Abstract
Pseudomonas putida CBB5 was isolated from soil by enrichment on caffeine. This strain used not only caffeine, theobromine, paraxanthine, and 7-methylxanthine as sole carbon and nitrogen sources but also theophylline and 3-methylxanthine. Analyses of metabolites in spent media and resting cell suspensions confirmed that CBB5 initially N demethylated theophylline via a hitherto unreported pathway to 1- and 3-methylxanthines. NAD(P)H-dependent conversion of theophylline to 1- and 3-methylxanthines was also detected in the crude cell extracts of theophylline-grown CBB5. 1-Methylxanthine and 3-methylxanthine were subsequently N demethylated to xanthine. CBB5 also oxidized theophylline and 1- and 3-methylxanthines to 1,3-dimethyluric acid and 1- and 3-methyluric acids, respectively. However, these methyluric acids were not metabolized further. A broad-substrate-range xanthine-oxidizing enzyme was responsible for the formation of these methyluric acids. In contrast, CBB5 metabolized caffeine to theobromine (major metabolite) and paraxanthine (minor metabolite). These dimethylxanthines were further N demethylated to xanthine via 7-methylxanthine. Theobromine-, paraxanthine-, and 7-methylxanthine-grown cells also metabolized all of the methylxanthines mentioned above via the same pathway. Thus, the theophylline and caffeine N-demethylation pathways converged at xanthine via different methylxanthine intermediates. Xanthine was eventually oxidized to uric acid. Enzymes involved in theophylline and caffeine degradation were coexpressed when CBB5 was grown on theophylline or on caffeine or its metabolites. However, 3-methylxanthine-grown CBB5 cells did not metabolize caffeine, whereas theophylline was metabolized at much reduced levels to only methyluric acids. To our knowledge, this is the first report of theophylline N demethylation and coexpression of distinct pathways for caffeine and theophylline degradation in bacteria.
Caffeine (1,3,7-trimethylxanthine) and related methylxanthines are widely distributed in many plant species. Caffeine is also a major human dietary ingredient that can be found in common beverages and food products, such as coffee, tea, and chocolates. In pharmaceuticals, caffeine is used generally as a cardiac, neurological, and respiratory stimulant, as well as a diuretic (3). Hence, caffeine and related methylxanthines enter soil and water easily through decomposed plant materials and other means, such as effluents from coffee- and tea-processing facilities. Therefore, it is not surprising that microorganisms capable of degrading caffeine have been isolated from various natural environments, with or without enrichment procedures (3, 10). Bacteria use oxidative and N-demethylating pathways for catabolism of caffeine. Oxidation of caffeine by a Rhodococcus sp.-Klebsiella sp. mixed-culture consortium at the C-8 position to form 1,3,7-trimethyluric acid (TMU) has been reported (8). An 85-kDa, flavin-containing caffeine oxidase was purified from this consortium (9). Also, Mohapatra et al. (12) purified a 65-kDa caffeine oxidase from Alcaligenes sp. strain CF8. Cells of a caffeine-degrading Pseudomonas putida strain (ATCC 700097) isolated from domestic wastewater (13) showed a fourfold increase in a cytochrome P450 absorption spectrum signal compared to cells grown on glucose. Recently, we reported a novel non-NAD(P)+-dependent heterotrimeric caffeine dehydrogenase from Pseudomonas sp. strain CBB1 (20). This enzyme oxidized caffeine to TMU stoichiometrically and hydrolytically, without producing hydrogen peroxide. Further metabolism of TMU has not been elucidated.
Several caffeine-degrading bacteria metabolize caffeine via the N-demethylating pathway and produce theobromine (3,7-dimethylxanthine) or paraxanthine (1,7-dimethylxanthine) as the initial product. Theophylline (1,3-dimethylxanthine) has not been reported to be a metabolite in bacterial degradation of caffeine. Subsequent N demethylation of theobromine or paraxanthine to xanthine is via 7-methyxanthine. Xanthine is further oxidized to uric acid by xanthine dehydrogenase/oxidase (3, 10). Although the identities of metabolites and the sequence of metabolite formation for caffeine N demethylation are well established, there is very little information on the number and nature of N-demethylases involved in this pathway.
The lack of adequate information on the metabolism and enzymology of theophylline, caffeine, and related methylxanthines prompted us to investigate the degradation of these compounds in detail. We isolated a unique caffeine-degrading bacterium, P. putida CBB5, from soil via enrichment with caffeine as the sole source of carbon and nitrogen. Here we describe a detailed study of the metabolism of theophylline, caffeine, and related di- and monomethylxanthines by CBB5. Our results indicate that CBB5 initially N demethylated caffeine to produce theobromine (major product) and paraxanthine (minor product) before the pathways converged to 7-methylxanthine and xanthine. Surprisingly, CBB5 was also capable of utilizing theophylline as a sole carbon and nitrogen source. CBB5 N demethylated theophylline to 1-methylxanthine and 3-methylxanthine, which were further N demethylated to xanthine. Theophylline N-demethylase activity was detected in cell extracts prepared from theophylline-grown CBB5 cells. 1-Methylxanthine and 3-methylxanthine were detected as products of this NAD(P)H-dependent reaction. To our knowledge, this is the first report of a theophylline degradation pathway in bacteria and coexpression of distinct caffeine and theophylline degradation pathways.
MATERIALS AND METHODS
Chemicals.
Caffeine, theobromine, paraxanthine, theophylline, 1-methylxanthine, 3-methylxanthine, 7-methylxanthine, xanthine, 1-methyluric acid, 1,3-dimethyluric acid, and uric acid were purchased from Sigma-Aldrich (St. Louis, MO). Yeast nitrogen base without amino acids and without ammonium sulfate (YNB) was obtained from ForMedium (Norfolk, United Kingdom), and soytone was purchased from Becton, Dickinson and Company (Sparks, MD). High-pressure liquid chromatography (HPLC)-grade methanol (J. T. Baker, Phillipsberg, NJ) was used in chromatographic studies.
Culture media.
Enrichment of caffeine-degrading bacteria was carried out with M9 mineral salts medium (15) containing 2.5 g·liter−1 caffeine as the sole source of carbon and nitrogen. A pure culture of strain CBB5 was also grown in M9 medium containing 1.0 g·liter−1 of theobromine, theophylline, paraxanthine, 7-methylxanthine, or 3-methylxanthine. Where indicated below, 4.0 g·liter−1 of soytone or YNB was added to M9 medium.
