Abstract
Nosocomial infections caused by antibiotic-resistant Klebsiella pneumoniae are emerging as a major health problem worldwide, while community-acquired K. pneumoniae infections present with a range of diverse clinical pictures in different geographic areas. In particular, an invasive form of K. pneumoniae that causes liver abscesses was first observed in Asia and then was found worldwide. We are interested in how differences in gene content of the same species result in different diseases. Thus, we sequenced the whole genome of K. pneumoniae NTUH-K2044, which was isolated from a patient with liver abscess and meningitis, and analyzed differences compared to strain MGH 78578, which was isolated from a patient with pneumonia. Six major types of differences were found in gene clusters that included an integrative and conjugative element, clusters involved in citrate fermentation, lipopolysaccharide synthesis, and capsular polysaccharide synthesis, phage-related insertions, and a cluster containing fimbria-related genes. We also conducted comparative genomic hybridization with 15 K. pneumoniae isolates obtained from community-acquired or nosocomial infections using tiling probes for the NTUH-K2044 genome. Hierarchical clustering revealed three major groups of genomic insertion-deletion patterns that correlate with the strains' clinical features, antimicrobial susceptibilities, and virulence phenotypes with mice. Here we report the whole-genome sequence of K. pneumoniae NTUH-K2044 and describe evidence showing significant genomic diversity and sequence acquisition among K. pneumoniae pathogenic strains. Our findings support the hypothesis that these factors are responsible for the changes that have occurred in the disease profile over time.
Klebsiella pneumoniae is a gram-negative bacterium that belongs to the gamma subdivision of the class Proteobacteria and exhibits relatively close genetic relatedness to other genera of the Enterobacteriaceae, including Escherichia, Salmonella, Shigella, and Yersinia (2). The conspicuous difference between K. pneumoniae and the other enterobacteria is the presence of a thick polysaccharide capsule, which is thought to be a significant virulence factor and to help the bacterium avoid phagocytosis (13). Infections caused by K. pneumoniae are seen throughout the world. This organism is a major cause of urinary tract infection and an important source of nosocomial infection (39). Moreover, K. pneumoniae is emerging worldwide as a major cause of bacteremia and drug-resistant infections (25, 38).
The clinical pattern of K. pneumoniae infection in humans has changed since this organism was discovered (19, 20) more than 100 years ago. Until the 1960s, K. pneumoniae was an important cause of community-acquired pneumonia in the United States (8) and elsewhere. However, the incidence of this type of infection has dropped to 1 to 3% in the United States and Europe, and hospital-acquired K. pneumoniae infection now predominates (22, 39, 48). The global pattern of community-acquired K. pneumoniae bacteremia varies with geographical area (25). In the United States, Europe, Australia, and Argentina, this condition is associated with urinary tract infection, vascular catheters, and cholangitis. In Asia and South Africa, classic K. pneumoniae pneumonia still exists (25) and has remained important over the past two decades. At the same time, an invasive form of K. pneumoniae infection, which presents as primary bacteremic liver abscesses, endophthalmitis, and meningitis, has been reported almost exclusively in Asia (21), especially in Taiwan (47, 50). Although the reasons for the preponderance of this severe invasive K. pneumoniae infection in Asia are unknown, they are likely to involve both host and microbial factors.
Recent studies by several groups have investigated and debated the major virulence factors of K. pneumoniae, including the magA (16) and rmpA (53) genes, capsular serotype K1 or K2 (11, 52), and even hypermucoviscosity (16, 53). In principle, other determinants may also contribute to pyogenic K. pneumoniae infection. To gather sufficient DNA sequence information for a systematic analysis of the genetic features that underlie the diverse clinical manifestations of K. pneumoniae infections, we undertook complete genome sequencing of a pathogenic strain, NTUH-K2044, which had been isolated from a Taiwanese liver abscess case (16). NTUH-K2044 is an appropriate strain because it possesses the magA and rmpA genes, belongs to capsular serotype K1, and has high virulence and hypermucoviscosity; these factors make this isolate very suitable as a model strain for genomic studies. We additionally used a genomic shotgun array (GSA) protocol developed in our laboratory (27) to compare the genome contents of NTUH-K2044 and multiple clinical isolates. The microarray data allowed us to examine the genome evolution of K. pneumoniae and to relate the various genomic signatures to the clinical patterns seen in K. pneumoniae infections.
MATERIALS AND METHODS
Bacteriological studies.
Clinical K. pneumoniae isolates were collected in the National Taiwan University Hospital (Taipei, Taiwan) and the National Cheng Kung University Hospital (Tainan, Taiwan). Each isolate was subcultured on 5% sheep blood agar and MacConkey agar plates (BBL, Becton Dickinson Microbiology Systems, Cockeysville, MD) to check its purity. Species identification was carried out by using a combination of standard conventional biochemical tests (BBL) (34) and Vitek Plus gram-negative identification cards (bioMérieux Vitek, Hazelwood, MO). MICs were determined using the broth microdilution method by following the Clinical and Laboratory Standards Institute guidelines (see the supplemental material). For virulence testing, groups of three female C57BL/6 mice (Charles River Japan, Inc., Atsugi, Japan) were given intraperitoneal injections consisting of 100 μl of bacteria diluted in saline, and the 100% lethal dose for mice was determined for each isolate. The animals were observed daily for 7 days. Mice that survived for more than 7 days were observed for another month to confirm that they survived after challenge.
Genome sequencing.
The complete sequence of NTUH-K2044 was determined by using a whole-genome shotgun approach (17, 18) and a combination of genomic libraries with two small inserts (1 to 4 kb and 2 to 3 kb). An additional 30- to 40-kb large-insert library was constructed using a CopyControl fosmid library production kit (Epicentre Biotechnologies, Madison, WI). The NTUH-K2044 genome was sequenced with 10-fold coverage using ABI3730xl automated capillary electrophoresis sequencers (Applied Biosystems, Foster City, CA) and was assembled using the Phred/Phrap/Consed software package (15, 23). Sequence gaps between contigs were closed by primer walking with linking clones and by sequencing PCR products of genomic DNA or specific fosmid clones. Low-quality reads or regions were eliminated by resequencing specific clones or by sequencing specific PCR products. The final sequencing error rates for the chromosome and plasmid were estimated to be 0.0016 and 0.0059 per 10 kb, respectively. To validate the finished chromosomal sequence, digestion with the I-CeuI enzyme, which recognizes the rRNA operons, was performed.
