Abstract
In Bacillus subtilis a null mutation of the relA gene, whose gene product is involved in the synthesis and/or hydrolysis of (p)ppGpp, causes a growth defect that can be suppressed by mutation(s) of yjbM and/or ywaC coding for small (p)ppGpp synthetases. All 35 suppressor mutations newly isolated were classified into two groups, either yjbM or ywaC, by mapping and sequencing their mutations, suggesting that there are no (p)ppGpp synthetases other than RelA, YjbM, and YwaC in B. subtilis. In order to understand better the relation between RelA and rRNA synthesis, we studied in the relA mutant the transcriptional regulation of seven rRNA operons (rrnO, -A, -J, -I, -E, -D, or -B) individually after integration of a promoter- and terminatorless cat gene. We identified the transcriptional start sites of each rrn operon (a G) and found that transcription of all rrn operons from their P1 promoters was drastically reduced in the relA mutant while this was almost completely restored in the relA yjbM ywaC triple mutant. Taken together with previous results showing that the intracellular GTP concentration was reduced in the relA mutant while it was restored in the triple mutant, it seems likely that continuous (p)ppGpp synthesis by YjbM and/or YwaC at a basal level causes a decrease in the amounts of intracellular GTP.
Guanosine 5′-diphosphate 3′-diphosphate (ppGpp) and guanosine 5′-triphosphate 3′-diphosphate (pppGpp), generally referred to as (p)ppGpp, are produced in cells of many bacteria and plants when they encounter adverse environmental conditions such as amino acid starvation (4, 5). As a global regulator, (p)ppGpp is known to control several cellular processes including transcription, translation, nucleotide metabolism, and DNA replication (1, 12, 22, 36). In Escherichia coli two homologous enzymes, RelA and SpoT, are involved in the regulation of intracellular (p)ppGpp levels. RelA is a ribosome-associated (p)ppGpp synthetase responding mainly to uncharged tRNAs that accumulate as a result of amino acid limitation (4). SpoT is a bifunctional (p)ppGpp synthetase and hydrolase and regulates (p)ppGpp levels in response to limitation of carbon source, fatty acid, or iron (3, 32, 35, 40). In contrast to E. coli, many other bacteria were assumed to possess only one gene encoding a RelA-SpoT homolog, Rel or RelA, considered to be a bifunctional (p)ppGpp synthetase and hydrolase (23, 38). We have recently found that two small RelA homologues, YjbM and YwaC, are capable of synthesizing (p)ppGpp in Bacillus subtilis, one of the best-characterized gram-positive bacteria (26). The putative homologues of RelA, YjbM and YwaC, are found in Streptococcus mutans and many gram-positive bacteria (20, 26), suggesting that intracellular (p)ppGpp levels in these bacteria are controlled by these three enzymes although the detailed regulatory mechanisms remain unclear.
During the course of characterizing a relA null mutant of B. subtilis, we found that this mutant strain grew more slowly than wild-type cells in LB medium (26). This growth defect could be suppressed by introduction of the yjbM and/or ywaC null mutation(s) or by expression of relA(D264G), encoding a RelA protein with a D264G mutation that abolishes (p)ppGpp synthetase activity (26). As this mutant RelA protein has normal hydrolase activity (26), these results suggest that the slow-growth characteristics of the relA null mutant could result from a slightly enhanced basal level of (p)ppGpp, which, however, was below the level of detection by our high-performance liquid chromatography system (26). (p)ppGpp binds directly to RNA polymerase and thereby inhibits the transcription of rRNA (rrn) operons, resulting in growth arrest (1, 2, 5, 6, 9, 29). Furthermore, it has been shown that increased levels of ppGpp caused by the relA mutation likely lead to a decrease in GTP pools, which inhibits rRNA operon promoter activity due to the reduced availability of initiating GTP (17, 30). Therefore, we studied the regulation of transcription for each individual rRNA operon in the relA mutant. Seven novel strains were constructed, each carrying a promoter- and terminatorless cat gene within either of the rRNA operons rrnO, -A, -J, -I, -E, -D, or -B to monitor their transcription activity. Using these strains, we experimentally determined all transcription start sites from promoters of seven individual rRNA operons and assessed the effects of the relA gene disruption and its suppressor mutations on the transcription activity of these rrn operons in B. subtilis.
MATERIALS AND METHODS
Strain construction.