Enrichment, isolation, and identification of caffeine-degrading bacteria.
Strain CBB5 was isolated from soil samples obtained from Coralville, IA, via enrichment in M9-caffeine medium at 29°C with rotary shaking at 200 rpm. Aliquots were periodically removed from the enrichment culture for analysis of caffeine utilization by HPLC. Growth of the enrichment culture was monitored by measuring the optical density at 600 nm (OD600). After three subcultures in M9-caffeine medium, an aliquot of the third subculture was serially diluted and plated on M9-caffeine agar. The plates were incubated at 29°C, and morphologically different colonies were isolated and screened for the ability to degrade caffeine. Subsequently, strain CBB5 was isolated and identified as P. putida by fatty acid methyl ester and 16S rRNA gene sequence analyses (MIDI Inc., Newark, DE).
Degradation assays.
Degradation of caffeine and related methylxanthines by CBB5 was investigated by adding these compounds to M9 medium as growth substrates. A seed culture of CBB5 was first grown in M9-caffeine medium supplemented with soytone. The seed culture was harvested by centrifugation (8,000 × g for 15 min at 4°C) and washed twice with M9 medium containing neither caffeine nor soytone. The cells were then suspended in M9 medium and inoculated into 50 ml soytone-free or soytone-supplemented M9 medium containing a test compound. All cultures were incubated at 29°C with rotary shaking at 200 rpm. Growth on the test compounds was monitored by measuring the OD600 of each culture. Concomitant degradation of the test compounds and accumulation of metabolites in the media (spent media) were monitored by HPLC. Identities of the metabolites were established by comparing the metabolites' retention times (Rts), absorption spectra, and electrospray ionization (ESI) mass spectra with those of authentic standards.
Degradation of caffeine and related methylxanthines by CBB5 was also demonstrated by performing resting cell assays. CBB5 grown in soytone-supplemented M9 medium containing either caffeine or a related methylxanthine was harvested by centrifugation (8,000 × g for 15 min at 4°C) when the OD600 reached 2.0 to 2.4. The cells were washed once with 50 mM potassium phosphate (KPi) buffer (pH 7.5) and suspended in 5 ml KPi buffer at a final OD600 of 4.0. Caffeine and related methylxanthines were added to the cell suspensions at a final concentration of 1 mM. The resting cell suspensions were incubated at 29°C with shaking at 200 rpm. Aliquots were removed from the cell suspensions periodically to monitor the degradation of test compounds and the formation of metabolites by HPLC.
Partial purification of xanthine-oxidizing enzymes.
CBB5 grown in soytone-supplemented M9 medium with 2.5 g·liter−1 theophylline was harvested in late log phase by centrifugation (13,800 × g for 10 min at 4°C). About 3.9 g (wet weight) of cells was suspended in 10 ml 50 mM KPi buffer (pH 7.5) with 10 μg·ml−1 DNase I. The cells were broken by passage through a chilled French press cell twice at 138 MPa. Unbroken cells and cell debris were removed from the lysate by centrifugation (20,400 × g for 20 min at 4°C). The clear supernatant was designated the cell extract. All purification procedures were performed at 4°C using an automated fast protein liquid chromatography system (ÄKTA Purifer; Amersham Pharmacia Biotech, Piscataway, NJ). A 4.0 M ammonium sulfate solution was added to cell extracts to a final concentration of 0.5 M. After 1 h, each mixture was centrifuged at 16,000 × g for 15 min. The supernatant was loaded onto a 120-ml (bed volume) phenyl Sepharose high-performance column (Amersham) preequilibrated with 50 mM KPi buffer (pH 7.5) containing 0.5 M ammonium sulfate. Unbound proteins were washed from the column with 120 ml 50 mM KPi buffer containing 0.5 M ammonium sulfate. Bound proteins were eluted with a 360-ml reverse gradient of ammonium sulfate (0.5 to 0 M in KPi buffer) at a flow rate of 1 ml·min−1. The column was then washed with 120 ml KPi buffer at the same flow rate, which was followed by a final wash with 120 ml deionized water. Fractions containing xanthine-oxidizing enzyme activities were identified by a spectrophotometric enzyme activity assay.
Enzyme activity assays.
To assay xanthine-oxidizing enzyme activity, a 1-ml reaction mixture consisting of 50 mM KPi buffer (pH 7.5), an appropriate amount of partially purified enzyme, 0.5 mM xanthine, and either 0.5 mM nitroblue tetrazolium (NBT) or 0.5 mM NAD+ as the electron acceptor was incubated at 30°C. When NBT was used, enzyme activity was determined by monitoring the increase in absorbance at 566 nm (Δɛ566 = 15,500 M−1·cm−1) due to formazan production with a UV-visible spectrophotometer (Shimadzu UV-2450). When NAD+ was used, enzyme activity was determined by monitoring the increase in absorbance at 340 nm (Δɛ340 = 6,220 M−1·cm−1) due to NADH production. Substrate preference was tested by replacing xanthine with 0.5 mM caffeine, theophylline, theobromine, or 3-methylxanthine.
To assay for theophylline N-demethylase activity in cell extracts prepared from theophylline-grown CBB5 cells, a 2-ml reaction mixture containing 50 mM KPi buffer (pH 7.5), an appropriate amount of cell extract, and 0.5 mM theophylline was incubated at 30°C. Aliquots were removed from the reaction mixture periodically to monitor consumption of theophylline and formation of metabolites by HPLC.
Analytical procedures.
Identification and quantification of caffeine and related methylxanthines and their metabolites were conducted with a Shimadzu LC-10AD HPLC system equipped with a photodiode array detector and a Shimadzu LCMS-2010EV single-stage quadruple mass analyzer. Compounds were separated on a Hypersil BDS C18 column (4.6 by 50 mm). Methanol-water-acetic acid (25:75:0.5, vol/vol/vol) was used as the mobile phase with a flow rate of 0.5 ml·min−1. For analysis of theophylline metabolites, the mobile phase was changed to methanol-water-acetic acid (7.5:92.5:0.5, vol/vol/vol) for better resolution. The metabolites resolved by the C18 column first passed through the photodiode array detector, during which UV-visible absorption spectra were recorded. Then molecules were ionized by ESI in positive-ion mode. The following selected ions corresponding to the M+1 molecular ions of compounds were monitored: caffeine, m/z 195; theobromine, theophylline, and paraxanthine, m/z 181; 1-methylxanthine, 3-methylxathine, and 7-methylxanthine, m/z 167; 1,3-dimethyluric acid, m/z 197; 1-methyluric acid and 3-methyluric acid, m/z 183; uric acid, m/z 169; and xanthine, m/z 153.
