Abstract
Azotobacter vinelandii is a soil bacterium related to the Pseudomonas genus that fixes nitrogen under aerobic conditions while simultaneously protecting nitrogenase from oxygen damage. In response to carbon availability, this organism undergoes a simple differentiation process to form cysts that are resistant to drought and other physical and chemical agents. Here we report the complete genome sequence of A. vinelandii DJ, which has a single circular genome of 5,365,318 bp. In order to reconcile an obligate aerobic lifestyle with exquisitely oxygen-sensitive processes, A. vinelandii is specialized in terms of its complement of respiratory proteins. It is able to produce alginate, a polymer that further protects the organism from excess exogenous oxygen, and it has multiple duplications of alginate modification genes, which may alter alginate composition in response to oxygen availability. The genome analysis identified the chromosomal locations of the genes coding for the three known oxygen-sensitive nitrogenases, as well as genes coding for other oxygen-sensitive enzymes, such as carbon monoxide dehydrogenase and formate dehydrogenase. These findings offer new prospects for the wider application of A. vinelandii as a host for the production and characterization of oxygen-sensitive proteins.
Azotobacter vinelandii is a free-living nitrogen-fixing bacterium of the gammaproteobacteria. It is found in soils worldwide, with features of nitrogen and energy metabolism relevant to agriculture (41, 42). This organism has been studied for more than 100 years by numerous scientists throughout the world. Prior to Joshua Lederberg's discovery of sexuality in Escherichia coli (47), A. vinelandii was the experimental organism of choice for many investigators during the emergence of biochemistry as a dominant discipline within the life sciences. Examples include the classical Lineweaver-Burk kinetic parameters, developed using enzymes from A. vinelandii (51), and the isolation by Severo Ochoa of polynucleotide phosphorylase from A. vinelandii, which was used in studies that contributed to the elucidation of the genetic code (62).
A. vinelandii is able to adapt its metabolism to diverse sources of nutrients. If no carbon source is present, A. vinelandii will undergo a differentiation process to form cysts that are resistant to desiccation and other chemical and physical challenges (74). While the process of encystment has been known for many years and studied at the physiological and morphological levels, there is little knowledge about the unique biosynthetic pathways that are involved and how they are regulated. Previous work has implicated the alternative sigma factors AlgU and RpoS in the differentiation process (13, 57, 64). Alginate polymers with different monomer compositions are an important structural component of the cyst, and at the end of exponential growth, A. vinelandii cells accumulate poly-beta-hydroxybutyrate (PHB) as a reserve carbon and energy source (81). The physiology of PHB formation has been well studied in a variety of different systems, and the PHB biosynthetic operon has been described (67, 77). A. vinelandii can also produce copolymers of hydroxybutyrate and hydroxyvalerate, known to improve the flexibility and stretch of bioplastics (63).
A. vinelandii has long served as a model for biochemical and genetic studies of biological nitrogen fixation, the conversion of N2 into NH3 by a nitrogenase enzyme. The best-studied nitrogenase consists of two oxygen-sensitive metalloproteins that, in the case of the molybdenum nitrogenase, are denominated the Fe protein and the MoFe protein. A. vinelandii is unusual in that it is one of the few bacteria that contain three nitrogenases with different subunit and metal cofactor compositions, namely, the molybdenum nitrogenase, the vanadium nitrogenase, and the iron-only nitrogenase. Expression of these nitrogenases is differentially regulated by metal availability from the medium (27).
Here we present the complete genome sequence of A. vinelandii DJ and discuss what the genome has revealed about the organism's ability to protect oxygen-sensitive processes. A. vinelandii has been cited as having one of the highest respiratory rates of any known bacterium (10). Diazotrophic growth under aerobic conditions is possible because A. vinelandii can adjust oxygen consumption rates to help maintain low levels of cytoplasmic oxygen, which is otherwise detrimental not only to nitrogenase but also to other oxygen-sensitive enzymes expressed by A. vinelandii. This phenomenon is called respiratory protection. In this work, we identify unique features of the A. vinelandii genome that help to explain the coexistence of oxygen-sensitive reactions and strict aerobic metabolism. The genome sequence and annotation allowed identification of the genes involved in respiration, including key players in respiratory protection. In addition, we have identified unexpected gene clusters encoding a carbon monoxide dehydrogenase (CODH), a formate dehydrogenase (FDH), and a second hydrogenase, all of which are also oxygen-sensitive enzymes.
MATERIALS AND METHODS
Strain description.
A. vinelandii O is a strain that forms gummy, slimy colonies of pale color. The earliest report of this strain dates from 1952 (92). Azotobacter agilis (later named Azotobacter vinelandii strain O) was part of the bacterial collection at the University of Wisconsin, Madison. In 1959, Bush and Wilson (11) reported the isolation of a nongummy chromogenic isolate of A. vinelandii O, which they named A. vinelandii OP. This new strain has a fluorescent color and well-defined colony shape. A. vinelandii strain OP has also been referred to as UW and CA. A. vinelandii strain DJ is a high-frequency transforming variant of A. vinelandii UW generated in 1984 through multiple rounds of transformation, using chromosomal DNAs from rifampin (rifampicin)-resistant and -sensitive strains (Dennis Dean, personal communication), and is available from the American Type Culture Collection under number ATCC BAA-1303.
Genome sequencing and assembly.
A total of 87,000 reads were generated by the Joint Genome Institute for a draft sequence released prior to this project. These reads came from two DNA libraries (insert sizes of 2 to 4 kb and 4 to 8 kb) obtained using mechanical shearing of DNA and cloning into pUC18, followed by a shotgun sequencing approach. The reads and clones were sent to Monsanto, where finishing occurred. The genome was then assembled and edited using Phred, Phrap, and Consed (23, 24, 32). Finishing was completed by generating an optical map (46) cut with the restriction enzymes BamHI and BsiWI and aligning the assembled sequences to the map. Gaps were closed by sequencing specific products. All rRNA operons were amplified with specific flanking primers, sequenced, and assembled individually. All positions with Phred scores of <40 were resequenced using an independent PCR fragment as a template. The error rate is estimated to be less than 1:10,000 bp.
Annotation and comparative genomics.
Genome annotation was done by supervised teams of undergraduate students using a web-based system over a preliminary automated annotation, both developed by J. C. Setubal. Comparison to other genomes was done using MUMmer (45) for alignments and orthoMCL (50) for protein families.
Phylogenetic analysis.
A total of 18 genomes (see Table S1 in the supplemental material) were used to build the phylogenetic tree. Chromohalobacter selexigens, Hahella chejuensis, and Marinomonas sp. strain MWYL1 were used as outgroup species. OrthoMCL (50) provided 1,445 protein families containing only one representative member of each ingroup genome. These protein sequences were aligned with Muscle (20), and the noninformative columns of the resulting (concatenated) alignment were removed by Gblocks (14). The tree was built using RAxML (79) with the PROTGAMMAWAGF model. Individual gene phylogenies were obtained by aligning protein sequences from A. vinelandii and Pseudomonas species, using the same method as that described above, but using MrBayes (35) for tree building.
Prediction of σ54- and NifA-binding sites.
Multiple alignments of known σ54- and NifA-binding-site DNA sequences were constructed from the literature (3). Using HMMER 1.8.5 (25), we generated profile hidden Markov models representing the binding sequence motifs from the alignments and used HMMER to search the genome for matches.
CydR binding site prediction.
We used the sequence proposed by Wu et al. (93) as a query against the genome in a BLASTN (1) search.
Nucleotide sequence accession number.
The A. vinelandii DJ sequence and its annotation are available at GenBank under accession number CP001157 and at www.azotobacter.org.
RESULTS
General features and phylogeny.
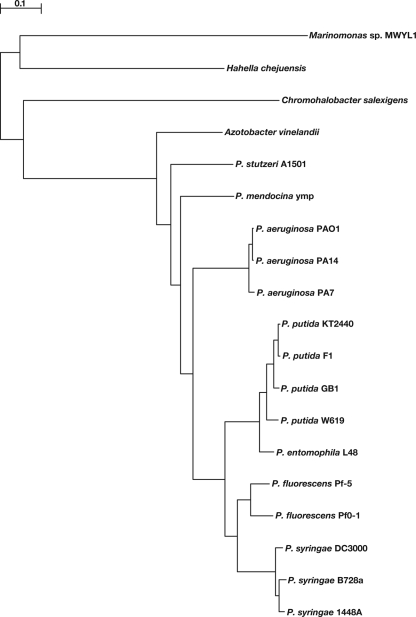
The general features of the A. vinelandii genome are shown in Table 1. Based on the sequence and its annotation, we have obtained a phylogeny for A. vinelandii (Fig. 1) that indicates that it groups most closely with the pseudomonads (family Pseudomonadaceae). Among the pseudomonads, the closest relative is the nitrogen-fixing strain Pseudomonas stutzeri A1501 (96). Table 1 shows that A. vinelandii and P. stutzeri A1501 share 46% and 56% of their respective protein coding gene complements. Figure S1 in the supplemental material presents a whole-chromosome alignment between A. vinelandii DJ and P. stutzeri A1501 in which large-scale conservation of gene order can readily be observed. The “X” pattern seen in this comparison is likely due to inversions around the origin of replication (22) and suggests that many such inversions have taken place since these two species diverged.