All B. subtilis strains used in this study were isogenic with B. subtilis strain 168 and are listed in Table S1 in the supplemental material. Strain RIK350 (trpC2 rrnA2+::catpt1) in which catpt1, a cat gene lacking any promoter or Rho-independent terminator sequence (27), is fused downstream of the rrnA P2 promoter to monitor transcription activity by primer extension analysis, was constructed as follows. Oligonucleotide primers (see Table S2 in the supplemental material) were used to amplify the upstream (primers rrnA-catF1 and rrnA-catR1) and downstream (primers rrnA-catF2 and rrnX-catR2) region of the rrnA promoter and 16S rRNA, respectively. Next, the chloramphenicol resistance gene of pCBB31 (13) was amplified by PCR using primers CAT-F2 and CAT-R. The three fragments obtained were used simultaneously as the template for PCR amplification with primers rrnA-catF1 and rrnX-catR2. The resulting fragment was transformed into B. subtilis 168, and chloramphenicol-resistant transformants were selected on LB plates. Proper integration was confirmed by PCR and DNA sequencing. Strains with catpt1 fused to the promoter region of other rrn operons, RIK351 to RIK356 were constructed analogously with the primers listed in Table S2 in the supplemental material. Primer rrnX-catR2 could be used for the generation of each integration cassette due to conservation of the 16S rRNA genes it anneals to.
Disruption of the relA gene in strain RIK350 to RIK356 was achieved by transformation of chromosomal DNA extracted from strain RIK900 (trpC2 relA::erm) (26), followed by selection of erythromycin-resistant transformants, yielding strains RIK901 to RIK907. In a similar manner, strains RIK908 (trpC2 ywaC::spc) (26) and RIK1000 (trpC2 ΔyjbM) (26) with a catpt1-tagged rrn operon (conferring chloramphenicol resistance) yielded RIK1023 to RIK1029 and RIK1030 to RIK1036, respectively, by subsequent disruption of the relA locus by transformation of RIK900 chromosomal DNA. Triple deletion mutants RIK1044 to RIK1050 with rrn operons containing catpt1 were constructed analogously from RIK1002 (trpC2 ΔyjbM ywaC::spc) (26). In all cases, proper integration was verified by PCR and DNA sequencing.
Medium.
B. subtilis strains were grown in LB medium or on LB agar (31). When required, antibiotics were added at the following concentrations: chloramphenicol, 5 μg ml−1; erythromycin, 0.5 μg ml−1; and spectinomycin, 100 μg ml−1.
Sucrose density gradient sedimentation analysis of ribosomes.
Cells grown in LB medium to an early exponential phase (optical density at 600 nm [OD600] of 0.2) at 37°C with shaking were collected and then disrupted by passage through a French pressure cell (Aminco) at 8,000 lb/in2, after which cell debris was removed by centrifugation as previously described (27). Supernatants were used as crude cell extracts. Aliquots of extract equivalent to 3.09 OD600 units of the culture were layered onto 10 to 40% sucrose density gradients and centrifuged at 4°C for 17.5 h at 65,000 × g (Hitachi P40ST rotor). Absorbance profiles were monitored at 254 nm using a Piston Gradient Fractionator (Bio ComP) and Bio-mini UV Monitor (ATTO Japan).
Primer extension analysis.
RNA was extracted from cells grown to an early exponential phase (OD600 of 0.2) in LB medium at 37°C with shaking. Thirty micrograms of total RNA and 1 pmol of infrared dye (IRD)-labeled oligonucleotide (rrn-in-cat2) complementary to the 5′-terminal region of the cat gene were mixed, and reverse transcription reactions were carried out using SuperScript II reverse transcriptase (Invitrogen) as previously described (24, 25, 27). The products were run on 5% polyacrylamide-6 M urea gels alongside a sequencing ladder generated by PCR cycle sequencing with rrn-in-cat2, which facilitated mapping of 3′ ends of the reverse transcripts (corresponding to the 5′ end of the RNAs) IRD-labeled reverse transcription and sequencing products were detected by a Li-Cor DNA analyzer, models 4200 and 4300 (Aloka). Quantification of each reverse transcript was performed by Scion image software (Scion Corporation).
RESULTS
Isolation and identification of two types of suppressor mutations from the relA null mutant.
The effects of relA mutation on cell physiology have been predominantly studied in B. subtilis using a relA gene containing a point mutation (14, 21) whose gene product appears to exhibit some biological activity, especially (p)ppGpp hydrolase activity, but lacks the synthetase activity. We therefore constructed a B. subtilis strain in which relA was replaced with an erythromycin resistance gene and found that the resultant relA null mutant grew more slowly than the wild type in LB medium (26). Interestingly, during cultivation of the relA null mutant on LB agar plates, we frequently observed the appearance of two types of larger colonies distinguishable from one another by colony morphology and growth characteristics. We isolated 35 spontaneous suppressor mutants and first examined whether these suppressor mutations were linked to either the yjbM or ywaC gene by transformation with the chromosomal DNAs carrying the cat genes inserted into the sites adjacent to either the yjbM or ywaC gene. Of the 35 suppressor mutants we characterized, 16 had mutations in yjbM, and the other 19 mapped within ywaC (Table 1). A variety of mutations including point mutations and deletion and addition mutations were observed among the suppressor mutations although deletion mutations were found only in yjbM, and addition mutations were found only in ywaC. More interestingly, neither suppressor completely restored the growth defect of the relA null mutant, which is in good agreement with our previous work (26). Furthermore, as these were the only suppressors found, it is most likely that there are no (p)ppGpp synthetases other than RelA, YjbM, and YwaC in B. subtilis although we cannot exclude the possibility that there is another minor (p)ppGpp synthetase in B. subtilis. Srivatsan and coworkers have recently reached a conclusion similar to ours regarding the identification of the suppressor mutations in the relA mutant (33).