Genome annotation.
The protein-encoding genes were predicted by using Glimmer 2.13 (12), GeneMark 2.4 (3), and GeneMark.hmm 2.1 (30), and the genes encoding at least 30 amino acids long were included. The name, description, probable function, and COG group of each predicted gene were assigned based on the results of BLASTP (E-value, <10−5; identity, >30%; matched length, >30%) using the RefSeq.microbial, nonredundant protein, and COG databases of NCBI (http://www.ncbi.nlm.nih.gov) and subsequent manual inspections. Ribosomal binding sites were located using RBSfinder (http://www.tigr.org). tRNAs were predicted by using tRNAscan-SE (29). rRNAs were identified by BLASTN analysis with known K. pneumoniae rRNA sequences.
Microarray analysis.
The methods used for preparation and fabrication of PCR products of the NTUH-K2044 genomic DNA on slides have been described previously (27), and details are provided in the supplemental material.
Resequencing by the 454 method.
Genome sequencing of the NK5 strain was performed using methods that have been described previously (31). Genomic DNA (5 μg) was used for the library preparation and titration steps. Following an emulsion PCR and two sequencing runs with a GS20 instrument (454 Life Sciences Corporation, Branford, CT), 586,208 reads with average size of 100 bp were obtained, and the random sequences were assembled using the Newbler software provided by the manufacturer.
Nucleotide sequence accession numbers.
The K. pneumoniae NTUH-K2044 chromosome and plasmid sequences have been deposited in the DDBJ database (http://www.ddbj.nig.ac.jp/index-e.html) under accession numbers AP006725 and AP006726, respectively.
RESULTS
General features of the NTUH-K2044 genome.
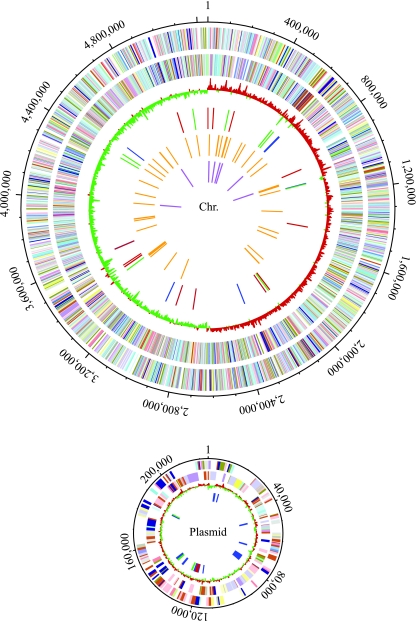
A clinical isolate, NTUH-K2044 (16), was selected for whole-genome sequencing. This strain came from the blood of a previously healthy individual who was diagnosed with a community-acquired primary liver abscess and metastatic meningitis. Using 88,196 reads, we assembled the shotgun sequences into two circular replicons: a 5,248,520-bp chromosome and a 224,152-bp plasmid. The two replicons contain about 5,006 and 281 protein-encoding genes with average lengths of 940 and 695 bp, respectively. The average G+C content of the chromosome is about 57.7%, which is the highest G+C content for a species in the family Enterobacteriaceae, while the average G+C content of the plasmid is 50.2%. There are eight rRNA operons (one has an extra 5S rRNA gene with the order 16S-23S-5S-5S, while the others consist of a single 16S-, 23S- 5S rRNA cluster) and 86 tRNA genes in the chromosome. The sequence of the NTUH-K2044 plasmid, pK2044, is highly similar to that of the large virulence plasmid pLVPK of K. pneumoniae CG43 (9). pK2044 is 4,767 bp longer than pLVPK, and the differences involve four major insertion-deletion events (see Fig. S1 in the supplemental material). The general features of the K. pneumoniae NTUH-K2044 genome are summarized in Table 1 and are shown in Fig. 1.
TABLE 1.
Global features of the K. pneumoniae NTUH-K2044 genome
| Genetic element | Size (bp) | G+C content (%) | No. of predicted coding genes | Avg gene length (bp) | Coding % | No. of rRNAs (16S-23S-5S) | No. of tRNAs |
|---|---|---|---|---|---|---|---|
| Chromosome | 5,248,520 | 57.68 | 5,006 | 939 | 89.52 | 8-8-9 | 86 |
| Plasmid | 224,152 | 50.17 | 281 | 695 | 87.10 | 0 | 0 |
FIG. 1.
Genomic maps of the K. pneumoniae NTUH-K2044 chromosome and plasmid. From the outside in, the first and second circles show the predicted protein-encoding regions on the plus and minus strands, by role, using the colors for the COG functional categories (http://www.ncbi.nlm.nih.gov/COG/grace/fiew.cgi). The third circle shows the GC skew. The fourth circle shows the transposases/transposons (blue), integrases/recombinases (green), and insertion sequences (red). The fifth and sixth circles show tRNAs and rRNAs, respectively.
Comparison with the strain MGH 78578 genome.
While this work was in progress, we became aware that the genome sequence of another K. pneumoniae strain, MGH 78578, which was isolated from a nosocomial pneumonia case, was available at the Genome Sequencing Center of Washington University (http://genome.wustl.edu/sub_genome_group.cgi?GROUP=3&SUB_GROUP=3). We compared the salient features of the coding sequences and identified gene clusters that are unique to each of the two genomes. As shown in Table 2, homolog sequences predicted for yersiniabactin synthesis (7), the virulence-associated vagCD operon (9), the siderophore transport iroNBCD cluster (42), the mucoid phenotype regulator rmpA gene (26), the type IV secretory pilX system (35), and the plasmid mobilization operon (36) are present only in NTUH-K2044. These genes belong to an integrative and conjugative element (designated ICEKp1) flanked by 17-bp direct repeat ends. The gene organization of ICEKp1 (see Fig. S2 in the supplemental material) is similar to that reported for the Yersinia high-pathogenicity island (7) and Escherichia coli ICEEc1 (41). The excision and integration abilities of ICEKp1 have been shown to be functional (28). In contrast, the genes that are unique to MGH 78578 include a citrate fermentation cluster (32), a fimbrial operon (stbABCDE) (46), and a group of associated genes encoding membrane proteins, which are similar to genes in Salmonella enterica. Additionally, there is an NTUH-K2044-specific sequence that is attributable to a phage insertion event (23,870 bp, comprising 27 genes), as well as at least two other chromosomal segments (58,275 bp and 31,336 bp, comprising 70 genes and 38 genes, respectively) that are present only in MGH 78578. Specifically, these two segments were confirmed to have phage origins by Prophage Finder (http://bioinformatics.uwp.edu/∼phage/ProphageFinder.php) (see Table S1 in the supplemental material). Notably, the lipopolysaccharide (LPS) and capsular polysaccharide (CPS) gene clusters of strains NTUH-K2044 and MGH 78578 are very different. The LPS gene cluster of NTUH-K2044 belongs to the KLEPN LPS O-Ag 1 type (GenBank accession no. L31775 and L31762), whereas that of MGH 78578 is more similar, but not identical, to the Serratia marcescens O4-antigen gene cluster (GenBank accession no. AF038816). The CPS gene clusters of NTUH-K2044 and MGH 78578 are responsible for the two distinct strain serotypes (serotypes K1 and K52, respectively). Thus, the genomes of these two K. pneumoniae clinical isolates are distinguished by the presence of unique sequences at multiple loci, some of which may represent key steps in the evolution of strain-specific features at the levels of metabolism, cell adhesion, and virulence.