TABLE 1.
General features of Azotobacter vinelandii DJ and Pseudomonas stutzeri A1501 genomesa
| Parameter | Value
|
|
|---|---|---|
| Azotobacter vinelandii DJ | Pseudomonas stutzeri A1501 | |
| Chromosome features | ||
| Size (bp) | 5,365,318 | 4,567,418 |
| % GC | 65.7 | 63.9 |
| Protein-encoding genes | ||
| Total no. | 5,051 | 4,128 |
| No. (%) with functional assignment | 3,561 (70.5) | 3,180 (77.0) |
| No. (%) of conserved hypothetical protein genes | 739 (14.6) | 0 (0.0) |
| No. (%) of hypothetical protein genes | 751 (14.9) | 948 (23.0) |
| No. of pseudogenes | 66 | 11 |
| No. (%) shared protein-encoding genes | 2,332 (44.1) | 2,298 (55.7) |
| RNAs | ||
| No. of rRNA operons | 6 | 4 |
| No. of tRNAs | 64 | 61 |
| No. of other RNAs | 18 | 0 |
FIG. 1.
Phylogenetic tree for A. vinelandii, all fully sequenced members of the Pseudomonas genus, and three outgroups. Accession numbers for the genomes used are given in Table S1 in the supplemental material. Bootstrap support for all branches was 100%.
Respiration and respiratory protection genes.
A. vinelandii is an obligately aerobic bacterium that uses the electron transport chain, with molecular oxygen as the final electron acceptor. This strict aerobic metabolism is supported by our analysis of the genome, which lacks genes for complete systems involved in anaerobic respiration using alternative terminal electron acceptors or fermentation processes. At the interface of catabolism and respiration, NADH and succinate are the major electron carriers that feed the electron transport chain, via complex I and complex II, respectively. The genome encodes four NADH-ubiquinone oxidoreductases, including one ATP-coupled NADH oxidoreductase of the Nuo type (Avin28440 to Avin28560) and three ATP-uncoupled NADH-ubiquinone oxidoreductases. The latter are Ndh (Avin12000) and two membrane-associated enzymes involved in the transport of cations across the membrane, i.e., the Nqr type (Avin14590 to Avin14640) and the Sha type (Avin19530 to Avin19580). While there is no association of Nuo, Nqr, and Sha with oxygen protection, the ndh gene product has been shown to be important for aerobic nitrogen fixation (5). It is possible that NADH-driven protection of nitrogenase is dependent on Ndh and CydAB oxidase via a ubiquinol-dependent electron transfer pathway. Despite its apparent involvement in nitrogen fixation (5), a copy of ndh is found in several Pseudomonas species, some of which are not known to fix nitrogen. In addition, A. vinelandii carries two copies of the Rnf system (Rnf1 [Avin50930 to Avin50980] and Rnf2 [Avin19220 to Avin19270]), which shows sequence similarity to a sodium-dependent NADH-ubiquinone oxidoreductase (18). Although there is no change in respiration upon inactivation of either one or both Rnf systems, Rnf1 is required for accumulation of nitrogenase Fe protein (18). An ortholog of Rnf1 exists in the diazotroph P. stutzeri A1501 (YP_001171857 to YP_00117852) but not in any other members of the Pseudomonas genus.
Ubiquinol-oxygen oxido-reduction can occur either through a two-step pathway, via cytochrome c reductase (complex III; Avin13060 to Avin13080) followed by a cytochrome terminal oxidase (complex IV), or in a single-step process, via a ubiquinol-dependent cytochrome terminal oxidase. The genome annotation identified the catalytic and biosynthetic genes of the following five terminal oxidases: cytochrome c oxidase (Cdt oxidase; Avin00950 to Avin01020), cytochrome o oxidase (Cox; Avin11170 to Avin11180), cytochrome bd copy I (CydAB I; Avin19880 to Avin19890), cytochrome bd copy II (CydAB II; Avin11040 to Avin11050), and cytochrome cbb3 (Cco; Avin19940 to Avin20010). While the Cox, CydAB II, and Cco oxidases are encoded in all Pseudomonas species analyzed to date, the Cdt oxidase gene is found only in P. stutzeri A1501 and Pseudomonas mendocina ymp.
The presence of two CydAB oxidases in A. vinelandii was unexpected. Phylogenetic analysis revealed that CydAB oxidase I (encoded by cydAB gene copy I) has its closest orthologs in P. stutzeri A1501 but groups with similar genes from members of the Acinetobacter and Shewanella genera rather than with those from other members of the Pseudomonas genus. Copy II, on the other hand, groups with similar genes from several members of the Pseudomonas genus (see Fig. S2 to S5 in the supplemental material). CydAB oxidase I has been characterized extensively (21, 39, 40, 94). The precise function of CydAB oxidase II remains to be elucidated.
Oxygen consumption by the aforementioned terminal oxidases is not the only factor responsible for oxygen protection of nitrogenase. The FeSII protein, known as the Shethna protein and encoded by Avin01520, forms a protective complex with nitrogenase when the enzyme is exposed to oxygen (58). In addition, it has been determined that cellular levels of ATP also contribute to protection of nitrogenase against oxygen damage (53). High concentrations of ATP directly correlate with high electron flux to nitrogenase, which influences the dissociation rate constant of the nitrogenase components (84) and, consequently, the susceptibility of the Fe protein to oxygen damage (83). The A. vinelandii genome encodes two sets of ATP synthase machineries. The first complex (Avin52150 to Avin52230) is the ortholog of the Pseudomonas ATP synthase and is located close to the putative origin of replication. The second copy (Avin19670 to Avin19750) is located 8 genes upstream of the sha operon (complex I-like) and 10 genes downstream of cydAB I (complex IV). This copy does not seem to be related phylogenetically to ATP synthases from the Pseudomonas genus, and the only orthologs are found in Burkholderia species, most of which are obligate aerobes, although very few can fix nitrogen.
Genomic analyses of A. vinelandii and microaerobic diazotrophs such as P. stutzeri show that they have similar respiratory complexes, suggesting that the regulation of these machineries, especially at the transcriptional level, is very important for adjusting rates of oxygen consumption in order to protect oxygen-sensitive processes. The transcriptional regulator CydR (Avin19910) seems to regulate various physiological processes associated with respiratory protection. Wu et al. (94) showed that CydR can coordinate an oxygen-labile [Fe-S] cluster, which provides a mechanism for CydR to sense subtle changes in the intercellular oxygen concentration and to regulate the expression of respiratory genes. During nitrogen fixation, CydR repression is presumably relieved, resulting in increased expression of uncoupled NADH dehydrogenase (Ndh) and CydAB I. A series of reports have suggested a role for CydR in several other metabolic processes, including synthesis of PHB (95) and flagellar motility (48). Although the role of CydR in respiration is not completely defined, its direct or indirect participation in other cellular processes is even more obscure.
The genomic location of cydR (Avin19910) downstream of the cco region (containing the cco genes [Avin19920 to Avin20010]) suggests that it is involved in the regulation of this system. In support of this, a putative CydR binding site was identified upstream of the cco genes, and expression of a ccoN::lacZ fusion is CydR dependent (data not shown). Similar regulation of Cco by FNR (a CydR ortholog) is seen in other organisms (16). CydR also binds upstream of the CydAB oxidase I promoter and is known to repress transcription of this operon (93). While cydR in A. vinelandii is adjacent to the cydAB operon (Avin19890 to Avin19880) encoding CydAB oxidase I, this organization is not seen in other organisms. Our promoter analysis has further identified a CydR binding site immediately upstream of Avin11170, suggesting that CydR also regulates the cox genes in A. vinelandii.
A putative CydR binding site is also located upstream of the iron-sulfur cluster biosynthetic genes (isc genes; Avin40380 to Avin40410). Cross regulation of Fe-S cluster biosynthesis and respiration makes biochemical sense for at least the following two reasons: (i) a large number of Fe-S proteins are involved in respiration, and (ii) elevated oxygen concentrations result in increased damage to Fe-S proteins under conditions in which the respiratory proteins are more highly expressed. Therefore, under conditions of high oxygen concentrations, CydR regulation would increase the expression of respiratory components and increase the capacity of the Fe-S cluster biosynthetic apparatus to supply the temporary higher demand for Fe-S clusters.
Nitrogen fixation.
