 mM sodium phosphate, 0.5 M NaCl, 30 mM imidazole [pH 7.4]) plus 1 mM phenylmethylsulfonyl fluoride (PMSF), and broken by sonication. After centrifugation, the supernatant was filtered through a 0.22-μl filter (Millipore), loaded in a 5-ml His-trap HP Ni2+ column (Amersham Biosciences), and washed with five times the column volume of wash buffer (20 mM sodium phosphate, 0.5 M NaCl, 500 mM imidazole [pH 7.4]); the column was then eluted with five times the column volume of elution buffer (20 mM sodium phosphate, 0.5 M NaCl, 80 mM imidazole [pH 7.4]). The protein was dialyzed against dialysis buffer (40% glycerol, 100 mM NaCl, 50 mM Tris-HCl [pH 7.5], 1 mM dithiothreitol [DTT]). The purified protein was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the protein concentration was measured by a Bradford assay.
The purification procedure of natural Orf4 recombinant protein, which was expressed from pET-11a harboring orf4 through the NdeI and BamHI sites, followed similar lines. Briefly, E. coli strain BL21(DE3) was transformed with the constructed plasmid, and overexpression of natural Orf4 was achieved by the addition of 1 mM IPTG to the culture medium when the culture had attained an optical density of 0.6 at 600 nm. After harvesting, cells from one liter of culture were resuspended in 30 ml of cell lysis buffer (50 mM Tris-HCl [pH 8.5], 10 mM EDTA, 5 mM DTT, 1 mM PMSF) and broken with a French press (1,250 lb/in2). Soluble proteins were separated from the cell debris by centrifugation (29,000 × g, 60 min), and the filtered supernatant was loaded in a 30-ml DEAE-Sepharose column (Amersham Biosciences) equilibrated with buffer A (50 mM Tris-HCl [pH 8.5], 1 mM DTT). The bound proteins were eluted with a 100-ml linear gradient of buffer A plus 1 M NaCl, and the fractions containing Orf4 were identified by 15% SDS-PAGE. The proteins were concentrated either by ultrafiltration (Millipore) before further purification by gel filtration with a Sephacryl 200 column or using a Superose 12 gel filtration column preequilibrated in gel filtration buffer (50 mM Tris-HCl [pH 7.5], 200 mM NaCl, 1 mM DTT).
Detection of Orf4 under stresses by Western blotting.
Overnight cultures of B. cereus in LB medium were subcultured into fresh LB medium, and cells were exposed to 42°C heat, 2.5% NaCl, and 4% ethanol for 0, 5, 10, 20, 30, 40, and 60 min at an optical density of 0.5 at 600 nm. Cell pellets were harvested by centrifugation, washed three times with phosphate buffer, and immediately frozen in liquid nitrogen for 30 min; they were then resuspended in lysis buffer (50 mM Tris [pH 7.5], 1 mM EDTA, 1 mM DTT, 1 mM PMSF), broken by sonication, and centrifuged at 13,000 × g for 20 min. Protein samples were separated by SDS-PAGE, and the resolved proteins transferred electrophoretically onto a nitrocellulose membrane using a semidry apparatus for 90 min. The blots were subsequently incubated in blocking buffer containing 5% milk in 1% Tween-20 in PBS at room temperature for 1 h, then incubated with 1/4,000 diluted primary antibody at 4°C overnight. Following three 10-min washes with 1% Tween-20 in PBS, the blots were then incubated with secondary antibody at room temperature for an hour. The membrane was washed as described above and developed using the Immobilon Western chemiluminescent horseradish peroxidase substrate (Millipore).
Real-time RT-PCR.
Real-time reverse transcription (RT)-PCR was employed to assay the orf4 expression level under distinct stress. Briefly, one-step quantitative RT-PCR was performed by incubating DNase I-treated RNA with SYBR Premix Ex Taq (perfect real time) (TaKaRa Bio Europe, France) using the LightCycler 1.5 instrument (Roche Applied Science, Mannheim, Germany). The cDNA was subjected to real-time PCR using the primer pairs listed in Table 1. Cycling conditions were 48°C for 30 min and 95°C for 15 min, followed by 40 cycles of 95°C for 15 s, 60°C for 1 min, and a dissociation step at 95°C for 15 s, 60°C for 30 s, and 95°C for 15 s. The standard curve method was used for extrapolating quantitative information for orf4 expression using a constructed plasmid, pET14b-orf4, as a reference standard.
Gel retardation assay.
A DNA retardation assay was performed as described previously (27). Briefly, 5, 10, 30, and 45 μg of rOrf4 were added to 200 ng of plasmid pGEX-6p-3 in TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]) to achieve a final volume of 20 μl, and the DNA-protein mixture was then incubated in a 37°C water bath for 30 min. The complex was resolved on 1% agarose gel in Tris-acetate-EDTA buffer, and the gel was stained with ethidium bromide.
DNA protection assay.
A DNA protection assay was performed as described previously (54), with modification. DNA protection from oxidative damage in vitro was assessed using pGEX-6p-3 plasmid DNA (4,983 bp, 10 mM), which was isolated from E. coli DH5α using a plasmid miniprep kit (Qiagen). Plasmid DNA pGEX-6p-3 was incubated with various amounts of rOrf4 in a buffer (20 mM Tris-HCl, 1 mM EDTA [pH 7.5]) for 30 min prior to the addition of 300 μM FeSO4 and 10 mM H2O2. Plasmid DNA was resolved by electrophoresis on 1% agarose gel in Tris-acetate-EDTA buffer, and the gel was stained with ethidium bromide.
Iron oxidation assay.
Ferrous ammonium sulfate was dissolved in deoxygenated water before the experiment in order to freshly prepare the reagent. Formation of ferric iron with O2 was assessed as described previously (13). The iron oxidation kinetics were monitored spectrophotometrically at 310 nm at room temperature after the addition of 300 μM Fe(II) to 1 μM Orf4 solution in 50 mM MOPS (morpholinepropanesulfonic acid) and 200 mM NaCl, pH 7.8. As a control, the rate of Fe(II) autoxidation without rOrf4 was also measured.
Sensitivity of B. cereus strains to t-BOOH.
