Abstract
The negative elongation factor NELF is a key component of an early elongation checkpoint generally located within 100 bp of the transcription start site of protein-coding genes. Negotiation of this checkpoint and conversion to productive elongation require phosphorylation of the carboxy-terminal domain of RNA polymerase II (pol II), NELF, and DRB sensitivity-inducing factor (DSIF) by positive transcription elongation factor b (P-TEFb). P-TEFb is dispensable for transcription of the noncoding U2 snRNA genes, suggesting that a NELF-dependent checkpoint is absent. However, we find that NELF at the end of the 800-bp U2 gene transcription unit and RNA interference-mediated knockdown of NELF causes a termination defect. NELF is also associated 800 bp downstream of the transcription start site of the β-actin gene, where a “late” P-TEFb-dependent checkpoint occurs. Interestingly, both genes have an extended nucleosome-depleted region up to the NELF-dependent control point. In both cases, transcription through this region is P-TEFb independent, implicating chromatin in the formation of the terminator/checkpoint. Furthermore, CTCF colocalizes with NELF on the U2 and β-actin genes, raising the possibility that it helps the positioning and/or function of the NELF-dependent control point on these genes.
The negative elongation factors (N-TEFs) DSIF and NELF are key components of a polymerase II (pol II) checkpoint that occurs early in the transcription cycle of the human immunodeficiency virus (HIV) genome and many protein-coding genes (7, 21, 46, 62). Release from an N-TEF-dependent block requires the activity of the cyclin-dependent kinase 9 (CDK9) subunit of positive transcription elongation factor b (P-TEFb), which phosphorylates serine (Ser) 2 in the YSPTSPS heptapeptide repeat of the pol II carboxy-terminal domain (CTD) and subunits of DSIF and NELF (7, 21, 46, 62). Accordingly, CDK9 inhibitors effectively inhibit the elongation of pol II transcription both in vitro and in vivo (13, 40). Once pol II has negotiated the early block, productive elongation can occur (37). Phosphorylation of Ser2 of the pol II CTD by CDK9 also activates cotranscriptional processing of transcripts from protein-coding genes and the mammalian noncoding small nuclear RNA (snRNA) genes (4, 17, 39, 42), presumably through interactions between phospho-CTD and processing factors (17). In many Drosophila protein-coding genes, the NELF-dependent checkpoint is located within 100 bp downstream of the transcription start site, where paused polymerase is also located (30). These genes generally have a short promoter-proximal region of nucleosome depletion, with the first nucleosome mapping close to where NELF is found (31, 38).
We have previously reported differences in elongation control between the short intronless pol II-transcribed human U2 snRNA genes with a transcription unit of less than 1 kb (40) and the β-actin protein-coding gene with a transcription unit of 5 kb (24). CDK9 inhibitors drastically affect the elongation of transcription of the β-actin gene but have little effect on transcription of the U2 snRNA genes (39). However, P-TEFb is recruited to the U2 genes, pol II transcribing these genes is phosphorylated on Ser2 of the CTD, and CDK9 inhibitors abolish recognition of the snRNA gene-specific 3′ box RNA 3′-end-processing signal (39). It is therefore unclear why P-TEFb plays no role in the elongation of transcription.
In order to understand the molecular basis of the requirement for P-TEFb function for transcription of human genes, we have carried out a comparative analysis of the U2 snRNA gene and protein-coding genes in HeLa cells, with regard to chromatin structure and the association of N-TEFs and elongation factors.
On the three protein-coding genes tested and the U2 snRNA genes, NELF recruitment occurs at the end of a promoter-proximal region of nucleosome depletion, implicating a nucleosomal roadblock in NELF-dependent transcriptional pausing in vivo. Like Drosophila protein-coding genes (31, 38), the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and γ-actin genes have a short promoter-proximal region of nucleosome depletion. In contrast, the P-TEFb-dependent checkpoint on the β-actin gene occurs at the end of a promoter-proximal region of nucleosome depletion that extends for approximately 800 bp. Thus, an elongation checkpoint is not always located close to the start of transcription in human protein-coding genes. The U2 gene is also nucleosome depleted for approximately 800 bp downstream from the promoter, which, in this case, comprises the entire transcribed region. NELF is therefore located at the end of the U2 transcription unit, circumventing the requirement for P-TEFb for transcription elongation. Interestingly, NELF is required for termination of U2 gene transcription rather than for attenuation.
