Abstract
The mammalian target of rapamycin (mTOR) interacts with raptor to form the protein complex mTORC1 (mTOR complex 1), which plays a central role in the regulation of cell growth in response to environmental cues. Given that glucose is a primary fuel source and a biosynthetic precursor, how mTORC1 signaling is coordinated with glucose metabolism has been an important question. Here, we found that the glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase (GAPDH) binds Rheb and inhibits mTORC1 signaling. Under low-glucose conditions, GAPDH prevents Rheb from binding to mTOR and thereby inhibits mTORC1 signaling. High glycolytic flux suppresses the interaction between GAPDH and Rheb and thus allows Rheb to activate mTORC1. Silencing of GAPDH or blocking of the Rheb-GAPDH interaction desensitizes mTORC1 signaling to changes in the level of glucose. The GAPDH-dependent regulation of mTORC1 in response to glucose availability occurred even in TSC1-deficient cells and AMPK-silenced cells, supporting the idea that the GAPDH-Rheb pathway functions independently of the AMPK axis. Furthermore, we show that glyceraldehyde-3-phosphate, a glycolytic intermediate that binds GAPDH, destabilizes the Rheb-GAPDH interaction even under low-glucose conditions, explaining how high-glucose flux suppresses the interaction and activates mTORC1 signaling. Taken together, our results suggest that the glycolytic flux regulates mTOR's access to Rheb by regulating the Rheb-GAPDH interaction, thereby allowing mTORC1 to coordinate cell growth with glucose availability.
The mTOR complex 1 (mTORC1) signal transduction pathway acts as a central controller of cell growth in mammals (20, 23, 29). mTORC1 integrates a wide range of intracellular and extracellular signals, including insulin, availability of nutrients (glucose and amino acids), cellular energy status, and hypoxia, to regulate protein synthesis and cell growth (11, 12, 17, 36, 46). Many of these environmental cues are integrated into tuberous sclerosis complex (TSC1-TSC2), the major upstream regulator of mTORC1. In response to the absence of insulin and to the low-energy status of cells, the TSC1-TSC2 complex stimulates the GTPase function of Rheb, a small GTPase that acts as a proximal key activator of mTORC1, which leads to the inhibition of Rheb-mediated mTORC1 activation. In contrast, inactivation of the TSC1-TSC2 complex results in the accumulation of GTP-bound Rheb and thus activation of mTORC1 (3, 13, 21, 27, 32, 39). For this reason, both the loss of TSC proteins and the overexpression of Rheb cause hyperactivation of mTORC1 signaling, which is frequently observed in many common human cancers (2, 5, 19, 25, 33). Therefore, a tight regulation of Rheb activity is critical for the proper operation of the mTORC1 pathway in response to environmental cues.
Rheb is an atypical member of the Ras superfamily of GTPases (1, 10, 47). As with other small GTPases, the activity of Rheb is regulated by its guanine nucleotide binding status. However, the negative control of GTP-bound Rheb by the TSC1-TSC2 complex has only recently been investigated, and the regulation of the nucleotide binding status of Rheb is not fully understood. A recent study proposed that translationally controlled tumor protein may function as a guanine nucleotide exchange factor for Rheb that causes the accumulation of GTP-bound Rheb (18). GTP-bound Rheb is essential for activating mTOR kinase (21, 28, 38). However, the interaction between Rheb and mTOR does not depend on the GTP binding status of Rheb (30), raising questions regarding the mechanism by which Rheb activates mTORC1. Recently, FKBP38 (immunophilin FK506-binding protein, 38 kDa) was found to be a direct binding partner of Rheb and an inhibitor of mTORC1 (4). GTP-bound Rheb binds FKBP38 and releases FKBP38 from mTORC1, resulting in activation of the mTORC1 pathway. However, there have been conflicting results regarding the effects of nutrient availability on Rheb activity (31, 37, 42, 50) and the effect of these newly identified regulators of Rheb function (44, 45). Thus, the precise molecular mechanisms underlying Rheb regulation and Rheb-mediated mTORC1 activation have remained unclear.
In this study, we identified glyceraldehyde-3-phosphate (Gly-3-P) dehydrogenase (GAPDH) as a novel Rheb binding protein and a negative regulator of Rheb. We found that the interaction between GAPDH and Rheb is induced when the glycolytic flux is suppressed under low-glucose conditions to inhibit mTORC1. Here, we provide a molecular mechanism underlying the cross talk between the glycolytic flux and the mTORC1 signaling.
MATERIALS AND METHODS
Cell culture, transfection, and sample preparation.
HEK293, TSC1+/+, and TSC1−/− mouse embryo fibroblasts (MEFs) were maintained in Dulbecco's modified Eagle's medium (DMEM) (Cambrex) containing 10% fetal bovine serum (Cambrex). Transfection was performed using Lipofectamine reagent (Invitrogen), according to the manufacturer's instructions. For RNA interference experiments, the medium for cells was supplemented with 1 mM sodium pyruvate, and cells were transfected with 100 nM of GAPDH (Bionner and GenePharma), AMPKα1 (siGENOME SMARTpool M-005027-02-0005; Dharmacon), or control luciferase small interfering RNA (siRNA) (Dharmacon) using Lipofectamine reagent. Target sequences for human GAPDH_#1 and GAPDH_#2 were 5′-GTGTGACCATGAGAAGTA-3′ and 5′-GTATGACAACAGCCTCAAGTT-3′, respectively. To deprive cells of glucose, cells were starved in serum-free DMEM for 1 h and then incubated in low-glucose DMEM containing 5 mg/ml d-glucose (Gibco BRL). To resupply cells with normal glucose, the medium was replaced with serum-free DMEM containing 25 mg/ml d-glucose (Gibco BRL). After treatment, cells were washed twice with phosphate-buffered saline and then harvested with CHAPS lysis buffer (40 mM HEPES, pH 7.5, 0.5% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate [CHAPS], 120 mM NaCl, 1 mM EDTA, 10 mM pyrophosphate, 50 mM NaF, 1.5 mM Na2VO3, 10 mM β-glycerophosphate, 1 mM phenylmethylsulfonyl fluoride [PMSF], 10 mM MgCl2, and 10 μg/ml leupeptin). The cell extracts generated were spun at 14,000 rpm for 15 min and then stored at −80°C.
Chemicals and materials.
All chemicals were purchased from Sigma unless stated otherwise. Anti-Myc 9E10 and antihemagglutinin (anti-HA) 12CA5 were harvested from the supernatants of corresponding hybridoma cell lines. Anti-Rheb (C19) antibodies were purchased from Santa Cruz, and anti-mTOR, Rheb, Raptor, pS6K1 (pThr389), S6K1, p4EBP1 (pThr37/46), 4EBP1, TSC1, pAMPK (pThr147), pERK (pThr202/Tyr204), extracellular signal-regulated kinase, pAKT (Ser473), AKT, ACC antibodies, and rapamycin were from Cell Signaling. Anti-GAPDH monoclonal antibodies were purchased from Biogenesis (Kingstone, NH), and pACC (Ser79) antibodies were purchased from Upstate Biotechnology. Compound C was obtained from Merck. Protein A-Sepharose and protein G-Sepharose beads were from RepliGen (Needham, MA) and Pierce, respectively. Horseradish peroxidase-conjugated goat anti-mouse immunoglobulin A (IgA)-IgM-IgG, peroxidase-conjugated goat anti-rabbit IgG, and peroxidase-conjugated rabbit anti-goat IgG antibodies were purchased from Kirkegaard and Perry Laboratories (Gaithersburg, MD). The enhanced chemiluminescence kit was purchased from Amersham Biosciences International (Buckinghamshire, United Kingdom).
Identification of Rheb binding proteins.