The orf4 gene was inserted into the expression vector pHY300PLK downstream of the original orf4 promoter for pRF304 or under the control of the constitutive promoter tufA for pRF305. t-BOOH resistance testing of vegetative B. cereus was performed as described previously (35). Briefly, overnight cultures of B. cereus strains in LB medium were subcultured into fresh LB medium. Wild-type B. cereus, the orf4-null mutant, the orf4 complementary strain harboring pRF304, and orf4 overexpression strains harboring pRF305 at the mid-logarithmic phase (optical density at 600 nm of 0.5) were exposed with or without 1.5 mM t-BOOH for 0, 1, 2, 3, 4, 5, and 6 h at 37°C, with shaking. Cell survival was determined by plating triplicate 20-μl samples of each diluted sample on LB agar and incubating them overnight at 37°C. The relative growth was calculated by comparing colonies of treated cultures with those of untreated cultures. Three independent experiments were conducted for all sets of t-BOOH exposures, and samples were plated in triplicate for each indicated time.
RESULTS
Orf4 was induced by environmental stresses and energy stress but did not regulate σB activity.
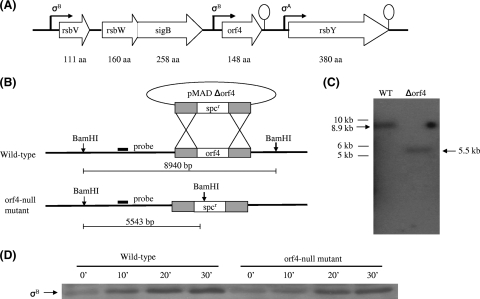
Whether or not Rsb regulators encoded by the sigB operon control σB activity in B. subtilis has been studied extensively (18, 23, 52). With a similar genetic organization, proteins encoded by the sigB cluster in B. cereus (Fig. 1A) have also been found to be involved in σB activation, with the exception that the function of orf4 remains unclear (46). Therefore, we were interested in investigating whether orf4 participates in σB regulation. For this purpose, an orf4 deletion strain, the Δorf4::Spr mutant, named WT708, was constructed using homologous recombination (Fig. 1B) according to the procedure of Arnaud et al. (8). Chromosomal DNAs extracted from WT708 and the parental strain were digested with BamHI and hybridized with a probe of 339 bp amplified from upstream of the orf4 gene. As shown in Fig. 1C, a signal representing the DNA fragment of 5.5 kb was visualized in the wild-type strain, whereas the expected DNA fragment of 8.9 kb appeared in WT708. Thus, Southern blotting hybridization confirmed the completion of orf4 deletion in WT708. To explore the possibility of orf4 being involved in σB regulation, both the wild-type strain and WT708 were treated with 42°C heat stress, which is frequently used to activate σB, and cells were harvested at different times in order to monitor σB activation by Western blotting using anti-σB antibody. No difference in σB activation was detected between the wild-type strain and WT708 (Fig. 1D), and we therefore conclude that orf4 is not involved in σB regulation.
FIG. 1.
Confirmation of orf4 deletion and orf4 not being involved in σB regulation. (A) Schematic diagram of genetic organization of sigB cluster. (B) Construction of the orf4 mutant by allelic exchange. The orf4 gene was replaced by a 1.2-kb spectinomycin resistance cassette. DNA was introduced into B. cereus by electroporation. BamHI restriction enzyme sites were present in the spectinomycin resistance cassette. The thick line indicates the fragment from the orf4 upstream region used as the probe for Southern blotting. The predicted hybridization sizes are shown. (C) Southern blotting confirming the disruption of orf4. Chromosomal DNA from the wild-type strain and the orf4 mutant were digested with BamHI and probed with the DNA upstream of orf4; the expected DNA sizes are indicated by the arrow symbol. Lane 1, wild-type (WT) genomic DNA; lane 2, orf4 mutant genomic DNA. The sizes of the molecular weight markers are indicated on the left. (D) Orf4 deletion did not affect σB activation. Cell lysates harvested from the wild-type and orf4 mutant strains at the indicated times were measured by Western blotting using anti-σB antibody.
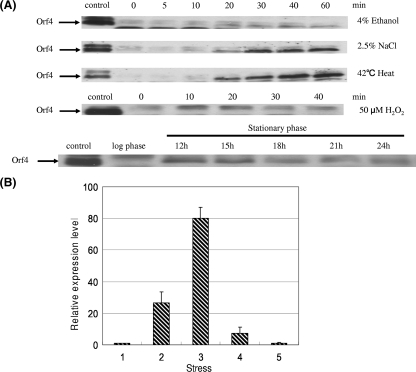
The sigB operon in B. subtilis can be activated by environmental stress, such as 4% ethanol, 2.5% NaCl, or 42°C heat stress during the exponential phase or energy stress during the stationary phase (23, 48). Northern blot analysis in a previous study suggested that orf4 in B. cereus was transcribed in a σB-dependent manner or cotranscribed with rsbV, rsbW, and sigB (45). Hence, in this study the B. cereus cell extract was analyzed by Western blotting using anti-Orf4 polyclonal antibody after the imposition of the above-described stresses and 50 μM H2O2 in order to examine whether the induction of orf4 is related to environmental stress, oxidative stress, or energy stress. Orf4 was found to be significantly induced by 2.5% NaCl treatment and 42°C heat stress, and slightly increased Orf4 expression was observed upon 4% ethanol exposure, but H2O2 treatment failed to activate Orf4. On the other hand, energy depletion resulted in a slight increase in Orf4 expression in the early stationary phase (12 h) and thereafter a decline (Fig. 2A). To confirm the results obtained by Western blotting, we employed real-time RT-PCR to estimate the amount of transcript of the orf4 gene in the wild-type strain without stress or in cells treated with heat, salt, ethanol, and H2O2, and cells were collected after treatment for 10 min. With exposure to 42°C heat, 2.5% NaCl, and 4% ethanol, the transcript level of the orf4 gene increased about 27-fold, 80-fold, and 7-fold, respectively; however, the mRNA level of the orf4 gene was not increased by H2O2 treatment (Fig. 2B). The results were comparable with what was observed by Western blotting. Our data suggest that orf4 responds to environmental stress diversely and that energy stress moderately induces orf4, but specific oxidative stress cannot activate orf4.
FIG. 2.
