Abstract
A 3-hydroxypropionate/4-hydroxybutyrate cycle operates in autotrophic CO2 fixation in various Crenarchaea, as studied in some detail in Metallosphaera sedula. This cycle and the autotrophic 3-hydroxypropionate cycle in Chloroflexus aurantiacus have in common the conversion of acetyl-coenzyme A (CoA) and two bicarbonates via 3-hydroxypropionate to succinyl-CoA. Both cycles require the reductive conversion of 3-hydroxypropionate to propionyl-CoA. In M. sedula the reaction sequence is catalyzed by three enzymes. The first enzyme, 3-hydroxypropionyl-CoA synthetase, catalyzes the CoA- and MgATP-dependent formation of 3-hydroxypropionyl-CoA. The next two enzymes were purified from M. sedula or Sulfolobus tokodaii and studied. 3-Hydroxypropionyl-CoA dehydratase, a member of the enoyl-CoA hydratase family, eliminates water from 3-hydroxypropionyl-CoA to form acryloyl-CoA. Acryloyl-CoA reductase, a member of the zinc-containing alcohol dehydrogenase family, reduces acryloyl-CoA with NADPH to propionyl-CoA. Genes highly similar to the Metallosphaera CoA synthetase, dehydratase, and reductase genes were found in autotrophic members of the Sulfolobales. The encoded enzymes are only distantly related to the respective three enzyme domains of propionyl-CoA synthase from C. aurantiacus, where this trifunctional enzyme catalyzes all three reactions. This indicates that the autotrophic carbon fixation cycles in Chloroflexus and in the Sulfolobales evolved independently and that different genes/enzymes have been recruited in the two lineages that catalyze the same kinds of reactions.
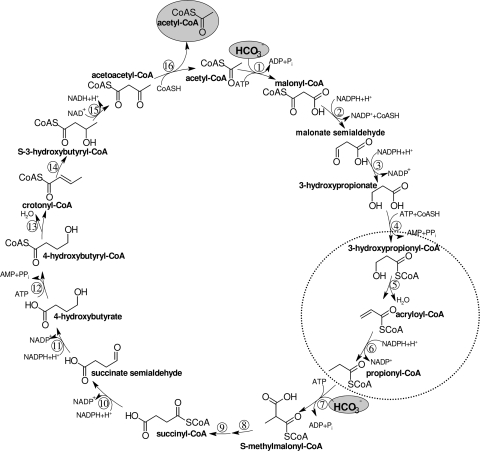
In the thermoacidophilic autotrophic crenarchaeum Metallosphaera sedula, CO2 fixation proceeds via a 3-hydroxypropionate/4-hydroxybutyrate cycle (8, 23, 24, 28) (Fig. 1). A similar cycle may operate in other autotrophic members of the Sulfolobales and in mesophilic Crenarchaea (Cenarchaeum sp. and Nitrosopumilus sp.) of marine group I. The cycle uses elements of the 3-hydroxypropionate cycle that was originally discovered in the phototrophic bacterium Chloroflexus aurantiacus (11, 16, 17, 19, 20, 32, 33). It involves the carboxylation of acetyl-coenzyme A (CoA) to malonyl-CoA by the biotin-dependent acetyl-CoA carboxylase. Malonyl-CoA is reduced via malonate semialdehyde to 3-hydroxypropionate (1), which is further reductively converted to propionyl-CoA (3). Propionyl-CoA is carboxylated to (S)-methylmalonyl-CoA by a propionyl-CoA carboxylase that is similar or identical to acetyl-CoA carboxylase. In fact, only one copy of the genes for the acetyl-CoA/propionyl-CoA carboxylase subunits is present in most Archaea, suggesting that this is a promiscuous enzyme that acts on both acetyl-CoA and propionyl-CoA (24). (S)-Methylmalonyl-CoA is epimerized to (R)-methylmalonyl-CoA, followed by carbon rearrangement to succinyl-CoA by coenzyme B12-dependent methylmalonyl-CoA mutase.
FIG. 1.
Proposed 3-hydroxypropionate/4-hydroxybutyrate cycle in M. sedula and other members of the Sulfolobales. Enzymes are the following: 1, acetyl-CoA carboxylase; 2, malonyl-CoA reductase (NADPH); 3, malonate semialdehyde reductase (NADPH); 4, 3-hydroxypropionyl-CoA synthetase (3-hydroxypropionate-CoA ligase, AMP forming); 5, 3-hydroxypropionyl-CoA dehydratase; 6, acryloyl-CoA reductase (NADPH); 7, propionyl-CoA carboxylase; 8, methylmalonyl-CoA epimerase; 9, methylmalonyl-CoA mutase; 10, succinyl-CoA reductase (NADPH); 11, succinate semialdehyde reductase (NADPH); 12, 4-hydroxybutyryl-CoA synthetase (4-hydroxybutyrate-CoA ligase, AMP-forming); 13, 4-hydroxybutyryl-CoA dehydratase; 14, crotonyl-CoA hydratase; 15, (S)-3-hydroxybutyryl-CoA dehydrogenase (NAD+); 16, acetoacetyl-CoA β-ketothiolase. The two steps of interest are highlighted.
In Chloroflexus succinyl-CoA is converted to (S)-malyl-CoA, which is cleaved by (S)-malyl-CoA lyase to acetyl-CoA (thus regenerating the CO2 acceptor molecule) and glyoxylate (16). Glyoxylate is assimilated into cell material by a yet not completely resolved pathway (37). In Metallosphaera succinyl-CoA is converted via 4-hydroxybutyrate to two molecules of acetyl-CoA (8), thus regenerating the starting CO2 acceptor molecule and releasing another acetyl-CoA for biosynthesis. Hence, the 3-hydroxypropionate/4-hydroxybutyrate cycle (Fig. 1) can be divided into two parts. The first part transforms one acetyl-CoA and two bicarbonates into succinyl-CoA, and the second part converts succinyl-CoA to two acetyl-CoA molecules.
The reductive conversion of 3-hydroxypropionate to propionyl-CoA requires three enzymatic steps: activation of 3-hydroxypropionate to its CoA ester, dehydration of 3-hydroxypropionyl-CoA to acryloyl-CoA, and reduction of acryloyl-CoA to propionyl-CoA. In C. aurantiacus these three steps are catalyzed by a single large trifunctional enzyme, propionyl-CoA synthase (2). This 200-kDa fusion protein consists of a CoA ligase, a dehydratase, and a reductase domain. Attempts to isolate a similar enzyme from M. sedula failed. Rather, a 3-hydroxypropionyl-CoA synthetase was found (3), suggesting that the other two reactions may also be catalyzed by individual enzymes.
Here, we purified the missing enzymes 3-hydroxypropionyl-CoA dehydratase and acryloyl-CoA reductase from M. sedula, identified the coding genes in the genome of M. sedula and other members of the Sulfolobales, produced recombinant enzymes as proof of function, and studied the enzymes in some detail. A comparison with the respective domains of propionyl-CoA synthase from C. aurantiacus indicates that the conversion of 3-hydroxypropionate to propionyl-CoA via the 3-hydroxypropionate route has evolved independently in these two phyla.
MATERIALS AND METHODS
Materials.
Chemicals and biochemicals were obtained from Roche Diagnostics (Mannheim, Germany), Fluka (Neu-Ulm, Germany), Merck (Darmstadt, Germany), Roth (Karlsruhe, Germany), Sigma-Aldrich (Deisenhofen, Germany), Bio-Rad (Munich, Germany), and Genaxxon (Biberach, Germany). Gases were obtained from Sauerstoffwerke Friedrichshafen (Friedrichshafen, Germany), radioisotopes were from American Radiolabeled Chemicals Inc./Biotrend Chemikalien GmbH (Cologne, Germany). Enzymes and primers were obtained from MBI Fermentas (St. Leon-Rot, Germany) and Genaxxon Biosciences GmbH (Biberach, Germany). Materials and equipment for protein purification were obtained from GE Healthcare (Munich, Germany), Millipore (Bedford, MA), Sigma-Aldrich (Deisenhofen, Germany), and Whatman Biosystems Ltd. (Madistone, Great Britain). Plasmids were obtained from Invitrogen (Karlsruhe, Germany), Novagene (Darmstadt, Germany), Fermentas (St. Leon-Rot, Germany), and Stratagene (La Jolla, CA).