A. vinelandii expresses three oxygen-sensitive nitrogenase enzymes with different structural subunits and metal cofactor dependencies. Genes encoding the well-studied molybdenum-dependent nitrogenase and its assembly machinery and regulation (nif) are located in two regions of the chromosome, adjacent to and equidistant from the origin of replication. Proximity to the origin might result in a higher gene dosage during active growth, which can contribute to the high expression levels of the Nif components. The major nif region (comprising genes Avin01360 to Avin01710, oriented away from the putative origin, on the plus strand) encodes the structural subunits and the majority of the assembly machinery. The minor nif region (genes Avin50990 to Avin51060, also on the plus strand) contains the regulatory genes nifL and nifA and genes required for Mo trafficking and nitrogenase cofactor biosynthesis. Adjacent to the minor nif region are the rnf1 region (upstream) and genes for a putative rhodanese (rhdnif; Avin51050) and a monothiol glutaredoxin (grxnif; Avin51060) (downstream). The rnf1 gene products are associated with the accumulation of active nitrogenase component 2, also referred to as Fe protein (18). Likewise, the inactivation of the nif-associated glutaredoxin resulted in a 50% loss of Fe protein activity (data not shown). The dramatic effect on Fe protein activity and the less pronounced effect on nitrogenase component 1, the MoFe protein, could be attributed to an inherent susceptibility to oxygen damage and/or deficiency in repair of the [Fe-S] cofactor in strains lacking these genes.
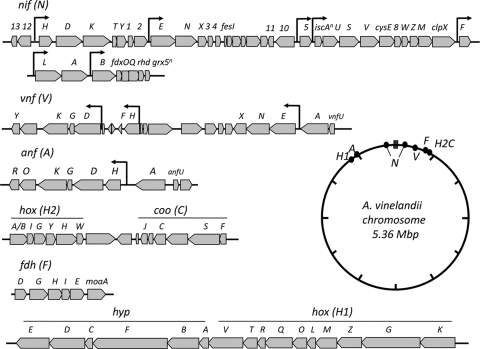
The genome sequence reveals that the region containing the structural genes for the vanadium nitrogenase (Avin02650 to Avin02660) is in close proximity to the gene cluster carrying genes (Avin02740 to Avin02780) involved in its assembly and regulation (Fig. 2). However, these two vnf clusters are separated by three genes predicted to participate in molybdopterin biosynthesis (Avin02700 to Avin02720) and a gene, pcaK, encoding a hydroxybenzoate transporter (Avin02690). In contrast, the anf genes, encoding the iron-only nitrogenase, are located in a single gene region (Avin48950 to Avin49000), which is regulated by the AnfA transcriptional activator (Avin49020) (Fig. 2).
FIG. 2.
Oxygen-sensitive protein-encoding genes in A. vinelandii and their genomic locations.
Although no additional nitrogenase genes were identified in the genome sequence, genes with sequence similarity to the nif, vnf, and anf genes were found scattered throughout the genome. Whether or not these other genes are involved in nitrogen fixation remains to be determined. Interestingly, a gene with sequence similarity to nifD was identified (Avin39870), but its inactivation did not affect growth under standard diazotrophic or nondiazotrophic conditions (data not shown). Phylogenetic analysis of the NifD-like sequence in comparison with its paralogs in A. vinelandii and P. stutzeri reinforced the idea that the three structural and accessory nitrogenase genes were derived from a common ancestor and that the nifD-like sequence resembles the ancestral gene (see Fig. S6 in the supplemental material).
A paralog of nifA, Avin26490, was identified as encoding a protein that contains an almost identical DNA recognition helix in the C-terminal DNA-binding domain, implying that this protein binds similar enhancer sequences to those recognized by NifA (Avin51000). A search for σ54 promoters and NifA upstream activator sequences detected the experimentally determined NifA-dependent promoters upstream of the nif and rnf genes and additional putative NifA-dependent σ54 promoters upstream of genes encoding ModE (Avin50680), a putative molybdate binding protein, ModA3 (Avin50730), and a putative McbC-like oxidoreductase (Avin48680). In view of the similarity in DNA recognition motifs, it is possible that these promoters are activated by either NifA or its paralog, Avin26490.
The genome sequence revealed two genes (Avin33440 and Avin47100) with 78% and 71% identity to vnfA (Avin02780), respectively. These VnfA paralogs have two of the three cysteines present in the proposed metal-binding cluster in the amino-terminal GAF domain of VnfA, similar to that of AnfA, which also has two cysteine residues implicated in metal or redox sensing (37, 69).
The nfuA gene (Avin28760) encodes a protein with a NifU-like C-terminal domain that shares sequence similarity with the products of vnfU (Avin02790) and anfU (Avin49030), which are located adjacent to vnfA (Avin02780) and anfA (Avin49020), respectively. Biochemical experiments suggest that NfuA represents an intermediate [Fe-S] cluster carrier involved in [Fe-S] protein maturation (2). It is likely that VnfU and AnfU play a role in the maturation of clusters present in vanadium- and iron-only nitrogenases.
In addition to paralogs of nitrogen fixation genes, other systems critical to nitrogen fixation appear to have been duplicated. Positioned 35 kb upstream of the minor nif gene cluster is the high-affinity molybdate transport system, encoded by modG and modEA1B1C1 (Avin50650 to Avin50690), which supports the expression of Mo-dependent nitrogenase under molybdenum-limiting conditions (65). The second known molybdate transport locus, modA2B2C2 (Avin01280 to Avin01300), is located 10 kb upstream of the major nif gene cluster. Interestingly, we identified a third putative molybdate transport system (Avin50700 to Avin50730), located directly next to the modEA1B1C1 operon.
Other oxygen-sensitive processes. (i) CODH.
The genome of A. vinelandii DJ contains genes with sequence similarity to membrane-bound Ni-dependent anaerobic CODH genes. CODH is an α2 homodimer of the cooS product (Avin04490) that carries out the reversible oxidation of CO to CO2 (71). Electrons extracted from CO are transferred to CooF (Avin04500), a hydrophobic FeS protein similar to the electron transfer subunits of oxidoreductases, such as the FDH beta subunit FdhH (Avin03820).
CODH contains a NiFe4S4 center, known as the C cluster, at its active site. The assembly of the CODH active site, which has been well studied in Rhodospirillum rubrum, requires the activities of three proteins, encoded by the cooJ, cooC, and cooT genes (43), involved in Ni storage and insertion (36, 91). Whereas one gene similar to cooC is present in the A. vinelandii genome (Avin04470), the genome does not code for proteins similar to CooT or CooJ. Avin04460, located downstream of cooC, encodes a 73-amino-acid histidine-rich protein with sequence similarity to the N-terminal portion (first 20%) of HypB proteins in R. rubrum (E value, 10−7) and Ralstonia eutropha (10−5), which are involved in Ni processing for hydrogenase maturation. Thus, it is likely that the product of Avin04460 substitutes for the role of CooJ in the maturation of CODH. There is a gene similar to hupE/ureJ genes (Avin04450) downstream of Avin04460. HupE/UreJ proteins are secondary Ni transporters, and their coding genes are widespread among bacteria, normally clustered with urease or hydrogenase genes. There is another gene in the hupE/ureJ family (Avin50400), located near the hypEDCFB gene cluster, that probably encodes a HupE protein. We hypothesize that Avin04450 and Avin50400 may encode two different Ni transport systems, specific for CODH and hydrogenase maturation, respectively.
The A. vinelandii coo gene cluster includes a gene for a flavin adenine dinucleotide-dependent pyridine nucleotide-disulfide oxidoreductase (Avin04480) inserted between the cooS and cooC genes. A similar protein is also encoded downstream of cooFS in Carboxydothermus hydrogenoformans. Proteins of this family utilize flavin adenine dinucleotide to shuttle electrons from NADH to a redox-active disulfide bridge. The product of Avin04480 might be involved in CODH maturation. It has been proposed that Ni insertion into the C cluster involves reversible Ni binding to an Fe3S4 center followed by coordination to a specific cysteinyl residue (Cys531 in R. rubrum CooS) (36). It is also noteworthy that in vitro Ni insertion into CODH requires dithionite as a reductant. Thus, the Avin04480 gene product could be involved in reduction of a cysteine residue in A. vinelandii apo-CODH to facilitate Ni insertion.
The genome sequence reveals the presence of a cooA gene (Avin47010), as previously reported (98). The cooA gene encodes a transcriptional activator distantly related to the CRP family. CooA is a heme-containing homodimeric protein that functions as a CO sensor and controls the expression of the coo genes (72). In R. rubrum, CooA activates transcription of two contiguous gene clusters, namely, one that encodes CODH and accessory proteins (cooFSCTJ) and another encoding a membrane-bound CO-tolerant NiFe hydrogenase and its accessory proteins (cooMKLXUH). Although A. vinelandii lacks the genes encoding the CO-tolerant hydrogenase, it contains a gene cluster encoding a chimerical soluble NiFe hydrogenase in the corresponding locus (see below).
(ii) FDH.
FDHs combine heterogeneous groups of enzymes found in both prokaryotes and eukaryotes that catalyze the oxidation of formate to CO2 and H+. In aerobic organisms, the FDHs are mostly NAD+-dependent FDHs. Many prokaryotes, however, thrive in anoxic environments, where FDHs are NAD+-independent enzymes containing a variety of redox centers with oxygen-sensitive active sites composed of molybdenum or tungsten cofactors. These organisms utilize formate (produced from pyruvate) as a main electron donor for a variety of inducible anaerobic respiratory pathways (49).
