Abstract
Burkholderia spp. that degrade phthalate have an ABC transporter-type phthalate transport system (OphFGH) and a specific porin (OphP) in addition to a permease-type phthalate transporter (OphD). OphFGH has a lower Km and higher Vmax than OphD, which affects how the bacteria grow. OphP is involved in both mechanisms of transport.
The ophD gene, encoding a permease-type transporter for phthalate, was cloned from Burkholderia multivorans ATCC 17616 (formerly B. cepacia ATCC 17616) (3). An ophD knockout mutant of strain ATCC 17616 grows slightly slower on phthalate but is still able to take up phthalate at a rate equivalent to that of the wild-type strain. This implies that strain ATCC 17616 must have a second phthalate-inducible phthalate uptake system. Another closely related phthalate-degrading strain, B. cepacia DBO1, carries a dysfunctional ophD gene and is able to transport phthalate at a very high rate (3). This also implies that there are two phthalate transport systems in Burkholderia spp.
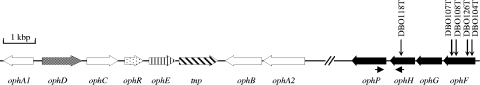
In order to locate the second phthalate transport system, random plasposon mutagenesis of B. cepacia DBO1 was performed by introducing pTnMod-Okm (8) into cells by triparental mating as described previously (2). Kanamycin-resistant transformants were screened for growth on phthalate as the sole source of carbon and energy on mineral salts basal medium (11). Eleven mutants that cannot grow on phthalate were obtained. Sequencing analysis revealed that four mutants (DBO104T, DBO107T, DBO108T, and DBO126T) have plasposon insertions in a gene encoding a substrate binding protein (SBP) of a putative ABC transporter system, and one mutant (DBO118T) has a plasposon insertion in the nucleotide binding domain (NBD) gene of the same ABC transporter system (Fig. 1). A third gene encoding a transmembrane domain (TMD) protein is located between the SBP and NBD genes. The SBP, TMD, and NBD genes were designated ophF, ophG, and ophH. A porin gene is present in the area downstream of the ophH gene and was designated ophP. The lengths of the ophFGHP genes are 984, 813, 783, and 1,065 nucleotides, respectively. OphF does not show high levels of similarity to other SBPs in the database. The highest score is 28% identity and 41% similarity to the SBP of an ABC-type transporter (accession no. ABH04832) from Heliobacillus mobilis. OphG shows 31% identity and 52% similarity to the inner membrane subunit of an ABC transporter (accession no. ABE33631) from B. xenovorans LB400. OphH shows 47% identity and 63% similarity to the NBD subunit of an ABC transporter (accession no. ABS28049) from Anaeromyxobacter sp. strain Fw109-5. OphP shows 77% identity and 86% similarity to a porin (accession no. EDN40637) from Ralstonia pickettii 12D. OphP belongs to the general bacterial porin family (TC no. 1.B.1) according to the transporter classification system of Saier et al.(10).
FIG. 1.
Map of the oph operons of B. cepacia DBO1, B. multivorans ATCC 17616, and B. vietnamiensis G4. The plasposon insertion sites of the phthalate-degrading mutants of B. cepacia DBO1 are indicated by arrows. There is a frameshift mutation in the ophD gene, which encodes the permease-type phthalate transporter, in strain DBO1. OphF (SBP), OphG (TMD), and OphH (NBD) constitute an ABC-type phthalate transporter. OphP is a phthalate-specific porin, which works with both phthalate transport systems. The nucleotide sequences of the ophFGHP genes of strains DBO1 (accession no. FJ790778), ATCC 17616 (accession no. BAG45600 to BAG45603), and G4 (accession no. ABO57274 to ABO57277) are 100% identical except for a silent mutation in the ophP gene of G4. The ophFGHP genes are located 17.3 kb and 21.9 kb from the other oph genes in strain ATCC 17616 (accession no. BAG45576 to BAG45583) and G4 (accession no. ABO57246 to ABO57253), respectively. The other oph genes are ophA1 (phthalate dioxygenase reductase), ophA2 (phthalate dioxygenase), ophB (4,5-dihydro-4,5-diohydroxyphthalate dehydrogenase), ophC (4,5-dihydroxyphthalate decarboxylase), ophE (quinolinate phosphoribosyl transferase), and ophR (regulator).