RESULTS
Isolation and identification of caffeine-degrading bacteria.
An enrichment culture of soil bacteria completely consumed 2.5 g·liter−1 of caffeine in 8 days when it was incubated in M9-caffeine medium. This culture was further enriched by successive transfers to fresh M9-caffeine medium. After three subcultures, several morphologically different colonies were isolated from M9-caffeine agar. One of these colonies, designated strain CBB5, was identified as P. putida biotype B (similarity index, 0.81) by fatty acid methyl ester analysis. Phylogenetic analysis based on the 16S rRNA gene sequence also indicated that CBB5 was a member of the genus of Pseudomonas, and the closest phylogenetic relative was P. putida strain AJ (GenBank accession no. AY391278; 99% identity). Based on these results, CBB5 was identified as a P. putida strain.
Growth and metabolism of caffeine by CBB5.
CBB5 utilized caffeine as a sole source of carbon and nitrogen. After 72 h of incubation, CBB5 reached stationary phase with an OD600 of ca. 0.4 (see Fig. S1 in the supplemental material). The caffeine concentration also decreased from 2.5 g·liter−1 to 0.2 g·liter−1. Supplementation of M9-caffeine medium with YNB or with soytone significantly increased the growth rate and final cell density of CBB5 (see Fig. S1 in the supplemental material). CBB5 completely utilized 2.5 g·liter−1 caffeine in 53 h when it was grown in YNB-supplemented medium. However, it took only 20 h to consume the same amount of caffeine in soytone-supplemented medium. Although YNB contains higher concentrations of various vitamins than soytone, 18 amino acids are present only in soytone. These differences in composition might account for the increased growth with soytone. A similar effect of soytone was also observed in resting cell suspension experiments. CBB5 resting cell suspensions prepared from cells cultivated in soytone-supplemented medium completely degraded 1 mM caffeine in 45 min, while a cell suspension having a similar density prepared from CBB5 grown in soytone-free medium took 80 min to completely degrade 1 mM caffeine. These results clearly showed that addition of soytone enhanced CBB5 growth without compromising caffeine degradation. Therefore, CBB5 was cultivated in soytone-supplemented M9-caffeine medium except where noted below.
Identification of metabolites resulting from caffeine degradation in growth media.
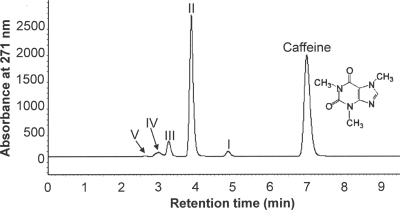
HPLC analyses of spent media removed at different times from CBB5 growing in soytone-supplemented M9-caffeine medium detected several metabolites (Fig. 1). The Rts and absorption spectra of major metabolites II and III (Table 1) were identical to those of theobromine and 7-methylxanthine, respectively. When metabolites II and III were ionized by ESI in positive-ion mode, their M+1 molecular ions had m/z values of 181 and 167, respectively, which were identical to the molecular weights of protonated theobromine and 7-methylxanthine. Likewise, metabolites I, IV, and V were identified as paraxanthine, xanthine, and uric acid based on Rts, absorption spectra, and ESI mass spectra (Table 1). When CBB5 was grown in soytone-free M9-caffeine medium, theobromine was still the major metabolite, followed by 7-methylxanthine and xanthine. These results suggested that CBB5 degraded caffeine via N demethylation to theobromine (major metabolite) and paraxanthine (minor metabolite), similar to previous reports for other caffeine-degrading bacteria (3, 10, 11, 19).
FIG. 1.
HPLC of spent medium of P. putida CBB5 grown in soytone-supplemented M9-caffeine medium. A sample was collected 20 h postinoculation. The identities of the metabolites are as follows: metabolite I, paraxanthine; metabolite II, theobromine; metabolite III, 7-methylxanthine; metabolite IV, xanthine; and metabolite V, uric acid. Physical characteristics of these metabolites are shown in Table 1.
TABLE 1.
UV absorption spectra and ESI mass spectral properties of metabolites formed from the degradation of caffeine, theophylline, and 3-methylxanthine by P. putida CBB5
| Growth substrate | Metabolite | UV λmax (nm) | m/z value of protonated molecular iona | Identity of metabolite |
|---|---|---|---|---|
| Caffeine (UV λmax, 273 nm; m/z 195) | I | 271 | NDb | Paraxanthine |
| II | 271 | 181 | Theobromine | |
| III | 269 | 167 | 7-Methylxanthine | |
| IV | 196, 267 | 153 | Xanthine | |
| V | 240, 284 | 169 | Uric acid | |
| Theophylline (UV λmax, 271 nm; m/z 181) | VI | 231, 287 | 197 | 1,3-Dimethyluric acid |
| VII | 231, 284 | 183 | 1-Methyluric acid | |
| VIII | 234, 287 | 183 | 3-Methyluric acid | |
| IX | 198, 267 | 167 | 1-Methylxanthine | |
| X | 198, 271 | 167 | 3-Methylxanthine | |
| IV | 196, 267 | 153 | Xanthine | |
| V | 240, 284 | 169 | Uric acid | |
| 3-Methylxanthine (UV λmax, 198 and 271 nm; | VIII | 234, 287 | 183 | 3-Methyluric acid |
| m/z 167) | IV | 196, 267 | 153 | Xanthine |
| V | 240, 284 | 169 | Uric acid |
m/z values were determined by ESI mass spectrometry operating in the positive-ion mode.
ND, not detected. No ESI mass spectrum was detected for paraxanthine, possibly because of the low concentration of this metabolite in spent media. Metabolite I was determined to be paraxanthine by comparing its HPLC Rt and UV spectrum with those of an authentic standard.
Catabolism of caffeine and related methylxanthines by CBB5 resting cells.