Rat brains were homogenized in lysis buffer (20 mM Tris-HCl, pH 7.8, 150 mM NaCl, 1 mM MgCl2, 1 mM EGTA, 1% Triton X-100) and cleared by centrifugation for 15 min at 14,000 rpm. Supernatants were spun at 67,000 rpm for 30 min, and brain extracts were incubated with immobilized glutathione S-transferase (GST)-tagged Rheb proteins for 2 h at 4°C. Proteins unique to Rheb were excised from the gel and digested with trypsin. Tryptic peptides were analyzed with a 4700 proteomics analyzer (Applied Biosystems, Framingham, MA). The measured spectrum was analyzed using an in-house version of Mascot (version 2.0) for peptide mass fingerprinting. The NCBInr database was used, and the Mascot score for a significant hit (P < 0.05) was greater than 67.
Plasmids.
HA-tagged Rheb clones and pGEX-2T-RhebWT were kindly provided by Ariel F. Castro (Indiana University School of Medicine), and Myc-mTOR was kindly provided by David M. Sabatini (Massachusetts Institute of Technology). Full-length Rheb cDNA obtained by PCR was subcloned into pEGFP-C1 (BD Biosciences) and pRSETB (Invitrogen) with an N-terminal green fluorescent protein (GFP) tag or His tag, respectively. To generate the GST-tagged Rheb fragment, the fragment of rat Rheb cDNA obtained by PCR was cloned into pGEX-4T-1 (Amersham Pharmacia Biotech). cDNAs encoding human GAPDH were obtained by PCR and subcloned into N-terminal pFLAG-CMV (Sigma).
Immunoprecipitation.
For immunoprecipitation, 0.5- to 1-mg aliquots of cell extracts were incubated with 2 μg of the indicated antibodies for 6 h at 4°C under gentle agitation. Immunocomplexes were collected with protein A- (RepliGen) or protein G-Sepharose beads (Pierce). Whole-cell lysates or immunoprecipitates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting. All immunoblots were detected by enhanced chemiluminescence (ECL system; Amersham).
In vitro binding assay.
GST-tagged Rheb proteins were immobilized on glutathione-Sepharose beads (GE Healthcare) and resuspended in binding buffer (40 mM HEPES, pH 7.5, 0.5% Triton X-100, 120 mM NaCl, 1 mM EDTA, 1 mM PMSF, and 10 mM MgCl2). To obtain soluble proteins, GST-tagged GBR was eluted off the beads with 50 mM glutathione in 50 mM Tris-HCl (pH 7.5). To prepare His-tagged Rheb proteins, cell lysates were incubated with nickel-nitrilotriacetic acid (Ni-NTA) agarose beads (Qiagen) and washed with buffer containing 20 mM, 50 mM, and 100 mM imidazole, in sequence. The beads were subsequently resuspended in binding buffer. For charging GST-Rheb with nucleotides in vitro, immobilized GST or GST-Rheb was washed with nucleotide-free buffer (50 mM Tris-HCl, pH 7.5, 250 mM NaCl, 0.5% Triton X-100, 1 mM dithiothreitol, 1 mM PMSF, and 20 mM EDTA). Beads were washed with loading buffer (40 mM HEPES, pH 7.5, 120 mM NaCl, 5 mM EDTA, and 2 mM MgCl2) and nucleotide preloaded with 200 μl of loading buffer containing 100 μM GDPβS or 100 μM GTPγS for 1 h at 4°C, and the process was then terminated by adding 5 μl of 1 M MgCl2. For nucleotide-free Rheb, beads were resuspended in nucleotide-free buffer. A 50 nM concentration of purified rabbit muscle GAPDH (Sigma) was incubated with 1 μg of immobilized GST-Rheb on glutathione-Sepharose or GST-glutathione-Sepharose for 2 h at 4°C in binding buffer. For in vitro competitive assays using GST-tagged GBR, GAPDH was mixed with immobilized His-tagged full-length Rheb on Ni-NTA agarose. Purified GST-GBR was then added in increasing amounts (0, 1, and 5 μg) and incubated for 2 h at 4°C.
RESULTS
Glycolytic flux regulates the interaction between Rheb and GAPDH.
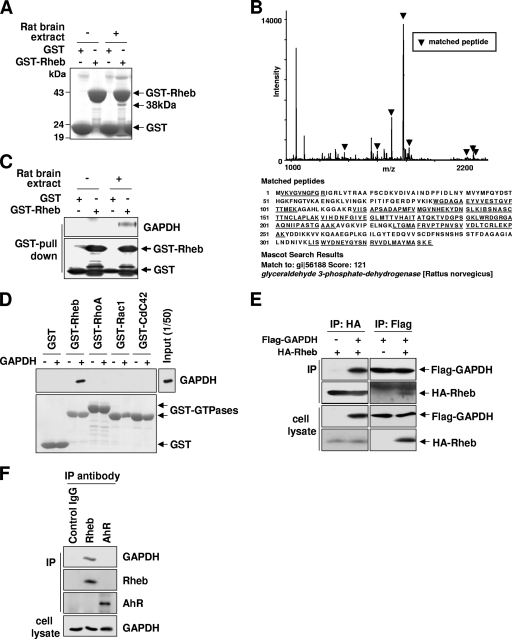
To gain a clearer understanding of the mechanisms that regulate Rheb GTPase, we sought to identify Rheb-interacting proteins. GST-tagged Rheb was incubated with rat brain extracts, and proteins specifically bound to GST-Rheb were resolved by SDS-PAGE and identified by mass spectrometry. One of the proteins identified was GAPDH, a glycolytic enzyme that converts Gly-3-P to 1,3-bisphosphoglycerate (Fig. 1A to C). Because GAPDH is a highly abundant protein, we analyzed the specificity of the interaction by using several control proteins and antibodies. We purified GST-tagged Rheb and other small GTPases from Escherichia coli and incubated them with purified GAPDH in vitro. Supporting the specificity of the Rheb-GAPDH interaction, GAPDH precipitated with Rheb but not with the other small GTPases (Fig. 1D). To confirm the interaction between GAPDH and Rheb in intact cells, we examined the association between transfected HA-Rheb and Flag-GAPDH in HEK293 cells. We found that immunoprecipitation with either Flag or HA antibodies revealed a specific interaction between Rheb and GAPDH (Fig. 1E). Next, we tested whether endogenous GAPDH interacts with endogenous Rheb. Immunoprecipitates obtained using an antibody specific to Rheb, but not control antibodies, contained endogenous GAPDH (Fig. 1F).
FIG. 1.
Identification of p38 as GAPDH. (A) Identification of GAPDH as a Rheb-associated protein. Precipitates of GST-Rheb from rat brain extracts were separated by SDS-PAGE and stained with Coomassie blue. The 38-kDa band was identified as GAPDH. (B) In-gel digestion and mass spectrometry analysis were done for identification of p38. Peptide mass fingerprinting showed that eight peptides were matched to the deduced sequence of a rat protein in the database, GAPDH. Matched peaks in the spectrum are indicated by arrowheads, and amino acid sequences of rat GAPDH are shown. Matched peptide sequences are shown by underlining. (C) Confirmation of identification of GAPDH as Rheb-associated protein. Precipitates of GST-Rheb from rat brain extracts were separated by SDS-PAGE and immunoblotted with anti-GAPDH antibodies. The GST immunoblot shows the relative amounts of GST proteins used. (D) GAPDH directly interacts with Rheb but not other small GTPases. GST, GST-Rheb, GST-RhoA, GST-Rac1, or GST-Cdc42 was incubated with purified rabbit muscle GAPDH. After GST pulldown assays were performed, bound GAPDH was analyzed by immunoblotting with anti-GAPDH antibodies (upper panel). The amounts of GST-small GTPases used were shown by Ponceau S staining (lower panel). (E) Coimmunoprecipitation of GAPDH and Rheb in intact cells. HEK293 cells were transfected with either HA-Rheb or Flag-tagged GAPDH alone or together with both. Cell lysates were immunoprecipitated with anti-HA antibodies or anti-Flag antibodies and then analyzed by Western blotting with indicated antibodies. (F) Confirmation of the interaction between endogenous GAPDH and Rheb. Immunoprecipitates were isolated from HEK293 cells by using Rheb-specific or control antibodies (anti-aryl hydrocarbon receptor [AhR] antibody and control goat IgG antibody). The amount of GAPDH isolated with Rheb was detected by immunoblotting with anti-GAPDH antibody.