The A. vinelandii genome contains a gene cluster that encodes a NAD+-independent FDH (fdhDGHIE; genes Avin03800 to Avin03840). While best BLAST (1) hits of fdhD are against Pseudomonas spp., including P. stutzeri A1501 (E value, 10−84), the catalytic subunit of FDH encoded by fdhGHI shows significant similarity to the alpha, beta, and gamma subunits of the well-studied nitrate-inducible Fdh-N enzyme from enterobacteria, including E. coli (85). Similarity E values against E. coli O157 were as follows: for fdhG, 0.0; for fdhH, 10−129; and for fdhI, 10−48. In the catalytic site of the alpha subunit (FdhG), the molybdopterin-guanine dinucleotide cofactor extracts electrons from formate. These electrons are transferred first to the [FeS]-containing beta subunit (FdhH) and finally to the heme-containing gamma subunit (FdhI). Although in E. coli Fdh-N function is associated with the activity of a respiratory nitrate reductase complex (NarGHI) located in the inner membrane, narGHI genes were not found in the genome of A. vinelandii. Interestingly, FdnG from Klebsiella pneumoniae was suggested to participate in relieving NifL inhibition of NifA (33), since K. pneumoniae strains carrying null mutations of fdnG (or the NADH-ubiquinone oxidoreductase genes nuoCD) showed significantly reduced nif induction under nitrogen-fixing conditions. The moaA gene (Avin03850), which encodes a SAM-dependent radical enzyme involved in the first step of the biosynthesis of the molybdenum cofactor (34), is located directly downstream of the fdhDGHIE gene cluster. Avin30330 encodes another MoaA homolog.
(iii) Hydrogenases.
Prokaryotes, mostly from anaerobic ecosystems, have the ability to use H2 by employing uptake hydrogenases or to produce H2 by the activity of H2-evolving hydrogenases. About one thousand hydrogenase sequences have been identified, many by genome sequencing, and more than 100 have been characterized genetically and/or biochemically (89). Three phylogenetically distinct classes of hydrogenases have been described, namely, [NiFe] hydrogenases, [FeFe] hydrogenases, and [Fe] hydrogenases ([Fe-S] cluster-free hydrogenases) (89, 90). Hydrogenases are usually sensitive to oxygen. For example, while [FeFe] hydrogenases are irreversibly destroyed by oxygen, the catalytic function of [NiFe] hydrogenases is reversibly inactivated by oxygen, but their structural integrity remains stable (8). The A. vinelandii genome contains genes encoding two [NiFe] hydrogenases, in membrane-bound and water-soluble forms (see below). Genes predicted to encode [FeFe] or [Fe] hydrogenases were not found.
The membrane-bound [NiFe] hydrogenase of A. vinelandii has been characterized extensively at both the biochemical (44, 75) and genetic (15, 31, 56) levels. The structural genes for this enzyme (hoxKG) (56) are clustered together with genes coding for hydrogenase accessory proteins, hoxKGZMLOQRTV (Avin50500 to Avin50590) and hypABFCDE (Avin50440 to Avin50480), encoding the components of the hydrogenase electron transport chain and proteins for the biosynthesis of the hydrogenase cofactors and hydrogenase maturation (15, 31, 56). Mutational analysis has shown the requirement of these genes for H2 oxidation activity (15, 31, 56).
It has generally been assumed that the presence of an uptake hydrogenase in nitrogen-fixing bacteria provides the advantage of recycling H2 produced by nitrogenase, thereby yielding extra reductant and/or ATP for N2 reduction and also providing respiratory protection for nitrogenase. Although competitive fitness experiments under carbon-limited conditions supported that notion for the related strain Azotobacter chroococcum (97), no significant difference was observed in ATP accumulation or in H2-dependent O2 uptake in a hoxKG-deficient mutant of A. vinelandii (52). Thus, the physiological role of the uptake hydrogenase in N2 fixation is not completely understood.
Close to hypE (Avin50440) there is a gene, Avin50400, which codes for a protein that belongs to the HupE/UreJ family of proteins involved in Ni transport or processing. Two adjacent open reading frames (Avin50410 and Avin50420), encoding putative membrane proteins, are unique to the A. vinelandii genome. Most pseudomonads possess a single copy of this membrane protein-encoding gene, but 12 copies are predicted for the A. vinelandii genome (Avin00560, Avin00570, Avin01720, Avin04580, Avin28060, Avin31780, Avin32050, Avin33410, Avin35450, Avin38580, Avin50420, and Avin50410). Whether these genes encode previously unknown accessory proteins for the A. vinelandii uptake hydrogenase and/or display an A. vinelandii-specific trait needs to be addressed.
A soluble NAD+-reducing [NiFe] hydrogenase is predicted to be encoded by Avin04360 to Avin04410. This putative multisubunit enzyme shares characteristics with both the soluble sensing hydrogenase HoxFUYHWI from Ralstonia eutropha (9) and the soluble H2-evolving, S0-reducing sulfhydrogenase ShyBCDA from Pyrococcus furiosus (54). There is weak but still significant similarity (E value, 10−4) between Avin04410 and R. eutropha hoxW (CAA63575), encoding the hydrogenase peptidase, and between Avin04370 and hoxI (NP_942732), encoding the NADPH-binding subunit (E value, 10−6) (9). These two A. vinelandii genes appear to be interspersed among the P. furiosus shyBCDA-like genes (Avin04360 and Avin04370 to Avin04390) for the hydrogenase structural genes and the accessory subunits encoded by shyBC. Thus, A. vinelandii would encode a putative soluble [NiFe] hydrogenase of chimerical nature among other previously characterized hydrogenases. A similar arrangement of genes to those of A. vinelandii is present in the genomes of other bacteria, such as the aerobic, nitrogen-fixing alphaproteobacterium Beijerinckia indica, the aerobic ammonia-oxidizing gammaproteobacterium Nitrosococcus mobilis Nb-231, and others, which have been uncovered by genome sequencing projects and represent the most similar relatives to the putative A. vinelandii soluble hydrogenase. The corresponding amino acid sequence identities to A. vinelandii shyB-hoxI-shyCDA-hoxW-like genes of these putative hydrogenase subunits are 66%, 75%, 63%, 73%, 76%, and 52% for B. indica and 58%, 60%, 65%, 71%, 65%, and 45% for N. mobilis, respectively. Interestingly, this gene cluster appears to have undergone extensive rearrangements during the evolution of hydrogenases. While this cluster is adjacent to the putative CODH gene cluster in A. vinelandii (Avin04450 to Avin04490), in N. mobilis it is adjacent to the hypBAEDCF-like genes for the maturation of the [NiFe] hydrogenase, and in B. indica they do not appear to be linked physically to hydrogen metabolism genes. Mutagenesis studies of these genes would shed light on hydrogen metabolism in A. vinelandii and other bacteria.
Alginate.
There are two possible strategies for keeping the cytoplasm anaerobic: either the bacterium can remove the oxygen after it has entered the cell, or it can create a barrier to impede O2 transfer into the cell. Sabra et al. showed that the alginate capsule of A. vinelandii was affected by the oxygen tension (73). Alginate is a linear copolymer of 1→4-linked β-d-mannuronic acid and α-l-guluronic acid, where some of the mannuronic acid residues may be acetylated. The alginate biosynthetic gene set consists of 12 genes (Avin10860 to Avin10970). The physical organization of this cluster is highly conserved in A. vinelandii and Pseudomonas species able to produce alginate, including Pseudomonas aeruginosa, Pseudomonas fluorescens, Pseudomonas syringae, Pseudomonas putida, Pseudomonas entomophila, and P. mendocina (see Fig. S7 in the supplemental material). In addition, A. vinelandii encodes a set of seven secreted mannuronan C-5 epimerases (AlgE1-7) that modify the polymer outside the cells (algE1, Avin51190; algE2, Avin51180; algE3, Avin51170; algE4, Avin51200; algE5, Avin33710; algE6, Avin51230; and algE7, Avin51250). Some of these epimerases are responsible for introducing consecutive guluronic acid residues into the polymer, and this structural feature is necessary for forming a gel with divalent cations. Close homologs of the epimerase genes algE1-7 are not described for other species except for P. syringae, which encodes a single bifunctional alginate epimerase/deacetylase, PsmE (7).
Wild-type strains of Pseudomonas usually do not produce alginate under laboratory conditions, while most strains of A. vinelandii produce alginate constitutively and use the polymer both for making the vegetative state capsule and for the cyst coat (see below). This probably explains why the regulation of alginate synthesis in A. vinelandii differs from that in Pseudomonas. Alginate production in P. aeruginosa is controlled by a complex regulatory system that includes AlgR, AlgB, AlgQ, AmrZ, and AlgP (30, 59). Orthologs of the genes encoding these regulators are present in the genome of A. vinelandii DJ, as follows: algR (Avin47610), algB (Avin11120), algP (Avin47540), algQ (Avin47550), and amrZ (Avin34410). While in Pseudomonas AlgR is required for alginate production, algR mutants of A. vinelandii produce alginate but are unable to form mature cysts (60). The participation of AlgB, AlgP, AlgQ, and AmrZ in the regulation of alginate synthesis has not been studied in A. vinelandii. Orthologs of these genes are present, however, in P. stutzeri A1501, which does not produce alginate.