Finally, binding sites for the boundary protein CTCF are located close to the checkpoint/terminator in the β-actin and U2 genes, implicating this factor together with chromatin in setting up a “late” NELF-dependent control point.
MATERIALS AND METHODS
Expression constructs.
To generate pFlag-NELF-E, pFlag-SSRP1, and pFlag-TFIIS, NELF-E, SSRP1, and TFIIS cDNAs were inserted into pFLAG-CMV-2 (Sigma). The Cdk9-myc construct was made by adding three myc tags at the C terminus of the Cdk9 construct used by Medlin et al. (39). Expression of each protein was verified by Western blot analysis.
Antibodies.
The following antibodies were used for chromatin immunoprecipitation (ChIP): anti-H2A (ab18255; Abcam), anti-H2B (ab1790; Abcam), anti-H3 (ab1791; Abcam), anti-H4 (ab31827; Abcam), anti-AcH3 (06-599; Upstate), anti-AcH4 (06-598; Upstate), anti-H3K4(Me)3 (ab8580; Abcam), anti-H3K36(Me)3 (ab9050; Abcam), anti-H3K9(Me)3 (ab8898; Abcam), anti-Flag M2 (F1804; Sigma), anti-Spt16 (sc-28734; Santa Cruz), anti-PTFγ (59), anti-pol II (sc-899X; Santa Cruz), anti-Ser2P (H5; Covance), anti-Ser5P (H14; Covance), anti-Ser7P (12), anti-c-Myc (sc-764X; Santa Cruz), anti-CTCF (07-729; Upstate) and anti-DSIF (D80020; Transduction Laboratories).
ChIP.
HeLa cells were either transfected and treated with DRB (100 μM; Sigma) or not treated for 4 h before being subjected to ChIP analysis (39). ChIP samples were analyzed by quantitative PCR using QuantiTect SYBR green PCR (Qiagen). Regions analyzed are described in Figure S10 in the supplemental material.
Nuclear run-on analysis.
Nuclear run-on analyses were carried out as described by Medlin et al. (40) with 80-nucleotide oligonucleotide probes complementary to RNA transcribed from the U2 gene. The 3′ ends of the probes correspond to positions −130 (proximal sequence element [PSE]), +48 (probe R1), +208 (probe R2), +288 (probe R3), +368 (probe R4), +448 (probe R5), +528 (probe R6), and +608 (probe R7), relative to the site of transcription initiation. The 3′ end of the 80-nucleotide 5S RNA probe corresponds to position +32, relative to the site of transcription initiation. Hybridization signals were quantified by densitometry, corrected with the background level, and normalized to probe R1.
Small interfering RNA-mediated knockdown.
Small interfering RNAs targeting NELF-E were purchased from Dharmacon and transfected using Lipofectamine 2000 (Invitrogen).
Western blot analysis.
Western blotting was carried out as described previously (39), using antibodies against NELF-E (58) and Int11 (16).
RESULTS
Human U2 snRNA and β-actin genes have an extended region of promoter-proximal nucleosome depletion.