The ophFGHP genes were shown to be present in B. multivorans ATCC 17616 and B. vietnamiensis G4 by using PCR primers based on the sequence of the ophFGHP genes (data not shown). The genome sequences show that strains ATCC 17616 (accession no. AP009386) and G4 (accession no. CP000615) have not only the ophFGHP genes but also an intact ophD gene. It is likely that both phthalate transport systems are functional in strains ATCC 17616 and G4. Strains DBO1 and ATCC 17616 were cultured on mineral salts basal medium containing phthalate, 4-hydroxybenzoate, or succinate. RNA, extracted from cells in the mid-log phase using an RNeasy mini kit (Qiagen), was used as the template for reverse transcription-PCRs (RT-PCRs) (Qiagen OneStep RT-PCR kit). The data show that the basal levels of expression of the ophA2, ophD, ophFGH, and ophP genes are low when the bacteria are cultured on 4-hydroxybenzoate or succinate (data not shown). The data additionally show that the ophP gene is cotranscribed with the ophFGH genes because a 0.5-kb PCR product was obtained using primers located inside the ophH and ophP genes (the locations of the primers are shown in Fig. 1). The relative quantification data obtained from real-time PCR show that the levels of expression of the ophD, ophA2, and ophP genes are 80 ± 47, 41 ± 2, and 237 ± 4 times greater when strain ATCC 17616 is cultured on phthalate instead of succinate (the rpoB gene was used as a control gene). The ratio of expression of the ophD, ophA2, and ophP genes is 0.8 ± 0.4 to 0.7 ± 0.2 to 2.0 ± 1.3 when bacteria grown on 4-hydroxybenzoate are compared to bacteria grown on succinate. Transcription of the genes for both types of phthalate transport systems is thus induced in the presence of phthalate.
An association between ABC transporter systems and specific porins has been observed previously. For example, BtuFCD (ABC transporter) and BtuB (porin) are transporters for the uptake of vitamin B12 (1), and GanFGK2 (ABC transporter) and GanL (porin) are transporters for galactan (6). The permease-type transporters for aromatic compounds are more often accompanied by nearby specific porins. For example, PhaJ (permease) and PhaK (porin) from Psedomonas putida U are essential for the uptake of phenylacetate (9). Disruption of either phaJ or phaK resulted in an inability of the mutants to utilize phenylacetate. BenP (porin) and BenK (permease) from Acinetobacter sp. strain ADP1 were proposed to play a role in the transport of aromatic compounds since the benPK operon was regulated in concert with other genes in the regulon (5). However, disruption of benP did not result in an obvious phenotype of the mutant strain under the laboratory conditions tested.
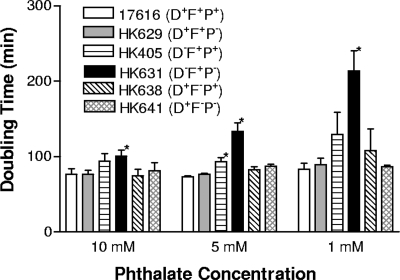
B. multivorans ATCC 17616 was chosen for further study because both of its phthalate transport systems are functional. Several B. multivorans ATCC 17616 mutants were created by allelic exchange mutagenesis, including HK405 (ΔophD), HK629 (ΔophP), HK631 (ΔophD ΔophP), HK638 (ΔophF), HK641 (ΔophF ΔophP), and HK642 (ΔophD ΔophF). The Kmr cassette from p34S-Km and the Tpr cassette from p34S-Tp2 were used to replace the target genes (7). All of the mutant strains can still grow on phthalate, except for HK642, which has lost both phthalate transport systems. The growth rates of these mutants were compared using basal medium (13) containing 10 mM, 5 mM, or 1 mM phthalate (Fig. 2). The average doubling time of the wild-type strain increased slightly from 76.0 ± 7.5 min to 83.4 ± 7.7 min when the phthalate concentration was decreased from 10 mM to 1 mM. The doubling times of mutant strains HK629, HK638, and HK641, each with an intact ophD gene, were about the same as that of the wild-type strain. However, HK405 and HK631, both with a disrupted ophD gene, grew slower than the wide-type strain. The doubling time of HK405 increased from 93.9 ± 10.2 min to 129.4 ± 29.4 min and the doubling time of HK631 increased from 100.8 ± 8.1 min to 213 ± 26.8 min when the phthalate concentration was decreased from 10 mM to 1 mM. RT-PCR showed that transcription of the ophC gene (downstream of the ophD gene) in mutants HK405 and HK631 is not affected by the ophD knockout (data not shown). This suggests that the increases in the doubling times of HK405 and HK631 were not due to a polar effect on OphC. The data imply that the OphD transport system is more important than the OphFGH transport system for growth of the bacteria with the substrate concentrations tested. The data also show that disruption of OphP affects bacterial growth only when OphD is also disrupted, as observed for mutant HK631. The effect was more obvious at low phthalate concentrations. As seen in other cases, the contribution of an individual specific porin sometimes could not be seen in nutrient-sufficient environments, as other porins present in the outer membrane could serve as nonspecific diffusion channels (12).