TABLE 2.
Differences in gene clusters between K. pneumoniae NTUH-K2044 and K. pneumoniae MGH 78578
| Strain | No. of genomic regions (no. of genes involved)
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Integrative and conjugative element ICEKp1a
|
Citrate fermentation | Fimbrial and membrane proteins | LPS synthesis | CPS synthesis | Phage insertion | |||||||
| ICEKp1 | Yersiniabactin biosynthesis | Viirulence-associated operon | Siderophore transport | Regulator of the mucoid phenotype | Type IV secretion system | Plasmid mobilization | ||||||
| NTUH-K2044 | 1 (60) | (11) | (2) | (4) | (1) | (9) | (3) | 0 | 0 | 1 (6) | 1 (15) | 1 (27) |
| MGH 78578 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (8) | 1 (15) | 1 (8) | 1 (18) | 2 (70, 38) |
Comparative analysis of various K. pneumoniae clinical strains.
To examine the genomic contents of various bacterial isolates that have different infection patterns, more than 50 strains of K. pneumoniae covering the years from 1990 to 2002 were collected. Clinical information was obtained about the place where the infection was acquired (community or hospital), the site of infection, and the presence of any underlying medical conditions. A detailed bacteriological analysis of all isolates will be reported elsewhere; here only the relevant information for the 15 isolates analyzed by using the GSA is summarized (Table 3). Six of these isolates were collected from patients who had nosocomial infections, and nine were community acquired. Liver abscesses were seen in five patients, while the urinary tract was the site of infection in another four patients. All liver abscess cases were community acquired. Notably, three strains (NK25, NK27, and NK29) were retrospectively identified as strains that were collected consecutively within a 2-week period in 1999 from one hospital.
TABLE 3.
Clinical and bacteriological features of the K. pneumoniae isolates
| Group | Strain | Clinical features
|
Virulence test
|
Resistance to antimicrobial agentsd
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β-Lactams
|
Non-β-lactams
|
|||||||||||||||
| Infectiona | Origin of infectionb | Date of isolation (mo/yr) | Mortality | Dose (CFU/mouse)c | Survival (days) | First-generation cephalosporins | Second-generation cephalosporins | Third-generation cephalosporins | Cephamycin | Chloramphenicol | Gentamicin | Quinolones | Tetracycline | TMP/SMX | ||
| 1 | NK1 | 1, 3 | C | 5/1993 | Y | 20,000 | 7 | |||||||||
| NK9 | 2 | C | 11/1996 | Y | 50 | 3 | ||||||||||
| NK252 | 2 | C | 10/2002 | Y | 500 | 5 | ||||||||||
| NK6 | 1, 3 | C | 10/1995 | Y | 2,000 | 2 | ||||||||||
| NK7 | 2 | C | 11/1993 | Y | 500 | 4 | ||||||||||
| 2 | NK2 | 1, 5 | H | 5/1993 | N | NA | >7 | R | R | R | R | |||||
| NK245 | 3 | H | 1/2002 | N | NA | >7 | R | R | R | R | R | R | R | |||
| NK25 | 4 | H | 1/1999 | N | NA | >7 | R | R | R | R | R | |||||
| NK29 | 3 | H | 2/1999 | N | NA | >7 | R | R | R | R | R | |||||
| NK27 | 6 | H | 2/1999 | N | NA | >7 | R | R | R | R | R | |||||
| NK3 | 1, 4 | C | 7/1993 | N | NA | >7 | ||||||||||
| NK4 | 1 | H | 8/1993 | N | NA | >7 | ||||||||||
| 3 | NK8 | 2 | C | 9/1996 | N | NA | >7 | R | R | R | ||||||
| NK10 | 2 | C | 12/1996 | N | NA | >7 | ||||||||||
| NK5 | 1 | C | 3/1994 | Y | 50 | 3 | ||||||||||
1, bacteremia; 2, liver abscess; 3, urinary tract infection; 4, pneumonia; 5, aspiration pneumonia; 6, surgical wound infection.
C, community acquired; H, hospital acquired.
NA, not available.
R, resistant. The first-generation cephalosporins were cephalothin and cefazolin; the second-generation cephalosporin was cefuroxime; the third-generation cephalosporins were ceftriaxone and either cefotaxime or ceftazidime; and the quinolones were nalidixic acid and the fluoroquinolones ciprofloxacin and levofloxacin. TMP/SMX, trimethoprim-sulfamethoxazole. The three isolates resistant to cephamycin (cefoxitin) in group 2 are also resistant to amoxicillin-clavulanic acid and/or ticarcillin-clavulanic acid. All isolates were resistant to ampicillin, and all isolates were susceptible to amikacin, cefepime, and imipenem.
All of the isolates were analyzed for susceptibility to various classes of antimicrobials by determining the MICs. As expected, they were all resistant to ampicillin due to intrinsic resistance. For four isolates (NK25, NK27, NK29, and NK 245) either the MIC of one or more of the third-generation cephalosporins was higher (>2 μg/ml) or the isolates were resistant to one or more of these cephalosporins. One of these four isolates, NK245, tested positive for the presence of an extended-spectrum β-lactamase (ESBL) when ESBL confirmatory testing was used (10). The other isolates did not show a >3 twofold decrease in the MIC for ceftazidime and cefotaxime in the presence of clavulanic acid and thus did not meet the ESBL producer criterion.