TABLE 1.
Location and identities of the mutation in the yjbM and ywaC
| Mutant | Position of mutationa | Amino acid substitution or description of mutation |
|---|---|---|
| yjbM mutants | ||
| yjbM4 | 316G → T | Asp106 → Tyr |
| yjbM8 | 241C → T | Gln81 → stop codon |
| yjbM27, -28 | 415G → T | Glu139 → stop codon |
| Δ(424A—485G) | 62-bp deletion | |
| yjbM31, -33, -34, -35 | Δ(363T—382C) | 20-bp deletion |
| yjbM32 | 368T → A | Val123 → Glu |
| yjbM43 | 319T → A | Tyr107 → Asn |
| yjbM44 | 283G → A | Ala95 → Thr |
| 460G → A | Glu154 → Lys | |
| yjbM46 | 514A → G | Arg172 → Gly |
| yjbM47 | Δ29T | 1-bp deletion |
| yjbM49 | Δ(462A—472A) | 11-bp deletion |
| yjbM50 | 239G → A | Cys80 → Tyr |
| yjbM52 | 29T → A | Leu10 → stop codon |
| ywaC mutants | ||
| ywaC1,- 2, -5, -6, -7, -11, -25 | 631T → A | Stop codon 210 → Lys |
| ywaC3 | 3G → T | Stop codon → Lys |
| ywaC9 | 575G → T | Ala192 → Val |
| ywaC10 | A511-514 → addition of A | Frameshift |
| ywaC12 | Addition of GCGCTCGAT after T90 | 9-bp addition |
| ywaC22 | 197A → T | Lys60 → Ile |
| ywaC23 | 328C → T | Gln110 → stop codon |
| ywaC24 | Addition of CC after A437 | Frameshift |
| ywaC26 | 572C → A | Ala191 → Asp |
| ywaC30 | 527T → A | Leu176 → stop codon |
| ywaC37 | 149A → CC | Frameshift |
| ywaC39 | 268G → C | Gly90 → Arg |
| ywaC40 | 575C → A | Ala192 → Glu |
Numbering from the start codon (ATG) of the open reading frame.
Effects of relA and its suppressor mutations on 70S ribosome formation.
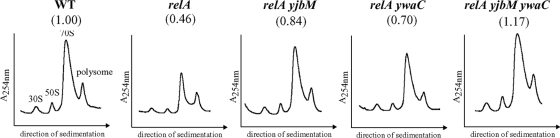
The results described above indicate that the presence of yjbM and ywaC in the relA null mutant background partially reverses inhibitory effects on growth of the relA null mutant. To explore the possibility that, as outlined in the introduction, the growth defect of the relA null mutant could be due to poor transcription from promoters in rrn operons, we first examined the formation of the 70S ribosome in relA null strains carrying a deletion of either or both of the yjbM and ywaC genes by 10% to 40% sucrose density gradient centrifugation and compared the result with that in the wild-type strain (Fig. 1). The amount of 70S ribosome formed in the relA mutant was apparently small compared with that in wild-type cells (Fig. 1). As the peaks corresponding to the 30S, 50S, and 70S particles were found in the profiles of the relA null mutant, it is most likely that the small amount of 70S ribosome in the relA null mutant is not caused by the inhibition of the normal processing pathways for ribosome formation but by the reduction of overall pre-rRNA synthesis, presumably due to the poor transcription of rrn operons. In contrast, the amount of 70S ribosome in the relA null mutant was partially restored by the introduction of the yjbM or ywaC mutation and completely restored in the triple mutant (Fig. 1), suggesting that the presence of yjbM and ywaC in the relA null background has inhibitory effects on growth and on the transcription activity in rrn operons.
FIG. 1.
Effects of the relA null mutation, the relA yjbM and relA ywaC double deletions, and the relA yjbM ywaC triple deletion on 70S ribosome formation. Crude cell extracts were sedimented through a 10 to 40% sucrose gradient as described in Materials and Methods. Peak values for 70S ribosomes in each strain are relative to the maximum value of the 70S peak in wild-type cells, set at 1.0.
RNAs transcribed from all promoters of each rrn operon are initiated with GTP in B. subtilis.