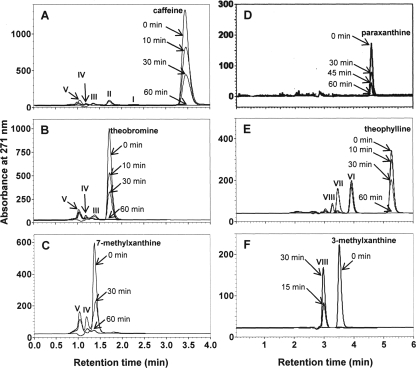
The degradation of caffeine and related methylxanthines by resting cell suspensions of CBB5 grown in soytone-supplemented M9-caffeine medium is shown in Fig. 2. CBB5 resting cell suspensions (OD600, 4) degraded 1 mM caffeine and most related methylxanthines within 60 min without any lag period (Fig. 2A to E). However, 1 mM 3-methylxanthine was completely degraded in 15 min (Fig. 2F). When caffeine was added to a cell suspension, theobromine, 7-methylxanthine, xanthine, and uric acid were detected as metabolites (Fig. 2A), suggesting that caffeine is degraded by the N-demethylation pathway, as was observed using spent media of CBB5 cultures (Fig. 1). When theobromine was added to a cell suspension, only 7-methylxanthine, xanthine, and uric acid were detected as metabolites (Fig. 2B). The same cell suspension at OD600 of 4 and 2 also metabolized 2 mM paraxanthine, but it did not accumulate significant amounts of downstream metabolites (Fig. 2D). However, small amounts of xanthine and uric acid were detected (data not shown). Thus, paraxanthine appears to be metabolized more rapidly than theobromine. Xanthine and uric acid were the only detectable metabolites when 7-methylxanthine was added to a CBB5 resting cell suspension (Fig. 2C). These results indicate that theobromine, paraxanthine, 7-methylxanthine, and xanthine are sequential downstream metabolites of caffeine. This conclusion was cross checked by growing CBB5 in the presence of theobromine, paraxanthine, and 7-methylxanthine with or without soytone. The metabolites formed in spent media and by resting cell suspensions further confirmed the N-demethylation sequence described above.
FIG. 2.
HPLC analyses of metabolites produced by P. putida CBB5 resting cell suspensions. CBB5 was cultivated in soytone-supplemented M9-caffeine medium. The cells were washed and resuspended at an OD600 of 4.0. The following compounds (1 mM) were then added to cell suspensions: (A) caffeine, (B) theobromine, (C) 7-methylxanthine, (D) paraxanthine, (E) theophylline, and (F) 3-methylxanthine. The identities of the metabolites produced are as follows: metabolite I, paraxanthine; metabolite II, theobromine; metabolite III, 7-methylxanthine; metabolite IV, xanthine; metabolite V, uric acid; metabolite VI, 1,3-dimethyluric acid; metabolite VII, 1-methyluric acid; and metabolite VIII, 3-methyluric acid. Physical characteristics of these metabolites are shown in Table 1.
Catabolism of theophylline by caffeine-grown CBB5.
When theophylline was added to a CBB5 resting cell suspension prepared from cells grown on soytone-supplemented M9-caffeine medium, theophylline was metabolized immediately without any lag period (Fig. 2E). Three new metabolites (metabolites VI, VII, and VIII) accumulated in the cell suspension. The Rts and absorption spectra of these metabolites (Table 1) were similar to those of uric acid, with a λmax at ca. 287 nm, suggesting that they could be dimethyluric acid or monomethyluric acid. When preparations were subjected to HPLC-ESI mass spectrometry analyses, the Rt and the M+1 molecular ion of metabolite VI (m/z 197) were identical to those of authentic 1,3-dimethyluric acid. Metabolites VII and VIII had M+1 molecular ions with m/z values of 183, suggesting that they are protonated monomethyluric acid. Metabolite VII was further confirmed to be 1-methyluric acid based on a comparison of its Rt to that of an authentic standard. Unfortunately, an authentic standard is not available commercially for confirmation that metabolite VIII is 3-methyluric acid. Only metabolite VIII was detected when 1 mM 3-methylxanthine was added to a CBB5 resting cell suspension prepared from cells grown on caffeine; 3-methylxanthine was completely degraded within 15 min (Fig. 2F). This further supports the conclusion that metabolite VIII is 3-methyluric acid.
These data suggest that caffeine-grown CBB5 can oxidize theophylline to 1,3-dimethyl-, 1-, and 3-methyluric acids and 3-methylxanthine to 3-methyluric acid. However, these methyluric acids were not metabolized further by resting cell suspensions (Fig. 2E and F). Crude cell extracts prepared from caffeine-grown CBB5 also did not metabolize these methyluric acids (data not shown). Since the expected theophylline metabolites (i.e., 1- and/or 3-methylxanthines) were not detected, degradation of theophylline was reexamined in soytone-free medium.
Growth and metabolism of theophylline when CBB5 is grown on theophylline.
CBB5 was grown in soytone-free M9 medium with 1.0 g·liter−1 and 2.5 g·liter−1 theophylline. After 72 h of incubation, the culture grown with 1.0 g·liter−1 theophylline reached stationary phase with an OD600 of 0.56 (see Fig. S1 in the supplemental material) and had consumed about 60% of the theophylline. The growth of CBB5 was almost completely inhibited by 2.5 g·liter−1 theophylline, unless M9 medium was supplemented with soytone. When grown in soytone-supplemented medium, CBB5 completely utilized 2.5 g·liter−1 theophylline in 30 h, and the final OD600 was 2.5 (see Fig. S1 in the supplemental material).