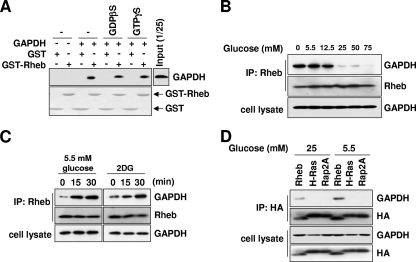
To further confirm the interaction between GAPDH and Rheb, we asked whether the interaction might be regulated by the state of the cell. First, we tested whether the interaction is regulated by the GTP-GDP-bound state of Rheb. The binding of GAPDH to Rheb did not depend on the guanyl nucleotide charge status of Rheb (Fig. 2A), indicating that GAPDH is unlikely to be either a guanine nucleotide exchange factor or a GTPase-activating protein. Next, we tested whether the interaction is regulated by glucose availability. Given that GAPDH is involved in glucose metabolism (8, 16) and that the GAPDH-mediated reaction is determined by extracellular glucose availability (6, 43), we reasoned that glucose availability might regulate the interaction of GAPDH to Rheb. The binding of endogenous GAPDH to Rheb was strongly enhanced in the absence of glucose, and this binding gradually decreased as extracellular glucose levels were elevated (Fig. 2B). The binding of Rheb to GAPDH was increased within 15 min of switching cells to low-glucose-containing medium (Fig. 2C). Moreover, treatment with the glucose analog 2-deoxyglucose (2DG), which inhibits glycolysis (7), strongly induced the interaction between Rheb and GAPDH (Fig. 2C). We confirmed that low-glucose conditions induce the binding of GAPDH to Rheb but not to other small GTPases such as H-Ras and Rap2A (Fig. 2D). These results support the idea that GAPDH specifically interacts with Rheb and that the glycolytic flux regulates the Rheb-GAPDH interaction.
FIG. 2.
GAPDH interacts with Rheb, and the interaction is regulated by glycolytic flux. (A) The interaction of Rheb with GAPDH does not depend on its nucleotide binding status. GST or GST-Rheb was charged with 100 μM GDPβS or 100 μM GTPγS. Each species of GST or GST-Rheb was incubated with purified rabbit muscle GAPDH. After GST pulldown assays were performed, bound GAPDH was analyzed by immunoblotting with anti-GAPDH antibodies (upper panel). The amount of GST-Rheb used was shown by Ponceau S staining (lower panel). (B) Interaction between Rheb and GAPDH is regulated by extracellular glucose availability. HEK293 cells were incubated in media containing the indicated concentrations of glucose for 30 min. Lysates prepared from these cells were immunoprecipitated with anti-Rheb antibodies. GAPDH bound to Rheb was analyzed by anti-GAPDH immunoblotting. (C) The inhibition of glycolytic flux induces Rheb-GAPDH interaction. HEK293 cells were treated with 25 mM 2DG or incubated in 5.5 mM glucose medium for the indicated times. Lysates prepared from these cells were immunoprecipitated with anti-Rheb antibodies. GAPDH bound to Rheb was analyzed by anti-GAPDH immunoblotting. (D) GAPDH specifically interacts with Rheb but not other Ras GTPases. HEK293 cells were transfected with wild-type HA-Rheb, HA-H-Ras, or HA-Rap2A and incubated in media containing the indicated concentration of glucose for 30 min. Lysates prepared from these cells were immunoprecipitated with anti-HA antibodies. GAPDH bound to Rheb was analyzed by anti-GAPDH immunoblotting.
Gly-3-P, an intermediate of the glycolytic pathway, destabilizes the interaction between GAPDH and Rheb.
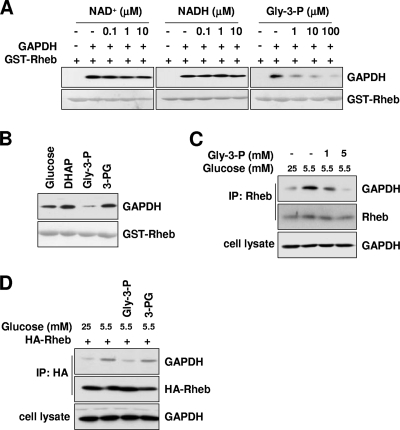
Knowing that glycolytic flux regulates the GAPDH-Rheb interaction, we wondered whether any glycolytic metabolite might regulate this interaction. GAPDH converts Gly-3-P to 1,3-bisphosphoglycerate while reducing NAD+ to NADH (16, 35, 49). Therefore, we asked whether any of the metabolites capable of binding GAPDH might regulate the interaction between GAPDH and Rheb. Neither NAD+ nor NADH had any effect on the Rheb-GAPDH interaction, but Gly-3-P showed an inhibitory effect on the interaction (Fig. 3A). We also tested other glycolytic metabolites such as glucose, dihydroxyacetone phosphate, and 3-phosphoglycerate (3-PG) without seeing any significant effects on the interaction (Fig. 3B). Furthermore, we found that the Rheb-GAPDH interaction under low-glucose conditions was reduced by Gly-3-P in a dose-dependent manner (Fig. 3C). In contrast, 3-PG (which is both a structural analog and a metabolic product of Gly-3-P) had no effect on the Rheb-GAPDH interaction (Fig. 3D). These results suggest that the GAPDH-Rheb interaction is negatively regulated by the binding of the glycolytic pathway intermediate Gly-3-P to GAPDH.
FIG. 3.
Gly-3-P, a glycolytic intermediate, destabilizes the Rheb-GAPDH interaction. (A) Gly-3-P, a glycolytic intermediate that binds GAPDH, inhibits Rheb-GAPDH interaction. GST-Rheb was incubated with purified rabbit muscle GAPDH in the presence of the indicated amounts of NAD+, NADH, or Gly-3-P. (B) The Rheb-GAPDH interaction is specifically inhibited by the glycolytic intermediate Gly-3-P. A 100 μM concentration of glycolytic intermediates was coincubated with purified rabbit muscle GAPDH and GST-Rheb in in vitro binding assays. DHAP, dihydroxyacetone phosphate. (A and B) After GST pulldown assays were performed, bound GAPDH was analyzed by immunoblotting with anti-GAPDH antibodies (upper panels). The amounts of GST-Rheb used were shown by Ponceau S staining (lower panels). (C) Glucose depletion-induced Rheb-GAPDH interaction is restored by Gly-3-P. HEK293 cells were incubated with 5.5 mM glucose medium in the absence or presence of the indicated concentrations of Gly-3-P for 30 min. Cell lysates were incubated with Rheb antibodies. Immunoprecipitates were blotted with indicated antibodies for GAPDH and Rheb. (D) Glucose depletion-induced Rheb-GAPDH binding is recovered by Gly-3-P but not another metabolite, 3-PG. HEK293 cells transfected with HA-Rheb were incubated with 5.5 mM glucose medium in the presence of either Gly-3-P or 3-PG for 30 min. Cell lysates prepared from these cells were immunoprecipitated with anti-HA antibodies. GAPDH bound to Rheb was analyzed by anti-GAPDH immunoblotting. (C and D) Cells were treated with glycolytic metabolites as previously described (34).
GAPDH negatively regulates mTORC1.