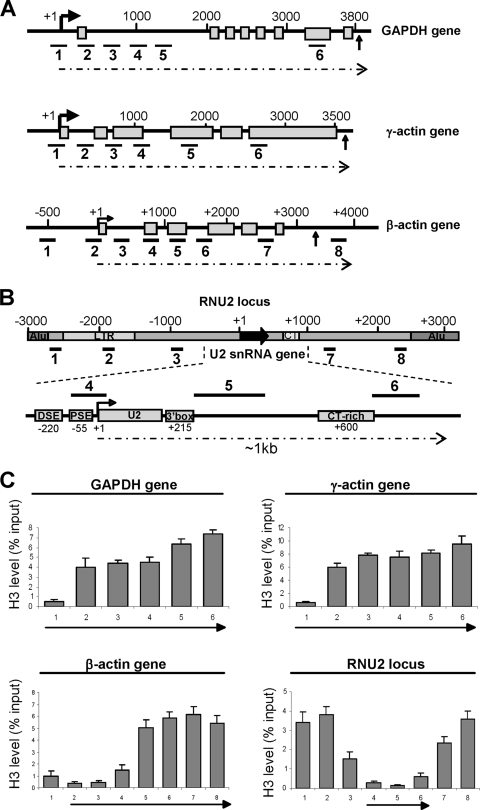
In vivo, transcription generally takes place in the context of chromatin, which comprises DNA associated with the histone proteins H2A, H2B, H3, and H4 in the form of nucleosomes, which can constitute a physical barrier to elongating pol II (5, 27, 32). A recent study indicates that a large proportion of active genes in Drosophila embryos have a nucleosome-free region at the transcription start site, followed by a nucleosome at position +1 with an upstream border at position +62, which correlates closely to the position of paused pol II (38). To determine the chromatin structure of the promoter-proximal region of a range of human genes transcribed by pol II, we analyzed the association of the H3 core histone with the human GAPDH, γ-actin, and β-actin protein-coding genes and the U2 snRNA genes by high-resolution ChIP, coupled with quantitative real-time PCR (qRT-PCR) analysis (Fig. 1). The protein-coding genes are all single-copy genes, with transcription units of >3 kb, and encode transcripts that are spliced and polyadenylated (Fig. 1A). The genes encoding human U2 snRNA are organized as a nearly perfect tandem array of an approximately 6-kb region repeated 10 to 20 times per haploid genome (the RNU2 locus) (Fig. 1B) (56). U2 snRNA genes have specialized, TATA-less promoters containing one essential PSE and an upstream enhancer-like distal sequence element (DSE) (26). The 3′ box, the snRNA gene-specific RNA 3′ processing element, is located immediately downstream of the snRNA-encoding region (61). Transcription of U2 snRNA genes extends for up to ∼1 kb from the start site (14, 40). It is unclear whether all repeats are active in all cells, but most must be active to ensure the high level of U2 snRNA detected in HeLa cells (36). ChIP localizes the PSE-binding complex, PTF, also known as SNAPc and PBP (26) at the PSE of the U2 genes (primer pair 4) as expected, and much lower signals are detected 200 bp upstream and 150 bp downstream, demonstrating that the resolution of our ChIP analysis is within 200 bp (see Fig. S1 in the supplemental material).
FIG. 1.
Human U2 snRNA and β-actin genes have an extended region of promoter-proximal nucleosome depletion. (A) Diagrams of the GAPDH, γ-actin, and β-actin genes, with the exons depicted as open boxes and the polyadenylation signals indicated with an arrow. The positions of primer pairs used for ChIP are shown below each diagram. Arrows indicate the start of transcription. (B) The 6-kb repeat of the human RNU2 locus, with U2 gene promoter and RNA processing elements noted in the expanded diagram below. The repeat also contains a CT microsatellite, two tandem Alu repeats, and a long-terminal-repeat element. The positions of the pairs of primers used for ChIP are shown below the 6-kb repeat and above the expanded gene diagram. (C) qRT-PCR of histone H3 ChIP analysis on the GAPDH, γ-actin, β-actin, and RNU2 loci. The y axes represent the percentages of input DNA immunoprecipitated, and the arrows under the x axes indicate the transcribed regions of the loci.
Like many Drosophila protein-coding genes (38), the GAPDH and γ-actin genes have a short promoter-proximal region of nucleosome depletion, and nucleosome occupancy is relatively high 200 bp downstream from the start of transcription (Fig. 1C). However, the β-actin and U2 snRNA genes do not conform to this pattern and instead have a more extensive region of promoter-proximal nucleosome depletion extending for approximately 800 bp (Fig. 1C; also see Fig. S2 in the supplemental material). A high density of nucleosomes is detected within the rest of the coding region of the β-actin gene (Fig. 1C; see primers 5 to 8 in Fig. S2 in the supplemental material) and the rest of the U2 repeat (Fig. 1C; see primer pairs 1-3 and 7-8 in Fig. S2 in the supplemental material). Importantly, all four core histones have the same pattern of depletion on β-actin and U2 genes (Fig. 1; also see Fig. S2 in the supplemental material). Thus, the extent of promoter-proximal nucleosome depletion can vary in human genes. Transcription of snRNA genes is highly efficient in dedifferentiated tissue culture cells in vivo, with initiation taking place as frequently as every 2 to 4 s (36). Nucleosomes can be transiently displaced from eukaryotic genes when transcription levels are very high (29, 51). However, in α-amanitin-treated cells, the histone H3 occupancy over the RNU2 locus remains the same, although the level of pol II is drastically reduced (see Fig. S3 in the supplemental material), arguing that nucleosomal depletion of the U2 gene is not simply due to a high level of transcription. Our results argue against the presence of a stable nucleosome between the PSE and DSE of the U2 genes as previously suggested (6, 45), but they are in accordance with the finding that regions within the U2 transcription unit are accessible to DNase I digestion (45). These results also indicate that transcribing pol II will encounter a high density of nucleosomes on protein-coding genes before completing a functional transcript, but not on the U2 genes, which could account for the differential requirement for P-TEFb activity for transcription elongation.