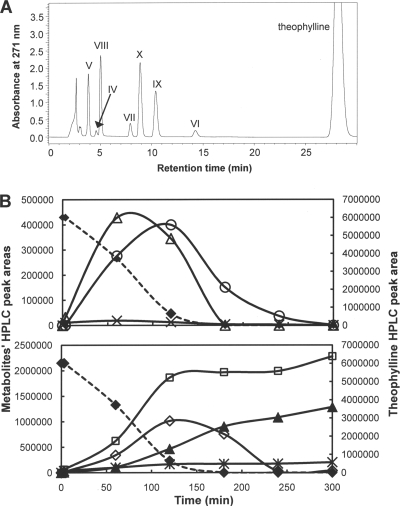
A resting cell suspension (OD600, 4) prepared from CBB5 cells grown on soytone-free M9-theophylline medium degraded 1 mM theophylline in ca. 2 h (Fig. 3B). Seven metabolites were detected by HPLC (Fig. 3A). The absorption spectra, Rts, and M+1 molecular ions of five of the seven metabolites were identical to those of xanthine (metabolite IV), uric acid (metabolite V), 1,3-dimethyluric acid (metabolite VI), 1-methyluric acid (metabolite VII), and very likely 3-methyluric acid (metabolite VIII [an authentic standard was not available for confirmation]). The remaining two metabolites (metabolites IX and X) were not detected previously in theophylline-metabolizing resting cells prepared from CBB5 grown on caffeine plus soytone (Fig. 2E). The physical properties of metabolites IX and X (Table 1) were identical to those of 1-methylxanthine and 3-methylxanthine, respectively, indicating that theophylline was also N demethylated by CBB5. More importantly, 1-methylxanthine, 3-methylxanthine, xanthine, and uric acid did not accumulate in the resting cell suspension and were completely utilized by CBB5 cells within 5 h (Fig. 3B). Meanwhile, 1,3-dimethyluric acid and 1- and 3-methyluric acids accumulated in the cell suspension, as observed for metabolism of theophylline by resting cell suspensions prepared from CBB5 grown on soytone-supplemented M9-caffeine medium (Fig. 2E). The methyluric acids formed from theophylline were estimated to account for 20 to 25% of the total metabolite pool, based on HPLC analysis with standards.
FIG. 3.
(A) HPLC analyses of metabolites produced from theophylline by a resting cell suspension of P. putida CBB5 grown in soytone-free M9-theophylline medium. The identities of metabolites are shown in Table 1. (B) Degradation of theophylline and various metabolites by a resting cell suspension of P. putida CBB5 grown in soytone-free M9-theophylline medium. Symbols: ⧫, theophylline; ○, 1-methylxanthine; ▵, 3-methylxanthine; ×, xanthine; *, 1,3-dimethyluric acid; ▴, 1-dimethyluric acid; □, 3-dimethyluric acid; ⋄, uric acid.
When 3-methylxanthine was added to the resting cell suspension prepared from CBB5 cells grown on soytone-free M9-theophylline medium, this compound was consumed completely in 3 h. Xanthine (metabolite IV), uric acid (metabolite V), and 3-methyluric acid (metabolite VIII) were the only detectable metabolites (data not shown). Xanthine and uric acid accumulated transiently and were completely degraded within 4 h. 3-Methyluric acid (metabolite VIII) accumulated in the medium and was not degraded further, which is consistent with the results for caffeine- or theophylline-grown CBB5 with or without the soytone supplement (Fig. 2F and Fig. 3B). When xanthine was added to the same resting cell suspension, it was completely consumed in 1 h. Uric acid was the only metabolite detected, and it was promptly degraded (data not shown). These data suggest that 3-methylxanthine, xanthine, and uric acid are sequential downstream metabolites of theophylline.
When theophylline was added to resting cell suspensions (OD600, 4) prepared from CBB5 cells grown on soytone-supplemented M9-theophylline medium, 1- and 3-methylxanthines were not detected. Instead, the di- and monomethyluric acids accumulated in the medium, consistent with the results for CBB5 resting cells grown on soytone-supplemented caffeine medium (Fig. 2E). However, small amounts of 1- and 3-methylxanthine were transiently detected if the OD600 of the resting cell suspension was reduced to 2 (data not shown), suggesting that CBB5 cells grown on soytone-supplemented media also metabolized theophylline via N demethylation. This suggestion is supported by the fact that theophylline N-demethylase activity was detected in crude extracts of CBB5 cells grown on soytone-supplemented media. Both 1- and 3-methylxanthines were detected when theophylline was used as the substrate (Fig. 4). These results suggest that addition of soytone did not alter theophylline metabolism via N-demethylase activity in CBB5 cells. Moreover, addition of NAD(P)H to the reaction mixture enhanced the production of 1- and 3-methylxanthines from theophylline (Fig. 4), indicating that theophylline N-demethylase is likely an NAD(P)H-dependent enzyme.
FIG. 4.
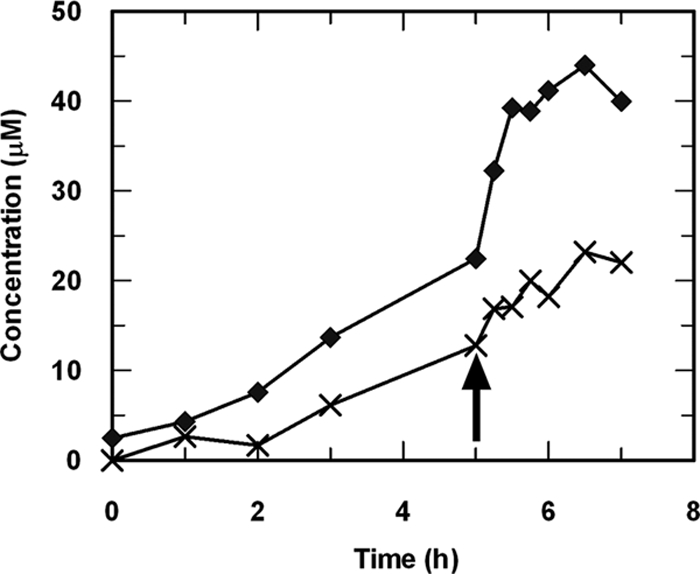
Production of 1-methylxanthine (⧫) and 3-methylxanthine (×) from theophylline by cell extracts prepared from P. putida CBB5 grown in soytone-supplemented M9-theophylline medium. After 5 h, 0.15 mM NADH was added to the enzyme reaction mixture (indicated by an arrow). Addition of NADPH at the same time also enhanced 1- and 3-methyxanthine production (data not shown).
Collectively, these data suggest that theophylline degradation occurred via initial N demethylation to 1-methylxanthine and 3-methylxanthine. Both of the latter compounds were further N demethylated to xanthine, which was subsequently oxidized to uric acid. 1,3-Dimethyluric and 1- and 3-methyluric acids formed from theophylline were not metabolized further by either resting cell suspensions or cell extracts prepared from CBB5 grown on theophylline or caffeine.
Coexpression of theophylline and caffeine metabolism in CBB5.