The clinical presentation of K. pneumoniae infection is complicated by host factors, such as age, gender, and underlying disease. To investigate the virulence behavior of the isolates under controlled host conditions, we conducted in vivo virulence testing with mice, and the viability of the animals was observed for up to 1 month. Bacterial strains that did not induce mortality within 1 week were scored as nonvirulent strains. Two distinct groups of bacteria were identified. As shown in Table 3, six isolates (NK1, NK6, NK7, NK9, NK252, and NK5) were lethal to the mice at doses of 50 to 20,000 CFU/mouse. In contrast, infection with the other nine isolates did not result in mortality within 7 days. All of the virulent strains were obtained from patients who had community-acquired infections.
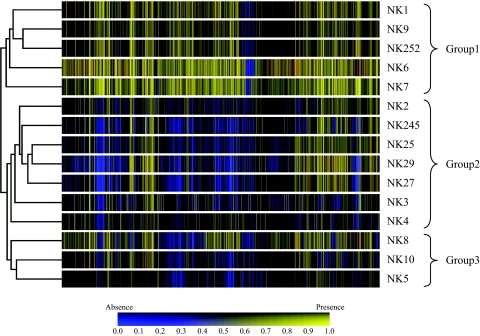
To analyze further the genomic contents of the various different K. pneumoniae isolates relative to the genomic content of NTUH-K2044 and to determine the genetic variations that are characteristic of two infection patterns, we conducted a comparative genomic hybridization analysis (24) using the GSA procedure (27). Briefly, DNA fragments for whole-genome shotgun sequencing were used to generate the probes for the microarray, and a total of 2,847 clones forming a tiling path covered the entire genome. When labeled DNA from 15 clinical isolates were hybridized with the NTUH-K2044 reference sequences, a total of 813 probes showed significantly reduced hybridization signals for at least one clinical isolate compared to NTUH-K2044. Hierarchical clustering analysis of the experimental data set based on these 813 probes revealed three major groups (Fig. 2).
FIG. 2.
Hierarchical cluster analysis of clinical isolates based on GSA with 813 genomic DNA probes. Three major groups of K. pneumoniae can be discerned based on the absence of hybridization signals for shotgun subclone probes. Group 1 strains gave signals for almost all the probes, suggesting that their sequences are nearly identical to that of the reference strain, NTUH-K2044. On the other hand, group 2 strains displayed a loss of signals for various chromosomal and plasmid probes. Three isolates (NK8, NK10, and NK5) are separated from group 1 and group 2; these isolates were classified in group 3.
Correlation of the microarray clustering patterns with the clinical data showed that group 1 isolates, which were the isolates that were most similar to NTUH-K2044 and had the fewest differences in genetic content, were invasive and caused mortality in the mouse model just like the reference strain, NTUH-K2044. In contrast, group 2 isolates, which were significantly different from NTUH-K2044 genetically in both chromosomal DNA and plasmid DNA, were not virulent when the same criteria were used. Within group 2, there is heterogeneity in the microarray patterns and antibiotic resistance profiles (Table 3 and Fig. 2). Five isolates, NK2, NK245, NK25, NK27, and NK29, were collected from patients with different episodes of nosocomial infection in the same hospital between May 1993 and January 2002. Together, NK2 and NK245 form a branch which is distinct from NK25, NK27, and NK29, and both NK2 and NK245 were resistant to quinolones, chloramphenicol, and trimethoprim-sulfamethoxazole; however, they differed in susceptibility to cephalosporins and other β-lactams. In contrast, NK25, NK27, and NK29 were resistant to chloramphenicol and nearly all cephalosporins but were susceptible to quinolones and trimethoprim-sulfamethoxazole (Table 3). Unlike the other group 2 strains, which showed resistance to multiple antimicrobials, NK3 and NK4 are susceptible to all antimicrobials tested except ampicillin. The three group 3 strains, NK5, NK8, and NK10, have a clinical presentation similar to that of the group 1 strains. The infections were acquired from the community and caused liver abscesses or bacteremia, and both groups of bacteria were susceptible to most antibiotics tested; however, group 3 isolates exhibit GSA patterns distinct from those of the group 1 or group 2 strains (Fig. 2). These isolates differed from group 1 strains by the absence of signals for specific chromosomal sequences and from group 2 strains by the presence of signals for specific plasmid sequences. Except for NK5, the group 3 strains did not cause mortality in the mouse virulence test. When the group 1 and group 3 strains were combined, there were five strains that caused community-acquired liver abscesses (NK7, NK9, NK252, NK8, and NK10), and these isolates can be distinguished from the nosocomial strains by the presence of common plasmid sequences (Fig. 3).
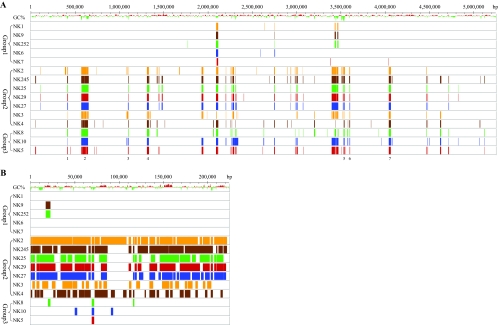
FIG. 3.
Linear representation of the NTUH-K2044 genome and hybridization patterns of K. pneumoniae isolates. Deviation from the average G+C content of the sequence is shown for the chromosomal (A) and plasmid (B) sequences. The absence of a signal (deletion) for each probe is indicated by a colored vertical line. The locations of the seven INDELs in the chromosome, whose sequences are not present in the group 2 and group 3 strains, are indicated.