Other regulators of alginate synthesis, including the algU-mucABCD operon (Avin13660 to Avin13730), the protease genes algW (Avin12950), mucP (Avin38920), and prc (Avin35170), and the diguanylate cyclase gene mucR (Avin49140), are also conserved in A. vinelandii and Pseudomonas species. A. vinelandii does not carry any close homolog to mucE, which in P. aeruginosa is necessary for the proteolytic events that degrade the anti-sigma factor MucA enabling alginate production (70). Strain DJ does not produce alginate due to the presence of an insertion sequence that splits algU (Avin13660) in two (55).
Polymer production and encystment.
A distinctive characteristic of A. vinelandii is the formation of a desiccation-resistant cell, described as a cyst, upon encountering adverse growth conditions or upon induction of vegetative cells with specific reagents. Only a few bacterial species have been observed to make cysts. Initial studies of cyst formation in Rhodospirillum centenum and Azospirillum brasiliense identified a few genes (4, 88) that are involved in the regulation of encystment in these bacteria. Genes similar to these regulators, however, have not been found in the A. vinelandii genome. Alginate, in addition to its role in respiratory protection, is a major component of the cyst capsule, and alginate with consecutive guluronic residues is essential for the formation of mature cysts (12, 80). During encystment, A. vinelandii was shown to incorporate phenolic lipids (alkylresorcinols and alkylpyrones) into the membrane. The proteins involved in the biosynthesis of these lipids have been characterized in A. vinelandii (29), and it has been shown that these lipids play a structural role in the cyst capsule, although they are not essential for desiccation resistance (78).
Even though A. vinelandii seems unable to utilize alginate as an energy source, it carries at least six enzymes with alginate lyase activities, including AlyA1-3 (Avin31810, Avin23960, and Avin13810), AlgE7 (Avin51250), Avin46500 (82; our unpublished data), and AlgL (Avin10900), the alginate lyase encoded in the alginate biosynthetic gene cluster that is also present in Pseudomonas species. One of these, AlyA3, is required for cyst germination, probably helping the germinating cells to escape from the cyst coat by depolymerizing the alginate (H. Ertesvåg, personal communication). In contrast, AlgL is not required for cyst germination (86).
Abundant PHB granules accumulate during encystment and are a major component of the central body of the cyst. Conditions or strains favoring greater polymer accumulation also produce more mature cysts (81), although PHB synthesis is not essential for encystment (76). The PHB biosynthetic genes phbB (Avin23650), phbA (Avin23640), and phaC (Avin23630), the regulatory genes phbR (Avin23660) and phbF (Avin23680), and the phasin gene phbP (Avin23670) were previously identified (66, 76, 77). The biosynthetic genes seem to be under CydR control, since the PhbB and PhbA proteins are increased in a cydR mutant (94) and a putative CydR binding site is present upstream of phbB (66). Genome analysis revealed additional genes probably involved in PHB metabolism, including genes similar to phbB and phbA and genes encoding putative phasins, PHB synthases, PHB depolymerases, and PHB oligomer hydrolases (see Table S6 in the supplemental material).
PHB is the polyhydroxyalkanoate (PHA) usually produced by A. vinelandii. However, the addition of valerate, heptanoate, or nonanoate to a culture of A. vinelandii UWD grown in glucose allows the synthesis of a copolymer, poly-(hydroxybutyrate-cohydroxyvalerate) (PHB-co-HV) (63). In the pseudomonads belonging to rRNA homology group I, an (R)-specific enoyl-coenzyme A hydratase, the product of the phaJ gene, is responsible for the channeling of β-oxidation intermediates to PHA synthesis (28). In accordance with the PHB-co-HV synthetic capacity of A. vinelandii, a phaJ gene (Avin30160) is present in its genome.
DISCUSSION
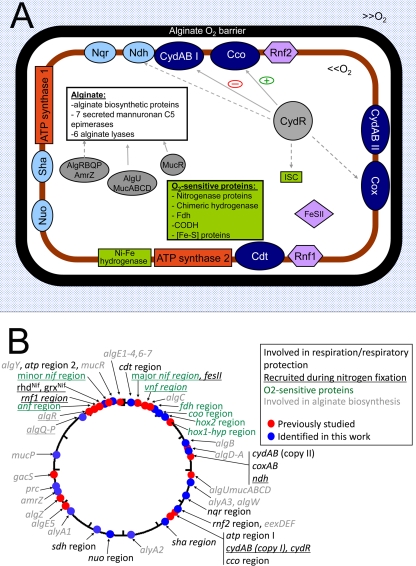
While P. stutzeri is the closest relative to A. vinelandii among fully sequenced prokaryotes, there are marked physiological and metabolic differences between these organisms. Unlike A. vinelandii, P. stutzeri can fix nitrogen only under microaerobic conditions, but in contrast to A. vinelandii, this organism can grow under anaerobic conditions by utilizing nitrate as a terminal electron acceptor. The classical high rates of respiration exhibited by A. vinelandii are apparently bestowed by the provision of five terminal oxidases (Fig. 3), supported by a number of NADH- and other substrate-driven respiratory chains. Unexpectedly, the genome encodes two cytochrome bd-type terminal oxidases. One of these, CydAB I, is the well-studied low-affinity bd oxidase known to be involved in respiratory protection of nitrogenase (68). This terminal oxidase appears to be essential for aerotolerant nitrogen fixation in A. vinelandii. The second bd-type terminal oxidase, CydAB II, resembles the cyanide-tolerant Cio terminal oxidases, which differ in their heme complement from the canonical cytochrome bd (17).
FIG. 3.
Respiratory protection in Azotobacter vinelandii. (A) Illustration of A. vinelandii cell and the key proteins involved in respiratory protection. A number of alginate biosynthetic proteins are produced that direct the formation of an alginate coating around the cell, acting as a physical O2 barrier (black lining). Respiratory proteins, including five terminal oxidases (dark blue), four NADH-ubiquinone oxidoreductases (light blue), two ATP synthases (orange), and other proteins involved in respiratory protection (lavender), provide the second line of defense against O2. Proteins sensitive to O2 exposure are shown in green boxes. Solid line arrows represent regulatory events for which experimental evidence exists. Dashed line arrows represent putative regulation on the basis of promoter binding regions. Regulatory proteins are depicted in gray. (B) Locations of respiratory and respiratory protection genes in the A. vinelandii genome. Proteins are color coded according to their role in respiration, respiratory protection, and nitrogen fixation. Genes whose protein products have been identified previously are represented by red dots. The genes identified in this work (represented by blue dots) encode the following proteins: cytochrome o oxidase (Avin11160 to Avin11180), Rnf2 complex (Avin19220 to Avin19270), Sdh complex (Avin29780 to Avin29810), Sha complex (Avin19530 to Avin19580), Nuo complex (Avin28440 to Avin28560), Nqr complex (Avin14590 to Avin14640), RhdNif (Avin51050), GrxNif (Avin51060), cytochrome c oxidase (Avin00940 to Avin01020), chimeric soluble Ni-Fe hydrogenase (Avin04360 to Avin04410), Fdh (Avin03820), CODH (Avin04450 to Avin04490), ATP synthase 2 complex (Avin52150 to Avin52230), AlgB (Avin11120), AlgQ (Avin47550), AmrZ (Avin34410), AlgP (Avin47540), AlgW (Avin12950), MucP (Avin38920), Prc (Avin35170), AlyA1-3 (Avin13810, Avin23960, and Avin31810), AlgY (Avin49400), and the alginate lyase (Avin 46500).
The distinctive ability of A. vinelandii to carry out nitrogen fixation under aerobic conditions is subject to regulation by the oxygen-responsive transcriptional regulator CydR, which like Fnr contains a [4Fe-4S]2+ cluster and negatively regulates expression of the cydAB genes that encode the low-affinity CydAB I enzyme (93). CydR also appears to control expression of the uncoupled NADH-ubiquinol dehydrogenase (Ndh) (6), which is thought to supply electrons to CydAB I oxidase and is also essential for aerotolerant nitrogen fixation (5). Our studies suggest that CydR activates expression of Cco, which may explain why CydR is required for growth under microaerobic conditions (94). Overall, CydR may have a role analogous to that of Anr in controlling the expression of terminal oxidases in Pseudomonas (87), but its involvement in the regulation of PHB metabolism (95) implies a wider role in the integration of carbon source and oxygen availability (Fig. 3).
Mechanisms for respiratory protection are complemented by the barrier to oxygen diffusion into the cell provided by the alginate capsule, which appears to play a major role in protecting nitrogenase from oxygen damage, particularly under phosphate-limiting conditions (73). It is noteworthy that alginate biosynthesis is constitutive in A. vinelandii and that a large number of secreted mannuronan C-5 epimerases are encoded in the genome by algE1-7. These enzymes may be required to increase the l-guluronic acid content of the alginate when the organism is grown at high oxygen tensions (73). Some of the six alginate lyase enzymes encoded by the genome may be responsible for altering the composition of alginate in response to oxygen availability (Fig. 3). P. stutzeri does not possess genes for alginate biosynthesis and therefore is not able to take advantage of this mechanism for reducing oxygen diffusion into the cell.