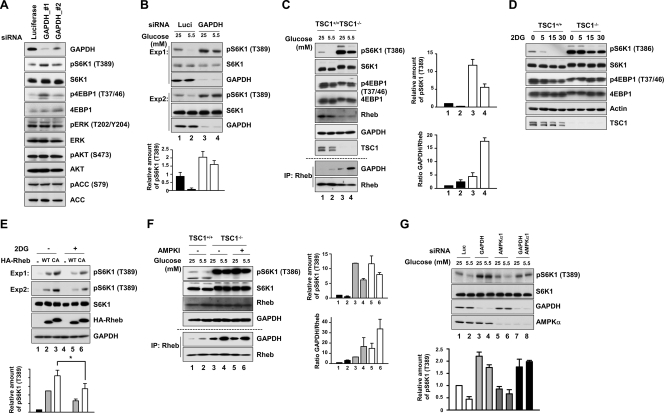
Knowing that GAPDH interacts with Rheb and that the interaction is regulated by the glycolytic pathway, we next asked whether this interaction affects the activity of mTORC1. We first analyzed the effect of GAPDH knockdown on the activity of mTORC1 by examining the phosphorylation states of S6K1 and 4EBP1. For this experiment, cells were supplemented with 1 mM sodium pyruvate to prevent cell death due to metabolic defects (15, 51). Knockdown of GAPDH using two distinct nonoverlapping siRNAs against GAPDH increased the phosphorylation states of both S6K1 and 4EBP1 but had no effect on other signaling molecules such as AKT, extracellular signal-regulated kinase, and ACC (Fig. 4A), indicating that GAPDH specifically regulates the mTORC1 pathway. The phosphorylation level inversely correlated with the amount of GAPDH remaining in the cell (Fig. 4A). This result indicates that GAPDH basally modulates mTORC1 activity under normal growth condition (25 mM glucose). We next tested the effect of GAPDH silencing under low-glucose conditions (5.5 mM glucose). HEK293 cells transfected with either control luciferase siRNA or GAPDH siRNA were cultured in low- and high-glucose medium, and the phosphorylation state of S6K1 was analyzed. Knockdown of GAPDH increased the basal phosphorylation of S6K1 under the high-glucose condition (as in Fig. 4A), and it significantly suppressed the inhibitory effects of low glucose levels on S6K1 phosphorylation (Fig. 4B). This result supports the idea that GAPDH is important for the glucose-responsive regulation of mTORC1 activity.
FIG. 4.
GAPDH has a negative effect on the mTORC1 activity. (A) GAPDH knockdown increases mTORC1 activity. HEK293 cells were transfected with GAPDH_#1, GAPDH_#2, or luciferase siRNA and then cultured in 25 mM glucose medium for 48 h. Cell lysates were prepared and analyzed by immunoblotting for the indicated proteins and phosphorylation states. (B) The negative effect of glucose depletion on mTORC1 activity is abolished in GAPDH-silenced cells. HEK293 cells expressing GAPDH_#1 or luciferase siRNA were incubated with media with the indicated concentrations of glucose for 30 min. Two examples of S6K1 phosphorylation obtained from three independent experiments are shown. The bar graph at the bottom shows the quantification of S6K1 phosphorylation. (C) mTORC1 signaling remains responsive to glucose depletion in TSC1-deficient cells, and the Rheb-GAPDH interaction is increased under glucose depletion conditions. TSC1+/+ and TSC1−/− MEFs were incubated with media with the indicated concentrations of glucose for 30 min. The phosphorylation of endogenous S6K1 and 4EBP1 was determined using phospho-specific antibodies for T389 and T37/46, respectively. The same cell lysates were immunoprecipitated with anti-Rheb antibodies. Immunoprecipitates were blotted with the indicated antibodies for GAPDH and Rheb. The bar graph shows the quantification of S6K1 phosphorylation and GAPDH/Rheb ratios in immunoprecipitations from four different experiments. (D) TSC1+/+ and TSC1−/− MEFs were incubated with 25 mM 2DG for the indicated times. The phosphorylation of endogenous S6K1 and 4EBP1 was determined using phospho-specific antibodies for T389 and T37/46, respectively. (E) Inhibition of glycolysis flux reduces the constitutively active mutant of Rheb-induced S6K1 phosphorylation. Cells transfected with either HA-Rheb WT or HA-Rheb CA were treated with 25 mM 2DG for 30 min. Two examples of S6K1 phosphorylation obtained from three independent experiments are shown. The bar graph at the bottom shows the quantification of S6K1 phosphorylation (*, P < 0.05). (F) The mTORC1 pathway was still responsive to glucose depletion under AMPK-inhibited conditions in TSC1-deficient MEFs. TSC1−/− MEFs were pretreated with the AMPK inhibitor (AMPKI) compound C (10 μM) for 30 min and then were incubated with media with the indicated concentration of glucose for 30 min. The phosphorylation of endogenous S6K1 was determined using phospho-specific antibodies for T389. The same cell lysates were immunoprecipitated with anti-Rheb antibodies. Immunoprecipitates were blotted with the indicated antibodies for GAPDH and Rheb. The bar graph shows the quantification of S6K1 phosphorylation and GAPDH/Rheb ratios in immunoprecipitations from three different experiments. (G) mTORC1 signaling remains responsive to glucose depletion in AMPKα1-silenced cells, and silencing of both AMPKα1 and GAPDH completely desensitizes mTORC1 signaling in response to glucose depletion. HEK293 cells expressing GAPDH_#1, AMPKα1, or luciferase siRNA were incubated with media with the indicated concentrations of glucose for 30 min. The phosphorylation of endogenous S6K1 was determined using phospho-specific antibodies for T389. The levels of expression of S6K1, GAPDH, and AMPKα1 were analyzed using anti-S6K1, anti-GAPDH, and anti-AMPKα antibodies. For quantitation of total protein, we used equal amounts of the same cell lysates that were used for detecting phosphorylated proteins by Western blotting separately. The bar graph shows the quantification of S6K1 phosphorylation from three different experiments.
Glucose availability has been known to regulate mTORC1 via the AMPK-TSC pathway and AMPK-mediated phosphorylation of raptor (14, 22). Our findings suggest that a novel additional signaling axis regulates mTORC1 in response to glucose availability. Supporting the idea that the AMPK-TSC axis is not the sole glucose-responsive pathway of mTORC1 activation, mTORC1 activity was still inhibited by glucose deprivation and 2DG in both TSC1-deficient MEFs and TSC1+/+ wild-type (WT) cells (Fig. 4C and D). In addition, incubation of cells with 2DG suppressed the phosphorylation of S6K1 induced either by WT or by a constitutively active Rheb mutant (Fig. 4E). To exclude a role for the AMPK-raptor axis in response to glucose depletion in TSC1-deficient cells, we monitored the effect of the specific AMPK inhibitor compound C on mTORC1 activity after glucose deprivation in TSC1-deficient cells. We found that mTORC1 was still inhibited by glucose depletion in AMPK inhibitor-treated TSC1-deficient cells (Fig. 4F). We also found that the interaction between GAPDH and Rheb was induced by glucose depletion in the absence of TSC1 (Fig. 4C) and even in AMPK-inhibited TSC1-deficient cells (Fig. 4F). This result suggests that the GAPDH-Rheb interaction is regulated by glucose availability independently of both the AMPK-TSC axis and the AMPK-raptor axis. We also found that AMPKα1-silenced HEK293 cells remain responsive to glucose deprivation (Fig. 4G, lanes 5 and 6), further supporting this idea. More importantly, the silencing of both AMPKα1 and GAPDH completely abolished mTORC1 signaling in response to glucose depletion (Fig. 4G, lanes 7 and 8). These results suggest that the GAPDH-Rheb axis constitutes a new pathway that regulates mTORC1 in response to glucose availability independently of the AMPK-TSC axis and the AMPK-raptor axis.