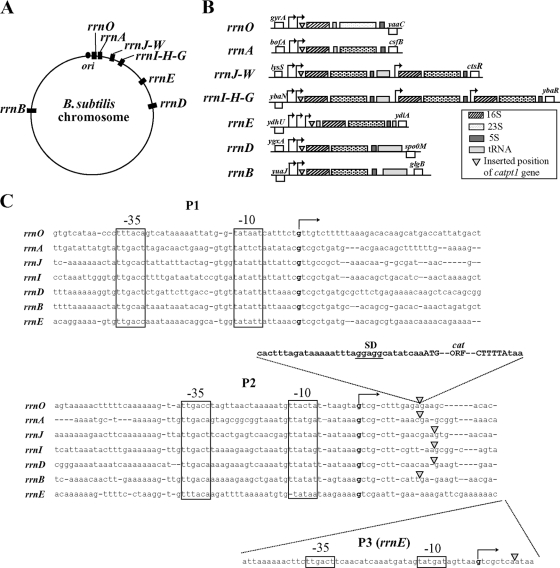
There are 10 rrn operons (rrnO, -A, -J, -W, -I, -H, -G, -E, -D, and -B), including the rrnJ-rrnW and rrnI-rrnH-rrnG clusters, in the B. subtilis genome (11, 19, 39). Two obvious σA consensus promoters (P1 and P2), have been found in the transcriptional regulatory regions of all the rrn operons, with the exception of rrnE, which has three promoters (P1, P2, and P3) (Fig. 2) (16, 17, 19, 28, 34, 37). Clusters of tRNA genes are located downstream of the 5S rRNA genes in rrnJ, -I, -E, -D, and -B; upstream of the 16S rRNA gene in rrnE; and in the spacer regions between the 16S rRNA and 23S rRNA genes in rrnO and rrnA (Fig. 2). Some laboratory strains of B. subtilis contain only nine rrn operons due to spontaneous deletions occurring within either rrnW, -H, or -G (39), indicating that these operons are dispensable for growth in B. subtilis. We therefore focused on the seven essential rrn operons, i.e., the first operons of the contiguous rrnJ-rrnW and rrnI-rrnH-rrnG clusters, rrnJ and -I, and the noncontiguous operons rrnO, -A, -E, -D, and -B to analyze the regulation of transcription of each rrn operon (Fig. 3). The highly conserved rRNA genes within each rrn operon, however, made it difficult to study individual rRNA operons. To overcome this problem in E. coli, Condon and coworkers (7) introduced a reporter gene encoding chloramphenicol acetyltransferase (CAT) into each rrn operon and could thus monitor the expression of each operon individually by measuring CAT activity or by a quantitative S1 protection assay. We adopted this strategy to assess transcriptional activity of individual rrn operons in B. subtilis. A truncated chloramphenicol resistance gene (catpt1), lacking its promoter as well as a rho-independent transcriptional terminator, was inserted downstream of the known P2 or P3 promoter regions in seven rrn operons (rrnO, -A, -J, -I, -E, -D, and -B) and upstream of sites where the primary transcript is cleaved during the posttranscriptional processing events that generate mature 16S, 23S, and 5S rRNAs (Fig. 2). The resultant strains were resistant to chloramphenicol as the cat gene was cotranscribed as an integral part of each rrn operon. Primer extension analyses on RNA isolated from these strains with an IRD-labeled reverse primer complementary to the 5′ region of the inserted cat gene were carried out to determine the transcriptional start sites of each rrn operon promoter. It has been reported that all promoters of rrnB and rrnO (17) and rrnA and rrnI (16) initiate transcription with GTP. As shown in Fig. 2 and 3, we found that all promoters examined initiated transcription with GTP. Our results are consistent with the previous reports (16, 17). Additional start sites in P1 promoters of rrnA, -J, -I, -E, and -D as well as P2 promoters in rrnO, -A, -D, and -J were observed one base upstream of the canonical start sites (Fig. 3 and 4).
FIG. 2.
Location (A) and structure (B) of the rrn operons in B. subtilis. (C) Sequence alignment of the promoter regions of seven rrn operons (rrnO, -A, -J, -I, -D, -B, and -E) in B. subtilis. The −35 and −10 regions are shown by boxes, and the transcriptional start sites, as determined by the results shown in Fig. 4, are indicated in boldface. The positions of the catpt1 gene, a chloramphenicol resistance gene containing only Shine-Dalgarno (SD) and open reading frame (ORF) sequences, are indicated by closed triangles. The sequence of the catpt1 gene is also shown.
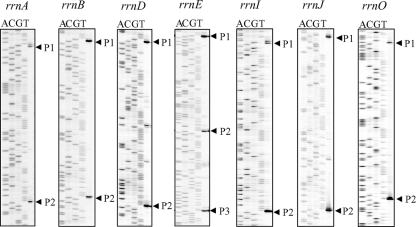
FIG. 3.