Expression of Orf4 in B. cereus. (A) Detection of Orf4 expression with Western blotting. Cells were harvested at different times under various stresses or at different growth stages. Cell extracts (20 μg) were used for SDS-PAGE and then Western blotting with anti-Orf4 polyclonal antibody. Orf4 is depicted by the arrows. (B) Real-time PCR quantification of orf4 transcript levels. Cultures of wild-type strain B. cereus were grown until the mid-exponential growth phase and exposed to the stresses indicated in panel A for 10 min. RNA was extracted for real-time PCR. Relative expression levels of orf4 are shown in comparison with that of mid-exponential phase, which is shown as onefold. Means and error bars of three independently stress-treated cultures are displayed. The numbers 1, 2, 3, 4, and 5 represent mid-exponential phase, 42°C heat, 2.5% NaCl, 4% ethanol, and 50 μM H2O2, respectively.
rOrf4 forms a filament structure.
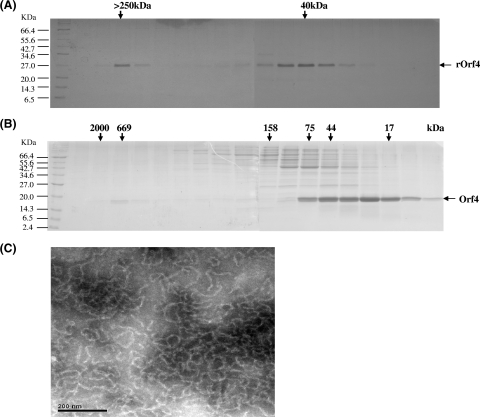
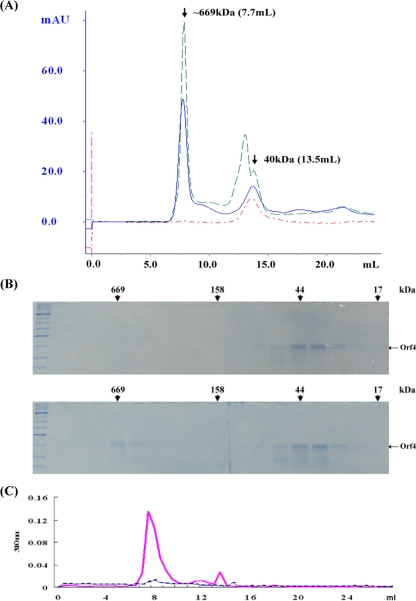
To characterize Orf4, the overexpressed recombinant protein rOrf4 was purified using a nickel column. Size exclusion analysis with a Sephacryl 200 column showed that rOrf4 was eluted mainly in a dimeric form of approximately 40 kDa, but a portion of rOrf4 was eluted in the void volume, showing the tendency of autoassembly (Fig. 3A). Although Orf4 is homologous with bacterioferritins (45), the phenomenon of a predominant rOrf4 dimeric form is in contrast to the characteristics of bacterioferritins, which are known to form a structure of high molecular weight containing 24 identical subunits (11, 29). To evaluate the perturbation of the 2.5-kDa N-terminal fusion His6 tag encoded in pET-14b for rOrf4 assembly, native Orf4 without a fusion tag in the vector pET-11a was utilized as a comparison. After DEAE chromatography purification, native Orf4 was filtered using a Superose 12 gel filtration column. The dimeric form was apparently predominant, despite the similarities with other cellular proteins, but relatively less natural Orf4 was eluted in fractions of high molecular weight (Fig. 3B). This result implied that the His6 tag did not influence rOrf4 assembly and that the newly synthesized native Orf4 was produced chiefly in dimeric form. Of note, the autoassembly tendency of rOrf4 is at least homologous with that of bacterioferritins of high molecular weight. Therefore, we attempted to investigate whether the structure of assembled rOrf4 resembles that of bacterioferritins. The transmission electron microscopy results revealed that the assembled rOrf4 appears as a polymer with a filament structure of the same thickness (Fig. 3C), which is extraordinarily different from the typical roughly spherical structure of bacterioferritins (1). However, it must be considered whether the His6 tag alters the conformation of rOrf4 from the spherical structure to the filament structure. If native Orf4 forms a spherical structure like that of bacterioferritins, the molecular mass should be around ∼480 kDa. In fact, the natural autoassembled Orf4 was found to exhibit the same molecular weight as assembled His6 tag rOrf4 by Superose 12 gel filtration analysis (Fig. 3B); thus, the His6 tag appears to have no influence on rOrf4 assembly in terms of either molecular weight or structure.
FIG. 3.
Characterization of the rOrf4 protein. (A) His6-tag rOrf4 was purified using a nickel column followed by Sephacryl 200 gel filtration chromatography. The fractions were analyzed by 12% SDS-PAGE. M represents protein markers and the labels on the left indicate the molecular masses (in thousands). (B) Native Orf4 was analyzed using Superose 12 following DEAE chromatography. The molecular masses (in thousands) of the gel filtration fractions are denoted by the arrows on the top of the SDS-PAGE results. (C) Negatively stained electron micrographs of the automated six-His-tagged rOrf4 polymer. The sample was placed on a carbon film mounted on a 300-mesh copper grid, immediately removed, and stained with 2% phosphotungstic acid (pH 7.2). Micrographs were taken using a Hitachi 7100 electron microscope operating at 75 kV. The scale bar corresponds to 200 nm.
Ferroxidase activity assay and incorporation of iron into rOrf4.
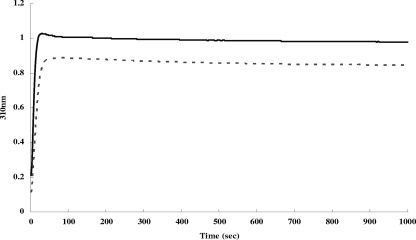
As bacterioferritins possess ferroxidase activity, which can convert iron ions from the ferrous to the ferric state (11), we employed spectrometric analysis at a λ310 nm to identify whether rOrf4 also has ferroxidase activity. The kinetics of absorption at a λ310 nm began at the onset of the addition of 300 μM FeCl2 to 1 μM dimeric rOrf4 in MOPS buffer. Consequently, an absorption at a λ of 310 nm, which was higher than that of the aero-oxidation of ferrous iron, reached a plateau within 40 s, indicating further ferrous iron oxidation exerted by rOrf4 (Fig. 4). Notwithstanding the excess ferrous irons present in the solution, absorbance specific to ferrous iron at a λ190 nm could not be detected by the end of the reaction (data not shown). In turn, all ferrous irons were oxidized rapidly, in agreement with the reported property of ferroxidase activity of bacterioferritins (2).
FIG. 4.