Strains and culture conditions.
M. sedula TH2 (DSM 5348) was grown autotrophically at 75°C on a chemically defined medium (pH 2.0) under gassing with a mixture of 19% CO2, 3% O2, and 78% H2 (generation time, 8 h) (21). As a control, cells were grown aerobically and heterotrophically with 0.05% yeast extract (generation time, 16 h) (21). Sulfolobus tokodaii (DSMZ 16993) was grown aerobically and heterotrophically at 75°C on a chemically defined medium (pH 3.0) with 1 g of glucose per liter (generation time, 6 h) (34). Cells were stored in liquid nitrogen until use. Escherichia coli strain DH5α and E. coli strain Rosetta 2 (DE3) (Merck, Germany) were grown at 37°C in Luria-Bertani (LB) medium (30). Antibiotics were added to E. coli cultures to a final concentration of 100 μg ampicillin ml−1 and 34 μg chloramphenicol ml−1.
Preparation of cell extracts.
For the purification of the reductase, cells were suspended in 1 volume of 100 mM Tris-HCl, pH 8.0, containing 5 mM MgCl2 and 0.02 mg DNase I ml−1. For the dehydratase purification, cells were suspended in 1 volume of 100 mM Tris-HCl, pH 7.2, containing 10 mM MgCl2, 5 mM 1,4-dithioerythritol (DTE), and 0.05 mg DNase I ml−1. The cell suspension was passed through a French pressure cell at 137 MPa and ultracentrifuged (at 100,000 × g) at 4°C for 1 h. The cell extract was used immediately or kept frozen at −70°C.
Enzyme assays.
The enzyme activities were determined in coupled spectrophotometric assays, in which the oxidation of NAD(P)H was followed spectrophotometrically at 365 nm (ɛ365 for NAD(P)H of 3,400 M−1 cm−1) and 65°C. One unit corresponds to 1 μmol of substrate converted per min. The addition of cell extract or (partially) purified enzyme started the reaction. The pH of the buffers as indicated below was adjusted at room temperature, and the actual value at high temperature was calculated. Note that the pka values of the buffers may deviate dramatically at high temperatures, and consequently their pH values and buffer capacities may also (10, 12, 15, 31). The variation of the pKa value of the buffer substance, d(pKa) per temperature unit dT, d(pKa)/dT, is −0.011 for 2-(N-morpholino)ethanesulfonic acid (MES), −0.011 for 4-morpholinopropanesulfonic acid (MOPS), and −0.028 for Tris.
(i) Acryloyl-CoA reductase.
Acryloyl-CoA was synthesized from acrylate, MgATP, and CoA by added 3-hydroxypropionyl-CoA synthetase (3-hydroxypropionate-CoA ligase; AMP forming) from M. sedula or S. tokodaii (3). The acryloyl-CoA-dependent oxidation of NADPH was followed. The assay mixture (0.5 ml) contained 100 mM MES, pH 6, 10 mM MgCl2, 3 mM ATP, 0.1 mM CoA, 0.5 mM NADPH, 0.3 units of recombinant 3-hydroxypropionyl-CoA synthetase, and 10 mM acrylate. When the Km values for acryloyl-CoA and NADPH were determined, the concentration of one substrate was varied (NADPH, 0.0625 to 0.5 mM; acrylate, 0.001 to 10 mM), while the concentration of the other substrate was kept constant (acrylate, 0.1 mM; NADPH, 0.5 mM). The buffer used to determine the pH optimum was a mixture of sodium citrate, MES, 4-(2-hydroxyethyl)piperazine-1-ethylsulfonic acid (HEPES), N-tris(hydroxymethyl)methyl-4-aminobutanesulfonic acid, and glycine at a 100 mM concentration each. The pH was adjusted to 4.5 to 9.5 at room temperature, and 20 mM MgCl2 was added. To test the effect of metal chelating agents, protein fractions were incubated for 1 h at 30°C, 60°C, and 80°C with 0 and 100 mM EDTA. The incubated fractions were measured with the coupled assay for reductase activity.
(ii) 3-Hydroxypropionyl-CoA dehydratase.
3-Hydroxypropionyl-CoA was synthesized from 3-hydroxypropionate, MgATP, and CoA by the addition of 3-hydroxypropionyl-CoA synthetase from S. tokodaii (3). The formation of acryloyl-CoA by the dehydratase was followed by its NADPH-dependent reduction to propionyl-CoA by the addition of acryloyl-CoA reductase from S. tokodaii. The assay mixture (0.5 ml) contained 100 mM Tris-HCl, pH 8.6 (alternatively, 100 mM MES-NaOH, pH 6.0, during native purification of the enzyme), 20 mM MgCl2, 3 mM ATP, 0.1 mM CoA, 0.5 mM NADPH, 1 mM 3-hydroxypropionate, 0.1 units of recombinant 3-hydroxypropionyl-CoA synthetase, and 0.2 units of recombinant acryloyl-CoA reductase. The Km value of 3-hydroxypropionyl-CoA was determined by adding variable concentrations of 3-hydroxypropionyl-CoA (0.025 to 0.15 mM) to the coupled assay. The conversion of 3-hydroxybutyryl-CoA to crotonyl-CoA was measured at 40°C using recombinant crotonyl-CoA carboxylase/reductase from Rhodobacter sphaeroides (13). The assay mixture (0.5 ml) contained 100 mM MOPS-NaOH, pH 7.7, 20 mM MgCl2, 0.5 mM NADPH, 0.2 mM (R)- or (S)-3-hydroxybutyryl-CoA, 30 mM NaHCO3, and 5 units of recombinant crotonyl-CoA carboxylase/reductase. The addition of purified recombinant 3-hydroxypropionyl-CoA dehydratase started the reaction. Buffers used to determine the pH optimum were MES-NaOH (pH 5.9 to 7.0), MOPS-NaOH (pH 7.6 to 8.5), and Tris-Cl (pH 9.1 to 10.1).
(iii) Malate dehydrogenase.
The oxaloacetate-dependent oxidation of NADH was measured spectrophotometrically at 65°C. The reaction mixture (0.5 ml) contained 100 mM Tris-HCl, pH 8.0, 10 mM MgCl2, 0.5 mM NADH, and cell extract. In this case the reaction was started by the addition of 5 mM oxaloacetate.
Purification of acryloyl-CoA reductase from M. sedula.
All steps in the described purification protocols were performed at 4°C. The pH of buffers was adjusted at room temperature.
(i) Preparation of cell extract.
Autotrophically grown frozen cells (40 g, wet weight) were suspended in 40 ml of 100 mM Tris-HCl (pH 8.0)-5 mM MgCl2 containing 0.8 mg DNase I and passed twice through a chilled French pressure cell at 137 MPa. The cell lysate was centrifuged at 20,000 × g for 15 min, and the supernatant was centrifuged at 100,000 × g for 1 h.
(ii) Heat precipitation and dialysis.
Since acryloyl-CoA reductase activity gradually became inactivated at 90°C and above, the supernatant from centrifugation at 100,000 × g (100,000 × g supernatant) was incubated for 15 min at 85°C. The sample was then cooled on ice for 15 min to precipitate unwanted protein, lipids, and pigments, followed by centrifugation (100,000 × g) at 4°C for 60 min. The supernatant (42 ml) was dialyzed (exclusion size, 14 kDa) twice overnight against 2 liters of 20 mM Tris-HCl (pH 8.5) buffer (buffer A).
(iii) Q-Sepharose chromatography.
The extract from step ii (43 ml) was loaded at a flow rate of 1 ml min−1 onto a 40-ml Q-Sepharose column (GE Healthcare) equilibrated with buffer A. The active protein eluted with the flowthrough.
(iv) Carboxymethylcellulose chromatography.
The flowthrough from step iii (87 ml) was applied onto a 40-ml carboxymethylcellulose (CM52) column (Whatman) equilibrated with buffer A. The column was washed with buffer A and developed with a 60-ml linear gradient of 0 to 100 mM KCl in buffer A at 1 ml min−1. The active protein eluted at 30 mM KCl.