Identification of transcription start sites of each rrn promoter by primer extension on RNA isolated from wild-type cells (RIK350 to RIK356). Cells were grown in LB medium at 37°C and collected at an OD600 of approximately 0.20. Thirty micrograms of total RNA extracted from the cells was used for primer extension analysis as described in Materials and Methods. The sequence ladder was generated by PCR cycle sequencing with the same primers used in the primer extension reaction.
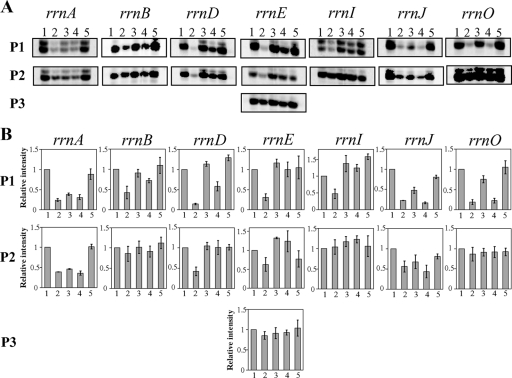
FIG. 4.
(A) Effects of the relA null mutation, the relA yjbM and relA ywaC double mutation, and the relA yjbM ywaC triple mutation on the transcription activity of each rrn operon. Primer extension products were generated with RNA isolated from the wild type (lane 1), the relA null mutant (lane 2), the relA yjbM (lane 3) or relA ywaC double mutant (lane 4), and the relA yjbM ywaC triple mutant (lane 5) as described in Materials and Methods. Representative results, obtained in three independent experiments, are shown. (B) Quantification of each reverse transcript shown in panel A was carried out as described in Materials and Methods, and relative signal intensity was calculated when the signal intensity of the product generated by reverse transcription of the RNA extracted from wild-type cells was set as 1.0. Each result is the average of three determinations. The error bars indicate standard deviations.
Transcription activity of rrn operons from P1 promoters is abolished in the relA null mutant and can be restored by its suppressor mutations.
Using the rrn operon transcription assay system stated above, we examined the effect of the relA null mutation on transcription activity of each rrn operon. Transcription activity from P1 promoters was drastically reduced in the relA null mutant. In addition, transcription from P2 promoters in the rrnA, -D, -E, and -J operons was also significantly decreased in the relA null mutant (Fig. 4). On the other hand, transcription from P2 promoters in the other operons, as well as from P3 in rrnE, although slightly decreased compared to wild type (Fig. 4), provided the major contribution to rRNA synthesis in relA null mutants.
Because all spontaneous suppressor mutations of the relA null mutant mapped to either yjbM or ywaC (Table 2), we next examined transcription activity of rrn operons in relA null strains carrying a deletion of either or both of these genes. Primer extension analyses showed that, compared to the relA null strain, transcription of each rrn operon was enhanced in the two double disruption mutants as well as in the triple disruption mutant although to different extents (Fig. 4). For each rrn operon we found that the transcription activity from P1 promoters was improved to a higher degree in relA yjbM than in relA ywaC, whereas the increase in transcription levels from P2 promoters were comparable for the two double disruption mutants. In the triple disruption mutant, transcription activity from both P1 and P2 promoters was almost completely restored to wild-type levels for each rrn operon compared to the relA null strain.
DISCUSSION
When the relA gene product was absent, transcription from all analyzed P1 promoters was abolished, thereby affecting the major pre-rRNA production pathway (Fig. 4). Thus, reduced ribosome synthesis may cause, at least in part, the growth defect observed for relA null mutant cells in LB medium (26). When two (p)ppGpp synthetase genes, yjbM and ywaC, were both inactivated in the relA mutant, growth (26) and ribosome formation (Fig. 1) were almost completely restored to wild-type levels. Therefore, as RelA is the only (p)ppGpp hydrolase in B. subtilis, it is likely that the basal level of (p)ppGpp maintained by these small (p)ppGpp synthetases repressed, directly or indirectly, transcription from P1 promoters in the relA mutant. However, transcription from the B. subtilis rrnB P1 promoter is not inhibited by the addition of ppGpp in vitro (17), suggesting that (p)ppGpp would not directly repress rRNA synthesis. Since the results were obtained with purified RNA polymerase (17), unknown cofactor(s), absent from the in vitro transcription reaction mixture, could be necessary for the downregulation of transcription by (p)ppGpp. In E. coli, (p)ppGpp decreases transcription by inhibition of the RNA polymerase via a cofactor, DksA (10, 30), but DksA homologues are not identified in B. subtilis (19), suggesting that there is a different mechanism for the regulation of rrn transcription by (p)ppGpp.