FIG. 2.
Doubling times of B. multivorans ATCC 17616 and mutants cultured on basal medium containing 1 mM, 5 mM, or 10 mM phthalate. Wild-type strain ATCC 17616, HK629 (ΔophP), HK405 (ΔophD), HK631 (ΔophD ΔophP), HK638 (ΔophF), and HK641 (ΔophF ΔophP) were tested. An asterisk indicates a statistically significant difference from the wild-type strain as determined by a t test.
The abilities of the mutant strains to transport phthalate at different concentrations were compared using established methods described in previous studies (3, 4). The initial rate of phthalate transport was determined at phthalate concentrations ranging from 1 to 26.5 μM and used to calculate the apparent Km and Vmax, assuming Michaelis-Menten kinetics (Table 1). The estimated Km values for the wild-type strain and HK629, which have both transport systems, are the lowest Km values. The Km values for HK405 and HK631, which have only the OphFGH transport system, are lower than those for HK638 and HK641, which have only the OphD transport system. The data suggest that the OphFGH transport system is more important when the phthalate concentration in the environment is low because of its higher affinity for the substrate. The Vmax values for the strains with the OphFGH transport system are higher than those for the strains with the OphD transport system. HK405, with the OphFGH system and OphP, has the highest Vmax, which is almost twice the Vmax of the wild-type strain. The presence of OphP does not affect the Km values for both systems significantly, but it does increase the Vmax values. This indicates that OphP plays a role in accelerating phthalate transport. It is surprising that the Vmax for HK629, which has both transport systems and no OphP, is lower than the Vmax values for HK631 and HK641, which have a single transport system and no OphP. It is not known if there is an interaction between the two transport systems that interferes with the transport process. Further investigation is needed to interpret this curious phenomenon.
TABLE 1.
Apparent Km and Vmax values for phthalate transport
| Strain | Genotype
|
Km (μM) | Vmax (nmol phthalate/min/mg protein) | ||
|---|---|---|---|---|---|
| ophD | ophF | ophP | |||
| ATCC 17616 | + | + | + | 9.6 ± 2.2 | 28.3 ± 6.1 |
| HK629 | + | + | − | 9.9 ± 0.7 | 3.1 ± 0.4 |
| HK405 | − | + | + | 16.5 ± 2.9 | 52.9 ± 5.5 |
| HK631 | − | + | − | 17.8 ± 3.8 | 31.2 ± 5.6 |
| HK638 | + | − | + | 28.3 ± 8.6 | 19.0 ± 4.2 |
| HK641 | + | − | − | 26.8 ± 10.0 | 9.4 ± 3.0 |
A number of compounds, including phthalate with either a hydroxyl, methyl, or chloro group substitution at the 4 position, were tested to determine their abilities to inhibit phthalate transport by mutants HK638 (with functional OphD and OphP) and HK405 (with functional OphFGH and OphP) (Table 2). The different profiles for substrate inhibition of phthalate transport for HK638 and HK405 indicate that the substrate specificities of the two phthalate transport systems are different. The OphFGH system has a narrower substrate range than the OphD system because 4-methylphthalate, quinolinate, and cinchomeronate could inhibit phthalate transport by HK638 but not phthalate transport by HK405. The profiles for substrate inhibition of phthalate transport for an Escherichia coli strain expressing OphD and HK638 with OphD and OphP are also different. All of the substrates except 4-methylphthalate and cinchomeronate have different abilities to inhibit phthalate transport by the E. coli strain expressing OphD and HK638. The differences could be due to the presence of OphP in HK638. Altogether, the two phthalate transport systems appear to function differently in the physiology of phthalate metabolism. The wild-type strain, having both transport systems, has the highest affinity for phthalate and a slightly compromised rate of phthalate transport. But it has the benefit of fast growth when there is plenty of substrate in the environment and a greater ability to transport phthalate into the cells when the substrate is less available.
TABLE 2.