(v) Ammonium sulfate precipitation.
To the active fraction of step iv (12.6 ml) saturated ammonium sulfate solution was added to a final saturation of 25%, and the mixture was stirred for 10 min on ice and centrifuged at 20,000 × g for 10 min.
(vi) Phenyl-Sepharose chromatography and dialysis.
The supernatant from step v (16 ml) was loaded onto a 40-ml phenyl-Sepharose column (GE Healthcare) equilibrated with 50 mM Tris-HCl, pH 7.9 (buffer B), containing 1 M (NH4)2SO4. After a wash with equilibration buffer, the column was developed with a 200-ml linear gradient at 1 ml min−1 from 1 M to 0 M (NH4)2SO4 in buffer B. The peak of activity eluted at around 450 mM ammonium sulfate. The pooled active fractions (20 ml) were dialyzed (exclusion size, 14 kDa) overnight twice against 2 liters of 10 mM MOPS-NaOH, pH 7.0, buffer (buffer C).
(vii) Resource-S chromatography.
The enzyme solution after dialyses (22 ml) was applied onto a 1-ml Resource-S column (GE Healthcare) equilibrated with buffer C. The column was washed with buffer C and developed with a 20-ml linear gradient of 0 to 1 M KCl in buffer C at 1.0 ml min−1. Activity eluted at around 30 mM KCl.
Heterologous expression of the acryloyl-CoA reductase gene from S. tokodaii and production of the enzyme in E. coli.
The gene encoding acryloyl-CoA reductase was amplified by PCR from S. tokodaii chromosomal DNA by using a forward primer (5′-TAAGTTTCATATGAAAGCAATTGTAGTTCC-3′) introducing an NdeI site (underlined) at the initiation codon and a reverse primer (5′-CTTCACAATAGGATCCTATTCAGTTAA-3′) introducing a BamHI site (underlined) after the stop codon. PCR conditions were as follows: 25 cycles of 1 min of denaturation at 94°C, 1 min of primer annealing at 47°C, and 2 min of elongation at 72°C. Pfu polymerase (Gennaxon) was used. The PCR products were isolated and cloned into pUC19 (Stratagene), resulting in plasmid pJK1. The sequence of the insert was determined for the control. Ligation of the NdeI/BamHI fragment of pJK1 into the similarly restricted pET16b (Novagen) resulted in plasmid pJK3. Introduction of the NdeI/BamHI fragment into the expression vector pET16b results in the expression of an extended 5′ coding region, resulting in a N-terminal 10-His tag for the recombinant protein. Competent E. coli Rosetta 2 (DE3) cells were transformed with pJK3, grown at 37°C in 2 liter of LB medium containing 100 μg ampicillin ml−1 and 34 μg chloramphenicol ml−1, and induced at an optical density of 0.6 with 0.4 mM isopropyl thiogalactopyranoside (IPTG). After additional growth for 3 h at 37°C, the cells (4.3 g, wet weight) were harvested and stored in liquid nitrogen until use.
Purification of recombinant acryloyl-CoA reductase from S. tokodaii. (i) Preparation of cell extract.
Frozen E. coli cells (2 g, wet weight) were suspended in 2 ml of 100 mM Tris-HCl (pH 8.0)-5 mM MgCl2 containing 0.04 mg of DNase I and passed through a chilled French pressure cell at 138 MPa. The cell lysate was centrifuged at 20,000 × g for 15 min.
(ii) Heat precipitation.
Cell extract was incubated for 15 min at 85°C and then cooled on ice for 15 min to remove precipitated protein, lipids, and pigments, followed by centrifugation (100,000 × g) at 4°C for 60 min.
(iii) Affinity chromatography.
For nickel affinity chromatography, 2 ml of heat-precipitated cell extract was applied onto a 7-ml Ni2+-chelating Sepharose affinity column (GE Healthcare) equilibrated with 20 mM Tris-HCl (pH 7.9)-250 mM KCl (buffer D). The column was washed with buffer D containing 100 mM imidazole and developed with a 30-ml step from 0.1 to 0.5 M imidazole in buffer D at 1.0 ml min−1. Active fractions (12 ml) were pooled, mixed with 50 ml of buffer A, and concentrated to 5.2 ml by ultrafiltration (Amicon YM 10 membrane; Millipore). The protein was stored at −20°C in 30% glycerol.
Purification of 3-hydroxypropionyl-CoA dehydratase from M. sedula. (i) Preparation of cell extract.
Autotrophically grown frozen cells (8.2 g, wet weight) were suspended in 8 ml of 10 mM Tris-HCl (pH 7.2)-5 mM DTE containing 0.05 mg of DNase I and passed twice through a chilled French pressure cell at 137 MPa. The cell lysate was centrifuged at 100,000 × g for 1 h.
(ii) Heat precipitation and dialysis.
The 100,000 × g supernatant (5 ml) was incubated for 15 min at 65°C and then cooled on ice for 15 min, followed by centrifugation (20,000 × g) at 4°C for 15 min. The supernatant (4.5 ml) was dialyzed (exclusion size, 14 kDa) twice overnight against 2 liters of 20 mM Tris-HCl (pH 8.5).
(iii) Q-Sepharose chromatography.
The extract from step ii (4.7 ml) was loaded at a flow rate 0.5 ml min−1 onto a 5-ml Q-Sepharose column (GE healthcare) equilibrated with 20 mM Tris-HCl, pH 8.5. The column was washed with 20 mM Tris-HCl, pH 8.5 (buffer E), and developed with a 90-ml linear gradient of 0 to 150 mM KCl in buffer E at 1.0 ml min−1. Activity eluted between 30 and 70 mM KCl. Active fractions were pooled and concentrated to 5 ml by ultrafiltration (Amicon YM 10 membrane; Millipore).
(iv) Dialysis.
The protein fraction of step iii (5 ml) was dialyzed (exclusion size, 14 kDa) twice overnight against 2 liters of 20 mM Tris-HCl, pH 6.5 (buffer F).
(v) Carboxymethylcellulose chromatography.
The dialyzed extract from step iv (5 ml) was applied onto a 5-ml carboxymethylcellulose (CM52) column (Whatman) equilibrated with buffer F at a flow rate of 0.5 ml min−1. The column was washed with buffer F and developed with a 50-ml linear gradient of 0 to 150 mM KCl in buffer F at 1 ml min−1. The active protein eluted between 50 and 140 mM KCl. Active fractions were pooled and concentrated to 5 ml by ultrafiltration (Amicon YM 10 membrane; Millipore).
(vi) Affinity chromatography.
Affinity chromatography was performed at room temperature. The concentrated protein solution obtained by step v (5 ml) was applied at a flow rate of 0.25 ml min−1 onto a 1-ml Cibacron blue 3GA agarose 3000 CL column (Sigma-Aldrich) that had been equilibrated with 20 ml of 20 mM MES-NaOH, pH 7.0 (buffer G). The column was washed with buffer G and developed with 25 mM KCl steps in buffer G up to 250 mM KCl. Activity eluted between 175 mM KCl and 250 mM KCl. Active fractions were pooled (2 ml). The protein concentration of this pool was too low to be determined.
Heterologous expression of 3-hydroxypropionyl-CoA dehydratase gene from M. sedula and production of the enzyme in E. coli.
The gene encoding 3-hydroxypropionyl-CoA dehydratase was amplified from M. sedula chromosomal DNA by PCR by using a forward primer (5′-CTCGTATCACATATGGAATTTGAAACAATAG-3′) introducing an NdeI site (underlined) at the initiation codon and a reverse primer (5′-CGAAAGCTTTAATTCTAACCAGATTAATC-3′) introducing an HindIII site (underlined) after the stop codon. PCR conditions were as follows: 33 cycles of 45 s of denaturation at 94°C, 45 s of primer annealing at 52°C, and 2 min of elongation at 72°C. A mixture of Pfu and Taq polymerase was used. The PCR products were isolated and cloned into pUC18, resulting in plasmid pUC18Msed_2001. Ligation of the NdeI/HindIII fragment of pUC18Msed_2001 into the similarly restricted pET16b resulted in plasmid pET16bMsed_2001. The sequence of the insert was determined as a control. Introduction of the NdeI/HindIII fragment into the expression vector pET16b results in the expression of an extended 5′ coding region, resulting in a N-terminal 10-His tag for the recombinant protein. Competent E. coli Rosetta 2 (DE3) cells were transformed with pET16Msed_2001, grown in a 12-liter fermentor at 37°C in LB medium containing 100 μg of ampicillin ml−1 and 34 μg of chloramphenicol ml−1, and induced at an optical density of 0.7 with 0.5 mM IPTG. After additional growth for 6 h at 30°C, the cells (28 g, wet weight) were harvested and stored at −20°C until use.