Kinetics of iron oxidation in B. cereus Orf4. Iron oxidation kinetics were monitored spectrophotometrically at 310 nm after the addition of 300 μM Fe(II) to 1 μM in deoxygenated MOPS buffer. Absorbance values with Fe(II) (solid line) or without Fe(II) (dashed line) are shown.
Bacterioferritins mediate the storage of irons in the cytoplasmic granular structure (51). In order to examine whether rOrf4 has the capacity to bind iron in the same way that bacterioferritins do, the FeCl2 and dimeric rOrf4 mixture was applied to a Superose 12 column and gel permeation fractions were monitored with UV at 280 nm. Most of the rOrf4 remained in dimer form, but a strong absorption appeared in the high-molecular-mass fractions at approximately 669 kDa, implying the occurrence of assembled rOrf4 induced by the presence of FeCl2 (Fig. 5A). Although the purification of native Orf4 dimer coincided with some cellular proteins, a similar profile was also observed when native Orf4 dimer was incubated with FeCl2 (Fig. 5A). All elution fractions were subsequently analyzed by SDS-PAGE, and surprisingly, only a small proportion of rOrf4 shifted from dimer to polymer form (Fig. 5B), corresponding to the strong absorption peak at a λ280 nm. However, the relatively small amount of assembled rOrf4 should not lead to such high absorption values. Interestingly, when all elution fractions were measured at an absorbance wavelength specific for ferric iron of 310 nm, a strong absorption peak was present, exactly as with the same fractions containing rOrf4 polymers (Fig. 5C). These results suggest that ferric irons were sequestered only in rOrf4 polymers and therefore contributed to the high absorption at a λ310 nm; this also explains the discrepancy of the high absorption at a λ280 nm produced by the small amount of rOrf4 due to cross-absorption between a λ280 nm and a λ310 nm. Hence, the incorporation of ferric irons into the rOrf4 assembly is consistent with the property of bacterioferritins, which form a spherical tetracosamer structure for iron accommodation (11).
FIG. 5.
Molecular mass determination of recombinant Orf4 proteins. (A) Size exclusion analysis. A total of 50 μM Orf4 was incubated with or without 500 μM FeCl2 and then subjected to Superose 12 gel permeation chromatography. Blue line and green line denote six-His-tagged rOrf4 dimer and native Orf4 dimer incubation with FeCl2, respectively. Red line represents six-His-tagged rOrf4 dimer incubation without FeCl2. Arrows indicate the molecular mass and corresponding elution volume for Orf4 dimer and iron-induced polymer. mAU, absorption units at 280 nm. (B) SDS-PAGE analysis. Fractions represented in panel A were analyzed by 12% PAGE. Six-His-tagged rOrf4 dimer was incubated without FeCl2 (top) or with FeCl2 (bottom). Molecular masses (in thousands) are shown. (C) Detection of ferric iron. The fractions represented in panel B were assessed at a λ310 nm. The solid line represents His6-tagged rOrf4 incubation with ferrous iron; the dashed line indicates incubation without ferrous iron.
Orf4 binds DNA and confers DNA protection against oxidative damage.
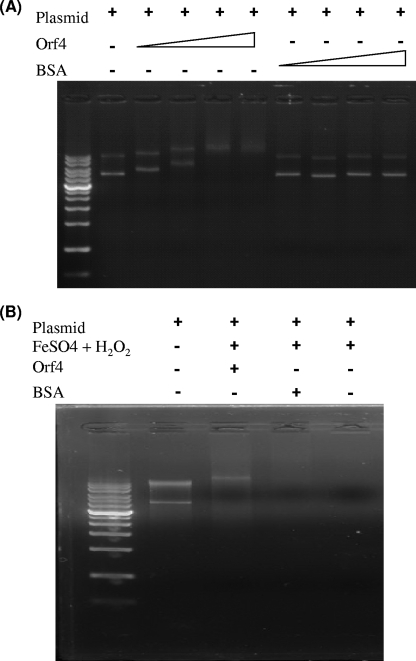
In addition to bacterioferritins, Orf4 is also homologous with Dps, which is induced under starvation conditions in a variety of bacteria to protect DNA from oxidative damage by iron sequestration and/or provide physical shielding of DNA through DNA binding in a nonspecific manner (38). To examine whether rOrf4 can bind DNA in the same way as Dps, plasmid pGEX-6p-3 (4,983 bp) was incubated with rOrf4 or bovine serum albumin (BSA) and then subjected to 1% agarose gel electrophoresis. The results clearly showed that migration of pGEX-6p-3 was retarded after incubation with rOrf4, and more rOrf4 caused cumulative DNA migration retardation until 30 μg and 40 μg of rOrf4 were added, presumably saturating the DNA binding sites (Fig. 6A). On the contrary, DNA migration retardation did not occur in the control BSA set, which revealed the capability of rOrf4 for DNA binding. To identify the manner in which rOrf4 DNA binding occurs, rOrf4 interacted with two separated DNA fragments with expected sizes of 3,187 bp and 1,796 bp, which were derived from pGEX-6p-3 restricted by BamHI and EcoRV. Incubation of rOrf4 with the separate DNA fragments led to electrophoretic retardation, suggesting that the DNA binding of rOrf4 occurs in a rather nonspecific manner. As rOrf4 can bind with DNA, we investigated whether rOrf4 confers DNA protection from oxidative damage, which is a vital biological function of Dps. To address this, plasmid pGEX-6p-3 was incubated with rOrf4 prior to H2O2 treatment in combination with FeSO4, which can enhance the production of hydroxyl radicals by the Fenton reaction. The complete breakdown of pGEX-6p-3 was visualized without rOrf4 or with BSA; however, DNA breakdown was inhibited after preincubation with rOrf4 (Fig. 6B). Taken together, these results show that rOrf4 is capable of protecting DNA against oxidative breakdown, not only by iron sequestration in the manner of bacterioferritins but also by direct DNA binding in the manner of Dps.
FIG. 6.
rOrf4 interaction with DNA. (A) DNA binding of rOrf4. Plasmid DNA pGEX-6p-3 (0.2 μg) was incubated with different amounts of rOrf4 and subsequently loaded onto 1% agarose gel for electrophoresis. Lane 1, 14-kb DNA marker; lane 2, pGEX-6p-3; lanes 3 to 6, pGEX-6p-3 incubation with 5, 10, 30, and 45 μg rOrf4; lanes 7 to10, pGEX-6p-3 incubation with 5, 10, 30, and 45 μg BSA. (B) DNA protection assay of rOrf4. Plasmid DNA pGEX-6p-3 (0.2 μg) was added to 10 mM H2O2 in the presence of FeSO4 after incubation with or without proteins. Lane 1, 14-kb DNA marker; lane 2, pGEX-6p-3; lanes 3 to 7, pGEX-6p-3 + 1 to 5 μg rOrf4+FeSO4 + H2O2; lane 8, pGEX-6p-3 + FeSO4 + H2O2.