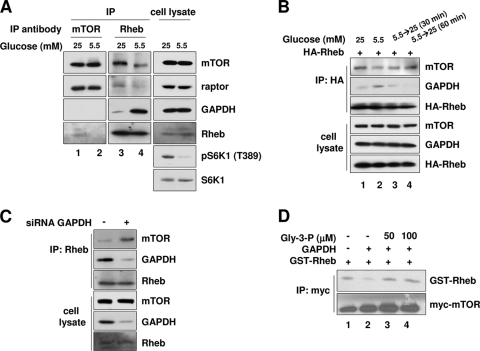
GAPDH dissociates Rheb from mTORC1 under glucose-depleted conditions.
Having characterized the inhibitory effect of GAPDH on mTORC1 signaling, we next sought to understand how the interaction between GAPDH and Rheb regulates mTORC1 signaling. Our results above showed that the binding of Rheb to GAPDH is enhanced under low-glucose conditions and that this correlates with the inhibition of mTORC1 signaling. We therefore reasoned that the interaction between GAPDH and Rheb somehow inhibits mTORC1's activity. We considered two possibilities for how this might occur: GAPDH might bind mTORC1 via Rheb and inhibit mTORC1 directly, or GAPDH could inhibit mTORC1 activity indirectly by sequestering Rheb and preventing it from binding to mTORC1. Our subsequent experiments supported this second mechanism. First, we found that glucose depletion stabilized the interaction between GAPDH and Rheb, whereas it destabilized the interaction between Rheb and mTOR. We confirmed these results by analyzing the interactions of both endogenous and recombinant proteins (Fig. 5A and B). Second, we could not detect GAPDH in mTORC1 immunoprecipitates, a result that excludes the first possibility (Fig. 5A, lanes 1 and 2). Third, when Rheb was isolated from GAPDH-depleted cells, more mTOR was found to be bound to it than when Rheb was isolated from control siRNA-transfected cells (Fig. 5C). These results suggest that GAPDH binds more extant Rheb under low-glucose conditions and thereby reduces the amount of Rheb bound to mTOR.
FIG. 5.
GAPDH causes Rheb to dissociate from mTORC1 under low-glucose conditions. (A) Coimmunoprecipitation of endogenous proteins. HEK293 cells were incubated with media with the indicated concentrations of glucose for 30 min and then immunoprecipitated with either anti-mTOR (lanes 1 and 2) or anti-Rheb (lanes 3 and 4) antibodies. Immunoprecipitates were blotted with the indicated antibodies for mTOR, raptor, Rheb, GAPDH, pS6K1, and S6K1. (B) Glucose depletion-induced Rheb-GAPDH binding reduces Rheb-mTOR association. HEK293 cells transfected with HA-Rheb were incubated in 5.5 mM glucose medium for 30 min and then incubated with 25 mM glucose medium for the indicated times. Cell lysates were immunoprecipitated with anti-HA antibodies and then immunoblotted with anti-mTOR and anti-GAPDH antibodies. (C) GAPDH knockdown increases Rheb-mTOR association. HEK293 cells were transfected with GAPDH_#1 or luciferase siRNA. Cell lysates were immunoprecipitated with Rheb antibodies. Immunoprecipitates were blotted with the indicated antibodies for mTOR, GAPDH, and Rheb. (D) GAPDH inhibits the association between Rheb and mTORC1 in vitro. The Myc-tagged mTOR immunocomplexes were prepared from HEK293 cells and incubated with soluble GST-Rheb in the presence or absence of purified rabbit muscle GAPDH, with or without Gly-3-P. GST-Rheb bound to the mTOR complex was analyzed by anti-Rheb immunoblotting. Amounts of Myc-mTOR immunocomplexes used are shown by anti-mTOR immunoblotting.
To further confirm if the alteration in the interaction between Rheb and mTOR depends on the interaction between GAPDH and Rheb, we incubated mTOR immunoprecipitates with purified GAPDH in vitro. This led to a decrease in the amount of Rheb bound to mTOR (Fig. 5D, compare lanes 1 and 2). Next, we included Gly-3-P, which is the metabolite capable of destabilizing the GAPDH-Rheb interaction, in the incubation. The addition of Gly-3-P increased the amount of Rheb bound to mTOR (Fig. 5D, lanes 3 and 4). All of these results are consistent with our observation that silencing GAPDH increases the basal activity of mTORC1 (Fig. 4A and B). Taken together, our results suggest that GAPDH negatively regulates mTORC1 by preventing Rheb from associating with mTOR under conditions of low glucose availability.
Inhibition of the Rheb-GAPDH interaction induces mTORC1 activation.
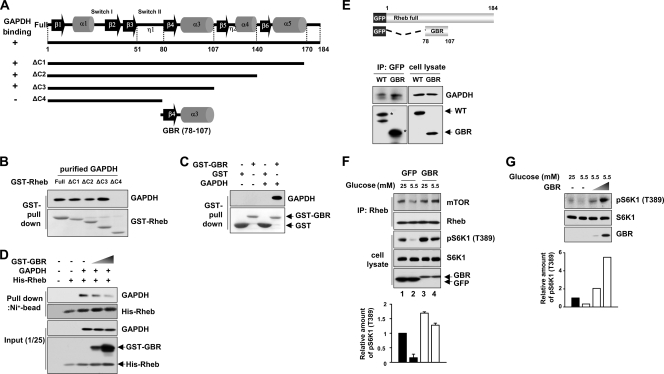
To further explore the negative roles of the GAPDH-Rheb interaction in mTORC1 signaling, we constructed Rheb truncation mutants (Fig. 6A) and analyzed the ability of each of the fragments to bind GAPDH (Fig. 6B). We identified a region of Rheb (residues 78 to 107) necessary for GAPDH binding and named it GBR (GAPDH binding region) (Fig. 6A to C). We next investigated whether GBR can interfere with the binding of Rheb to GAPDH. Increasing the amount of GBR inhibited the interaction between Rheb and GAPDH in vitro (Fig. 6D). Given that the GBR fragment interferes with the interaction between GAPDH and Rheb, we reasoned that GBR may enhance the interaction between Rheb and mTOR. To test this, we constructed a GFP-tagged GBR and confirmed that it binds to endogenous GAPDH by comparing its binding with that of the whole Rheb protein (Fig. 6E). We found that expression of GBR stabilized the Rheb-mTOR interaction even under low-glucose conditions (Fig. 6F, compare lanes 2 and 4). The stabilization of the Rheb-mTOR interaction in cells expressing the GBR was accompanied by an increase in S6K1 phosphorylation. Furthermore, the cells expressing GBR exhibited a dose-dependent increase in S6K1 phosphorylation even under low-glucose conditions (Fig. 6G). This result further supports the idea that the interaction between GAPDH and Rheb is inhibitory to mTORC1 signaling.
FIG. 6.