In addition to the oxygen diffusion barrier and respiratory removal of oxygen at the cell surface, a number of other strategies may be used by A. vinelandii to protect oxygen-sensitive enzymes in the cytoplasm. These include maintenance of a low redox state and efficient energy metabolism (61). In the latter context, it is interesting that A. vinelandii contains a second operon encoding an F-type ATP synthase (Avin19670 to Avin19750) that is not closely related to the ancestral operon, with its closest relative being present in the genus Burkholderia. The redundancy of oxygen protection mechanisms may explain why A. vinelandii DJ is able to fix nitrogen under standard atmospheric conditions in spite of its inability to produce alginate.
The A. vinelandii genome encodes several unexpected anaerobic enzymes in addition to the three well-characterized oxygen-sensitive nitrogenases and the membrane-bound [NiFe] hydrogenase (Fig. 3). The presence of NAD+-independent FDH is unusual for a strict aerobe that does not possess a respiratory nitrate reductase. Even more intriguing is the presence of genes predicted to encode a Ni-dependent CODH, although a homolog of the heme-containing CO sensor, CooA, has been found previously (98). These proteins are characteristic of anaerobic CO metabolism and contrast with the molybdopterin-containing CODHs found in aerobes (71). CO oxidation by A. vinelandii has not been reported previously, although it may be an important detoxification process given the CO sensitivity of hydrogenase and respiratory enzymes. The electron acceptor for CO oxidation could potentially be provided by the gene cluster encoding the soluble chimeric NiFe hydrogenase (Avin04360 to Avin04410), which intriguingly is positioned in the corresponding locus to that of the membrane-bound CO-tolerant NiFe hydrogenase in R. rubrum. It has been suggested that this energy-conserving hydrogenase produces a proton gradient to drive ATP synthesis by coupling CO-dependent H2 production and proton translocation in R. rubrum (26). Clearly, the role of CO in A. vinelandii metabolism requires further investigation.
The ability of A. vinelandii to reconcile an obligate aerobic lifestyle with the maintenance of fundamental oxygen-sensitive processes, such as nitrogen fixation, is a remarkable metabolic accomplishment that has important implications for biotechnological exploitation. A. vinelandii is a model organism for biochemical studies on the basis of the high yield and quality of enzymes that can be prepared from it. Moreover, powerful genetic approaches facilitated by homologous recombination are readily available, as are a stringent system for controlled expression of proteins encoded within the genome (38) and a means to achieve high-level protein expression (19). Given the high quality of proteins purified from A. vinelandii, these combined genetic and biochemical tools make this an ideal organism for the production of enzymes, particularly those that are oxygen sensitive.
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation grant 0523357 to D.W.W. and by M. J. Murdock Charitable Trust Life Sciences Program grants (2004262:JVZ and 2006245:JVZ) to D.W.W.
We thank Paul Rudnick, Nirav Merchant, and other contributors at the University of Arizona, the Joint Genome Institute, and the Oak Ridge National Laboratory who developed the first draft version of the A. vinelandii genome. We also thank the students at Hiram College (50 students), Virginia Tech (9 students), and Seattle Pacific University (94 students) who participated in the deep annotation of this genome as part of their undergraduate coursework.
This report is dedicated to the memory of Christina Kennedy (1945-2009). Christina made numerous contributions to our understanding of nitrogen fixation and Azotobacter biology over her nearly 4-decade-long career, culminating in the publication of its genome.
Footnotes
Published ahead of print on 8 May 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bandyopadhyay, S., S. G. Naik, I. P. O'Carroll, B. H. Huynh, D. R. Dean, M. K. Johnson, and P. C. Dos Santos. 2008. A proposed role for the Azotobacter vinelandii NfuA protein as an intermediate iron-sulfur cluster carrier. J. Biol. Chem. 28314092-14099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrios, H., B. Valderrama, and E. Morett. 1999. Compilation and analysis of sigma(54)-dependent promoter sequences. Nucleic Acids Res. 274305-4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berleman, J. E., B. M. Hasselbring, and C. E. Bauer. 2004. Hypercyst mutants in Rhodospirillum centenum identify regulatory loci involved in cyst cell differentiation. J. Bacteriol. 1865834-5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertsova, Y. V., A. V. Bogachev, and V. P. Skulachev. 2001. Noncoupled NADH:ubiquinone oxidoreductase of Azotobacter vinelandii is required for diazotrophic growth at high oxygen concentrations. J. Bacteriol. 1836869-6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertsova, Y. V., A. V. Bogachev, and V. P. Skulachev. 1998. Two NADH:ubiquinone oxidoreductases of Azotobacter vinelandii and their role in the respiratory protection. Biochim. Biophys. Acta 1363125-133. [DOI] [PubMed] [Google Scholar]
- 7.Bjerkan, T. M., C. L. Bender, H. Ertesvag, F. Drablos, M. K. Fakhr, L. A. Preston, G. Skjak-Braek, and S. Valla. 2004. The Pseudomonas syringae genome encodes a combined mannuronan C-5-epimerase and O-acetylhydrolase, which strongly enhances the predicted gel-forming properties of alginates. J. Biol. Chem. 27928920-28929. [DOI] [PubMed] [Google Scholar]
- 8.Buhrke, T., O. Lenz, N. Krauss, and B. Friedrich. 2005. Oxygen tolerance of the H2-sensing [NiFe] hydrogenase from Ralstonia eutropha H16 is based on limited access of oxygen to the active site. J. Biol. Chem. 28023791-23796. [DOI] [PubMed] [Google Scholar]
- 9.Burgdorf, T., E. van der Linden, M. Bernhard, Q. Y. Yin, J. W. Back, A. F. Hartog, A. O. Muijsers, C. G. de Koster, S. P. Albracht, and B. Friedrich. 2005. The soluble NAD+-reducing [NiFe]-hydrogenase from Ralstonia eutropha H16 consists of six subunits and can be specifically activated by NADPH. J. Bacteriol. 1873122-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burk, D. 1930. The influence of oxygen gas upon the organic catalysis of nitrogen fixation by Azotobacter. J. Phys. Chem. 341195-1209. [Google Scholar]
- 11.Bush, J. A., and P. W. Wilson. 1959. A non-gummy chromogenic strain of Azotobacter vinelandii. Nature 184381.13815135 [Google Scholar]
- 12.Campos, M., J. M. Martinez-Salazar, L. Lloret, S. Moreno, C. Nunez, G. Espin, and G. Soberon-Chavez. 1996. Characterization of the gene coding for GDP-mannose dehydrogenase (algD) from Azotobacter vinelandii. J. Bacteriol. 1781793-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castaneda, M., J. Sanchez, S. Moreno, C. Nunez, and G. Espin. 2001. The global regulators GacA and sigma(S) form part of a cascade that controls alginate production in Azotobacter vinelandii. J. Bacteriol. 1836787-6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castresana, J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17540-552. [DOI] [PubMed] [Google Scholar]
- 15.Chen, J. C., and L. E. Mortenson. 1992. Identification of six open reading frames from a region of the Azotobacter vinelandii genome likely involved in dihydrogen metabolism. Biochim. Biophys. Acta 1131199-202. [DOI] [PubMed] [Google Scholar]
- 16.Cosseau, C., and J. Batut. 2004. Genomics of the ccoNOQP-encoded cbb3 oxidase complex in bacteria. Arch. Microbiol. 18189-96. [DOI] [PubMed] [Google Scholar]
- 17.Cunningham, L., M. Pitt, and H. D. Williams. 1997. The cioAB genes from Pseudomonas aeruginosa code for a novel cyanide-insensitive terminal oxidase related to the cytochrome bd quinol oxidases. Mol. Microbiol. 24579-591. [DOI] [PubMed] [Google Scholar]
- 18.Curatti, L., C. S. Brown, P. W. Ludden, and L. M. Rubio. 2005. Genes required for rapid expression of nitrogenase activity in Azotobacter vinelandii. Proc. Natl. Acad. Sci. USA 1026291-6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dos Santos, P. C., D. C. Johnson, B. E. Ragle, M. C. Unciuleac, and D. R. Dean. 2007. Controlled expression of nif and isc iron-sulfur protein maturation components reveals target specificity and limited functional replacement between the two systems. J. Bacteriol. 1892854-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edgar, R. C. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edwards, S. E., C. S. Loder, G. Wu, H. Corker, B. W. Bainbridge, S. Hill, and R. K. Poole. 2000. Mutation of cytochrome bd quinol oxidase in reduced stationary phase survival, iron deprivation, metal toxicity, and oxidative stress in Azotobacter vinelandii. FEMS Microbiol. Lett. 18571-77. [DOI] [PubMed] [Google Scholar]