Purification of heterologously expressed 3-hydroxypropionyl-CoA dehydratase from M. sedula. (i) Preparation of cell extract.
Frozen E. coli cells (5.8 g, wet weight) were suspended in 6 ml of 10 mM Tris-HCl (pH 7.2)-5 mM DTE containing 0.05 mg of DNase I and passed twice through a chilled French pressure cell at 137 MPa. The cell lysate was centrifuged at 100,000 × g for 1 h.
(ii) Heat precipitation.
The supernatant (6 ml) was incubated for 15 min at 85°C and then cooled on ice for 15 min, followed by centrifugation (20,000 × g) at 4°C for 15 min.
(iii) Affinity chromatography.
For nickel affinity chromatography, the supernatant (4.8 ml) was applied onto a 1-ml His-Trap FF column (GE Healthcare) equilibrated with 20 mM Tris-HCl (pH 7.8)-100 mM KCl (buffer H). The column was washed with buffer H containing 100 mM imidazole and developed with 0.5 M imidazole in buffer H at 1.0 ml min−1. Active fractions (3.5 ml) were pooled and stored at −20°C in 30% glycerol.
Protein analyzing methods.
Protein was determined by the method of Bradford (9) using bovine serum albumin as the standard. Protein fractions were analyzed by sodium dodecyl sulfate (SDS)-12.5% polyacrylamide gel electrophoresis (PAGE) (26). Proteins were visualized by Coomassie brilliant blue R-250 staining (38). In-gel protein digestion with trypsin and matrix-assisted laser desorption ionization-mass spectrometry analysis of peptides was carried out by TopLab (Martinsried, Germany). Proteins were identified using ProFound (Proteometrics) software and the NCBI database. The native molecular mass was determined on a 300-ml Superdex 200 (GE Healthcare) gel filtration column calibrated with ovalbumin (43 kDa), bovine serum albumin (67 and 134 kDa), aldolase (158 kDa), and ferritin (440 kDa). Purified recombinant acryloyl-CoA reductase (1.0 mg) was analyzed for metals by inductively coupled plasma optical emission spectroscopy in the Chemical Analysis Laboratory of R. Auxier (University of Georgia, Athens, GA).
Computational analysis.
The BLAST searches were performed via the NCBI BLAST server (http://www.ncbi.nlm.nih.gov/BLAST/) (4, 7). The amino acid sequences were aligned using CLUSTAL W (35) implemented within BioEdit software (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). The phylogenetic trees were reconstructed using neighbor-joining algorithms (29) in the TREECONW program package (36).
RESULTS
Reductive conversion of 3-hydroxypropionate by cell extracts.
A spectrophotometric enzyme assay was developed that used excess of purified 3-hydroxypropionyl-CoA synthetase (3) for the synthesis of 3-hydroxypropionyl-CoA or acryloyl-CoA from 3-hydroxypropionate or acrylate in the presence of MgATP and CoA. This synthetase acts nearly equally well on either carboxylic acid. Cell extracts of M. sedula catalyzed the additional NADPH-dependent transformation of 3-hydroxypropionyl-CoA. The specific activity in autotrophically grown cells was 37 nmol min−1 mg−1 of protein (at 65°C, pH 7.5 at room temperature), whereas in heterotrophically grown cells this activity was 3 nmol min−1 mg−1 of protein. When acryloyl-CoA was tested as the substrate, the same results were obtained. This indicates that extracts contained a 3-hydroxypropionyl-CoA dehydratase, which formed acryloyl-CoA, and a NADPH-dependent acryloyl-CoA reductase forming propionyl-CoA. The reductase activity may limit the conversion rate and clearly is more than 10-fold downregulated in heterotrophically grown cells. Whether the dehydratase is also downregulated and whether a single enzyme or two enzymes catalyze the two reactions cannot be decided based on these experiments.
Enrichment of acryloyl-CoA reductase from M. sedula.
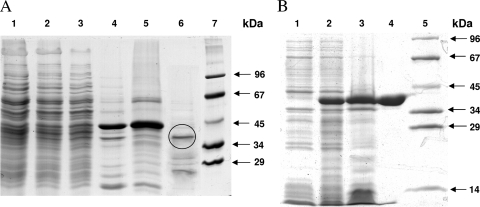
Based on the acryloyl-CoA-dependent oxidation of NADPH in the spectrophotometric assay, the enzyme that catalyzes acryloyl-CoA reduction to propionyl-CoA was enriched from extract of autotrophically grown cells. The enrichment after several purification steps was over 200-fold, and the yield was approximately 10% (Table 1). In 12.5% SDS-PAGE gels a prominent band of 40 kDa was seen (Fig. 2); it was analyzed by in-gel digestion with trypsin and peptide fingerprint mass spectrometry. At this stage of the work, the genome of the related S. tokodaii was available (25) but not that of M. sedula (5). Comparison with the database suggested that the 40-kDa protein was the putative acryloyl-CoA reductase. The best hit was found in S. tokodaii (100% identity and 3% coverage; 11 identical amino acids in a 334-amino-acid-containing protein; gene identifiers gi 15920695, NP_376364, and ST0480).
TABLE 1.
Partial purification of acryloyl-CoA reductase from M. sedula and heterologously produced acryloyl-CoA reductase from S. tokodaii
| Enzyme (accession no.) | Purification step | Volume (ml) | Total activity (μmol min−1)a | Protein (mg) | Specific activity (μmol mg−1 min−1) | Yield (%) | Purification (n-fold) |
|---|---|---|---|---|---|---|---|
| M. sedula reductase (Msed_1426) | Cell extract | 82.5 | 8,400 | ||||
| Heat precipitation (85°C) | 45.5 | 43 | 3,000 | 0.014 | 100 | 1 | |
| Dialysis | 43 | 40 | 1,800 | 0.018 | 94 | 2 | |
| Q-Sepharose | 86.5 | 42 | 470 | 0.09 | 98 | 6 | |
| Carboxymethylcellulose | 16.5 | 18 | 9.9 | 1.8 | 43 | 130 | |
| Phenylsepharose | 20 | 4.6 | 1.6 | 2.9 | 11 | 210 | |
| S. tokodaii reductase (ST0480) | Cell extract | 2.6 | 140 | ||||
| Heat precipitation (85°C) | 2 | 46 | 10 | 4.6 | 100 | 1 | |
| His-Trap | 7.8 | 32 | 1.7 | 18.7 | 70 | 4.1 |
Activities were determined using the respective photometric assay at 65°C.
FIG. 2.
SDS-PAGE (12.5%) of fractions obtained during purification of native and recombinant acryloyl-CoA reductase. Proteins were stained with Coomassie blue. (A) Enzyme fractions during purification of the native enzyme from M. sedula. Lane 1, cell extract of autotrophically grown cells (20 μg); lane 2, after heat precipitation (20 μg); lane 3, after ultracentrifugation (20 μg); lane 4, after Q-Sepharose chromatography (20 μg); lane 5, after carboxymethylcellulose chromatography (20 μg); lane 6, after phenyl-Sepharose chromatography (10 μg; after an additional purification step using a Resource S column the marked band was cut out and sequenced); lane 7, molecular mass standard proteins. (B) Heterologous expression of the acryloyl-CoA reductase gene from S. tokodaii in E. coli Rosetta 2 (DE3). Lane 1, whole cells before induction; lane 2, whole cells after 3 h of induced growth; lane 3, cell extract after heat precipitation (20 μg); lane 4, purified recombinant acryloyl-CoA reductase after Ni2+ affinity column (20 μg); lane 5, molecular mass standard proteins.