Blocking of the Rheb-GAPDH interaction increases the mTORC1 activity. (A and B) Identification of the GAPDH binding region in Rheb. Considering the secondary structure elements of Rheb (48), we generated GST-tagged serial deletion mutants of Rheb (ΔC1 to ΔC4). The recovery of GST-Rheb constructs with GAPDH is indicated on the left of panel A as + or −, and the immunoblot data supporting this summary are shown in panel B. (C) GAPDH specifically interacts with the Rheb (78-107) fragment in vitro. GST-tagged Rheb fragment 78 to 107 (GST-GBR) was incubated with purified rabbit muscle GAPDH. Bound GAPDH was analyzed by immunoblotting with anti-GAPDH antibodies (upper panel), and the amounts of GST or GST-GBR used were shown by Ponceau S staining (lower panel). (D) GST-GBR disrupts the interaction between GAPDH and full-length Rheb. For in vitro competition assays using GST-tagged GBR, GAPDH was mixed with immobilized His-tagged full-length Rheb on Ni-NTA agarose. Purified GST-GBR was then added in increasing amounts (0, 1, and 5 μg) and incubated for 2 h at 4°C. GAPDH remaining on the Ni-NTA agarose was analyzed by anti-GAPDH immunoblotting. The 1/25 input of the samples was immunoblotted using anti-Rheb antibodies to determine the relative amounts of His-Rheb and GST-GBR used. (E) GFP-tagged GBR specifically interacts with endogenous GAPDH. (Top panels) Schematic representation of GFP-tagged Rheb constructs. (Bottom panels) HEK293 cells transfected with GFP-tagged Rheb WT or GFP-tagged GBR were subjected to coimmunoprecipitation with anti-GFP antibodies. Immunoprecipitates were blotted with anti-GAPDH antibodies for GAPDH and with anti-GFP antibodies for GFP-tagged proteins. (F) Overexpression of GBR induces both the Rheb-mTOR interaction and the hyperactivation of mTORC1. HEK293 cells transfected with either enhanced GFP or enhanced GFP-tagged GBR were incubated with media with the indicated concentrations of glucose for 30 min. Cell lysates were immunoprecipitated with Rheb antibodies. Immunoprecipitates and cell lysates were blotted with the indicated antibodies. Representative results of immunoblotting from three independent experiments are shown. (G) HEK293 cells transfected with incremental doses (0, 0.2, and 2 μg) of enhanced GFP-tagged GBR were incubated in 5.5 mM glucose medium for 30 min. The phosphorylation of S6K1 was determined by immunoblotting. The bar graph at the bottom shows the increase in phosphorylation of S6K1 over that of the control. For quantitation of total protein, we used equal amounts of the same cell lysates that were used for detecting phosphorylated proteins by Western blotting separately.
DISCUSSION
In this study, we demonstrate that glycolysis is linked to the mTORC1 pathway via the direct binding of GAPDH to Rheb. The glycolytic flux regulates the interaction between GAPDH and Rheb, and this interaction inhibits mTORC1 signaling by preventing Rheb from binding to mTOR. We also found that the inhibitory effect of GAPDH on mTORC1 was restored by Gly-3-P, a glycolytic intermediate that binds GAPDH. Gly-3-P destabilizes the interaction between GAPDH and Rheb, suggesting that this glycolytic intermediate acts as a messenger to regulate mTORC1 signaling in response to changes in the glycolytic flux.
Although it is known that Rheb is necessary for the activation of mTOR, the exact regulatory function of Rheb has not been clearly elucidated. Unlike other Ras GTPases that bind their effectors only when they are charged with GTP, the GTP-GDP status of Rheb does not alter its ability to bind mTOR (30). In view of this atypical property of Rheb, it has been speculated that an unidentified factor may regulate the binding of Rheb to mTOR (3). In our study, we found that GAPDH regulates the binding of Rheb to mTOR in a manner that is dependent upon glucose levels and independent of the nucleotide-charged status of Rheb (Fig. 2). To our knowledge, this constitutes the first evidence that GAPDH regulates mTORC1 activity by binding Rheb and thereby modulating its interaction with mTOR.
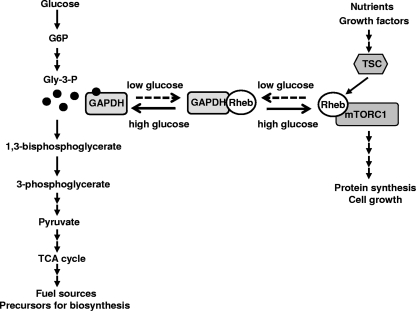
Although GAPDH is known as a glycolytic enzyme, recent studies have shown that it may be involved in diverse cellular processes, including membrane fusion, microtubule bundling, nuclear RNA export, DNA repair, transcriptional regulation, and apoptosis (9, 24, 41). Our findings further extend the multifunctional character of GAPDH. Although the Rheb-GAPDH interaction increases when the glycolytic flux is reduced, the interaction was still detected even under high-glucose conditions (Fig. 2). This implies that GAPDH shuttles between the glycolysis pathway and the mTORC1 pathway to facilitate the immediate response of mTORC1 signaling to the glycolytic flux (Fig. 7).
FIG. 7.
Model for the regulation of mTORC1 signaling in response to glycolytic flux via GAPDH-Rheb interaction. GAPDH acts as a glycolytic messenger that senses the glycolytic flow and signals to the mTORC1 pathway by interacting with Rheb. The extracellular glucose sufficiency increases the intracellular glycolytic flux, resulting in the accumulation of Gly-3-P and GAPDH charged with Gly-3-P, the form that is incapable of binding Rheb. Under low-glucose conditions, a decrease in the supply of extracellular glucose may result in the accumulation of GAPDH uncharged with Gly-3-P, which prevents Rheb from binding to mTORC1 and thereby inhibits mTORC1 signaling. TCA, tricarboxylic acid.
The glycolytic pathway is regulated in an allosteric manner by phosphofructokinase 1 (PFK-1), which functions to regulate the flow of glycolysis (26). The enzymatic activity of PFK-1 is regulated by the end product of glycolysis such as ATP. For this reason, the increased production of a glycolytic intermediate (such as fructose 6-phosphate) by upstream enzymes in the glycolytic pathway would not increase the glycolytic flux via PFK-1. On the other hand, the GAPDH-mediated reaction is substrate limited and determined by the instantaneous concentration of the substrate (26). Whereas PFK-1 is likely regulated by both the net glycolytic flux and the nonglycolytic processes that affect cellular ATP levels, GAPDH is likely to be entirely controlled by the influx of glucose. Therefore, we hypothesize that GAPDH is better suited than other enzymes in the glycolytic pathway to monitor the glycolytic flux that depends on the supply of extracellular glucose.
Previously, it was suggested that a link exists between AMPK and the mTORC1 pathway under conditions of energy starvation (22). In that study, treatment with 2DG or glucose starvation resulted in decreased cellular ATP levels and the subsequent activation of AMPK. AMPK was proposed to stimulate TSC2 to serve as a GTPase-activating protein for Rheb and thereby inhibit the mTORC1 pathway under conditions of glucose starvation. A recent report showed that activated AMPK regulates mTORC1 signaling not only through TSC1-TSC2 but also through direct phosphorylation of raptor (14). In our study, we found a new pathway in which the glycolytic enzyme GAPDH directly binds to Rheb and thereby inhibits mTORC1 signaling in response to glucose availability independently of the AMPK-TSC axis and the AMPK-raptor axis. Interestingly, we found that the GAPDH-Rheb interaction is dramatically increased in TSC1-deficient MEFs and AMPK-inhibited TSC1-deficient MEFs, even under high-glucose conditions (Fig. 4C and F). An increase in the GAPDH-Rheb interaction was also observed in AMPKα1-silenced HEK293 cells (data not shown). Although both the absence of TSC1 and the inhibition of AMPK activity increased basal GAPDH-Rheb interaction, the GAPDH-Rheb interaction was still responsive to glucose (Fig. 4C and F). However, at this time, it is unclear whether AMPK activity directly regulates the GAPDH-Rheb interaction. Interestingly, we observed that AMPK activity is significantly elevated in TSC1-deficient MEFs relative to WT MEFs (data not shown). We presume that TSC1-deficient cells have a highly activated mTORC1 pathway and thus consume much cellular energy to support cell growth. This idea is supported by the increased levels of both AMPK and ACC phosphorylation in these cells. One might expect this to result in energy depletion and eventual energy crisis-mediated cell death. However, we did not observe any kind of cell death in TSC1-deficient cells. Possibly, TSC1-deficient cells do not undergo energy starvation-induced apoptosis because they lack p53, as suggested in a previous model (25a ). We observed that the GAPDH-Rheb interaction is dramatically enhanced in TSC1-deficient MEFs and that this interaction is regulated by glucose availability in both TSC1-deficient MEFs and AMPK-inhibited cells. We hypothesize that the GAPDH-Rheb interaction may act as a cellular safety device to prevent uncontrolled mTOR activation and energy crisis-mediated cell death under abnormal cellular circumstances, such as TSC deficiency and/or loss of AMPK function. Further study would be needed to confirm such a protective role for GAPDH. Relevant questions would include whether the GAPDH-Rheb interaction is elevated in TSC patient tissue or when AMPK function is lost and whether the silencing of GAPDH severely induces energy crisis-mediated cell death under these circumstances.