We examined the chromosomal contents of the different K. pneumoniae strains in more detail, and it became evident that at least seven chromosomal regions were very different in the major groups (Fig. 3). We designated these regions INDEL1 to INDEL7, which refer to the insertion-deletion nature of the variations. The molecular features of the chromosomal sequences of these regions are summarized in Table 4, and Table S2 in the supplemental material provides a list of predicted genes for the INDEL regions. Briefly, a total of 144 predicted genes were not present in the group 2 and group 3 strains; 36 of these genes code for proteins that show similarity to known hypothetical proteins in other bacteria, while 22 of them have no known annotation. Remarkably, five INDEL regions (INDEL1, INDEL2, INDEL3, INDEL5, and INDEL6) have G+C contents much lower than the average chromosomal DNA G+C content. Moreover, pairwise dinucleotide covariation analysis of the INDELs revealed that the dinucleotide frequencies of INDEL1, INDEL3, and INDEL5 differ significantly from the overall dinucleotide frequencies of the genome. Together, the results obtained are consistent with the hypothesis that these genomic features distinguishing the major groups were acquired horizontally during evolution. Finally, we identified sequence elements that are characteristic of a pathogenicity island, such as tRNA genes, insertion sequences, and genes encoding integrases and transposases; these elements are clustered in INDEL2 and INDEL3. The presence of fimbria-pilus genes in INDEL2 suggests that this region could potentially make an important contribution to the virulence phenotype of the invasive strains.
TABLE 4.
Genomic features that distinguished the major groups of K. pneumoniae strains
| INDEL | Location in reference strain | Length (bp) | Predicted no. of genes | Features
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| tRNA | Insertion element | Integrase | Transposase | ΔG+C (%)a | ρb | ||||
| 1 | 410,975-418,760 | 7,786 | 6 | − | − | − | − | −12.00 | 0.5 |
| 2 | 566,263-644,763 | 78,501 | 69 | + | − | + | + | −6.20 | 0.8 |
| 3 | 1,096,872-1,103,609 | 6,738 | 8 | + | − | + | + | −13.19 | 0.4 |
| 4 | 1,308,978-1,331,600 | 22,623 | 19 | + | − | − | − | −3.18 | 0.9 |
| 5 | 3,529,235-3,543,692 | 14,458 | 13 | − | − | − | − | −20.48 | −0.5 |
| 6 | 3,597,896-3,604,737 | 6,842 | 6 | − | − | − | + | −5.03 | 0.9 |
| 7 | 4,040,733-4,065,069 | 24,337 | 23 | − | − | − | − | 3.95 | 1.0 |
Deviation from the average G+C content of chromosomal DNA.
Pairwise covariation of 3:1 dinucleotide frequencies of predicted genes in INDEL regions compared to the whole genome as assessed by the Spearman rank correlation coefficient (ρ).
DISCUSSION
In this study, genome sequencing and subsequent molecular analysis with GSA provided data for exposing the magnitude of the genetic diversity in clinical isolates of K. pneumoniae and allowed identification of the genomic signatures that are associated with various K. pneumoniae infection patterns. Based on the GSA results, 15 clinical isolates were clustered into three major groups according to their hybridization patterns, using the completely sequenced strain NTUH-K2044 as a reference (Fig. 2 and 3). Overall, the genomic signatures correlated well with the clinical features (community versus hospital infection; liver abscess versus other types of infection) and the virulence phenotypes observed in mice (Table 3). Therefore, we have developed methods that through comparative genomics are capable of identifying the genetic determinants characterizing different K. pneumoniae infections. Since we used NTUH-K2044 genomic fragments as probes for the GSA experiments, this approach detected only the loss of sequences in the genomes tested. For this reason, we designed a high-density oligonucleotide microarray based on all newly available K. pneumoniae genome sequences from our laboratory and used this microarray to analyze representative strains with different infection patterns. The grouping results for the oligonucleotide microarray experiments (data not shown) are consistent with the conclusions reported here.
Almost no strain in group 2 and group 3 was lethal for mice when the strains were injected intraperitoneally; NK5 was the exception, and it consistently caused mortality at a relatively low dose (Table 3). We therefore wondered whether the NK5 strain has unique genetic features that give rise to this virulence; hence, we determined this strain's entire genome sequence using 454 technology (31) (data not shown). We found that the CPS region of the NK5 strain was identical to that of a serotype K2 strain, Chedid, and that of another virulent strain, CG43 (H.-Y. Shu, unpublished), which suggests that the CPS associated with the K2 serotype (33) is the origin of virulence for mice. Thus, all the strains tested that are lethal to mice are either serotype K1 strains (NTUH-K2044, NK1, NK6, NK7, NK9, and NK252) or a serotype K2 strain (NK5) (Table 3). This finding is consistent with previous reports indicating that, among isolates belonging to 77 K. pneumoniae K serotypes (37, 39) distributed across different geographical areas, serotype K1 and K2 isolates are the isolates most virulent for humans and mice (1, 21, 33). The GSA data also indicate that the group 1 and group 3 strains contain almost identical versions of the 224-kb plasmid found in the NTUH-K2044 isolate (Fig. 3). Notably, the plasmid sequence is highly similar to that of the large virulence plasmid pLVPK of K. pneumoniae CG43 (9). Since pLVPK is essential for the virulence of CG43 (26) and since a common rmpA gene is present in CG43 and in all group 1 and group 3 strains, we suggest that RmpA could contribute to the unique clinical manifestation of liver abscesses caused by these strains. Consistent with this notion, a recent epidemiology study by Yu et al. revealed a statistical correlation between the rmpA gene and virulence for abscess formation (53). Given that RmpA functions as an activator of the cps genes (26, 49), its role in mediating the hypermucoviscous phenotype of the group 1 and group 3 strains deserves further investigation. The possibility that this plasmid sequence, which has now been implicated in liver abscesses, is distinct from the virulence factors identified by the mouse intraperitoneal injection assay is worth considering. Moreover, the results of this study support the hypothesis that although the K. pneumoniae strains isolated in different regions of Taiwan have distinct genomic backgrounds, the strains that cause liver abscesses share common genetic determinants and these determinants seem to be propagated through a plasmid.
The hypermucoid phenotype is a hallmark of K. pneumoniae pathogens (16). The CPS protects the invading bacteria against phagocytosis and complement-mediated serum killing. A cluster of genes required for K. pneumonia CPS synthesis is in INDEL5 on the chromosome (Table 4). The general organization of the genes is similar to that of the genes encoding E. coli group I CPS (1, 40). Notably, the central portion of the cps gene cluster contains genes that seem to be involved in specific and unique oligosaccharide repeat unit biosynthesis and polymerization for each of the sequenced isolates (NTUH-K2044, MGH 78578, and Chedid [1]), while the flanking sequences are conserved (H.-Y. Shu, unpublished). As indicated in Table S2 in the supplemental material, magA, which has been shown recently to be associated with the K1 serotype (11, 43) and to be significantly more prevalent in invasive strains (16), is present in all group 1 strains. Thus, it is likely that a genetic mechanism(s) resulting in variation in the K antigen (1, 14) may contribute to the association between serotype and infection pattern.