Sensitivity of B. cereus and orf4-null mutant to oxidative stress.
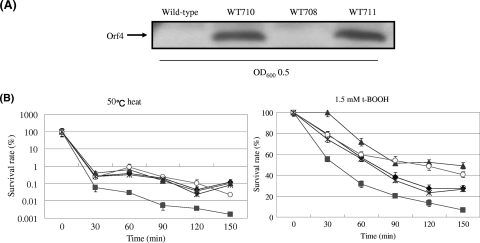
Although rOrf4 was shown to protect DNA from oxidative breakdown in vitro (Fig. 6B), the effect of orf4 on cell protection upon oxidative stress needed to be investigated. For the specific oxidative assay, wild-type B. cereus and the mutant strain WT708 were treated with 1.5 mM t-BOOH for 6 h, and cell cultures were harvested at specific time intervals for cell viability determination by plating count. The results showed that exposure to t-BOOH significantly lowered cell viability in both the wild-type and WT708 strains, although the survival rate of the wild type was approximately 1.5- to 2-fold higher than that of WT708 (Fig. 7B). Hence, deletion of orf4 caused B. cereus to be slightly sensitive to t-BOOH. In addition, we constructed a plasmid designated pRF304, which contained the orf4 gene controlled by the orf4 original promoter in the expression vector pHY300PLK. For orf4 complementation, pRF304 was introduced into WT708 by electroporation in order to generate another strain, WT709. As expected, the reduced cell viability observed in WT708 upon t-BOOH treatment was restored in WT709 (Fig. 7B).
FIG. 7.
Orf4 protects cells from oxidative stress. (A) Constitutive expression of Orf4. Four B. cereus strains including the wild-type strain, WT708 (orf4-null mutant), WT710 (wild-type strain harboring pRF305), and WT711 (orf4-null mutant harboring pRF305) were measured for Orf4 expression by Western blotting in the absence of environmental stress. The constitutive Orf4 expressions in strain WT710 and WT711 are clearly shown. OD600, optical density at 600nm. (B) Orf4 overexpression increases cell viability under the condition of 50°C heat stress or 1.5 mM t-BOOH treatment. A cell culture was harvested every 30 min to measure cell viability by plating count. The diamond, square, asterisk, triangle, and circle symbols denote the survival rate of wild-type B. cereus, WT708, WT709 (orf4-null mutant harboring pRF304), WT710, and WT711, respectively. The means of three replications are displayed for cell viability at each recorded time point.
Cell viability was determined upon treatment with 1.5 mM t-BOOH in order to investigate the effect of overexpressed Orf4 under the condition of specific oxidative stress followed by heat stress. To meet this end, another expression plasmid denoted pRF305, which was modified from pRF304 by replacing the orf4 original promoter with a constitutive B. cereus tufA promoter, was used. This plasmid pRF305 was then introduced into the wild-type and WT708 strains to generate two new strains, WT710 and WT711, respectively. The wild type with a barely detectable expression of orf4 during the exponential phase contrasts to WT710 and WT711, in that both strains constitutively expressed Orf4, even in the absence of stress (Fig. 7A). When these four bacterial strains were subjected to 1.5 mM t-BOOH heat stress, two strains, the wild type and WT711, exhibited comparable viabilities until 60 min; thereafter, WT711 showed a higher survival rate, homologous with that of strain WT710, than those of the wild type and WT708. WT710 showed a cell viability that was up to sevenfold higher than that of WT708 at the 150-min time point (Fig. 7B). Unlike WT708, which exhibited only 1.5- to 2-fold relative sensitivities to t-BOOH treatment, the strains with preloaded Orf4 resulted in more effective protection against specific oxidative damage compared with that of the wild type. Furthermore, the survival rates of these four bacterial strains were also examined, as cells were exposed to 50°C heat stress. The cell viabilities of three strains, the wild type, WT710 and WT711, as a result, were increased 25- to 27-fold in comparison with that of WT708 (Fig. 7B).
DISCUSSION
For B. subtilis, it has been well established that the alternative sigma factor σB governs the transcription of general stress proteins and the regulation of σB activity through a partner-switching mechanism (25). This mechanism is most likely employed in B. cereus, in which proteins encoded by the sigB cluster are known to be involved in the regulation of σB activity, with the exception that the role of orf4 is currently unclear (46). Our data showed indistinguishable σB activations upon 42°C heat stress for the wild-type strain and the orf4-null mutant, suggesting that orf4 is not involved in the regulation of σB activity (Fig. 2C). However, significant orf4 induction upon 42°C heat stress and 2.5% salt treatment implies orf4 induction relative to environmental stress (Fig. 1). This postulation is supported by the fact that orf4 is a known member of the B. cereus SigB regulon (47). Notably, 4% ethanol exposure caused only a slight increase in Orf4 expression (Fig. 2A and B). This result is consistent with a previous study (46) that showed less σB induction by ethanol exposure than by salt and heat stress.
Two miniferritin/Dps genes are found in B. subtilis and many other Bacillus spp. (44). Dps2/MrgA miniferritin is induced by peroxide via modification of the Per transcription factor by an Fe2+/H2O2-catalyzed reaction (31), and Dps1/DpsA miniferritin is induced by general stress (heat, salt, and ethanol stress, and glucose starvation), which is σB dependent during exponential growth (7). The fact that Orf4 is induced by general stress but not by H2O2 treatment indicates that the regulation of orf4 is similar to that of Dps1/DpsA (Fig. 1).