Taken together, mTORC1 inhibition under low-glucose conditions seems to occur as a result of multiple pathways: (i) the AMPK-TSC axis, (ii) the AMPK-raptor axis, and (iii) the GAPDH-Rheb axis. Cells may have developed these multiple ways of regulating mTORC1 signaling in response to glucose levels to deal with a wider diversity of energy and stress conditions. In this light, it is important that the two AMPK-regulated pathways depend on the AMP/ATP ratio. Therefore, these pathways will respond not only to glucose-deprived conditions but also to many other stress conditions that reduce ATP levels. In contrast, the GAPDH-Rheb pathway is more directly linked to glucose metabolism. Thus, the GAPDH-Rheb axis may be responsible for more intimate cross talk between the glycolytic pathway and the mTORC1 pathway, and the AMPK-dependent pathways may be responsive to other cellular energy states and/or stress conditions that alter the AMP/ATP ratio. Further studies are needed to address the relative contributions of the three pathways to the regulation of mTORC1 in response to glucose availability. From an evolutionary point of view, whether GAPDH is involved in the regulation of TORC1 in yeast will also be an interesting question. Given that the GAPDH-Rheb axis can inhibit mTORC1 signaling even when there is a TSC1 deficiency or Rheb is constitutively active, our study provides important insight into the development of therapeutic approaches to human diseases caused by defects in TSC.
Acknowledgments
We thank A. F. Castro for providing the Rheb constructs and D. M. Sabatini for Myc-mTOR. We also thank D. J. Kwiatkowski for the TSC1 MEFs.
This work was supported by FPR08B1-160 of the 21C Frontier Functional Proteomics Project and by the Global Research Network Program from the Korean Ministry of Education, Science and Technology.
Footnotes
Published ahead of print on 18 May 2009.
REFERENCES
- 1.Aspuria, P. J., and F. Tamanoi. 2004. The Rheb family of GTP-binding proteins. Cell. Signal. 161105-1112. [DOI] [PubMed] [Google Scholar]
- 2.Astrinidis, A., and E. P. Henske. 2005. Tuberous sclerosis complex: linking growth and energy signaling pathways with human disease. Oncogene 247475-7481. [DOI] [PubMed] [Google Scholar]
- 3.Avruch, J., K. Hara, Y. Lin, M. Liu, X. Long, S. Ortiz-Vega, and K. Yonezawa. 2006. Insulin and amino-acid regulation of mTOR signaling and kinase activity through the Rheb GTPase. Oncogene 256361-6372. [DOI] [PubMed] [Google Scholar]
- 4.Bai, X., D. Ma, A. Liu, X. Shen, Q. J. Wang, Y. Liu, and Y. Jiang. 2007. Rheb activates mTOR by antagonizing its endogenous inhibitor, FKBP38. Science 318977-980. [DOI] [PubMed] [Google Scholar]
- 5.Basso, A. D., A. Mirza, G. Liu, B. J. Long, W. R. Bishop, and P. Kirschmeier. 2005. The farnesyl transferase inhibitor (FTI) SCH66336 (lonafarnib) inhibits Rheb farnesylation and mTOR signaling: role in FTI enhancement of taxane and tamoxifen anti-tumor activity. J. Biol. Chem. 28031101-31108. [DOI] [PubMed] [Google Scholar]
- 6.Bonini, B. M., C. Van Vaeck, C. Larsson, L. Gustafsson, P. Ma, J. Winderickx, P. Van Dijck, and J. M. Thevelein. 2000. Expression of Escherichia coli otsA in a Saccharomyces cerevisiae tps1 mutant restores trehalose 6-phosphate levels and partly restores growth and fermentation with glucose and control of glucose influx into glycolysis. Biochem. J. 350261-268. [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, J. 1962. Effects of 2-deoxyglucose on carbohydrate metabolism: review of the literature and studies in the rat. Metabolism 111098-1112. [PubMed] [Google Scholar]
- 8.Canback, B., S. G. E. Andersson, and C. G. Kurland. 2002. The global phylogeny of glycolytic enzymes. Proc. Natl. Acad. Sci. USA 996097-6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chuang, D.-M., C. Hough, and V. V. Senatorov. 2005. Glyceraldehyde-3-phosphate dehydrogenase, apoptosis, and neurodegenerative diseases. Annu. Rev. Pharmacol. Toxicol. 45269-290. [DOI] [PubMed] [Google Scholar]
- 10.Clark, G. J., M. S. Kinch, K. Rogers-Graham, S. M. Sebti, A. D. Hamilton, and C. J. Der. 1997. The Ras-related protein Rheb is farnesylated and antagonizes Ras signaling and transformation. J. Biol. Chem. 27210608-10615. [DOI] [PubMed] [Google Scholar]
- 11.Corradetti, M. N., K. Inoki, and K.-L. Guan. 2005. The stress-inducted proteins RTP801 and RTP801L are negative regulators of the mammalian target of rapamycin pathway. J. Biol. Chem. 2809769-9772. [DOI] [PubMed] [Google Scholar]
- 12.Fingar, D. C., and J. Blenis. 2004. Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene 233151-3171. [DOI] [PubMed] [Google Scholar]
- 13.Garami, A., F. J. Zwartkruis, T. Nobukuni, M. Joaquin, M. Roccio, H. Stocker, S. C. Kozma, E. Hafen, J. L. Bos, and G. Thomas. 2003. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol. Cell 111457-1466. [DOI] [PubMed] [Google Scholar]
- 14.Gwinn, D. M., D. B. Shackelford, D. F. Egan, M. M. Mihaylova, A. Mery, D. S. Vasquez, B. E. Turk, and R. J. Shaw. 2008. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell 30214-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hara, M. R., N. Agrawal, S. F. Kim, M. B. Cascio, M. Fujimuro, Y. Ozeki, M. Takahashi, J. H. Cheah, S. K. Tankou, L. D. Hester, C. D. Ferris, S. D. Hayward, S. H. Snyder, and A. Sawa. 2005. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat. Cell Biol. 7665-674. [DOI] [PubMed] [Google Scholar]
- 16.Harris, J. I., and M. Water. 1976. Glyceraldehyde-3-phosphate dehydrogenase, p. 1-50. In P. D. Boyer (ed.), The enzyme XIII, 3rd ed. Academic Press, New York, NY.