The comparison of the genome sequences of NTUH-K2044 and MGH 78578 (Table 2) supports the idea that gene acquisition and perhaps gene loss play an important role in strain evolution in an environment where the pathogen is under selection pressure from the host. The very distinct LPS and CPS gene clusters that were found when these two strains were compared emphasizes the conclusion that the serotype differences reflect major genetic differences. However, the strain differences also include an integrative and conjugative element similar to one found in E. coli, as well as a fimbrial gene cluster from S. enterica. Furthermore, the fact that the NTUH-K2044 and MGH 78578 strains contain different prophage sequences reflects the impact of phage lysogeny on prokaryotic diversity (5, 6), and these sequences can also be treated as evolutionary markers for separation of the different lineages of clinical K. pneumoniae. Thus, the genome variation markers, together with the INDEL results, suggest that gene transfer between the various closely related enterobacterial species that inhabit the human body may be one of the more important evolutionary mechanisms acting on K. pneumoniae. Clearly, further sequence information for different Klebsiella species is needed to help us understand which genes are central to a specific strain and how a strain fits into a particular pathogenic niche.
The fact that strains NK25, NK27, and NK29 (Fig. 2), which have highly similar genomic signatures and antimicrobial profiles and form a tight cluster, turned out to be from a hospital outbreak suggests strongly that GSA can be a useful tool for tracing epidemics as well as reconstructing the phylogeny of K. pneumoniae isolates. It is noteworthy that a microarray approach provides more detailed genome-wide information on strain variation than sequence-based single-gene analysis, pooled analysis of a number of conserved gene sequences, PCR-based techniques like randomly amplified polymorphic DNA (RAPD) analysis (4), or analysis of chromosomal DNA restriction patterns by pulsed-field gel electrophoresis (44). In fact, we were able to detect minor sequence variations in the three isolates by GSA (Fig. 3), and this suggests that these isolates represent independent clones from the same lineage. We concluded that this approach should be able to provide a fast and highly accurate way to identify and trace the origin of a nosocomial outbreak of K. pneumoniae infections.
Finally, genetic diversity and dynamic genome organization appear to be general characteristics of Enterobacteriaceae species. Touchon et al. (45) examined the evolutionary genome dynamics of E. coli and suggested an important adaptive role for metabolic diversification in virulence when E. coli and Shigella species are compared. It seems likely that a similar situation exists with Klebsiella species. One possibility is that enzymes of the methionine salvage pathway (see Fig. S3 in the supplemental material), which are not present in E. coli but whose genes are present in all Klebsiella genomes, including other genomes sequenced by our group and unpublished genomes, might have a role in pathogenesis via oxidative stress (51). In this context, this pathway also seems to be present in other pathogenic enterobacteria, including Serratia, Erwinia, Enterobacter, and Citrobacter species and some Yersinia species. In contrast, this group of genes is not present in Escherichia, Salmonella, and Shigella (see Table S3 in the supplemental material). In addition, there is genetic variation in the methionine salvage pathway among sequenced Yersinia species (Y. enterocolitica, Y. pestis, and Y. pseudotuberculosis). Together, the data support the idea that this pathway and perhaps other pathways vary in enterobacteria and provide diversity in virulence mechanisms.
In conclusion, the rapid evolution of enteric bacteria, including Klebsiella, is a constant medical problem. Direct control of such evolution is difficult, but the results presented here provide insights into how clinically relevant K. pneumoniae strains are evolving and support the hypothesis that such strains gain new genetic features from other strains and species by horizontal transfer and perhaps interstrain recombination. The combined use of genome sequencing and GSA when the clinically relevant evolution of pathogenic bacteria is studied provides a simple approach that should increase our understanding of the changes that occur when bacteria sidestep modern medicine.
Supplementary Material
Acknowledgments
We express our gratitude to Yan-Hwa Wu Lee for enthusiastic support of the microbial genomics study of K. pneumoniae, to Monto Ho for critical reading and discussion of the manuscript, and to Ming-Wei Lin for advice on statistical analysis. We are grateful to the staff of the National DNA Sequencing Core at the National Yang-Ming University for genome sequencing of the K. pneumoniae strains.
This project was supported by NRPGM of NSC (S.F.T. and J.T.W.), by institutional funds from NYMU (S.F.T.) and NHRI (S.F.T., T.F.L., and I.J.S.), and by a grant from the Ministry of Education, Aim for the Top University Plan.
Footnotes
Published ahead of print on 15 May 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Arakawa, Y., R. Wacharotayankun, T. Nagatsuka, H. Ito, N. Kato, and M. Ohta. 1995. Genomic organization of the Klebsiella pneumoniae cps region responsible for serotype K2 capsular polysaccharide synthesis in the virulent strain Chedid. J. Bacteriol. 1771788-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balows, A., and B. Duerden. 1998. Topley & Wilson's microbiology and microbial infections, 9th ed., vol. 2. Oxford University Press, Oxford, United Kingdom.