Substrate inhibition of phthalate uptake
| Competing substrate | % Inhibition of phthalate uptakea
|
||
|---|---|---|---|
| E. coli (OphD)b | HK638 (OphD+ OphF− OphP+) | HK405 (OphD− OphF+ OphP+) | |
| Phthalate | 91 ± 1 | >95 | >95 |
| 4-Hydroxyphthalate | 86 ± 2 | >95 | 34 ± 13 |
| 4-Chlorophthalate | 66 ± 4 | >95 | >95 |
| 4-Methylphthalate | 32 ± 4 | 31 ± 7 | <5 |
| Quinolinate | <10 | 20 ± 7 | <5 |
| Cinchomeronate | 27 ± 6 | 28 ± 9 | <5 |
| Salicylate | <10 | 51 ± 7 | >95 |
Inhibition of phthalate uptake was calculated as follows: % inhibition = [100 − (14C-labeled phthalate transport rate with competing substrate/14C-labeled phthalate transport rate without competing substrate)] × 100. The concentrations of 14C-labeled phthalate and competing substrate used were 50 μM and 1 mM, respectively (3).
Data from reference 3.
Acknowledgments
We thank K. Lam for isolating and studying the mutant strains. We thank L. Seliger and M. M. Murillo for sequencing and PCR facility maintenance.
Footnotes
Published ahead of print on 8 May 2009.
REFERENCES
- 1.Cadieux, N., C. Bradbeer, E. Reeger-Schneider, W. Koster, A. K. Mohanty, M. C. Wiener, and R. J. Kadner. 2002. Identification of the periplasmic cobalamin-binding protein BtuF of Escherichia coli. J. Bacteriol. 184706-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang, H. K., P. Mohseni, and G. J. Zylstra. 2003. Characterization and regulation of the genes for a novel anthranilate 1,2-dioxygenase from Burkholderia cepacia DBO1. J. Bacteriol. 1855871-5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang, H. K., and G. J. Zylstra. 1999. Characterization of the phthalate permease OphD from Burkholderia cepacia ATCC 17616. J. Bacteriol. 1816197-6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, H. K., and G. J. Zylstra. 1998. Novel organization of the genes for phthalate degradation from Burkholderia cepacia DBO1. J. Bacteriol. 1806529-6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark, T. J., C. Momany, and E. L. Neidle. 2002. The benPK operon, proposed to play a role in transport, is part of a regulon for benzoate catabolism in Acinetobacter sp. strain ADP1. Microbiology 1481213-1223. [DOI] [PubMed] [Google Scholar]
- 6.Delangle, A., A. F. Prouvost, V. Cogez, J. P. Bohin, J. M. Lacroix, and N. H. Cotte-Pattat. 2007. Characterization of the Erwinia chrysanthemi Gan locus, involved in galactan catabolism. J. Bacteriol. 1897053-7061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dennis, J. J., and G. J. Zylstra. 1998. Improved antibiotic-resistance cassettes through restriction site elimination using Pfu DNA polymerase PCR. BioTechniques 25772-774, 776. [DOI] [PubMed] [Google Scholar]
- 8.Dennis, J. J., and G. J. Zylstra. 1998. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl. Environ. Microbiol. 642710-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olivera, E. R., B. Minambres, B. Garcia, C. Muniz, M. A. Moreno, A. Ferrandez, E. Diaz, J. L. Garcia, and J. M. Luengo. 1998. Molecular characterization of the phenylacetic acid catabolic pathway in Pseudomonas putida U: the phenylacetyl-CoA catabolon. Proc. Natl. Acad. Sci. USA 956419-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saier, M. H., Jr., C. V. Tran, and R. D. Barabote. 2006. TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 34D181-D186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stanier, R. Y., N. J. Palleroni, and M. Duodoroff. 1966. The aerobic pseudomonads: a taxonomic study. J. Gen. Microbiol. 43159-271. [DOI] [PubMed] [Google Scholar]
- 12.Tamber, S., M. M. Ochs, and R. E. Hancock. 2006. Role of the novel OprD family of porins in nutrient uptake in Pseudomonas aeruginosa. J. Bacteriol. 18845-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zylstra, G. J., R. H. Olsen, and D. P. Ballou. 1989. Cloning, expression, and regulation of the Pseudomonas cepacia protocatechuate 3,4-dioxygenase genes. J. Bacteriol. 1715907-5914. [DOI] [PMC free article] [PubMed] [Google Scholar]