Heterologous production of acryloyl-CoA reductase from S. tokodaii in E. coli.
The putative reductase gene ST0480 from S. tokodaii coded for a hypothetical 36-kDa member of the zinc-containing alcohol dehydrogenase family (clusters of orthologous groups of proteins family 1064). It was cloned into the expression vector pET16b and expressed in E. coli Rosetta 2(DE3). This strain carries a plasmid (pRARE2) with genes for rare tRNA species. The His10 tag at the N terminus allowed an efficient purification of the enzyme after heat precipitation of most of the E. coli protein (Fig. 2 and Table 1). The overproduced enzyme was soluble, and the obtained preparation was virtually pure and showed one single 39-kDa band (expected value, 38.6 kDa) on SDS-PAGE gels (Fig. 2). The recombinant His-tagged enzyme catalyzed the acryloyl-CoA-dependent NADPH oxidation and formation of propionyl-CoA, whereas it was inactive with 3-hydroxypropionyl-CoA, indicating that it was the wanted reductase. The enzyme could be stored for months in 30% glycerol at −20°C. In the meantime, the genome of M. sedula became available (5). A gene encoding a protein with 76% amino acid sequence identity to the identified acryloyl-CoA reductase of S. tokodaii is present in the genome of M. sedula (gi 146304192; Msed_1426). Reinterpretation of the fingerprint mass spectrometry data identified the same gene (Msed_1426) in M. sedula as a best hit (100% identity and 73% coverage). It is located upstream of a gene encoding succinic seminaldehyde reductase (gi 146304190; Msed_1424), an enzyme also involved in autotrophic CO2 fixation (D. Kockelkorn and G. Fuchs, unpublished data).
Characterization of the recombinant acryloyl-CoA reductase.
The native molecular mass determined by gel filtration was 43 kDa, suggesting that the enzyme existed as a monomer. The catalytic properties were analyzed at 65°C by the coupled spectrophotometric assay. Controls showed that the enzyme exhibited neither 3-hydroxypropionyl-CoA synthetase nor 3-hydroxypropionyl-CoA dehydratase activity. Its optimum pH was 6.0 (65°C); half-maximal activity was obtained at pH 7.5. The enzyme activity followed Michaelis-Menten kinetics, with apparent Km values for NADPH of 36 μM and for acryloyl-CoA of around 3 μM. The specific activity was 18.7 μmol min−1 mg−1, corresponding to a turnover number of 13 s−1. The enzyme did not act on either NADH or crotonyl-CoA. Incubation with 100 mM EDTA for 1 h at up to 80°C did not inactivate the enzyme. Addition of Zn2+ or other divalent metal ions to the enzyme assay did not stimulate or inactivate activity. However, a metal analysis (27 elements) of acryloyl-CoA reductase by plasma emission spectroscopy revealed the presence of 0.8 mol of Zn2+ per mol of enzyme monomer, whereas other metals could not be detected. The UV-visible spectrum showed only a protein absorption band near 280 nm.
Spectrophotometric assay for 3-hydroxypropionyl-CoA dehydratase.
The availability of recombinant 3-hydroxypropionyl-CoA synthetase and acryloyl-CoA reductase allowed the design of a spectrophotometric assay for 3-hydroxypropionyl-CoA dehydratase. Assays containing both auxiliary enzymes in excess allowed detection and quantification of dehydratase activity in cell extract and protein fractions of M. sedula. Low concentrations of CoA (0.1 mM) were optimal and necessary because higher concentrations inhibited the assay. 3-Hydroxypropionyl-CoA dehydratase activity in extracts of autotrophically grown cells was 2.4 μmol min−1 mg−1 of cell protein and was not downregulated in heterotrophically grown cells (3.1 μmol min−1 mg−1 of cell protein). As a control, malate dehydrogenase was measured in both types of extracts. Its specific activity in autotrophically grown cells was 1.9 μmol min−1 mg−1 versus 3.5 μmol min−1 mg−1 in heterotrophically grown cells. Note that the heterotrophic growth rate was three times higher.
Purification of 3-hydroxypropionyl-CoA dehydratase from M. sedula.
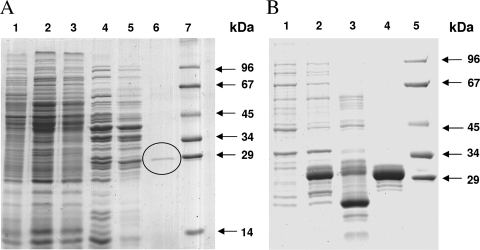
The enzyme was purified from autotrophically grown cells by six steps including use of a Cibacron blue affinity column with high-resolution power (Table 2 and Fig. 3). The enzyme was quite robust and was stored at 4°C. The preparation exhibited one dominant band of 29 kDa in 12.5% SDS-PAGE gels (Fig. 3). It was analyzed by in-gel digestion with trypsin and mass spectrometric peptide mass fingerprint analysis, followed by comparison with the database. At this stage of the work, the genome of M. sedula was available. The best hit was the Msed_2001 gene annotated as an enoyl-CoA hydratase gene (100% identity and 41% coverage) coding for a 28.3-kDa (259 amino acids) protein.
TABLE 2.
Partial purification of 3-hydroxypropionyl-CoA dehydratase from M. sedula (Msed_2001) and heterologously produced 3-hydroxypropionyl-CoA dehydratase from M. sedula
| M. sedula enzyme type | Purification step | Volume (ml) | Total activity (μmol min−1)a | Protein (mg) | Specific activity (μmol mg−1 min−1) | Yield (%) | Purification (n-fold) |
|---|---|---|---|---|---|---|---|
| Purified | Cell extract | 5 | 26.6 | 215 | 0.12 | 100 | 1 |
| Heat precipitation (65°C) | 4.5 | 21 | 71.1 | 0.3 | 79 | 2.5 | |
| Dialysis | 4.7 | 20.7 | 69 | 0.3 | 77.8 | 2.5 | |
| Q-Sepharose | 5 | 14.2 | 3.3 | 4.3 | 53 | 36 | |
| Carboxymethylcellulose | 5 | 3.6 | 0.5 | 7.2 | 13.5 | 58 | |
| Cibacron blue | 2 | ||||||
| Heterologously expressed | Cell extract | 6 | 234 | ||||
| Heat precipitation (65°C) | 4.8 | 271 | 28 | 9.9 | 100 | 1 | |
| His-Trap | 3.5 | 73 | 0.48 | 151 | 27 | 15 |
Activities were determined using the respective photometric assay at 65°C.
FIG. 3.
SDS-PAGE (12.5%) of fractions obtained during purification of native and recombinant 3-hydroxypropionyl-CoA dehydratase. Proteins were stained with Coomassie blue. (A) Enzyme fractions during purification of the native enzyme from M. sedula. Lane 1, cell extract of autotrophically grown cells (20 μg); lane 2, after heat precipitation (20 μg); lane 3, after dialysis overnight (20 μg); lane 4, after Q-Sepharose chromatography (10 μg); lane 5, after carboxymethylcellulose chromatography (10 μg); lane 6, after Cibacron blue chromatography (the marked band is 3-hydroxypropionyl-CoA dehydratase; protein amount not determined); lane 7, molecular mass standard proteins. (B) Heterologous expression of the 3-hydroxypropionyl-CoA dehydratase gene from M. sedula in E. coli Rosetta 2 (DE3). Lane 1, whole cells before induction; lane 2, whole cells after 3 h of induced growth; lane 3, cell extract after heat precipitation (20 μg); lane 4, purified recombinant 3-hydroxypropionyl-CoA dehydratase after Ni2+ affinity column (10 μg); lane 5, molecular mass standard proteins.
Heterologous production of 3-hydroxypropionyl-CoA dehydratase from M. sedula in E. coli and characterization of the recombinant enzyme.