Resting cell suspensions prepared from CBB5 grown in soytone-supplemented M9-theophylline medium N demethylated caffeine, theobromine, paraxanthine, and 7-methylxanthine without any lag period (see Fig. S2 in the supplemental material). Similarly, resting cell suspensions prepared from CBB5 grown on caffeine or its N-demethylated metabolites degraded theophylline without a noticeable lag (Fig. 2E; see Fig. S2 in the supplemental material). Thus, the enzymes involved in both the caffeine and theophylline N-demethylation pathways are coexpressed in CBB5 cells grown on either compound or the N-demethylated metabolites obtained from caffeine. The number of N-demethylases involved and their specificities with respect to caffeine and theophylline metabolism remain to be determined.
Growth and metabolism of 3-methylxanthine by CBB5.
Since resting cell suspension prepared from theophylline-grown CBB5 could metabolize 3-methylxanthine (Fig. 3), we tested the ability of CBB5 to utilize this compound as a sole carbon and nitrogen source. When grown in soytone-free and soytone-supplemented M9 media containing 1.0 g·liter−1 3-methylxanthine, the OD600 of CBB5 cultures reached 0.14 (after 72 h) and 2.5 (after 30 h), respectively. 3-Methyluric acid (metabolite VIII) was detected under both conditions and was not metabolized further. Resting cell suspensions prepared from CBB5 grown on 3-methylxanthine degraded this compound at rates comparable to the rates observed for resting cell suspensions prepared from CBB5 grown on caffeine, theophylline, theobromine, paraxanthine, and 7-methylxanthine (see Fig. S2 in the supplemental material). Interestingly, a resting cell suspension prepared from 3-methylxanthine-grown CBB5 also degraded theophylline without any lag period, but at a rate significantly lower than that of a resting cell suspension prepared from theophylline- or caffeine-grown CBB5 (see Fig. S2 in the supplemental material). However, only methyluric acids were produced from theophylline. Caffeine, theobromine, and 7-methylxanthine were not degraded by 3-methylxanthine-grown resting cells incubated for 60 min, while paraxanthine was degraded minimally. A resting cell suspension prepared from CBB5 cells grown in soytone-supplemented M9 medium did not degrade any methylxanthines except theophylline and 3-methylxanthine, which were degraded at significantly lower rates (see Fig. S2 in the supplemental material).
Origin of 1,3-dimethyluric acid and 1- and 3-methyluric acids from theophylline.
Yamaoka-Yano and Mazzafera (19) previously reported that caffeine-degrading P. putida L possessed a xanthine oxidase which also oxidized theophylline and 3-methylxanthine. In the present study we observed oxidation of theophylline and 1- and 3-methylxanthines at the C-8 position (Fig. 2E and 3). Whether the formation of these compounds was due to a xanthine oxidase with a broad substrate range, similar to the enzyme described by Yamaoka-Yano and Mazzafera, was investigated further. Cell extracts prepared from theophylline-grown CBB5 cells were fractionated on a phenyl Sepharose high-performance column. Enzyme activity assays detected two peaks of xanthine-oxidizing fractions, A and B, which were eluted from the column with 0.25 M and 0.18 M ammonium sulfate, respectively. Fraction A oxidized xanthine to uric acid, which was identified by HPLC, in the presence of NAD+ (data not shown). Fraction A was a highly specific xanthine dehydrogenase. It exhibited no activity with caffeine, theobromine, theophylline, and 3-methyxantine. In contrast, fraction B oxidized xanthine to uric acid with concomitant reduction of NBT. Ferricyanide, but not NAD+, could replace NBT as the electron acceptor. Fraction B was also active with theophylline and 3-methylxanthine, producing 1,3-dimethyluric acid and 3-methyluric acid, respectively. Caffeine and theobromine were not substrates for fraction B. For this enzyme fraction, the highest level of activity was observed with 3-methylxanthine (100%); the levels of activity with xanthine and theophylline were 83% and 13%, respectively. Thus, the production of 1,3-dimethyluric acid and 3- and 1-methyluric acids from theophylline and 3- and 1-methylxanthines, respectively (Fig. 2E and 3), is probably due to the activity of the broad-substrate-range xanthine-oxidizing enzyme in fraction B.
DISCUSSION
We isolated a unique caffeine-degrading bacterium, P. putida CBB5, which uses not only caffeine, theobromine, and 7-methylxanthine as sole carbon and nitrogen sources but also theophylline and 3-methylxanthine. While caffeine metabolism by bacteria has been studied to some extent, there is very little information on theophylline and 3-methylxanthine metabolism by bacteria. Theophylline has not been proposed to be an intermediate in the caffeine degradation pathways of bacteria (3, 10, 11, 19, 20). Caffeine-degrading Serratia marcescens was not able to utilize theophylline or 3-methylxanthine as a growth substrate (11). Woolfolk (18) reported that Pseudomonas isolates capable of growth on 0.1% caffeine also grew well on all dimethylxanthines, including theophylline. However, details of theophylline metabolism in these Pseudomonas isolates were not elucidated. In contrast, caffeine catabolism in plant species and in caffeine-degrading fungi has been reported to proceed via theophylline, which is further degraded by N demethylation to 3-methylxanthine (7, 10, 16).
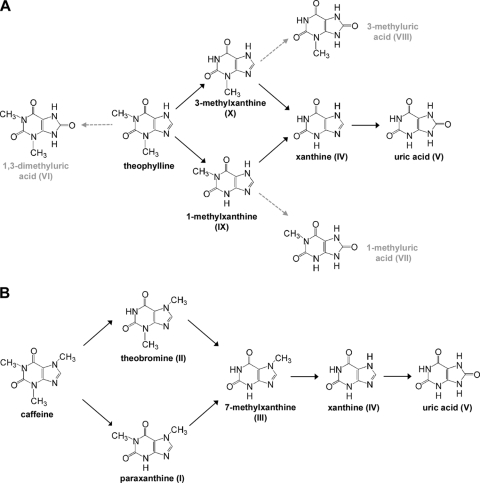
CBB5 metabolized theophylline by a previously unknown N-demethylation pathway, forming 1-methylxanthine and 3-methylxanthine (Fig. 3). These two intermediates were further N demethylated to xanthine, which was finally oxidized to uric acid. Metabolism of theophylline by this N-demethylation pathway was confirmed using CBB5 culture media, resting cell suspensions, and cell extracts. CBB5 also oxidized theophylline to 1,3-dimethyluric acid and 1- and 3-methyluric acids (Fig. 2E and 3). However, these methyluric acids were not metabolized further by resting cells or crude cell extracts (Fig. 3B). A broad-substrate-range xanthine-oxidizing enzyme that was partially purified from CBB5 appears to be involved in the oxidation of theophylline to the methyluric acids. This hypothesis is supported by the fact that 3-methylxanthine is the best substrate for this enzyme and by the fact that 3-methyluric acid was also produced from 3-methylxanthine (Fig. 2F). Based on these results, a new N-demethylation pathway is proposed for theophylline degradation in CBB5 (Fig. 5A). In this pathway, formation of methyluric acids is indicated by dead-end metabolites of theophylline and 1- and 3-methylxanthines.