Differential localization of NELF on snRNA and protein-coding genes.
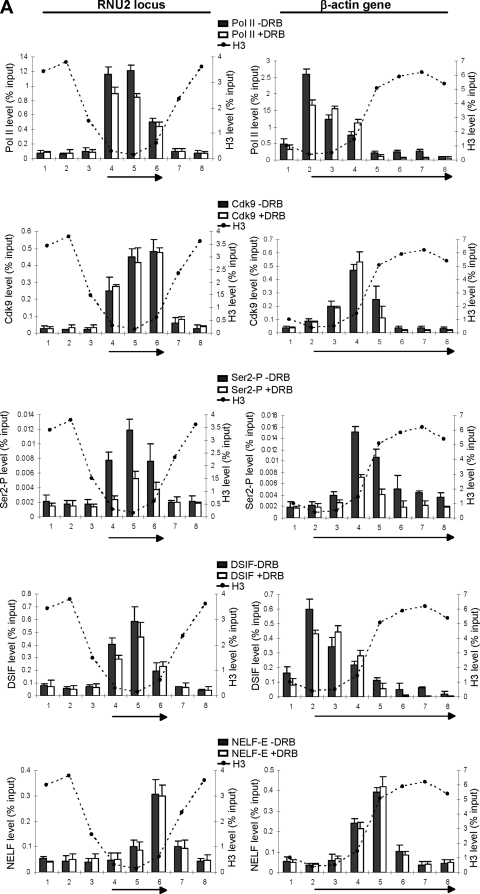
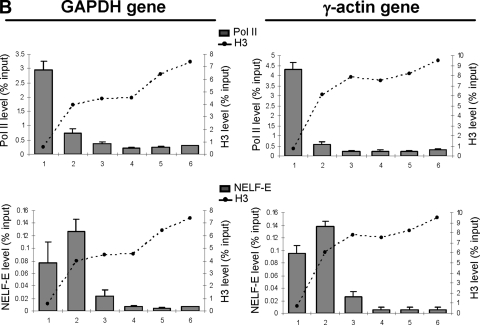
NELF is a complex comprising four subunits (NELF-A, -B, -C/D, and -E) which acts in collaboration with DSIF to induce promoter-proximal pausing of RNA pol II on protein-coding genes (46, 48). NELF/DSIF-induced arrest is implicated in an early elongation checkpoint, which may help ensure efficient capping of nascent mRNA through interaction between DSIF and components of the capping enzyme (35). The elongation block is released when P-TEFb phosphorylates the CTD of pol II and subunits of DSIF and NELF (46). Because P-TEFb activity is not required for efficient elongation of transcription of the U2 snRNA genes (39), we investigated whether there is a NELF/DSIF-induced block on these genes. The positions of pol II, P-TEFb, Ser2 phosphorylation, the Spt5 subunit of DSIF, and the NELF-E subunit of NELF on the U2 and β-actin genes were determined by ChIP with or without prior treatment of the cells with the CDK9 inhibitor, DRB (Fig. 2A). In untreated cells, pol II is detected across the whole transcription unit of the U2 and β-actin genes. The level of pol II is highest at the promoter region and decreases toward the 3′ end of the genes. CDK9 is detected across the whole transcription unit of the U2 genes, suggesting that P-TEFb is recruited early in the transcription cycle and stays associated with pol II during transcription. CDK9 is detected on the β-actin gene only after 800 bp, at the point where the histone occupancy increases. Ser2 phosphorylation follows the pattern of P-TEFb association on both genes and, therefore, occurs earlier in transcription of the U2 gene. The localization of DSIF correlates with pol II in both cases (Fig. 2A), in accordance with previous studies indicating that DSIF travels with pol II (22, 23). NELF is recruited to both genes later in the transcription cycle. On the β-actin gene, NELF accumulates just downstream of the region of nucleosome depletion, which in this case is within the transcribed region (Fig. 2A). On the U2 genes, NELF is also associated with the region of the gene where nucleosome occupancy increases, as confirmed by fine mapping (see Fig. S4 in the supplemental material), which occurs, in this case, at the very end of the transcription unit. Treatment of cells with DRB does not significantly alter the pattern of association of P-TEFb, pol II, DSIF, or NELF on the U2 genes (Fig. 2A). However, the already low levels of pol II and DSIF downstream from the position of NELF association with the β-actin gene are reduced significantly in response to DRB treatment, indicating that transcription is effectively blocked where NELF and nucleosome occupancy levels are high, if CDK9 is not active. Interestingly, the length of the nucleosome-depleted region corresponds closely to the region of both genes that continues to be transcribed in the presence of CDK9 inhibitors. These results are in accordance with nuclear run-on analysis indicating that pol II transcription of snRNA genes is not affected by inhibition of CDK9 while transcription of the β-actin gene is restricted to a few hundred base pairs at the beginning of the gene (40).