Although Orf4 shows approximately a 30% identity and 50% similarity with bacterioferritins (21), Orf4 has some distinctive features in comparison with bacterioferritins. For example, unlike findings for most of the bacterioferritin homologs, which readily form tetracosamers, even in the absence of iron (20, 54), our results showed that rOrf4 has a predominant dimeric form and a polymer form (Fig. 3A and B), but the proportions of the dimeric and polymer forms varied slightly in different preparations. Interestingly, a similar observation was reported in overproduced E. coli Bfr (bacterioferritin), which comprised a dimeric form and a tetracosameric form in various proportions (6). The physiological significance of the dimeric form in Bfr is assumed to be participation in the release mechanism of iron stores, or alternatively it may have a function distinct from that of the tetracosameric form, e.g., electron transfer (6). Consistent with these assumptions, our data showed a series of reactions, including ferrous oxidation and induction of rOrf4 assembly, in which conformational change must occur, and the entry of ferric iron into the nucleation core within the polymer after the addition of FeCl2 elucidates the necessity of iron for rOrf4 assembly and deaggregation through the interchangeable Fe state (Fig. 4 and 5).
Formation of the rOrf4 polymer can be facilitated rapidly by the addition of ferrous iron (Fig. 5A and B), and the amount of iron-induced rOrf4 polymer depends on the quantity of FeCl2. In contrast, the automated rOrf4 polymer can be formed at a relatively slow rate from dimeric rOrf4. The molecular mass of automated-assembly rOrf4 of ∼669 kDa is distinct from that of bacterioferritins, whose molecular masses are estimated to be around 450 to 480 kDa. In accordance with this observation, transmission electron microscopy imaging revealed a filament structure in the automated rOrf4 polymer, which is different from the typical roughly spherical structure of bacterioferritins (Fig. 3C). The automated-assembly rOrf4 still retained the capacity to incorporate iron (data not shown).
Ferric irons were detected at a λ310 nm only in the fractions corresponding to rOrf4 polymers, but no absorbance was detected in the dimeric rOrf4 fractions (Fig. 5C). This result implies that rOrf4 is able to oxidize ferrous iron efficiently, thereby incorporating mineralized ferric iron into the rOrf4 polymer, as do bacterioferritins. However, rOrf4 exhibited a reaction that was most likely monophasic (Fig. 4), as opposed to the biphasic kinetics of iron oxidation/incorporation in bacterioferritins (10, 13). Therefore, the rOrf4 polymer with the filament structure probably employed a distinct mechanism to coordinate storage of ferric irons within the intramolecular space compared to the storage of ferric iron in the nanocages of bacterioferritin homologs (10, 11, 13).
Organisms develop complex strategies to protect cells from injury from oxidants upon oxidative stress. Dps-like proteins play important roles in protecting DNA, usually by the formation of Dps-DNA complexes (12) and reduce the DNA degradation caused by iron-dependent free-radical generation by the Fenton reaction (53). Orf4 showed homology (approximately a 27% identity and 40% similarity) with DpsA from the Synechococcus strains PCC7947 and PCC6031 (21) and was found to be able to protect DNA from degradation by the oxidative damage caused by H2O2 treatment through direct DNA binding in a nonspecific manner (Fig. 6). Thus, Orf4 has a DNA binding property homologous with that of Dps. The DNA binding of Dps has been attributed to the presence of lysine-rich residues, resulting in a positively charged N terminus or requiring C-terminal extension (13, 22, 42). Three lysine residues (Lys10, Lys14, and Lys18) are situated at the N terminus of Orf4; however, evidence is still needed to identify whether these residues are involved in DNA binding.
Although our in vitro system demonstrated that rOrf4 can bind DNA and sequester iron, the cell protection provided by orf4 against oxidative stress resulted only in a 1.5- to 2-fold decrease in viability in the WT708 strain compared with that of the wild-type strain, which is not as significant as that of other essential antioxidation genes, including sigB, dps, and katA in B. cereus (24, 37, 45). We can interpret this result as wild-type B. cereus expressing a barely detectable level of Orf4 without environmental stress during the logarithmic phase and as the H2O2 treatment failing to induce Orf4; therefore, the Orf4 level in the wild-type strain would be expected to be only slightly higher than that in WT708, which did not express Orf4. As a certain Orf4 level is required for cell protection upon environmental stress, the relatively smaller difference in Orf4 expression satisfactorily explains the small difference in cell viability. Moreover, the cell survival rates of WT710 and WT711 were elevated upon 1.5 mM t-BOOH treatment or 50°C heat stress, as cells preloaded with Orf4 constitutively expressed pRF305; this result further supports that the Orf4 level is critical to cell protection under oxidative stress.
We demonstrated in this study that Orf4 possesses the properties of ferroxidase activity and iron sequestration and prevents DNA from oxidative degradation in vitro and that Orf4 displays the ability to increase cell viability during environmental stress in vivo. In conclusion, Orf4 was characterized as a Dps-like bacterioferritin that is regulated in much the same way as Dps1/DpsA. These results suggest that Orf4 may have a distinct role in iron metabolism in response to environmental stress, rather than exerting direct activity against specific oxidative stress.
Acknowledgments
We thank Michael Yudkin (Oxford University, Oxford, United Kingdom) for helpful discussions. We thank M. Débarbouillé (Pasteur Institute, Paris, France) for kindly providing pMAD. We are grateful to Yuan-Yuan Shih and Chia-Yuan Chang for technical assistance.
This work was supported by a grant of the National Science Council of Taiwan to C.C.C. (96-2320-B-017-001-MY3).
Footnotes
Published ahead of print on 1 May 2009.