Instead, it has been proposed that transcription of rRNA operons is regulated by the intracellular concentration of GTP in B. subtilis (17). This agrees with the observation that reduction of the intracellular GTP level in response to amino acid starvation causes downregulation of stringently controlled-promoters with +1G start sites (18). In Thermus thermophilus, the concentration of the intracellular GTP level also appears to play an important role in the regulation of rrn promoters (15). Taking these observations together, it is likely that reduction of the transcription from P1 promoters in the relA mutant is due to the limitation of initiating GTP. Interestingly, we found by high-performance liquid chromatography analysis that the intracellular GTP level in the relA null mutant was threefold lower than in the wild type and that both GTP concentration and growth were equally restored by suppressor mutations that reduced ongoing (p)ppGpp synthesis (26). It is conceivable that the basal level of (p)ppGpp present in the relA null mutant negatively regulates the activity of IMP dehydrogenase, the first enzyme of GTP biosynthesis (21), thus causing a reduction of the intracellular GTP level and the concomitant downregulation of the transcription from P1 promoters.
However, at present it is not known why the P1 promoters of each rrn operon tested, as well as the P2 promoters in rrnA, -J, -E, and -D, are selectively repressed in the relA null mutant. We speculate that the rrn P1 promoters, as well as P2 promoters in rrnA, -J, -E, and -D, are regulated in a manner distinct from other P2 promoters. It has been proposed that the P1 promoters of rrn operons are regulated more than their respective P2 promoters since the P2 promoters are less inhibited than the P1 promoters at slow growth rates (17). In this context, one possible explanation is that only the P1 promoters and the P2 promoters in rrnA, -J, -E, and -D are regulated by some unknown factor(s), which can activate or repress the transcription from these promoters. The activation of the unknown factor(s) may be regulated by the intracellular concentration of GTP and/or required for the function of RelA protein. However, the detailed mechanism explaining how RelA differentially regulates the transcription activity of P1 and P2 promoters in rrn operons remains unclear. Further investigations would be necessary to clarify the precise regulatory mechanism(s) by which (p)ppGpp causes a reduction in GTP levels and how this preferentially inhibits transcription from P1 promoters and from P2 promoters in rrnA, -J, -E, and -D.
The activity of P1 promoters, as assessed by primer extension, is higher in the relA yjbM disruption mutant than in relA ywaC (Fig. 4), mirroring the higher GTP levels in relA yjbM than in relA ywaC (26). We have previously reported that the (p)ppGpp synthesis activity of YjbM exceeds that of YwaC in vitro (26). In addition, the yjbM gene is predominantly transcribed in the logarithmic growth phase when ribosome synthesis is very high while the transcription of ywaC is dependent on σM, which is activated by a variety of stress conditions such as high salinity, ethanol, heat, acid stress, and exposure to several cell wall antibiotics (8), explaining the low expression level of ywaC during vegetative growth as detected by Northern blot analysis (26). On the basis of these results, YjbM would be mainly responsible for maintaining the basal (p)ppGpp level during growth in LB medium. However, deletion of yjbM in the relA mutant did not restore the transcription activity of all tested P1 promoters. As shown in Fig. 4, in the relA yjbM mutant, transcription from the P1 promoters of the rrnI, -E, -D, and -B operons was almost equal to that of the wild type, but that of the rrnA and -J operons was only partially restored. As the intracellular GTP concentration in the relA yjbM mutant was also partially recovered (about 60% of the wild-type level) (26), these results suggest that sensitivity to the intracellular concentration of GTP may be different for each P1 promoter although the detailed mechanisms remain to be clarified. In addition, the contribution of YwaC to the production of the basal (p)ppGpp level remains unclear. To address this question, it is necessary to understand in more detail the functions of YjbM and YwaC in vivo, including transcriptional regulation of these genes and characteristics of cell growth after induction of either gene in a triple null mutant background.
Supplementary Material
Acknowledgments
We are grateful to Roy H. Doi for helpful discussion and critical reading of the manuscript.
This work was supported by the Frontier Project “Adaptation and Evolution of Extremophile”; in part by a Grant-in-Aid for Scientific research (C) (to F.K.) and (A) (to H.N.) and for Young Scientist (B) (to H.N.); and in part by Creative Scientific Research grant (16GS0304 to F.K.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan. This work was partly supported by an NISR Research Grant from the Noda Institute for Scientific Research to F.K.