FIG. 5.
Proposed N-demethylation pathways for degradation of (A) theophylline and (B) caffeine by P. putida CBB5. The dashed arrows indicate fortuitous oxidation of theophylline and 1- and 3-methylxanthines to 1,3-dimethyluric acid and 1- and 3-methyluric acids, respectively, which were not metabolized further.
The formation of methyluric acids from theophylline and 1- and 3-methylxanthines is not unusual. A xanthine oxidase that was active with theophylline and 3-methylxanthine was identified in caffeine-degrading P. putida L cell extracts by activity staining of native polyacrylamide gel electrophoresis gels (19). This enzyme was subsequently purified, but theophylline and 3-methylxanthine products were not reported. An NAD+-dependent xanthine dehydrogenase activity was also identified in cell extracts of caffeine-degrading S. marcescens using a similar activity staining procedure (11). This xanthine dehydrogenase was also active with theophylline, 3-methylxanthine, and 1-methylxanthine. Apart from the reports described above, it appears that most xanthine-oxidizing enzymes exhibit high specificity with xanthine and monomethylxanthines. Dikstein et al. (5) studied xanthine oxidase activities in 10 different species of human intestinal bacteria. All of the enzymes were inactive with theophylline, while seven were active with 3-methylxanthine. At present, the physiological role of the broad-substrate-range xanthine-oxidizing enzyme in CBB5 is unclear since CBB5 could not metabolize methyluric acids. S. marcescens, which could oxidize theophylline, 3-methylxathine, and 1-methylxanthine (11), was also not capable of utilizing methyluric acids as growth substrates.
CBB5 also metabolized caffeine via successive N demethylation. Theobromine was the major product of the initial N demethylation, although paraxanthine was also detected as a minor metabolite. These two dimethylxanthines were further N demethylated to 7-methylxanthine and xanthine before oxidation to uric acid (Fig. 1). Based on these data, a sequential N-demethylation pathway is proposed here for caffeine (Fig. 5B). This pathway is further supported by the fact that CBB5 metabolized theobromine, paraxanthine, and 7-methylxanthine with the same pathway (Fig. 2). The pathway proposed for caffeine N demethylation in CBB5 is similar, but not identical, to that in caffeine-degrading P. putida L (19). In the latter organism, dimethylxanthines and methylxanthines produced from caffeine were also shown to be oxidized to the corresponding methyluric acids. Xanthine oxidase with broad substrate specificity was reported to be responsible for oxidizing these compounds. In our study, methyluric acids were not detected when caffeine, theobromine, paraxanthine, and 7-methylxanthine were used as growth substrates for CBB5 or in resting cell studies.
Multiple N-demethylases might be involved in N demethylation of caffeine and related methylxanthines. For example, during purification of a theobromine N-demethylase from caffeine-degrading P. putida No. 352 that was inhibited by Zn2+, Asano et al. (1) also detected caffeine N-demethylase activity in the cell extracts which was not inhibited by Zn2+. Glück and Lingens (6) partially purified a 7-methylxanthine N-demethylase from caffeine-degrading P. putida WS. This enzyme was specific for 7-methylxanthine and had no activity with caffeine and theobromine. Caffeine and theobromine also did not inhibit 7-methylxanthine N demethylation by this enzyme. A recent study by Dash and Gummadi (2) showed that caffeine and theobromine N-demethylases in Pseudomonas sp. strain NCIM5235 were inducible in nature, but the caffeine N-demethylase activity in theobromine-grown cells was 10-fold lower than that in caffeine-grown cells. These data implied that different N-demethylases were responsible for caffeine and theobromine N demethylation. Previous attempts to purify caffeine N-demethylase were unsuccessful because of the instability of the enzyme. However, NAD(P)H was required as a cosubstrate for caffeine N-demethylase (3, 10). The only known bacterial N-demethylase in the UniProt protein database (17) is N-methylproline demethylase (StcD) of Sinorhizobium meliloti, which is involved in stachydrine catabolism (14). The biochemical properties of StcD were not studied in detail, but its protein sequence is similar to those of α/β-barrel oxidoreductase flavoproteins. A caffeine N-demethylase nucleotide sequence (accession no. E07469) from P. putida strain IF-3 was also found in the GenBank database. The deduced protein sequence showed a high degree of similarity with Rieske (2Fe-2S) domain-containing oxygenase subunits of many putative vanillate O-demethylases, putative phenylpropionate dioxygenases, and HpxD of Klebsiella pneumoniae, which is involved in the oxidation of hypoxanthine to uric acid (4). We are currently purifying different N-demethylases from CBB5 for detailed biochemical characterization, including characterization of substrate specificity.
The caffeine and theophylline N-demethylation pathways proposed in Fig. 5 were coexpressed in CBB5 when caffeine, N-demethylated metabolites of caffeine, or theophylline was used as the growth substrate. However, CBB5 grown in 3-methylxanthine did not metabolize caffeine, theobromine, or 7-methylxanthine. Furthermore, there also was much less degradation of theophylline, with production of only methyluric acids (see Fig. S2 in the supplemental material). There are precedents for differential regulation of caffeine degradation by an alternative carbon or nitrogen source. For instance, when caffeine-degrading Pseudomonas sp. strain NCIM5235 was grown with theobromine as the sole source of carbon and nitrogen, the caffeine N-demethylase and theobromine N-demethylase activities in cell extracts were 10-fold lower and 3- to 4-fold higher, respectively, than those in cell extracts from caffeine-grown cells (2). Caffeine dehydrogenase activity in Pseudomonas sp. strain CBB1 was repressed when this organism was grown in the presence of soytone plus caffeine. However, addition of YNB did not suppress the activity of this enzyme (20). Expression of caffeine oxidase in Alcaligenes sp. strain CF8 was completely suppressed by starch, but glucose increased the caffeine oxidase expression twofold. Sodium nitrate also stimulated caffeine oxidase expression in Alcaligenes sp. strain CF8, while ammonium sulfate and ammonium chloride substantially reduced production of this enzyme (12).