REFERENCES
- 1.Abdul-Tehrani, H., A. J. Hudson, Y. S. Chang, A. R. Timms, C. Hawkins, J. M. Williams, P. M. Harrison, J. R. Guest, and S. C. Andrews. 1999. Ferritin mutants of Escherichia coli are iron deficient and growth impaired, and fur mutants are iron deficient. J. Bacteriol. 1811415-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aitken-Rogers, H., C. Singleton, A. Lewin, A. Taylor-Gee, G. R. Moore, and N. E. Le-Brun. 2004. Effect of phosphate on bacterioferritin-catalysed iron(II) oxidation. J. Biol. Inorg. Chem. 9161-170. [DOI] [PubMed] [Google Scholar]
- 3.Almirón, M., A. J. Link, D. Furlong, and R. Kolter. 1992. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 62646-2654. [DOI] [PubMed] [Google Scholar]
- 4.Andrews, S. C. 1998. Iron storage in bacteria. Adv. Microb. Physiol. 40281-351. [DOI] [PubMed] [Google Scholar]
- 5.Andrews, S. C., N. E. L. Brun, V. Barynin, A. J. Thomson, G. R. Moore, J. R. Guest, and P. M. Harrison. 1995. Site-directed replacement of the coaxial heme ligands of bacterioferritin generates heme-free variants. J. Biol. Chem. 27023268-23274. [DOI] [PubMed] [Google Scholar]
- 6.Andrews, S. C., J. M. A. Smith, C. Hawkins, J. M. Williams, P. M. Harrison, and J. R. Guest. 1993. Overproduction, purification and characterization of the bacterioferritin of Escherichia coli and a C-terminally extended variant. Eur. J. Biochem. 213329-338. [DOI] [PubMed] [Google Scholar]
- 7.Antelmann, H., S. Engelmann, R. Schmid, A. Sorokin, A. Lapidus, and M. Hecker. 1997. Expression of a stress- and starvation-induced dps/pexB-homologous gene is controlled by the alternative sigma factor σB in Bacillus subtilis. J. Bacteriol. 1797251-7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arnaud, M., A. Chastanet, and M. Débarbouillé. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 706887-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhattacharyya, G., and A. Grove. 2007. The N-terminal extensions of Deinococcus radiodurans Dps-1 mediate DNA major groove interactions as well as assembly of the dodecamer. J. Biol. Chem. 28211921-11930. [DOI] [PubMed] [Google Scholar]
- 10.Bozzi, M., G. Mignogna, S. Stefanini, D. Barra, C. Longhi, P. Valenti, and E. Chiancone. 1997. A novel non-heme iron-binding ferritin related to the DNA-binding proteins of the Dps family in Listeria innocua. J. Biol. Chem. 2723259-3265. [DOI] [PubMed] [Google Scholar]
- 11.Carrondo, M. A. 2003. Ferritins, iron uptake and storage from the bacterioferritin viewpoint. EMBO J. 221959-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ceci, P., S. Cellai, E. Falvo, C. Rivetti, G. L. Rossi, and E. Chiancone. 2004. DNA condensation and self-aggregation of Escherichia coli Dps are coupled phenomena related to the properties of the N-terminus. Nucleic Acids Res. 325935-5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ceci, P., A. Ilari, E. Falvo, and E. Chiancone. 2003. The Dps protein of Agrobacterium tumefaciens does not bind to DNA but proects it toward oxidative cleavage: x-ray crystal structure, iron binding, and hydroxyl-radical scavenging properties. J. Biol. Chem. 27820319-20326. [DOI] [PubMed] [Google Scholar]
- 14.Ceci, P., L. Mangiarotti, C. Rivetti, and E. Chiancone. 2007. The neutrophil-activating Dps protein of Helicobacter pylori, HP-NAP, adopts a mechanism different from Escherichia coli Dps to bind and condense DNA. Nucleic Acid Res. 352247-2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, C.-C., R. J. Lewis, R. Harris, M. D. Yudkin, and O. Delumeau. 2003. A supramolecular complex in the environmental stress signalling pathway of Bacillus subtilis. Mol. Microbiol. 491657-1669. [DOI] [PubMed] [Google Scholar]
- 16.Chiancone, E., P. Ceci, A. Ilari, F. Ribacchi, and S. Stefanini. 2004. Iron and proteins for iron storage and detoxification. Biometals 17197-202. [DOI] [PubMed] [Google Scholar]
- 17.de-Vries, Y. P., L. M. Hornstra, W. M. de-Vos, and T. Abee. 2004. Growth and sporulation of Bacillus cereus ATCC 14579 under defined conditions: temporal expression of genes for key sigma factors. Appl. Environ. Microbiol. 702514-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dufour, A., and W. G. Haldenwang. 1994. Interactions between a Bacillus subtilis anti-sigma factor (RsbW) and its antagonist (RsbV). J. Bacteriol. 1761813-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunn, A. K., A. K. Klimowicz, and J. Handelsman. 2003. Use of a promoter trap to identify Bacillus cereus genes regulated by tomato seed exudates and a rhizosphere resident, Pseudomonas aureofaciens. Appl. Environ. Microbiol. 691197-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dussurget, O., E. Dumas, C. Archambaud, I. Chafsey, C. Chambon, M. He′braud, and P. Cossart. 2005. Listeria monocytogenes ferritin protects against multiple stresses and is required for virulence. FEMS Microbiol. Lett. 250253-261. [DOI] [PubMed] [Google Scholar]
- 21.Fouet, A. S., O. Namy, and G. Lambert. 2000. Characterization of the operon encoding the alternative σB factor from Bacillus anthracis and its role in virulence. J. Bacteriol. 1825036-5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grant, R. A., D. J. Filman, S. E. Finkel, R. Kolter, and J. M. Hogle. 1998. The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nat. Struct. Biol. 5294-303. [DOI] [PubMed] [Google Scholar]
- 23.Hecker, M., J. Pané-Farré, and U. Völker. 2007. SigB-dependent general stress response in Bacillus subtilis and related gram-positive bacteria. Annu. Rev. Microbiol. 61215-236. [DOI] [PubMed] [Google Scholar]
- 24.Horsburgh, M. J., M. O. Clements, H. Crossley, E. Ingham, and S. J. Foster. 2001. PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect. Immun. 693744-3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Igoshin, O. A., M. S. Brody, C. W. Price, and M. A. Savageau. 2007. Distinctive topologies of partner-switching signaling networks correlate with their physiological roles. J. Mol. Biol. 3691333-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ilari, A., P. Ceci, D. Ferrari, G. L. Rossi, and E. Chiancone. 2002. Iron incorporation into Escherichia coli Dps gives rise to a ferritin-like microcrystalline core. J. Biol. Chem. 27737619-37623. [DOI] [PubMed] [Google Scholar]