- 22.Eisen, J. A., J. F. Heidelberg, O. White, and S. L. Salzberg. 2000. Evidence for symmetric chromosomal inversions around the replication origin in bacteria. Genome Biol. 1RESEARCH0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8186-194. [PubMed] [Google Scholar]
- 24.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8175-185. [DOI] [PubMed] [Google Scholar]
- 25.Finn, R. D., J. Tate, J. Mistry, P. C. Coggill, S. J. Sammut, H. R. Hotz, G. Ceric, K. Forslund, S. R. Eddy, E. L. Sonnhammer, and A. Bateman. 2008. The Pfam protein families database. Nucleic Acids Res. 36D281-D288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fox, J. D., Y. He, D. Shelver, G. P. Roberts, and P. W. Ludden. 1996. Characterization of the region encoding the CO-induced hydrogenase of Rhodospirillum rubrum. J. Bacteriol. 1786200-6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frazzon, J., and D. R. Dean. 2002. Biosynthesis of the nitrogenase iron-molybdenum-cofactor from Azotobacter vinelandii. Met. Ions Biol. Syst. 39163-186. [PubMed] [Google Scholar]
- 28.Fukui, T., N. Shiomi, and Y. Doi. 1998. Expression and characterization of (R)-specific enoyl coenzyme A hydratase involved in polyhydroxyalkanoate biosynthesis by Aeromonas caviae. J. Bacteriol. 180667-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Funa, N., H. Ozawa, A. Hirata, and S. Horinouchi. 2006. Phenolic lipid synthesis by type III polyketide synthases is essential for cyst formation in Azotobacter vinelandii. Proc. Natl. Acad. Sci. USA 1036356-6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galindo, E., C. Pena, C. Nunez, D. Segura, and G. Espin. 2007. Molecular and bioengineering strategies to improve alginate and polydydroxyalkanoate production by Azotobacter vinelandii. Microb. Cell Fact. 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garg, R. P., A. L. Menon, K. Jacobs, R. M. Robson, and R. L. Robson. 1994. The hypE gene completes the gene cluster for H2-oxidation in Azotobacter vinelandii. J. Mol. Biol. 236390-396. [DOI] [PubMed] [Google Scholar]
- 32.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8195-202. [DOI] [PubMed] [Google Scholar]
- 33.Grabbe, R., and R. A. Schmitz. 2003. Oxygen control of nif gene expression in Klebsiella pneumoniae depends on NifL reduction at the cytoplasmic membrane by electrons derived from the reduced quinone pool. Eur. J. Biochem. 2701555-1566. [DOI] [PubMed] [Google Scholar]
- 34.Hanzelmann, P., and H. Schindelin. 2004. Crystal structure of the S-adenosylmethionine-dependent enzyme MoaA and its implications for molybdenum cofactor deficiency in humans. Proc. Natl. Acad. Sci. USA 10112870-12875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huelsenbeck, J. P., and F. Ronquist. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17754-755. [DOI] [PubMed] [Google Scholar]
- 36.Jeon, W. B., J. Cheng, and P. W. Ludden. 2001. Purification and characterization of membrane-associated CooC protein and its functional role in the insertion of nickel into carbon monoxide dehydrogenase from Rhodospirillum rubrum. J. Biol. Chem. 27638602-38609. [DOI] [PubMed] [Google Scholar]
- 37.Joerger, R. D., M. R. Jacobson, and P. E. Bishop. 1989. Two nifA-like genes required for expression of alternative nitrogenases by Azotobacter vinelandii. J. Bacteriol. 1713258-3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson, D. C., M.-C. Unciuleac, and D. R. Dean. 2006. Controlled expression and functional analysis of iron-sulfur cluster biosynthetic components within Azotobacter vinelandii. J. Bacteriol. 1887551-7561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jünemann, S., and J. M. Wrigglesworth. 1995. Cytochrome bd oxidase from Azotobacter vinelandii. J. Biol. Chem. 27016213-16220. [DOI] [PubMed] [Google Scholar]
- 40.Kelly, M. J., R. K. Poole, M. G. Yates, and C. Kennedy. 1990. Cloning and mutagenesis of genes encoding the cytochrome bd terminal oxidase complex in Azotobacter vinelandii: mutants deficient in the cytochrome d complex are unable to fix nitrogen in air. J. Bacteriol. 1726010-6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kennedy, C., P. Rudnick, T. MacDonald, and T. Melton. 2005. Genus Azotobacter, p. 384-401. In G. M. Garrity (ed.), Bergey's manual of sytematic bacteriology, vol. 2, part B. Springer-Verlag, New York, NY. [Google Scholar]
- 42.Kennedy, C., and A. Toukdarian. 1987. Genetics of azotobacters: applications to nitrogen fixation and related aspects of metabolism. Annu. Rev. Microbiol. 41227-258. [DOI] [PubMed] [Google Scholar]
- 43.Kerby, R. L., P. W. Ludden, and G. P. Roberts. 1997. In vivo nickel insertion into the carbon monoxide dehydrogenase of Rhodospirillum rubrum: molecular and physiological characterization of cooCTJ. J. Bacteriol. 1792259-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kow, Y. W., and R. H. Burris. 1984. Purification and properties of membrane-bound hydrogenase from Azotobacter vinelandii. J. Bacteriol. 159564-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kurtz, S., A. Phillippy, A. L. Delcher, M. Smoot, M. Shumway, C. Antonescu, and S. L. Salzberg. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Latreille, P., S. Norton, B. S. Goldman, J. Henkhaus, N. Miller, B. Barbazuk, H. B. Bode, C. Darby, Z. Du, S. Forst, S. Gaudriault, B. Goodner, H. Goodrich-Blair, and S. Slater. 2007. Optical mapping as a routine tool for bacterial genome sequence finishing. BMC Genomics 8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lederberg, J., and E. L. Tatum. 1946. Gene recombination in Escherichia coli. Nature 158558. [DOI] [PubMed] [Google Scholar]
- 48.Leon, R., and G. Espin. 2008. flhDC, but not fleQ, regulates flagella biogenesis in Azotobacter vinelandii, and is under AlgU and CydR negative control. Microbiology 1541719-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leopoldini, M., S. G. Chiodo, M. Toscano, and N. Russo. 2008. Reaction mechanism of molybdoenzyme formate dehydrogenase. Chemistry 148674-8681. [DOI] [PubMed] [Google Scholar]
- 50.Li, L., C. J. Stoeckert, Jr., and D. S. Roos. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 132178-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lineweaver, H., and D. Burk. 1934. The determination of enzyme dissociation constants. J. Am. Chem. Soc. 56658-666. [Google Scholar]
- 52.Linkerhagner, K., and J. Oelze. 1995. Hydrogenase does not confer significant benefits to Azotobacter vinelandii growing diazotrophically under conditions of glucose limitation. J. Bacteriol. 1776018-6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Linkerhagner, K., and J. Oelze. 1997. Nitrogenase activity and regeneration of the cellular ATP pool in Azotobacter vinelandii adapted to different oxygen concentrations. J. Bacteriol. 1791362-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ma, K., R. Weiss, and M. W. Adams. 2000. Characterization of hydrogenase II from the hyperthermophilic archaeon Pyrococcus furiosus and assessment of its role in sulfur reduction. J. Bacteriol. 1821864-1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martinez-Salazar, J. M., S. Moreno, R. Najera, J. C. Boucher, G. Espin, G. Soberon-Chavez, and V. Deretic. 1996. Characterization of the genes coding for the putative sigma factor AlgU and its regulators MucA, MucB, MucC, and MucD in Azotobacter vinelandii and evaluation of their roles in alginate biosynthesis. J. Bacteriol. 1781800-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Menon, A. L., L. E. Mortenson, and R. L. Robson. 1992. Nucleotide sequences and genetic analysis of hydrogen oxidation (hox) genes in Azotobacter vinelandii. J. Bacteriol. 1744549-4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moreno, S., R. Najera, J. Guzman, G. Soberon-Chavez, and G. Espin. 1998. Role of alternative sigma factor AlgU in encystment of Azotobacter vinelandii. J. Bacteriol. 1802766-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moshiri, F., J. Kim, C. Fu, and R. Maier. 1994. The FeSII protein of Azotobacter vinelandii is not essential for aerobic nitrogen fixation but confers significant protection to oxygen-mediated inactivation of nitrogenase in vitro and in vivo. Mol. Microbiol. 14101-114. [DOI] [PubMed] [Google Scholar]
- 59.Muhammadi, and N. Ahmed. 2007. Genetics of bacterial alginate: alginate genes distribution, organization and biosynthesis in bacteria. Curr. Genomics 8191-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nunez, C., S. Moreno, G. Soberon-Chavez, and G. Espin. 1999. The Azotobacter vinelandii response regulator AlgR is essential for cyst formation. J. Bacteriol. 181141-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oelze, J. 2000. Respiratory protection of nitrogenase in Azotobacter species: is a widely held hypothesis unequivocally supported by experimental evidence? FEMS Microbiol. Rev. 24321-333. [DOI] [PubMed] [Google Scholar]