FIG. 2.
Differential localization of NELF on snRNA and protein-coding genes. (A) qRT-PCR of ChIP analysis of the RNU2 locus and β-actin gene, using antibodies against subunits of the protein complexes noted, with or without treatment of the cells with DRB (+DRB and −DRB, respectively). The H3 levels are indicated with dotted lines. (B) qRT-PCR of ChIP analysis of the GAPDH and γ-actin genes, using antibodies against NELF-E. The H3 levels are indicated with dotted lines.
We have also analyzed the distribution of pol II CTD Ser5 and Ser7 phosphorylation on the U2 and β-actin genes, since the CTD phosphorylation pattern may influence the behavior of pol II at a NELF-dependent block. Ser5 is at its highest, relative to pol II, at the promoters of both the U2 and β-actin genes (see Fig. S5 in the supplemental material). In contrast, Ser7 phosphorylation relative to pol II is highest at the promoter of the U2 gene, but at the end of the β-actin transcription unit. The timing of Ser7 phosphorylation may therefore influence P-TEFb function during transcription of protein-coding genes and snRNA genes.
Taken together, these results demonstrate that, in contrast to the β-actin gene, there is no transcriptional pausing by NELF in the transcribed region of U2 genes. Instead, NELF is recruited at the end of the transcription unit. This result emphasizes that the requirement for the elongation function of P-TEFb in vivo is dependent on the presence of a NELF-induced block to transcription in vivo as in vitro. On both the β-actin and snRNA genes, the nucleosome-depleted region can be efficiently transcribed by pol II in the presence of the CDK9 inhibitor, indicating that P-TEFb activity is critical only for transcription of nucleosomal templates in vivo.
Interestingly, NELF association maps close to the beginning of the GAPDH and γ-actin genes, demonstrating that this factor is not recruited at the same point of the transcription cycle on all protein-coding genes (Fig. 2B). However, in all cases, NELF association correlates with an increase in histone density, suggesting that nucleosomes play a central role in elongation checkpoint formation and/or function.
Knockdown of NELF causes a defect in termination of transcription of the U2 genes.
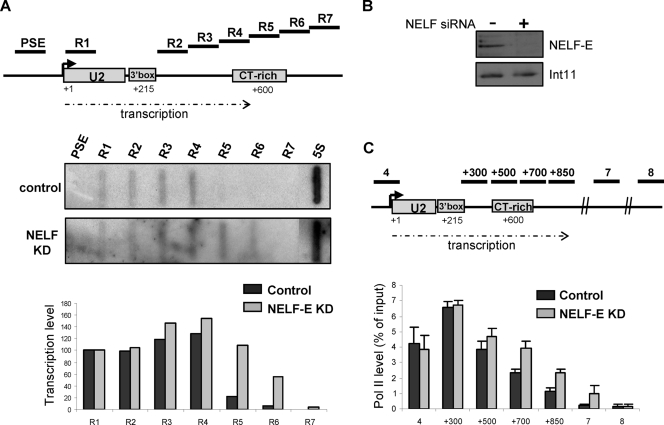
In order to determine whether NELF association with the end of the U2 gene transcription unit plays a functional role, we have knocked down the level of the NELF-E subunit by RNA interference (RNAi) (Fig. 3B) and assayed transcription by nuclear run-on analysis and pol II occupancy by ChIP (Fig. 3A and C). Knockdown of NELF-E has a marked effect on termination of transcription of the U2 genes, extending high levels of transcription downstream by at least 200 bp and increasing pol II levels downstream of the 3′ box. These results indicate that NELF plays a key role in termination of transcription of the U2 genes.