The enzyme was produced as an N-terminal His10-tagged protein in E. coli. Most of the protein was insoluble (>90%), but in the 100,000 × g supernatant of the E. coli cell extract, sufficient dehydratase activity was still present even after heat precipitation to allow enzyme purification. After Ni affinity chromatography, an almost pure enzyme was obtained (Table 2 and Fig. 3). A strong protein band at around 30 kDa (expected size, 30.8 kDa) was observed in 12.5% SDS-PAGE gels, with traces of minor bands. The purified enzyme catalyzed the expected reaction at a rate of 151 μmol min−1 mg−1 of protein at 65°C. The UV-visible spectrum showed an absorption maximum around 270 nm. The calculated absorption coefficient at 280 nm (22,560 M−1 cm−1) compared reasonably well with the experimentally determined value (27,600 M−1 cm−1). The native molecular mass determined by gel filtration resulted in 23 kDa, indicating a monomeric enzyme. This experimental value may be underestimated due to the very small amounts of enzyme used. The optimum pH was at 8.1 (at room temperature, corresponding to pH 7.5 at 65°C), with half-maximal activities at pH 6.5 and 9.5 (as determined at room temperature; pH 6 and pH 8.5, respectively, when extrapolated to 65°C). The extrapolated maximum rate (Vmax) at 65°C was 186 μmol min−1 mg−1, corresponding to a turnover number of 96 s−1; the apparent Km for 3-hydroxypropionyl-CoA was 60 μM.
It was also tested whether the enzyme acted on 3-hydroxybutyryl-CoA. In this assay crotonyl-CoA formation was followed spectrophotometrically at 40°C using crotonyl-CoA carboxylase/reductase from R. sphaeroides (13). This auxiliary enzyme catalyzes the following reaction: crotonyl-CoA + CO2 + NADPH + H+ → ethylmalonyl-CoA + NADP+. 3-Hydroxypropionyl-CoA dehydratase acted nearly equally as well on (S)-3-hydroxybutyryl-CoA as on 3-hydroxypropionyl-CoA. The enzyme did not convert the (R)-stereoisomer of 3-hydroxybutyryl-CoA. The apparent Km value for (S)-3-hydroxybutyryl-CoA was 75 μM; the extrapolated Vmax was 34 μmol min−1 mg−1. When extrapolated to 65°C assuming doubling of the rate per 10°C temperature increase, the Vmax value was nearly same as that determined at 65°C for 3-hydroxypropionyl-CoA.
DISCUSSION
Role of the enzymes.
We characterized the missing enzymes 3-hydroxypropionyl-CoA dehydratase and acryloyl-CoA reductase that are required for the reductive conversion of 3-hydroxypropionate to propionyl-CoA in the process of CO2 fixation in autotrophic members of the Sulfolobales. Three separate enzymes are required, 3-hydroxypropionyl-CoA synthetase (3), 3-hydroxypropionyl-CoA dehydratase, and acryloyl-CoA reductase. In M. sedula cell extract the reductase was the rate-limiting enzyme. The overall activity of 3-hydroxypropionate reduction as well as the 3-hydroxypropionyl-CoA synthetase and the acryloyl-CoA reductase activities were upregulated in autotrophically grown cells. These results are consistent with the proposed function in autotrophic CO2 fixation. In Chloroflexus, all three steps are catalyzed by a large fusion protein with three enzyme domains (2). These enzyme domains are only distantly related to the three separate enzyme entities in the Sulfolobales (see below).
The autotrophic 3-hydroxypropionate/4-hydroxybutyrate carbon fixation cycle leads from acetyl-CoA plus two bicarbonate molecules to succinyl-CoA and back to two molecules of acetyl-CoA. The conversion of acetyl-CoA to succinyl-CoA via 3-hydroxypropionate probably has evolved independently in two genera in the order Sulfolobales (e.g., Sulfolobus and Metallosphaera sp.) and in the green non-sulfur bacteria (e.g., Chloroflexus sp.). This is concluded from the finding that the Metallosphaera and Chloroflexus enzymes or enzyme domains have been derived from genes that are only distantly related. The suggestion of a convergent evolution is further corroborated by the completely different genes coding for malonyl-CoA reductase (1) and malonate semialdehyde reductase (Kockelkorn and Fuchs, unpublished). In support of this, two completely different solutions have been realized in these two lineages to regenerate the CO2 acceptor molecule acetyl-CoA from succinyl-CoA.
Enzyme properties and catalyzed reactions.
The properties of the two enzymes are summarized in Table 3. Acryloyl-CoA reductase (E.C. 1.3.1.x) catalyzes the following irreversible reaction: acryloyl-CoA + NADPH + H+ → propionyl-CoA + NADP+. The enzyme does not require cofactors and does not use NADH as an electron donor. Also it does not act on crotonyl-CoA. This finding is important because crotonyl-CoA is an intermediate in the conversion of succinyl-CoA to two molecules of acetyl-CoA (Fig. 1). Nonspecific reduction of crotonyl-CoA to butyryl-CoA would be disastrous, resulting in a dead-end product (butyryl-CoA) that traps CoA. An NADH-dependent, oxygen-sensitive acryloyl-CoA reductase has been studied in Clostridium propionicum that is involved in propionate fermentation (18). The complex enzyme contains an electron-transferring flavoprotein and does not exhibit any similarity to the enzyme of M. sedula or to other trans-2-enoyl-CoA reductases.
TABLE 3.
Molecular and catalytic properties of recombinant acryloyl-CoA reductase from S. tokodaii and 3-hydroxypropionyl-CoA dehydratase from M. sedula
| Organism and enzymea | Substrates | Products | Specific activity (U/mg)b | Apparent Km (μM) | Optimum pH | Turnover (s−1) | Native molecular mass (kDa [calculated])c | Composition | Specificity (%) |
|---|---|---|---|---|---|---|---|---|---|
| S. tokodaii acryloyl-CoA reductase | Acryloyl-CoA, NADPH, H+ | Propionyl-CoA, NADP+ | 18.7 | Acryloyl-CoA, <10; NADPH, 36 | 6 | 13 | 43 (36) | Monomer | Acryloyl-CoA 100; crotonyl-CoA, <1 |
| M. sedula 3-hydroxypropionyl-CoA dehydratase | 3-Hydroxypropionyl-CoA, (S)-3-hydroxybutyryl-CoA | Acryloyl-CoA, crotonyl-CoA, H2O | 151 | 3-Hydroxypropionyl-CoA, 60; (S)-3-hydroxybutyryl-CoA, 75 | 8.1 | 96 | 23 (31) | Monomer | 3-Hydroxypropionyl-CoA, 100; (S)-3-hydroxybutyryl-CoA, 100; (R)-3-hydroxybutyryl-CoA, <1 |
Accession numbers are ST0480 for the S. tokodaii reductase and Msed_2001 for the M. sedula dehydratase.
Activities were determined using the respective photometric assay at 65°C.
Determined by gel filtration.
3-Hydroxypropionyl-CoA dehydratase (E.C.4.2.1.17) catalyzes the following reversible reaction: 3-hydroxypropionyl-CoA → acryloyl-CoA + H2O. It also catalyzes the following reversible reaction: crotonyl-CoA + H2O → (S)-3-hydroxybutyryl-CoA. The (R)-stereoisomer is not accepted when tested in the dehydration reaction. Crotonyl-CoA hydration takes part in the conversion of succinyl-CoA to two molecules of acetyl-CoA (Fig. 1), which also involves the (S)-stereoisomer rather than the (R)-stereoisomer of 3-hydroxybutyryl-CoA. Therefore, the dehydratase may fulfill a dual function.
Comparison with the corresponding domains of propionyl-CoA synthase from C. aurantiacus, distribution, and phylogenetic trees.