In summary, CBB5 metabolized theophylline by a previously unknown N-demethylation pathway to 1- and 3-methylxanthines. The monomethylxanthines were further N demethylated to xanthine. In contrast, CBB5 N demethylated caffeine to theobromine (major metabolite) and paraxanthine (minor metabolite). These two dimethylxanthines were subsequently N demethylated to 7-methylxanthine and xanthine. The theophylline and caffeine N-demethylation pathways converge at xanthine, which is eventually oxidized to uric acid. The two pathways are coexpressed in the presence of caffeine, theophylline, and related methylxanthines. Curiously, 1,3-dimethyluric acid and 1- and 3-methyluric acids were also produced during theophylline metabolism, but they were not degraded further. A broad-substrate-range xanthine-oxidizing enzyme was responsible for the formation of these methyluric acids.
Supplementary Material
Acknowledgments
This research was supported by University of Iowa research funds.
We thank Kevin Erpelding for his assistance during protein purification and Shuvendu Das for his help with liquid chromatography-mass spectrometry experiments.
Footnotes
Published ahead of print on 15 May 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Asano, Y., T. Komeda, and H. Yamada. 1994. Enzymes involved in theobromine production from caffeine by Pseudomonas putida No. 352. Biosci. Biotechnol. Biochem. 582303-2304. [Google Scholar]
- 2.Dash, S. S., and S. N. Gummadi. 2008. Inducible nature of the enzymes involved in catabolism of caffeine and related methylxanthines. J. Basic Microbiol. 48227-233. [DOI] [PubMed] [Google Scholar]
- 3.Dash, S. S., and S. N. Gummadi. 2006. Catabolic pathways and biotechnological applications of microbial caffeine degradation. Biotechnol. Lett. 281993-2002. [DOI] [PubMed] [Google Scholar]
- 4.de la Riva, L., J. Badia, J. Aguilar, R. A. Bender, and L. Baldoma. 2008. The hpx genetic system for hypoxathine assimilation as a nitrogen source in Klebsiella pneumonia: gene organization and transcriptional regulation. J. Bacteriol. 1907892-7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dikstein, S., F. Bergmann, and Y. Henis. 1957. Studies on uric acid and related compounds. IV. The specificity of bacterial xanthine oxidases. J. Biol. Chem. 22467-77. [PubMed] [Google Scholar]
- 6.Glück, M., and F. Lingens. 1988. Heteroxanthinedemethylase, a new enzyme in the degradation of caffeine by Pseudomonas putida. Appl. Microbiol. Biotechnol. 2859-62. [Google Scholar]
- 7.Hakil, M., S. Denis, G. Viniegra-González, and C. Augur. 1998. Degradation and product analysis of caffeine and related dimethylxanthines by filamentous fungi. Enzyme Microb. Technol. 22355-359. [Google Scholar]
- 8.Madyastha, K. M., and G. R. Sridhar. 1998. A novel pathway for the metabolism of caffeine by a mixed culture consortium. Biochem. Biophys. Res. Commun. 249178-181. [DOI] [PubMed] [Google Scholar]
- 9.Madyastha, K. M., G. R. Sridhar, B. B. Vadiraja, and Y. S. Madhavi. 1999. Purification and partial characterization of caffeine oxidase—a novel enzyme from a mixed culture consortium. Biochem. Biophys. Res. Commun. 263460-464. [DOI] [PubMed] [Google Scholar]
- 10.Mazzafera, P. 2004. Catabolism of caffeine in plants and microorganisms. Front. Biosci. 91348-1359. [DOI] [PubMed] [Google Scholar]
- 11.Mazzafera, P., O. Olsson, and G. Sandberg. 1996. Degradation of caffeine and related methylxanthines by Serratia marcescens isolated from soil under coffee cultivation. Microb. Ecol. 31199-207. [DOI] [PubMed] [Google Scholar]
- 12.Mohapatra, B. R., N. Harris, R. Nordin, and A. Mazumder. 2006. Purification and characterization of a novel caffeine oxidase from Alcaligenes species. J. Biotechnol. 125319-327. [DOI] [PubMed] [Google Scholar]
- 13.Ogunseitan, O. A. 2002. Caffeine-inducible enzyme activity in Pseudomonas putida ATCC 700097. World J. Microbiol. Biotechnol. 18423-428. [Google Scholar]
- 14.Phillips, D. A., E. S. Sande, J. A. C. Vriezen, F. J. de Bruijn, D. L. Rudulier, and C. M. Joseph. 1998. A new genetic locus in Sinorhizobium meliloti is involved in stachydrine utilization. Appl. Environ. Microbiol. 643954-3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor laboratory Press, Cold Spring Harbor, NY.
- 16.Schwimmer, S., R. H. Kurtzman, Jr., and E. Heftmann. 1971. Caffeine metabolism by Penecillium roqueforti. Arch. Biochem. Biophys. 147109-113. [DOI] [PubMed] [Google Scholar]
- 17.The UniProt Consortium. 2008. The Universal Protein Resource (UniProt). Nucleic Acids Res. 36D190-D195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woolfolk, C. A. 1975. Metabolism of N-methylpurines by a Pseudomonas putida strain isolated by enrichment on caffeine as the sole source of carbon and nitrogen. J. Bacteriol. 1231088-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamaoka-Yano, D. M., and P. Mazzafera. 1999. Catabolism of caffeine and purification of a xanthine oxidase responsible for methyluric acids production in Pseudomonas putida L. Rev. Microbiol. 3062-70. [Google Scholar]
- 20.Yu, C. L., Y. Kale, S. Gopishetty, T. M. Louie, and M. Subramanian. 2008. A novel caffeine dehydrogenase in Pseudomonas sp. strain CBB1 oxidizes caffeine to trimethyluric acid. J. Bacteriol. 190772-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.