FIG. 3.
Knockdown of NELF causes a defect in termination of transcription of the U2 genes. (A) The relative position of single-stranded oligonucleotides used for nuclear run-on analysis is noted on the diagram of the U2 gene transcription unit. The angled arrow indicates the start of transcription. The extent of transcription is indicated by a broken arrow. The result of nuclear run-on analysis with and without RNAi-mediated knockdown (KD) of NELF-E is shown below. An oligonucleotide complementary to transcripts from the pol III-transcribed 5S RNA gene was used as a control of the level of transcription. Corrected hybridization signals (with R1 taken as 100%) are shown in the graph below. (B) Western blot analysis of NELF-E was carried out before and after RNAi-mediated knockdown. Western blot analysis of Int11 was used as a control for protein levels. (C) qRT-PCR of ChIP analysis of the RNU2 locus, using an antibody against pol II occupancy, after NELF-E knockdown. Additional primers (+300/+500/+700/+850) are noted above the diagram of the RNU2 locus. All other primers are as shown in Fig. 1.
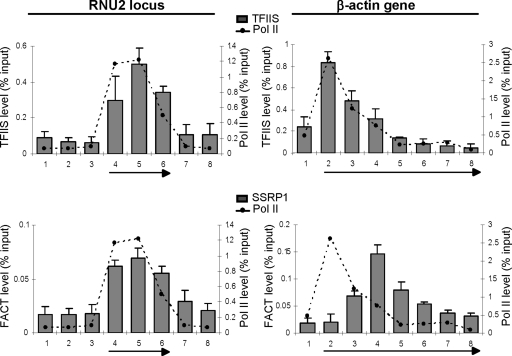
TFIIS and FACT are associated with both snRNA genes and protein-coding genes.
Several studies provide evidence that nucleosomes can efficiently pause transcribing pol II in vitro and in vivo and that histone acetyltransferases (HATs), chromatin remodellers (Swi/snf, FACT), TFIIS, and activation domains of transcription factors are all implicated in relief of the pause (5, 8-10, 27, 32, 47, 54). It is not clear why NELF participates in a P-TEFb-reversible elongation checkpoint on protein-coding genes but functions to halt transcription of the U2 snRNA genes. Since an increase in nucleosome density correlates with checkpoint or termination function, differential association of factors required for transcription of nucleosomal templates may underlie differences in NELF function. Accordingly, we have analyzed the association of TFIIS and FACT (facilitates transcription through chromatin) with the U2 and β-actin genes (Fig. 4). These factors have been shown to play roles in elongation of transcription on chromatin templates (25, 27, 43). TFIIS promotes productive elongation by stimulating cleavage of backtracked transcripts that arise as pol II stalls during early elongation (57). FACT, which comprises Spt16 and SSRP1 proteins (50), destabilizes nucleosome structure during passage of pol II (2). TFIIS is associated with both genes and correlates with pol II occupancy (Fig. 4). In addition, the ratio of TFIIS to pol II is approximately the same for both genes, suggesting that TFIIS travels with the polymerase in vivo regardless of chromatin structure. The two subunits of FACT are also found to be associated with both genes (Fig. 4; also see Fig. S6 in the supplemental material). On snRNA genes, FACT association largely correlates with pol II occupancy. In contrast, FACT appears to be recruited to the β-actin gene at the same position as NELF. Like Gomes et al. (23), we find that FACT association is drastically reduced after DRB treatment (S. Egloff and S. Murphy, unpublished observations), indicating that P-TEFb activity is required for FACT recruitment, possibly through Ser2 phosphorylation of the CTD. This is further emphasized by the close correlation between FACT localization and Ser2 phosphorylation on both types of genes. Thus, although the transcribed region of U2 genes is largely nucleosome-depleted, TFIIS and FACT are still recruited to the genes. These results therefore indicate that the failure of pol II to negotiate the NELF-dependent block at the end of the U2 snRNA genes is not due to the failure to recruit TFIIS and FACT. The role of FACT in elongation of transcription of snRNA genes is unclear, since DRB treatment abolishes FACT association (23) without affecting transcription of snRNA genes (40).