Footnotes
Published ahead of print on 15 May 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Artsimovitch, I., V. Patlan, S. Sekine, M. N. Vassylyeva, T. Hosaka, K. Ochi, S. Yokoyama, and D. G. Vassylyev. 2004. Structural basis for transcription regulation by alarmone ppGpp. Cell 117299-310. [DOI] [PubMed] [Google Scholar]
- 2.Barker, M. M., T. Gaal, C. A. Josaitis, and R. L. Gourse. 2001. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J. Mol. Biol. 305673-688. [DOI] [PubMed] [Google Scholar]
- 3.Battesti, A., and E. Bouveret. 2006. Acyl carrier protein/SpoT interaction, the switch linking SpoT-dependent stress response to fatty acid. Mol. Microbiol. 621048-1063. [DOI] [PubMed] [Google Scholar]
- 4.Braeken, K., M. Moris, R. Daniels, J. Vanderleyden, and J. Michiels. 2006. New horizons for (p)ppGpp in bacterial and plant physiology. Trends Microbiol. 1445-54. [DOI] [PubMed] [Google Scholar]
- 5.Cashel, M., D. R. Gentry, V. J. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. American Society for Microbiology, Washington, DC.
- 6.Chatterji, D., N. Fujita, and A. Ishihama. 1998. The mediator for stringent control, ppGpp, binds to the beta-subunit of Escherichia coli RNA polymerase. Genes Cells 3279-287. [DOI] [PubMed] [Google Scholar]
- 7.Condon, C., J. Philips, Z.-Y. Fu, C. Squires, and C. L. Squires. 1992. Comparison of the expression of the seven ribosomal RNA operons in Escherichia coli. EMBO J. 114175-4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eiamphungporn, W., and J. D. Helmann. 2008. The Bacillus subtilis σM regulon and its contribution to cell envelope stress responses. Mol. Microbiol. 67830-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gralla, J. D. 2005. Escherichia coli ribosomal RNA transcription: regulatory roles for ppGpp, NTPs, architectural proteins and a polymerase-binding protein. Mol. Microbiol. 55973-977. [DOI] [PubMed] [Google Scholar]
- 10.Haugen, S. P., W. Ross, and R. L. Gourse. 2008. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat. Rev. Microbiol. 6507-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henkin, T. M. 2002. Ribosomes, protein synthesis factors, and tRNA synthetases, p. 313-322. In A. L. Sonenshein J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to aells. American Society for Microbiology, Washington, DC.
- 12.Hou, Z., M. Cashel, H. J. Fromm, and R. B. Honzatko. 1999. Effectors of the stringent response target the active site of Escherichia coli adenylosuccinate synthetase. J. Biol. Chem. 27417505-17510. [DOI] [PubMed] [Google Scholar]
- 13.Imamura, D., K. Kobayashi, J. Sekiguchi, N. Ogasawara, M. Takeuchi, and T. Sato. 2004. spoIVH (ykvV), a requisite cortex formation gene, is expressed in both sporulating compartments of Bacillus subtilis. J. Bacteriol. 1865450-5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inaoka, T., and K. Ochi. 2002. RelA protein is involved in induction of genetic competence in certain Bacillus subtilis strains by moderating the level of intracellular GTP. J. Bacteriol. 1843923-3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kasai, K., T. Nishizawa, K. Takahashi, T. Hosaka, H. Aoki, and K. Ochi. 2006. Physiological analysis of the stringent response elicited in an extreme thermophilic bacterium, Thermus thermophilus. J. Bacteriol. 1887111-7122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koga, K., A. Ikegami, K. Nakasone, R. Murayama, G. Akanuma, Y. Natori, H. Nanamiya, and F. Kawamura. 2006. Construction of Bacillus subtilis strains carrying the transcriptional bgaB fusion with the promoter region of each rrn operon and their differential transcription during spore development. J. Gen. Appl. Microbiol. 52119-124. [DOI] [PubMed] [Google Scholar]
- 17.Krásný, L., and R. L. Gourse. 2004. An alternative strategy for bacterial ribosome synthesis: Bacillus subtilis rRNA transcription regulation. EMBO J. 234473-4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krásný, L., H. Tišerová, J. Jonák, D. Rejman, and H. Šanderová. 2008. The identity of the transcription +1 position is crucial for changes in gene expression in response to amino acid starvation in Bacillus subtilis. Mol. Microbiol. 6942-54. [DOI] [PubMed] [Google Scholar]
- 19.Kunst, F., N. Ogasawara, I. Moszer, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390249-256. [DOI] [PubMed] [Google Scholar]
- 20.Lemos, J. A., V. K. Lin, M. M. Nascimento, J. Abranches, and R. A. Burne. 2007. Three gene products govern (p)ppGpp production by Streptococcus mutans. Mol. Microbiol. 651568-1581. [DOI] [PubMed] [Google Scholar]