- 3.Borodovsky, M., and J. McIninch. 1993. GeneMark: parallel gene recognition for both DNA strands. Comput. Chem. 17123-133. [Google Scholar]
- 4.Brisse, S., and J. Verhoef. 2001. Phylogenetic diversity of Klebsiella pneumoniae and Klebsiella oxytoca clinical isolates revealed by randomly amplified polymorphic DNA, gyrA and parC genes sequencing and automated ribotyping. Int. J. Syst. Evol. Microbiol. 51915-924. [DOI] [PubMed] [Google Scholar]
- 5.Brüssow, H., C. Canchaya, and W. D. Hardt. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68560-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canchaya, C., G. Fournous, and H. Brussow. 2004. The impact of prophages on bacterial chromosomes. Mol. Microbiol. 539-18. [DOI] [PubMed] [Google Scholar]
- 7.Carniel, E., I. Guilvout, and M. Prentice. 1996. Characterization of a large chromosomal “high-pathogenicity island” in biotype 1B Yersinia enterocolitica. J. Bacteriol. 1786743-6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carpenter, J. L. 1990. Klebsiella pulmonary infections: occurrence at one medical center and review. Rev. Infect. Dis. 12672-682. [DOI] [PubMed] [Google Scholar]
- 9.Chen, Y. T., H. Y. Chang, Y. C. Lai, C. C. Pan, S. F. Tsai, and H. L. Peng. 2004. Sequencing and analysis of the large virulence plasmid pLVPK of Klebsiella pneumoniae CG43. Gene 337189-198. [DOI] [PubMed] [Google Scholar]
- 10.Chen, Y. T., H. Y. Shu, L. H. Li, T. L. Liao, K. M. Wu, Y. R. Shiau, J. J. Yan, I. J. Su, S. F. Tsai, and T. L. Lauderdale. 2006. Complete nucleotide sequence of pK245, a 98-kilobase plasmid conferring quinolone resistance and extended-spectrum-beta-lactamase activity in a clinical Klebsiella pneumoniae isolate. Antimicrob. Agents Chemother. 503861-3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chuang, Y. P., C. T. Fang, S. Y. Lai, S. C. Chang, and J. T. Wang. 2006. Genetic determinants of capsular serotype K1 of Klebsiella pneumoniae causing primary pyogenic liver abscess. J. Infect. Dis. 193645-654. [DOI] [PubMed] [Google Scholar]
- 12.Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 274636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Domenico, P., R. J. Salo, A. S. Cross, and B. A. Cunha. 1994. Polysaccharide capsule-mediated resistance to opsonophagocytosis in Klebsiella pneumoniae. Infect. Immun. 624495-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drummelsmith, J., and C. Whitfield. 1999. Gene products required for surface expression of the capsular form of the group 1 K antigen in Escherichia coli (O9a:K30). Mol. Microbiol. 311321-1332. [DOI] [PubMed] [Google Scholar]
- 15.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8186-194. [PubMed] [Google Scholar]
- 16.Fang, C. T., Y. P. Chuang, C. T. Shun, S. C. Chang, and J. T. Wang. 2004. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J. Exp. Med. 199697-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, et al. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269496-512. [DOI] [PubMed] [Google Scholar]
- 18.Fraser, C. M., and R. D. Fleischmann. 1997. Strategies for whole microbial genome sequencing and analysis. Electrophoresis 181207-1216. [DOI] [PubMed] [Google Scholar]
- 19.Friedlander, C. 1983. Die Mikrokokken der Pneumonie. Fortschr. Med. 1715-733. [Google Scholar]
- 20.Friedlander, C. 1882. Ueber die Schizomyceten bei der acuten fibrosen Pneumoniae. Virchows Arch. Pathol Anat. Physiol. Klin. Med. 87319-324. [Google Scholar]
- 21.Fung, C. P., F. Y. Chang, S. C. Lee, B. S. Hu, B. I. Kuo, C. Y. Liu, M. Ho, and L. K. Siu. 2002. A global emerging disease of Klebsiella pneumoniae liver abscess: is serotype K1 an important factor for complicated endophthalmitis? Gut 50420-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gastmeier, P., K. Groneberg, K. Weist, and H. Ruden. 2003. A cluster of nosocomial Klebsiella pneumoniae bloodstream infections in a neonatal intensive care department: identification of transmission and intervention. Am. J. Infect. Control 31424-430. [DOI] [PubMed] [Google Scholar]
- 23.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8195-202. [DOI] [PubMed] [Google Scholar]
- 24.Kallioniemi, A., O. P. Kallioniemi, D. Sudar, D. Rutovitz, J. W. Gray, F. Waldman, and D. Pinkel. 1992. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science 258818-821. [DOI] [PubMed] [Google Scholar]
- 25.Ko, W. C., D. L. Paterson, A. J. Sagnimeni, D. S. Hansen, A. Von Gottberg, S. Mohapatra, J. M. Casellas, H. Goossens, L. Mulazimoglu, G. Trenholme, K. P. Klugman, J. G. McCormack, and V. L. Yu. 2002. Community-acquired Klebsiella pneumoniae bacteremia: global differences in clinical patterns. Emerg. Infect. Dis. 8160-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lai, Y. C., H. L. Peng, and H. Y. Chang. 2003. RmpA2, an activator of capsule biosynthesis in Klebsiella pneumoniae CG43, regulates K2 cps gene expression at the transcriptional level. J. Bacteriol. 185788-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, L. H., J. C. Li, Y. F. Lin, C. Y. Lin, C. Y. Chen, and S. F. Tsai. 2004. Genomic shotgun array: a procedure linking large-scale DNA sequencing with regional transcript mapping. Nucleic Acids Res. 32e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin, T. L., C. Z. Lee, P. F. Hsieh, S. F. Tsai, and J. T. Wang. 2008. Characterization of integrative and conjugative element ICEKp1-associated genomic heterogeneity in a Klebsiella pneumoniae strain isolated from a primary liver abscess. J. Bacteriol. 190515-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lukashin, A. V., and M. Borodovsky. 1998. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 261107-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Margulies, M., M. Egholm, W. E. Altman, S. Attiya, J. S. Bader, L. A. Bemben, J. Berka, M. S. Braverman, Y. J. Chen, Z. Chen, S. B. Dewell, L. Du, J. M. Fierro, X. V. Gomes, B. C. Godwin, W. He, S. Helgesen, C. H. Ho, G. P. Irzyk, S. C. Jando, M. L. Alenquer, T. P. Jarvie, K. B. Jirage, J. B. Kim, J. R. Knight, J. R. Lanza, J. H. Leamon, S. M. Lefkowitz, M. Lei, J. Li, K. L. Lohman, H. Lu, V. B. Makhijani, K. E. McDade, M. P. McKenna, E. W. Myers, E. Nickerson, J. R. Nobile, R. Plant, B. P. Puc, M. T. Ronan, G. T. Roth, G. J. Sarkis, J. F. Simons, J. W. Simpson, M. Srinivasan, K. R. Tartaro, A. Tomasz, K. A. Vogt, G. A. Volkmer, S. H. Wang, Y. Wang, M. P. Weiner, P. Yu, R. F. Begley, and J. M. Rothberg. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437376-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyer, M., P. Dimroth, and M. Bott. 2001. Catabolite repression of the citrate fermentation genes in Klebsiella pneumoniae: evidence for involvement of the cyclic AMP receptor protein. J. Bacteriol. 1835248-5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mizuta, K., M. Ohta, M. Mori, T. Hasegawa, I. Nakashima, and N. Kato. 1983. Virulence for mice of Klebsiella strains belonging to the O1 group: relationship to their capsular (K) types. Infect. Immun. 4056-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murray, P. R., and E. J. Baron. 2003. Manual of clinical microbiology, 8th ed. ASM Press, Washington, DC.