FIG. 4.
TFIIS and FACT are associated with both snRNA and protein-coding genes. qRT-PCR of ChIP analysis of the RNU2 locus and β-actin genes, using antibodies against the proteins or subunits of the protein complexes noted. The levels of pol II occupancy are indicated with dotted lines.
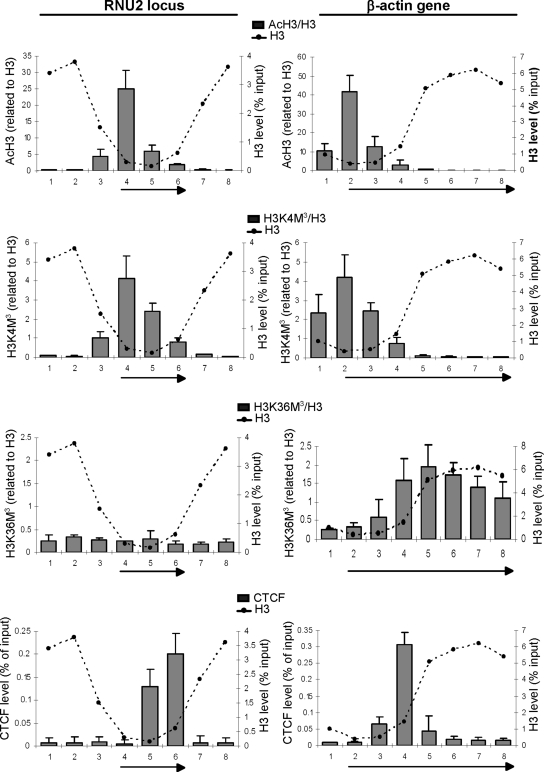
The chromatin landscape of the RNU2 locus and the β-actin gene.
The histones, particularly their N-terminal tails, are subject to a large number of dynamic posttranslational modifications which can influence all stages of the transcription cycle (28, 32). Some posttranslational modifications of histone tails, such as acetylation of histone H3 and H4 by HATs or trimethylation of H3K4 and H3K36 by methyltransferases (HMTs), are associated with active transcription (32). Others, such as H3K9 trimethylation, correlate mainly with gene repression. As the chromatin state could influence transcription, we have compared the histone modification landscapes across the RNU2 locus and the β-actin gene by ChIP. H3 and H4 acetylation is highly enriched in the promoter region of both genes, where it could facilitate nucleosome removal (Fig. 5; also see Fig. S7 in the supplemental material). Trimethylation of H3K4 is also detected surrounding the transcription start sites and mirrors the histone acetylation pattern (Fig. 5). These results are in accordance with the general picture emerging from genome-wide analysis that these modifications are enriched at the 5′ end of actively transcribed genes (1, 3). Even though core histones are highly depleted around the start sites of the U2 and β-actin genes, methylated and acetylated histones are still readily detectable, emphasizing that a very high proportion of the remaining histones are modified.
FIG. 5.
Chromatin modifications across the RNU2 locus and β-actin gene. qRT-PCR of ChIP analysis on the RNU2 locus and β-actin gene. The y axis is the ratio of the percentage of input DNA immunoprecipitated with the antibodies noted to the percentage of input DNA immunoprecipitated with an anti-H3 antibody. The H3 levels are indicated with dotted lines.
Interestingly, the levels of H3K36 di- and trimethylation are very low relative to those of H3 across the whole U2 repeat, whereas these marks are clearly detected in the coding region of the β-actin gene, specifically following the region of nucleosome depletion (Fig. 5 and data not shown). H3K36 di- and trimethylation are directed by the Set2 methyltransferase-containing complex (52, 53). Human Set2 directly binds pol II CTD phosphorylated on Ser5 and Ser2 (34), linking Ser2 kinase to H3K36 trimethylation. Accordingly, H3K36 trimethylation closely correlates with the appearance of Ser2 phosphorylation on the β-actin gene (Fig. 2A and 5), and treatment of cells with the CDK9 inhibitors DRB and KM05382 drastically affects the level of H3K36 trimethylation on the β-actin gene (see Fig. S8 in the supplemental material; also data not shown). The low level of signal detected on the U2 genes is instead unaffected, indicating that it likely represents nonspecific background levels. Thus, although Ser2 phosphorylation is detected on pol II transcribing both types of genes (39) (Fig. 2), it differentially influences the chromatin landscape. In Saccharomyces cerevisiae, H3K36 methylation helps to recruit deacetylases to suppress cryptic-gene internal initiation on long or infrequently transcribed genes (33). Set2 activity may therefore be unnecessary for the correct expression of the short and highly transcribed U2 genes.