- 17.Harris, T. E., and J. C. Lawrence, Jr. 2003. TOR signaling. Sci. STKE 2003re15. [DOI] [PubMed] [Google Scholar]
- 18.Hsu, Y.-C., J. J. Chern, Y. Cai, M. Liu, and K.-W. Choi. 2007. Drosophila TCTP is essential for growth and proliferation through regulation of dRheb GTPase. Nature 445785-788. [DOI] [PubMed] [Google Scholar]
- 19.Inoki, K., M. N. Corradetti, and K.-L. Guan. 2005. Dysregulation of the TSC-mTOR pathway in human disease. Nat. Genet. 3719-24. [DOI] [PubMed] [Google Scholar]
- 20.Inoki, K., and K.-L. Guan. 2006. Complexity of the TOR signaling network. Trends Cell Biol. 16206-212. [DOI] [PubMed] [Google Scholar]
- 21.Inoki, K., Y. Li, T. Xu, and K. L. Guan. 2003. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 171829-1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inoki, K., T. Zhu, and K.-L. Guan. 2003. TSC2 mediates cellular energy response to control cell growth and survival. Cell 115577-590. [DOI] [PubMed] [Google Scholar]
- 23.Jacinto, E., and M. N. Hall. 2003. TOR signalling in bugs, brain and brawn. Nat. Rev. Mol. Cell Biol. 4117-126. [DOI] [PubMed] [Google Scholar]
- 24.Kim, J.-W., and C. V. Dang. 2005. Multifaceted roles of glycolytic enzymes. Trends Biochem. Sci. 30142-150. [DOI] [PubMed] [Google Scholar]
- 25.Kwiatkowski, D. J., and B. D. Manning. 2005. Tuberous sclerosis: a GAP at the crossroads of multiple signaling pathways. Hum. Mol. Genet. 14R251-R258. [DOI] [PubMed] [Google Scholar]
- 25a.Lee, C. H., K. Inoki, M. Karbowniczek, E. Petroulakis, N. Sonenberg, E. P. Henske, and K. L. Guan. 2007. Constitutive mTOR activation in TSC mutants sensitizes cells to energy starvation and genomic damage via p53. EMBO J. 264812-4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lehninger, A. L., M. M. Cox, and D. L. Nelson. 2004. Principles of metabolic regulation, illustrated with the metabolism of glucose and glycogen, p. 560-600. In D. L. Nelson and M. M. Cox (ed.), Lehninger principles of biochemistry, 4th ed. W. H. Freeman & Co., New York, NY.
- 27.Li, Y., M. N. Corradetti, K. Inoki, and K.-L. Guan. 2004. TSC2: filling the GAP in the mTOR signaling pathway. Trends Biochem. Sci. 2932-38. [DOI] [PubMed] [Google Scholar]
- 28.Li, Y., K. Inoki, and K. L. Guan. 2004. Biochemical and functional characterizations of small GTPase Rheb and TSC2 GAP activity. Mol. Cell. Biol. 247965-7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loewith, R., and M. N. Hall. 2004. TOR signalling in yeast: temporal and spatial control of cell growth, p. 139-166. In M. N. Hall, M. Raff, and G. Thomas (ed.), Cell growth: control of cell size. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 30.Long, X., Y. Lin, S. Ortiz-Vega, K. Yonezawa, and J. Avruch. 2005. Rheb binds and regulates the mTOR kinase. Curr. Biol. 15702-713. [DOI] [PubMed] [Google Scholar]
- 31.Long, X., S. Ortiz-Vega, Y. Lin, and J. Avruch. 2005. Rheb binding to mammalian target of rapamycin (mTOR) is regulated by amino acid sufficiency. J. Biol. Chem. 28023433-23436. [DOI] [PubMed] [Google Scholar]
- 32.Manning, B. D., and L. C. Cantley. 2003. Rheb fills a GAP between TSC and TOR. Trends Biochem. Sci. 28573-576. [DOI] [PubMed] [Google Scholar]
- 33.Mavrakis, K. J., H. Zhu, R. L. A. Silva, J. R. Mills, J. Teruya-Feldstein, S. W. Lowe, W. Tam, J. Pelletier, and H.-G. Wendel. 2008. Tumorigenic activity and therapeutic inhibition of Rheb GTPase. Genes Dev. 222178-2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitro, N., P. A. Mak, L. Vargas, C. Godio, E. Hampton, V. Molteni, A. Kreusch, and E. Saez. 2007. The nuclear receptor LXR is a glucose sensor. Nature 445219-223. [DOI] [PubMed] [Google Scholar]
- 35.Nagradova, N. K. 2001. Study of the properties of phosphorylating D-glyceraldehyde-3-phosphate dehydrogenase. Biochemistry (Moscow) 661067-1076. [DOI] [PubMed] [Google Scholar]
- 36.Reiling, J. H., and D. M. Sabatini. 2006. Stress and mTORture signaling. Oncogene 256373-6383. [DOI] [PubMed] [Google Scholar]
- 37.Roccio, M., J. L. Bos, and F. J. T. Zwartkruis. 2006. Regulation of the small GTPase Rheb by amino acids. Oncogene 25657-664. [DOI] [PubMed] [Google Scholar]
- 38.Sancak, Y., C. C. Thoreen, T. R. Peterson, R. A. Lindquist, S. A. Kang, E. Spooner, S. A. Carr, and D. M. Sabatini. 2007. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol. Cell 25903-915. [DOI] [PubMed] [Google Scholar]
- 39.Saucedo, L. J., X. Gao, D. A. Chiarelli, L. Li, D. Pan, and B. A. Edgar. 2003. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat. Cell Biol. 5566-571. [DOI] [PubMed] [Google Scholar]
- 40.Reference deleted.
- 41.Sirover, M. A. 1999. New insights into an old protein: the functional diversity of mammalian glyceraldehyde-3-phosphate dehydrogenase. Biochim. Biophys. Acta 1432159-184. [DOI] [PubMed] [Google Scholar]
- 42.Smith, E. M., S. G. Finn, A. R. Tee, G. J. Browne, and C. G. Proud. 2005. The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses. J. Biol. Chem. 28018717-18727. [DOI] [PubMed] [Google Scholar]
- 43.Torralba, A. S., K. Yu, P. Shen, P. J. Oefner, and J. Ross. 2003. Experimental test of a method for determining causal connectivities of species in reactions. Proc. Natl. Acad. Sci. USA 1001494-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uhlenbrock, K., M. Weiwad, R. Wetzker, G. Fischer, A. Wittinghofer, and I. Rubio. 2009. Reassessment of the role of FKBP38 in the Rheb/mTORC1 pathway. FEBS Lett. 583965-970. [DOI] [PubMed] [Google Scholar]
- 45.Wang, X., B. D. Fonseca, H. Tang, R. Liu, A. Elia, M. J. Clemens, U.-A. Bommer, and C. G. Proud. 2008. Re-evaluating the roles of proposed modulators of mammalian target of rapamycin complex 1 (mTORC1) signaling. J. Biol. Chem. 28330482-30492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wullschleger, S., R. Loewith, and M. N. Hall. 2006. TOR signaling in growth and metabolism. Cell 124471-484. [DOI] [PubMed] [Google Scholar]
- 47.Yamagata, K., L. K. Sanders, W. E. Kaufmann, W. Yee, C. A. Barnes, D. Nathans, and P. F. Worley. 1994. rheb, a growth factor- and synaptic activity-regulated gene, encodes a novel Ras-related protein. J. Biol. Chem. 26916333-16339. [PubMed] [Google Scholar]
- 48.Yu, Y., S. Li, X. Xu, Y. Li, K. Guan, E. Arnold, and J. Ding. 2005. Structural basis for the unique biological function of small GTPase RHEB. J. Biol. Chem. 28017093-17100. [DOI] [PubMed] [Google Scholar]
- 49.Yun, M., C. G. Park, J. Y. Kim, and H. W. Park. 2000. Structural analysis of glyceraldehyde 3-phosphate dehydrogenase from Escherichia coli: direct evidence of substrate binding and cofactor-induced conformational changes. Biochemistry 3910702-10710. [DOI] [PubMed] [Google Scholar]
- 50.Zhang, Y., X. Gao, L. J. Saucedo, B. Ru, B. A. Edgar, and D. Pan. 2003. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat. Cell Biol. 5578-581. [DOI] [PubMed] [Google Scholar]
- 51.Zheng, L., R. G. Roeder, and Y. Luo. 2003. S phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell 114255-266. [DOI] [PubMed] [Google Scholar]