- 35.Núñez, B., P. Avila, and F. de la Cruz. 1997. Genes involved in conjugative DNA processing of plasmid R6K. Mol. Microbiol. 241157-1168. [DOI] [PubMed] [Google Scholar]
- 36.Núñez, B., and F. De La Cruz. 2001. Two atypical mobilization proteins are involved in plasmid CloDF13 relaxation. Mol. Microbiol. 391088-1099. [DOI] [PubMed] [Google Scholar]
- 37.Ørskov, I., and F. Ørskov. 1984. Serotyping of Klebsiella. Methods Microbiol. 14143-164. [Google Scholar]
- 38.Paterson, D. L., W. C. Ko, A. Von Gottberg, S. Mohapatra, J. M. Casellas, H. Goossens, L. Mulazimoglu, G. Trenholme, K. P. Klugman, R. A. Bonomo, L. B. Rice, M. M. Wagener, J. G. McCormack, and V. L. Yu. 2004. International prospective study of Klebsiella pneumoniae bacteremia: implications of extended-spectrum beta-lactamase production in nosocomial Infections. Ann. Intern. Med. 14026-32. [DOI] [PubMed] [Google Scholar]
- 39.Podschun, R., and U. Ullmann. 1998. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 11589-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rahn, A., J. Drummelsmith, and C. Whitfield. 1999. Conserved organization in the cps gene clusters for expression of Escherichia coli group 1 K antigens: relationship to the colanic acid biosynthesis locus and the cps genes from Klebsiella pneumoniae. J. Bacteriol. 1812307-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schubert, S., S. Dufke, J. Sorsa, and J. Heesemann. 2004. A novel integrative and conjugative element (ICE) of Escherichia coli: the putative progenitor of the Yersinia high-pathogenicity island. Mol. Microbiol. 51837-848. [DOI] [PubMed] [Google Scholar]
- 42.Sorsa, L. J., S. Dufke, J. Heesemann, and S. Schubert. 2003. Characterization of an iroBCDEN gene cluster on a transmissible plasmid of uropathogenic Escherichia coli: evidence for horizontal transfer of a chromosomal virulence factor. Infect. Immun. 713285-3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Struve, C., M. Bojer, E. M. Nielsen, D. S. Hansen, and K. A. Krogfelt. 2005. Investigation of the putative virulence gene magA in a worldwide collection of 495 Klebsiella isolates: magA is restricted to the gene cluster of Klebsiella pneumoniae capsule serotype K1. J. Med. Microbiol. 541111-1113. [DOI] [PubMed] [Google Scholar]
- 44.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 332233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Touchon, M., C. Hoede, O. Tenaillon, V. Barbe, S. Baeriswyl, P. Bidet, E. Bingen, S. Bonacorsi, C. Bouchier, O. Bouvet, A. Calteau, H. Chiapello, O. Clermont, S. Cruveiller, A. Danchin, M. Diard, C. Dossat, M. E. Karoui, E. Frapy, L. Garry, J. M. Ghigo, A. M. Gilles, J. Johnson, C. Le Bouguenec, M. Lescat, S. Mangenot, V. Martinez-Jehanne, I. Matic, X. Nassif, S. Oztas, M. A. Petit, C. Pichon, Z. Rouy, C. S. Ruf, D. Schneider, J. Tourret, B. Vacherie, D. Vallenet, C. Medigue, E. P. Rocha, and E. Denamur. 2009. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 5e1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Townsend, S. M., N. E. Kramer, R. Edwards, S. Baker, N. Hamlin, M. Simmonds, K. Stevens, S. Maloy, J. Parkhill, G. Dougan, and A. J. Baumler. 2001. Salmonella enterica serovar Typhi possesses a unique repertoire of fimbrial gene sequences. Infect. Immun. 692894-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsai, F. C., Y. T. Huang, L. Y. Chang, and J. T. Wang. 2008. Pyogenic liver abscess as endemic disease, Taiwan. Emerg. Infect. Dis. 141592-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsay, R. W., L. K. Siu, C. P. Fung, and F. Y. Chang. 2002. Characteristics of bacteremia between community-acquired and nosocomial Klebsiella pneumoniae infection: risk factor for mortality and the impact of capsular serotypes as a herald for community-acquired infection. Arch. Intern. Med. 1621021-1027. [DOI] [PubMed] [Google Scholar]
- 49.Wacharotayankun, R., Y. Arakawa, M. Ohta, K. Tanaka, T. Akashi, M. Mori, and N. Kato. 1993. Enhancement of extracapsular polysaccharide synthesis in Klebsiella pneumoniae by RmpA2, which shows homology to NtrC and FixJ. Infect. Immun. 613164-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, J. H., Y. C. Liu, S. S. Lee, M. Y. Yen, Y. S. Chen, S. R. Wann, and H. H. Lin. 1998. Primary liver abscess due to Klebsiella pneumoniae in Taiwan. Clin. Infect. Dis. 261434-1438. [DOI] [PubMed] [Google Scholar]
- 51.Wray, J. W., and R. H. Abeles. 1995. The methionine salvage pathway in Klebsiella pneumoniae and rat liver. Identification and characterization of two novel dioxygenases. J. Biol. Chem. 2703147-3153. [DOI] [PubMed] [Google Scholar]
- 52.Yeh, K. M., A. Kurup, L. K. Siu, Y. L. Koh, C. P. Fung, J. C. Lin, T. L. Chen, F. Y. Chang, and T. H. Koh. 2007. Capsular serotype K1 or K2, rather than magA and rmpA, is a major virulence determinant for Klebsiella pneumoniae liver abscess in Singapore and Taiwan. J. Clin. Microbiol. 45466-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu, W. L., W. C. Ko, K. C. Cheng, H. C. Lee, D. S. Ke, C. C. Lee, C. P. Fung, and Y. C. Chuang. 2006. Association between rmpA and magA genes and clinical syndromes caused by Klebsiella pneumoniae in Taiwan. Clin. Infect. Dis. 421351-1358. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.