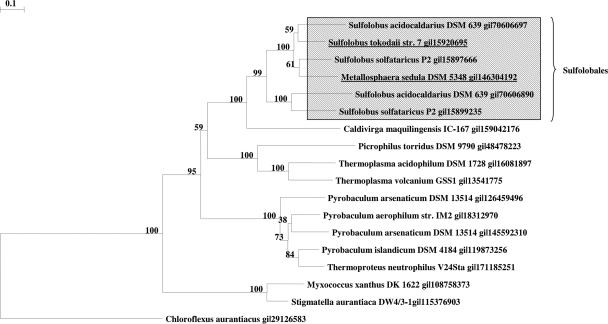
The genes coding for the two enzymes are not clustered on the Metallosphaera or Sulfolobus genome. The grouping of acryloyl-CoA reductase (Fig. 4) and of 3-hydroxypropionyl-CoA dehydratase (Fig. 5) with related proteins in the database reveals interesting aspects. Similar reductases exist in other autotrophic members of the Sulfolobales (Metallosphaera and Sulfolobus spp.) and probably fulfill the same function as in M. sedula. The function of similar enzymes in other Crenarchaea (Caldivirga, Picrophilus, Thermoplasma, Pyrobaculum, and Thermoproteus spp.) and Eubacteria (Myxobacteria) cannot be predicted. The reductase domain of propionyl-CoA synthase of C. aurantiacus shows only 28% amino acid sequence identity and 47% similarity over only one-fourth of the acryloyl-CoA reductase from M. sedula. In fact, only the NADPH binding site is highly conserved. Surprisingly, a similar gene product in the autotrophic marine group I Archaea (the Nitrosopumilus sp. Nmar_0523 and the Cenarchaeum sp. CENSYa_1407) shows only moderate similarity with the Metallosphaera enzyme (27% amino acid sequence identity and 45% similarity); these archaea have been proposed to use a similar autotrophic 3-hydroxypropionate/4-hydroxybutyrate cycle (8). Whether this discrepancy reflects large evolutionary distances between the Sulfolobales and the marine group I Crenarchaea, convergent evolution of the pathway in these groups, or eventually the operation of two different autotrophic pathways cannot be decided based on this study.
FIG. 4.
Phylogenetic tree of acryloyl-CoA reductase. The tree is based on amino acid sequence analysis and rooted with propionyl-CoA synthase from C. aurantiacus. Tree topography and evolutionary distances are given by the neighbor-joining method with Poisson correction. The scale bar represents a difference of 0.1 substitutions per site. Numbers at nodes indicate the percentage bootstrap values for the clade of this group in 1,000 replications. The enzyme group from the order Sulfolobales is indicated.
FIG. 5.
Phylogenetic tree of 3-hydroxypropionyl-CoA dehydratase. The tree is based on amino acid sequence analysis and rooted with propionyl-CoA synthase from C. aurantiacus. Tree topography and evolutionary distances are given by the neighbor-joining method with Poisson correction. The scale bar represents a difference of 0.1 substitutions per site. Numbers at nodes indicate the percentage bootstrap values for the clade of this group in 1,000 replications. The enzyme groups from the order Sulfolobales and from Clostridia are indicated. Fusion refers to the dehydratase domain of an archaeal fusion enzyme that contains an enoyl-CoA hydratase fused to a 3-hydroxyacyl-CoA dehydrogenase.
Also, similar dehydratases exist in other autotrophic members of the Sulfolobales and probably fulfill the same function as in Metallosphaera. Many members of the Thermoproteales (Thermoproteus, Pyrobaculum, and Caldivirga) and Desulfurococcales (Aeropyrum sp.) contain similar genes whose function is unknown. Furthermore, similar enzymes exist in Clostridiales and other Eubacteria. Again, the dehydratase domain of propionyl-CoA synthase of C. aurantiacus shows only 40% amino acid sequence identity and 57% similarity. A corresponding gene product in the autotrophic marine group I Archaea (Nitrosopumilus sp. Nmar_1308 and Cenarchaeum sp. CENSYa_0166) shows only moderate similarity with the Metallosphaera enzyme (42% amino acid sequence identity and 65 to 69% similarity for Nmar_1308 and 65% CENSYa_0166). It should be stressed, however, that the enoyl-CoA hydratase family and the Zn-containing alcohol dehydrogenase family comprise many only distantly related members and that the two reactions under study may well be catalyzed by members of other protein families.
Interestingly, Metallosphaera organisms as well as other autotrophic members of the Sulfolobales and Proteales contain another enzyme that catalyzes the (S)-3-hydroxybutyryl-CoA dehydratase/crotonyl-CoA hydratase reaction (Kockelkorn and Fuchs, unpublished). Work is in progress to characterize this second enzyme, which consists of an enoyl-CoA hydratase and a 3-hydroxyacyl-CoA dehydrogenase domain and which is twice as large as the enzymes under study here. In Fig. 5 the (de)hydratase domain of these fusion proteins is shown in the phylogenetic tree. Because the members of the Thermoproteales use a similar conversion of succinyl-CoA to two acetyl-CoA in their autotrophic dicarboxylate/4-hydroxybutyrate cycle (22), the role of this fusion protein is likely the same as in the Sulfolobales, i.e., the conversion of crotonyl-CoA via (S)-3-hydroxybutyryl-CoA to acetoacetyl-CoA. Cleavage of acetoacetyl-CoA by β-ketothiolase forms two molecules of acetyl-CoA, thus regenerating the CO2 acceptor and releasing another acetyl-CoA molecule for biosynthesis.
Related members of the Zn-containing alcohol dehydrogenase and enoyl-CoA hydratase families and possible active sites.
Acryloyl-CoA reductase belongs to the Zn-containing alcohol dehydrogenase enzyme family. It appears that the enzyme contains Zn2+ although it could not be inactivated by EDTA. A possible explanation for this apparent discrepancy could be an insufficient accessibility of zinc by EDTA and/or high thermal stability of the reductase (27). An alignment of the acryloyl-CoA reductase with the well-studied horse liver alcohol dehydrogenase shows the presence of the strictly conserved sequence motif GHE of the catalytic zinc binding side (amino acid position 59 to 61) with two flanking cysteines (amino acids 38 and 146). In addition, a conserved sequence for a structural zinc binding site DXCXXCXXXXXXXC (where X is any residue) can be found (amino acids 90, 93, 96, and 104). Here, amino acid 90 is changed from cysteine to aspartic acid, as already observed in Sulfolobus solfataricus and Aeropyrum pernix (6, 14). Whether the native and active reductases contain one or two zinc ions cannot be decided based on our data. 3-Hydroxypropionyl-CoA dehydratase belongs to the enoyl-CoA hydratase enzyme family. This class of enzymes catalyzes the syn-addition of a water molecule to a trans-α-β unsaturated CoA-activated acid. Since the OH− is added to the β-position and the H+ to the α-position from the 2-Re-side of crotonyl-CoA, the S-stereoisomer results. In the case of the best-studied crotonyl-CoA hydratase (crotonase) (S)-3-hydroxybutyryl-CoA is formed. The amino acids in the active sites of these enzymes are well conserved in the Metallosphaera enzyme.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie.
We thank Gabor Igloi, Freiburg, for DNA sequencing and Nasser Gad'on, Freiburg, for cell culturing.
Footnotes
Published ahead of print on 8 May 2009.
REFERENCES
- 1.Alber, B., M. Olinger, A. Rieder, D. Kockelkorn, B. Jobst, M. Hügler, and G. Fuchs. 2006. Malonyl-coenzyme A reductase in the modified 3-hydroxypropionate cycle for autotrophic carbon fixation in archaeal Metallosphaera and Sulfolobus spp. J. Bacteriol. 1888551-8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alber, B. E., and G. Fuchs. 2002. Propionyl-coenzyme A synthase from Chloroflexus aurantiacus, a key enzyme of the 3-hydroxypropionate cycle for autotrophic CO2 fixation. J. Biol. Chem. 27712137-12143. [DOI] [PubMed] [Google Scholar]
- 3.Alber, B. E., J. W. Kung, and G. Fuchs. 2008. 3-Hydroxypropionyl-coenzyme A synthetase from Metallosphaera sedula, an enzyme involved in autotrophic CO2 fixation. J. Bacteriol. 1901383-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. [DOI] [PubMed] [Google Scholar]
- 5.Auernik, K. S., Y. Maezato, P. H. Blum, and R. M. Kelly. 2008. The genome sequence of the metal-mobilizing, extremely thermoacidophilic archaeon Metallosphaera sedula provides insights into bioleaching-associated metabolism. Appl. Environ. Microbiol. 74682-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Auld, D. S., and T. Bergman. 2008. Medium- and short-chain dehydrogenase/reductase gene and protein families: the role of zinc for alcohol dehydrogenase structure and function. Cell. Mol. Life. Sci. 653961-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benson, D. A., I. Karsch-Mizrachi, D. J. Lipman, J. Ostell, and D. L. Wheeler. 2006. GenBank. Nucleic Acids Res. 34D16-D20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berg, I. A., D. Kockelkorn, W. Buckel, and G. Fuchs. 2007. A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Science 3181782-1786. [DOI] [PubMed] [Google Scholar]
- 9.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 10.Dawson, R. M. C., D. C. Elliott, W. H. Elliott, and K. M. Jones. 1986. Data for biochemical research, 3rd ed. Oxford University Press, New York, NY.