- 62.Ortiz, P. J., and S. Ochoa. 1959. Studies on polynucleotides synthesized by polynucleotide phosphorylase. IV. P32-labeled ribonucleic acid. J. Biol. Chem. 2341208-1212. [PubMed] [Google Scholar]
- 63.Page, W. J., J. Manchak, and B. Rudy. 1992. Formation of poly(hydroxybutyrate-co-hydroxyvalerate) by Azotobacter vinelandii UWD. Appl. Environ. Microbiol. 582866-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Page, W. J., A. Tindale, M. Chandra, and E. Kwon. 2001. Alginate formation in Azotobacter vinelandii UWD during stationary phase and the turnover of poly-beta-hydroxybutyrate. Microbiology 147483-490. [DOI] [PubMed] [Google Scholar]
- 65.Pau, R. N., and D. M. Lawson. 2002. Transport, homeostasis, regulation, and binding of molybdate and tungstate to proteins. Met. Ions Biol. Syst. 3931-74. [PubMed] [Google Scholar]
- 66.Peralta-Gil, M., D. Segura, J. Guzman, L. Servin-Gonzalez, and G. Espin. 2002. Expression of the Azotobacter vinelandii poly-beta-hydroxybutyrate biosynthetic phbBAC operon is driven by two overlapping promoters and is dependent on the transcriptional activator PhbR. J. Bacteriol. 1845672-5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pettinari, M. J., G. J. Vazquez, D. Silberschmidt, B. Rehm, A. Steinbuchel, and B. S. Mendez. 2001. Poly(3-hydroxybutyrate) synthesis genes in Azotobacter sp. strain FA8. Appl. Environ. Microbiol. 675331-5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Poole, R. K., and S. Hill. 1997. Respiratory protection of nitrogenase activity in Azotobacter vinelandii—roles of the terminal oxidases. Biosci. Rep. 17303-317. [DOI] [PubMed] [Google Scholar]
- 69.Premakumar, R., T. M. Loveless, and P. E. Bishop. 1994. Effect of amino acid substitutions in a potential metal-binding site of AnfA on expression from the anfH promoter in Azotobacter vinelandii. J. Bacteriol. 1766139-6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Qiu, D., V. M. Eisinger, D. W. Rowen, and H. D. Yu. 2007. Regulated proteolysis controls mucoid conversion in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 1048107-8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ragsdale, S. W. 2004. Life with carbon monoxide. Crit. Rev. Biochem. Mol. Biol. 39165-195. [DOI] [PubMed] [Google Scholar]
- 72.Roberts, G. P., R. L. Kerby, H. Youn, and M. Conrad. 2005. CooA, a paradigm for gas sensing regulatory proteins. J. Inorg. Biochem. 99280-292. [DOI] [PubMed] [Google Scholar]
- 73.Sabra, W., A. P. Zeng, H. Lunsdorf, and W. D. Deckwer. 2000. Effect of oxygen on formation and structure of Azotobacter vinelandii alginate and its role in protecting nitrogenase. Appl. Environ. Microbiol. 664037-4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sadoff, H. L. 1975. Encystment and germination in Azotobacter vinelandii. Bacteriol. Rev. 39516-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Seefeldt, L. C., and D. J. Arp. 1986. Purification to homogeneity of Azotobacter vinelandii hydrogenase: a nickel and iron containing alpha beta dimer. Biochimie 6825-34. [DOI] [PubMed] [Google Scholar]
- 76.Segura, D., T. Cruz, and G. Espin. 2003. Encystment and alkylresorcinol production by Azotobacter vinelandii strains impaired in poly-beta-hydroxybutyrate synthesis. Arch. Microbiol. 179437-443. [DOI] [PubMed] [Google Scholar]
- 77.Segura, D., E. Vargas, and G. Espin. 2000. Beta-ketothiolase genes in Azotobacter vinelandii. Gene 260113-120. [DOI] [PubMed] [Google Scholar]
- 78.Segura, D., O. Vite, Y. Romero, S. Moreno, M. Castaneda, and G. Espin. 2009. Isolation and characterization of Azotobacter vinelandii mutants impaired in alkylresorcinol synthesis: alkylresorcinols are not essential for cyst desiccation resistance. J. Bacteriol. 1913142-3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stamatakis, A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 222688-2690. [DOI] [PubMed] [Google Scholar]
- 80.Steigedal, M., H. Sletta, S. Moreno, M. Maerk, B. E. Christensen, T. Bjerkan, T. E. Ellingsen, G. Espin, H. Ertesvag, and S. Valla. 2008. The Azotobacter vinelandii AlgE mannuronan C-5-epimerase family is essential for the in vivo control of alginate monomer composition and for functional cyst formation. Environ. Microbiol. 101760-1770. [DOI] [PubMed] [Google Scholar]
- 81.Stevenson, L. H., and M. D. Socolofsky. 1966. Cyst formation and poly-beta-hydroxybutyric acid accumulation in Azotobacter. J. Bacteriol. 91304-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Svanem, B. I., W. I. Strand, H. Ertesvag, G. Skjak-Braek, M. Hartmann, T. Barbeyron, and S. Valla. 2001. The catalytic activities of the bifunctional Azotobacter vinelandii mannuronan C-5-epimerase and alginate lyase AlgE7 probably originate from the same active site in the enzyme. J. Biol. Chem. 27631542-31550. [DOI] [PubMed] [Google Scholar]
- 83.Thorneley, R. N., and G. A. Ashby. 1989. Oxidation of nitrogenase iron protein by dioxygen without inactivation could contribute to high respiration rates of Azotobacter species and facilitate nitrogen fixation in other aerobic environments. Biochem. J. 261181-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Thorneley, R. N., and D. J. Lowe. 1983. Nitrogenase of Klebsiella pneumoniae. Kinetics of the dissociation of oxidized iron protein from molybdenum-iron protein: identification of the rate-limiting step for substrate reduction. Biochem. J. 215393-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tishkov, V. I., and V. O. Popov. 2004. Catalytic mechanism and application of formate dehydrogenase. Biochemistry (Moscow) 691252-1267. [DOI] [PubMed] [Google Scholar]
- 86.Trujillo-Roldan, M. A., S. Moreno, D. Segura, E. Galindo, and G. Espin. 2003. Alginate production by an Azotobacter vinelandii mutant unable to produce alginate lyase. Appl. Microbiol. Biotechnol. 60733-737. [DOI] [PubMed] [Google Scholar]
- 87.Ugidos, A., G. Morales, E. Rial, H. D. Williams, and F. Rojo. 2008. The coordinate regulation of multiple terminal oxidases by the Pseudomonas putida ANR global regulator. Environ. Microbiol. 101690-1702. [DOI] [PubMed] [Google Scholar]
- 88.Valverde, A., Y. Okon, and S. Burdman. 2006. cDNA-AFLP reveals differentially expressed genes related to cell aggregation of Azospirillum brasilense. FEMS Microbiol. Lett. 265186-194. [DOI] [PubMed] [Google Scholar]
- 89.Vignais, P. M. 2008. Hydrogenases and H(+)-reduction in primary energy conservation. Results Probl. Cell Differ. 45223-252. [DOI] [PubMed] [Google Scholar]
- 90.Vignais, P. M., B. Billoud, and J. Meyer. 2001. Classification and phylogeny of hydrogenases. FEMS Microbiol. Rev. 25455-501. [DOI] [PubMed] [Google Scholar]
- 91.Wattt, R. K., and P. W. Ludden. 1999. Nickel-binding proteins. Cell. Mol. Life Sci. 56604-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wilson, P. W., and S. G. Knight. 1952. Experiments in bacterial physiology. Burgess Publishing Co., Minneapolis, MN.
- 93.Wu, G., H. Cruz-Ramos, S. Hill, J. Green, G. Sawers, and R. K. Poole. 2000. Regulation of cytochrome bd expression in the obligate aerobe Azotobacter vinelandii by CydR (Fnr). Sensitivity to oxygen, reactive oxygen species, and nitric oxide. J. Biol. Chem. 2754679-4686. [DOI] [PubMed] [Google Scholar]
- 94.Wu, G., S. Hill, M. J. Kelly, G. Sawers, and R. K. Poole. 1997. The cydR gene product, required for regulation of cytochrome bd expression in the obligate aerobe Azotobacter vinelandii, is an Fnr-like protein. Microbiology 1432197-2207. [DOI] [PubMed] [Google Scholar]
- 95.Wu, G., A. J. Moir, G. Sawers, S. Hill, and R. K. Poole. 2001. Biosynthesis of poly-beta-hydroxybutyrate (PHB) is controlled by CydR (Fnr) in the obligate aerobe Azotobacter vinelandii. FEMS Microbiol. Lett. 194215-220. [DOI] [PubMed] [Google Scholar]
- 96.Yan, Y., J. Yang, Y. Dou, M. Chen, S. Ping, J. Peng, W. Lu, W. Zhang, Z. Yao, H. Li, W. Liu, S. He, L. Geng, X. Zhang, F. Yang, H. Yu, Y. Zhan, D. Li, Z. Lin, Y. Wang, C. Elmerich, M. Lin, and Q. Jin. 2008. Nitrogen fixation island and rhizosphere competence traits in the genome of root-associated Pseudomonas stutzeri A1501. Proc. Natl. Acad. Sci. USA 1057564-7569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yates, M. G., and F. O. Campbell. 1989. The effect of nutrient limitation on the competition between H2-uptake hydrogenase positive (Hup+) recombinant strain of Azotobacter chroococcum and the Hup− mutant parent in mixed populations. J. Gen. Microbiol. 135221-226. [Google Scholar]
- 98.Youn, H., R. L. Kerby, M. Conrad, and G. P. Roberts. 2004. Functionally critical elements of CooA-related CO sensors. J. Bacteriol. 1861320-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.