- 27.Ishikawa, T., Y. Mizunoe, S.-I. Kawabata, A. Takade, M. Harada, S. N. Wai, and S.-I. Yoshida. 2003. The iron-binding protein Dps confers hydrogen peroxide stress resistance to Campylobacter jejuni. J. Bacteriol. 1851010-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jensen, G. B., B. M. Hansen, J. Eilenberg, and J. Mahillon. 2003. The hidden lifestyles of Bacillus cereus and relatives. Environ. Microbiol. 5631-634. [DOI] [PubMed] [Google Scholar]
- 29.Keren, N., R. Aurora, and H. B. Pakrasi. 2004. Critical roles of bacterioferritins in iron storage and proliferation of cyanobacteria. Plant Physiol. 1351666-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuo, S., S. Zhang, R. L. Woodbury, and W. G. Haldenwang. 2004. Associations between Bacillus subtilis σB regulators in cell extracts. Microbiology 1504125-4136. [DOI] [PubMed] [Google Scholar]
- 31.Lee, J. W., and J. D. Helmann. 2006. The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nat. Struct. Biol. 440363-367. [DOI] [PubMed] [Google Scholar]
- 32.Liu, X., K. Hintze, B. Lonnerdal, and E. C. Theil. 2006. Iron at the center of ferritin, metal/oxygen homeostasis and novel dietary strategies. Biol. Res. 39167-171. [DOI] [PubMed] [Google Scholar]
- 33.Liu, X., K. Kim, T. Leighton, and E. C. Theil. 2006. Paired Bacillus anthracis Dps (mini-ferritin) have different reactivities with peroxide. J. Biol. Chem. 28127827-27835. [DOI] [PubMed] [Google Scholar]
- 34.Liu, X., and E. C. Theil. 2005. Ferritins: dynamic management of biological iron and oxygen chemistry. Acc. Chem. Res. 38167-175. [DOI] [PubMed] [Google Scholar]
- 35.Loprasert, S., W. Whangsuk, R. Sallabhan, and S. Mongkolsuk. 2004. DpsA protects the human pathogen Burkholderia pseudomallei against organic hydroperoxide. Arch. Microbiol. 18296-101. [DOI] [PubMed] [Google Scholar]
- 36.Martinez, A., and R. Kolter. 1997. Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J. Bacteriol. 1795188-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nair, S., and S. E. Finkel. 2004. Dps protects cells against multiple stresses during stationary phase. J. Bacteriol. 1864192-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olsen, K. N., M. H. Larsen, C. G. M. Gahan, B. Kallipolitis, X. A. Wolf, R. Rea, C. Hill, and H. Ingmer. 2005. The Dps-like protein Fri of Listeria monocytogenes promotes stress tolerance and intracellular multiplication in macrophage-like cells. Microbiology 151925-933. [DOI] [PubMed] [Google Scholar]
- 39.Petersohn, A., M. Brigulla, S. Haas, J. D. Hoheisel, U. Volker, and M. Hecker. 2001. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 1835617-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ratledge, C., and L. G. Dover. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54881-941. [DOI] [PubMed] [Google Scholar]
- 41.Salvetti, S., E. Ghelardi, F. Celandroni, M. Ceragioli, F. Giannessi, and S. Senesi. 2007. FlhF, a signal recognition particle-like GTPase, is involved in the regulation of flagellar arrangement, motility behaviour and protein secretion in Bacillus cereus. Microbiology 1532541-2552. [DOI] [PubMed] [Google Scholar]
- 42.Schoeni, J. L., and A. C. Wong. 2005. Bacillus cereus food poisoning and its toxins. J. Food Prot. 68636-648. [DOI] [PubMed] [Google Scholar]
- 43.Stillman, T. J., M. Upadhyay, V. A. Norte, S. E. Sedelnikova, M. Carradus, S. Tzokov, P. A. Bullough, C. A. Shearman, M. J. Gasson, C. H. Williams, P. J. Artymiuk, and J. Green. 2005. The crystal structures of Lactococcus lactis MG1363 Dps proteins reveal the presence of an N-terminal helix that is required for DNA binding. Mol. Microbiol. 571101-1112. [DOI] [PubMed] [Google Scholar]
- 44.Theil, E. C. 2007. Coordinating response to iron and oxygen stress with DNA and mRNA promoters: the ferritin story. Biometals 20513-521. [DOI] [PubMed] [Google Scholar]
- 45.van-Schaik, W., M. H. Tempelaars, J. A. Wouters, W. M. de-Vos, and T. Abee. 2004. The alternative sigma factor σB of Bacillus cereus: response to stress and role in heat adaptation. J. Bacteriol. 186316-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van-Schaik, W., M. H. Tempelaars, M. H. Zwietering, W. M. de-Vos, and T. Abee. 2005. Analysis of the role of RsbV, RsbW, and RsbY in regulating σB activity in Bacillus cereus. J. Bacteriol. 1875846-5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van-Schaik, W., M. van der Voort, D. Molenaar, R. Moezelaar, W. M. de Vos, and T. Abee. 2007. Identification of the σB regulon of Bacillus cereus and conservation of σB-regulated genes in low-GC-content gram-positive bacteria. J. Bacteriol. 1894384-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vijay, K., M. S. Brody, E. Fredlund, and C. W. Price. 2000. A PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the σB transcription factor of Bacillus subtilis. Mol. Microbiol. 35180-188. [DOI] [PubMed] [Google Scholar]
- 49.Vilas-Bôas, G. T., A. P. Peruca, and O. M. Arantes. 2007. Biology and taxonomy of Bacillus cereus, Bacillus anthracis, and Bacillus thuringiensis. Can. J. Microbiol. 53673-687. [DOI] [PubMed] [Google Scholar]
- 50.Wai, S. N., K. Nakayama, K. Umene, T. Moriya, and K. Amako. 1996. Construction of a ferritin-deficient mutant of Campylobacter jejuni: contribution of ferritin to iron storage and protection against oxidative stress. Mol. Microbiol. 201127-1134. [DOI] [PubMed] [Google Scholar]
- 51.Waidner, B., S. Greiner, S. Odenbreit, H. Kavermann, J. Velayudhan, F. Stähler, J. Guhl, E. Bissé, A. H. van-Vliet, S. C. Andrews, J. G. Kusters, D. J. Kelly, R. Haas, M. Kist, and S. Bereswill. 2002. Essential role of ferritin Pfr in Helicobacter pylori iron metabolism and gastric colonization. Infect. Immun. 703923-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wise, A. A., and C. W. Price. 1995. Four additional genes in the sigB operon of Bacillus subtilis that control activity of the general stress factor σB in response to environmental signals. J. Bacteriol. 177123-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamamoto, Y., L. B. Poole, R. R. Hantgan, and Y. Kamio. 2002. An iron-binding protein, Dpr, from Streptococcus mutans prevents iron-dependent hydroxyl radical formation in vitro. J. Bacteriol. 1842931-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao, G., P. Ceci, A. Ilari, L. Giangiacomo, T. M. Laue, E. Chiancone, and N. D. Chasteen. 2002. Iron and hydrogen peroxide detoxification properties of DNA-binding protein from starved cells. A ferritin-like DNA-binding protein of Escherichia coli. J. Biol. Chem. 27727689-27696. [DOI] [PubMed] [Google Scholar]