- 11.Eisenreich, W., G. Strauss, U. Werz, G. Fuchs, and A. Bacher. 1993. Retrobiosynthetic analysis of carbon fixation in the phototrophic eubacterium Chloroflexus aurantiacus. Eur. J. Biochem. 215619-632. [DOI] [PubMed] [Google Scholar]
- 12.Ellis, K. J., and J. F. Morrison. 1982. Buffers of constant ionic strength for studying pH-dependent processes. Methods Enzymol. 87405-426. [DOI] [PubMed] [Google Scholar]
- 13.Erb, T. J., I. A. Berg, V. Brecht, M. Müller, G. Fuchs, and B. E. Alber. 2007. Synthesis of C5-dicarboxylic acids from C2-units involving crotonyl-CoA carboxylase/reductase: the ethylmalonyl-CoA pathway. Proc. Natl. Acad. Sci. USA 10410631-10636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esposito, L., F. Sica, C. A. Raia, A. Giordano, M. Rossi, L. Mazzarella, and A. Zagari. 2002. Crystal structure of the alcohol dehydrogenase from the hyperthermophilic archaeon Sulfolobus solfataricus at 1.85 Å resolution. J. Mol. Biol. 318463-477. [DOI] [PubMed] [Google Scholar]
- 15.Good, N. E., and S. Izawa. 1972. Hydrogen ion buffers. Methods Enzymol. 2453-68. [DOI] [PubMed] [Google Scholar]
- 16.Herter, S., J. Farfsing, N. Gad'On, C. Rieder, W. Eisenreich, A. Bacher, and G. Fuchs. 2001. Autotrophic CO2 fixation by Chloroflexus aurantiacus: study of glyoxylate formation and assimilation via the 3-hydroxypropionate cycle. J. Bacteriol. 1834305-4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herter, S., G. Fuchs, A. Bacher, and W. Eisenreich. 2002. A bicyclic autotrophic CO2 fixation pathway in Chloroflexus aurantiacus. J. Biol. Chem. 27720277-20283. [DOI] [PubMed] [Google Scholar]
- 18.Hetzel, M., M. Brock, T. Selmer, A. J. Pierik, B. T. Golding, and W. Buckel. 2003. Acryloyl-CoA reductase from Clostridium propionicum. An enzyme complex of propionyl-CoA dehydrogenase and electron-transferring flavoprotein. Eur. J. Biochem. 270902-910. [DOI] [PubMed] [Google Scholar]
- 19.Holo, H. 1989. Chloroflexus aurantiacus secretes 3-hydroxypropionate, a possible intermediate in the assimilation of CO2 and acetate. Arch. Microbiol. 151252-256. [Google Scholar]
- 20.Holo, H., and R. Sirevåg. 1986. Autotrophic growth and CO2 fixation of Chloroflexus aurantiacus. Arch. Microbiol. 148173-180. [Google Scholar]
- 21.Huber, G., C. Spinnler, A. Gambacorta, and K. O. Stetter. 1989. Metallosphaera sedula gen. and sp. nov. represents a new genus of aerobic, metal mobilizing, thermoacidophilic Archaebacteria. Syst. Appl. Microbiol. 1238-57. [Google Scholar]
- 22.Huber, H., M. Gallenberger, U. Jahn, E. Eylert, I. A. Berg, D. Kockelkorn, W. Eisenreich, and G. Fuchs. 2008. A dicarboxylate/4-hydroxybutyrate autotrophic carbon assimilation cycle in the hyperthermophilic archaeum Ignicoccus hospitalis. Proc. Natl. Acad. Sci. USA 1057851-7856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hügler, M., H. Huber, K. O. Stetter, and G. Fuchs. 2003. Autotrophic CO2 fixation pathways in archaea (Crenarchaeota). Arch. Microbiol. 179160-173. [DOI] [PubMed] [Google Scholar]
- 24.Hügler, M., R. S. Krieger, M. Jahn, and G. Fuchs. 2003. Characterization of acetyl-CoA/propionyl-CoA carboxylase in Metallosphaera sedula. Carboxylating enzyme in the 3-hydroxypropionate cycle for autotrophic carbon fixation. Eur. J. Biochem. 270736-744. [DOI] [PubMed] [Google Scholar]
- 25.Kawarabayasi, Y., Y. Hino, H. Horikawa, K. Jin-no, M. Takahashi, M. Sekine, S. Baba, A. Ankai, H. Kosugi, A. Hosoyama, S. Fukui, Y. Nagai, K. Nishijima, R. Otsuka, H. Nakazawa, M. Takamiya, Y. Kato, T. Yoshizawa, T. Tanaka, Y. Kudoh, J. Yamazaki, N. Kushida, A. Oguchi, K. Aoki, S. Masuda, M. Yanagii, M. Nishimura, A. Yamagishi, T. Oshima, and H. Kikuchi. 2001. Complete genome sequence of an aerobic thermoacidophilic crenarchaeon, Sulfolobus tokodaii strain7. DNA Res. 8123-140. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 27.Magonet, E., P. Hayen, D. Delforge, E. Delaive, and J. Remacle. 1992. Importance of the structural zinc atom for the stability of yeast alcohol dehydrogenase. Biochem. J. 287361-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Menendez, C., Z. Bauer, H. Huber, N. Gad'on, K. O. Stetter, and G. Fuchs. 1999. Presence of acetyl coenzyme A (CoA) carboxylase and propionyl-CoA carboxylase in autotrophic Crenarchaeota and indication for operation of a 3-hydroxypropionate cycle in autotrophic carbon fixation. J. Bacteriol. 1811088-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4406-425. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 31.Stoll, V. S., and J. S. Blanchard. 1990. Buffers: principles and practice. Methods Enzymol. 18224-38. [DOI] [PubMed] [Google Scholar]
- 32.Strauss, G., W. Eisenreich, A. Bacher, and G. Fuchs. 1992. 13C-NMR study of autotrophic CO2 fixation pathways in the sulfur-reducing Archaebacterium Thermoproteus neutrophilus and in the phototrophic Eubacterium Chloroflexus aurantiacus. Eur. J. Biochem. 205853-866. [DOI] [PubMed] [Google Scholar]
- 33.Strauss, G., and G. Fuchs. 1993. Enzymes of a novel autotrophic CO2 fixation pathway in the phototrophic bacterium Chloroflexus aurantiacus, the 3-hydroxypropionate cycle. Eur. J. Biochem. 215633-643. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki, T., T. Iwasaki, T. Uzawa, K. Hara, N. Nemoto, T. Kon, T. Ueki, A. Yamagishi, and T. Oshima. 2002. Sulfolobus tokodaii sp. nov. (f. Sulfolobus sp. strain 7), a new member of the genus Sulfolobus isolated from Beppu Hot Springs, Japan. Extremophiles 639-44. [DOI] [PubMed] [Google Scholar]
- 35.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van de Peer, Y., and R. De Wachter. 1994. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10569-570. [DOI] [PubMed] [Google Scholar]
- 37.Zarzycki, J., A. Schlichting, N. Strychalski, M. Müller, B. E. Alber, and G. Fuchs. 2008. Mesaconyl-coenzyme A hydratase, a new enzyme of two central carbon metabolic pathways in bacteria. J. Bacteriol. 1901366-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zehr, B. D., T. J. Savin, and R. E. Hall. 1989. A one-step, low background Coomassie staining procedure for polyacrylamide gels. Anal. Biochem. 182157-159. [DOI] [PubMed] [Google Scholar]