- 21.Lopez, J. M., A. Dromerick, and E. Freese. 1981. Response of guanosine 5′-triphosphate concentration to nutritional changes and its significance for Bacillus subtilis sporulation. J. Bacteriol. 146605-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milon, P., E. Tischenko, J. Tomsic, E. Caserta, G. Folkers, A. L. La Teana, M. V. Rodnina, C. L. Pon, R. Boelens, and C. O. Gualerzi. 2006. The nucleotide-binding site of bacterial translation initiation factor 2 (IF2) as a metabolic sensor. Proc. Natl. Acad. Sci. USA 10313962-13967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mittenhuber, G. 2001. Comparative genomics and evolution of genes encoding bacterial (p)ppGpp synthetases/hydrolases (the Rel, RelA and SpoT proteins). J. Mol. Microbiol. Biotechnol. 3585-600. [PubMed] [Google Scholar]
- 24.Nanamiya, H., E. Shiomi, M. Ogura, T. Tanaka, K. Asai, and F. Kawamura. 2003. Involvement of ClpX protein in the post-transcriptional regulation of a competence specific transcription factor, ComK protein, of Bacillus subtilis. J. Biochem. (Tokyo) 133295-302. [DOI] [PubMed] [Google Scholar]
- 25.Nanamiya, H., G. Akanuma, Y. Natori, R. Murayama, S. Kosono, T. Kudo, K. Kobayashi, N. Ogasawara, S. M. Park, K. Ochi, and F. Kawamura. 2004. Zinc is a key factor in controlling alternation of two types of L31 protein in the Bacillus subtilis ribosome. Mol. Microbiol. 52273-283. [DOI] [PubMed] [Google Scholar]
- 26.Nanamiya, H., K. Kasai, A. Nozawa, C. S. Yun, T. Narisawa, K. Murakami, Y. Natori, F. Kawamura, and Y. Tozawa. 2008. Identification and functional analysis of novel (p)ppGpp synthetase genes in Bacillus subtilis. Mol. Microbiol. 67291-304. [DOI] [PubMed] [Google Scholar]
- 27.Natori, Y., H. Nanamiya, G. Akanuma, S. Kosono, T. Kudo, K. Ochi, and F. Kawamura. 2007. A fail-safe system for the ribosome under zinc-limiting conditions in Bacillus subtilis. Mol. Microbiol. 63294-307. [DOI] [PubMed] [Google Scholar]
- 28.Ogasawara, N., S. Moriya, and H. Yoshikawa. 1983. Structure and organization of rRNA operons in the region of the replication origin of the Bacillus subtilis chromosome. Nucleic Acids Res. 116301-6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paul, B. J., W. Ross, T. Gaal, and R. L. Gourse. 2004. rRNA transcription in Escherichia coli. Annu. Rev. Genet. 38749-770. [DOI] [PubMed] [Google Scholar]
- 30.Paul, B. J., M. M. Barker, W. Ross, D. A. Schneider, C. Webb, J. W. Foster, and R. L. Gourse. 2004. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell 118311-322. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 32.Seyfzadeh, M., J. Keener, and M. Nomura. 1993. spoT-dependent accumulation of guanosine tetraphosphate in Escherichia coli. Proc. Natl. Acad. Sci. USA 9011004-11008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Srivatsan, A., Y. Han, J. Peng, A. K. Tehranchi, R. Gibbs, J. D. Wang, and R. Chen. 2008. High-precision, whole-genome sequencing of laboratory strains facilitates genetic studies. PLoS Genet. 4e1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stewart, G., C, and K. F. Bott. 1983. DNA sequence of the tandem ribosomal RNA promoter for B. subtilis operon rrnB. Nucleic Acids Res. 116289-6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vinella, D., C. Albrecht, M. Cashel, and R. D'Ari. 2005. Iron limitation induces SpoT-dependent accumulation of ppGpp in Escherichia coli. Mol. Microbiol. 56958-970. [DOI] [PubMed] [Google Scholar]
- 36.Wang, J. D., G. M. Sanders, and A. D. Grossman. 2007. Nutritional control of elongation of DNA replication by(p) ppGpp. Cell 128865-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wellington, S. R., and G. B. Spiegelman. 1993. The kinetics of formation of complexes between Escherichia coli RNA polymerase and the rrnB P1 and P2 promoters of Bacillus subtilis. Effects of guanosine tetraphosphate on select steps of transcription initiation. J. Biol. Chem. 2687205-7214. [PubMed] [Google Scholar]
- 38.Wendrich, T. M., and M. A. Marahiel. 1997. Cloning and characterization of a relA/spoT homologue from Bacillus subtilis. Mol. Microbiol. 2665-79. [DOI] [PubMed] [Google Scholar]
- 39.Widom, R. L., E. D. Jarvis, G. LaFauci, and R. Rudner. 1988. Instability of rRNA operons in Bacillus subtilis. J. Bacteriol. 170605-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiao, H., M. Kalma, K. Ikehara, S. Zemel, G. Glaser, and M. Cashel. 1991. Residual guanosine 3′,5′-bisphosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 2665980-5990. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.