H3K9 trimethylation is absent on snRNA genes, while it is detected on the β-actin gene (see Fig. S9 in the supplemental material). The role of this modification remains unclear as it is found on both highly expressed protein-coding genes and regions of heterochromatin (32).
In conclusion, a chromatin mark linked to P-TEFb activity is missing on snRNA genes, despite the presence of Ser2 phosphorylation (39) (Fig. 2), indicating that this CTD modification does not automatically lead to Set2 recruitment and/or activity. The differential effect of P-TEFb activity on the chromatin landscape further emphasizes differences in P-TEFb function in expression of snRNA and protein-coding genes.
CTCF binding sites are located close to the “late” NELF-dependent control point.
Finally, we scanned the nucleosome-depleted region of the U2 and β-actin genes for features that might influence NELF positioning and noted the presence of potential binding sites for the boundary protein CTCF close to the downstream border of nucleosome depletion in both the U2 gene and the β-actin gene (Fig. 5; also see Fig. S4 in the supplemental material). CTCF is a multifunctional protein found at insulators, where it prevents unwanted enhancer/promoter interactions (55), and has recently been implicated in positioning of nucleosomes (19). ChIP analysis indicates that CTCF is indeed associated with this region of the U2 and β-actin genes, raising the possibility that this factor is involved in the function and/or positioning of the NELF-dependent control point.
DISCUSSION
Does a nucleosomal roadblock regulate elongation of transcription?
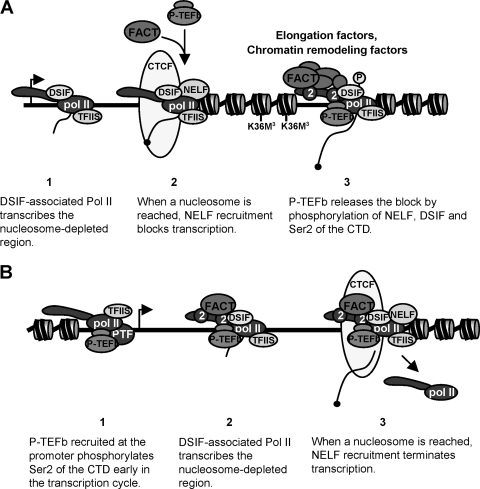
Our results, taken together, shed new light on the positioning and function of the NELF-dependent elongation checkpoint in vivo. P-TEFb independency closely correlates with nucleosomal depletion on both snRNA and protein-coding genes, suggesting that chromatin structure plays a major role in the elongation function of P-TEFb. The increase in nucleosome occupancy maps with the transcriptional block imposed by N-TEFs, suggesting that this checkpoint is directly regulated by chromatin structure. As P-TEFb activity is critical for the release of this block, and also for recruitment of chromatin-remodeling activities, we propose that the main function of P-TEFb in elongation of transcription is to promote pol II transcription through a nucleosomal checkpoint. This model explains why pol II can transcribe the U2 snRNA gene and the beginning of protein-coding genes without P-TEFb activity (Fig. 6). TFIIS, which travels with pol II, could help to release the block. However, DSIF/NELF has been reported to inhibit TFIIS in vitro (44). Our data support the notion that the function of NELF recruitment in this process is to avoid premature release of the block by TFIIS to provide a window of time for recruitment of capping enzymes and/or P-TEFb. Phosphorylation of Ser2 of the CTD by P-TEFb would allow the association of chromatin modification factors such as FACT and Set2, which in turn would augment pol II processivity on chromatinized templates to efficiently transcribe the rest of the gene. This model is supported by the fact that inhibition of P-TEFb activity leads to a loss of FACT association and Set2 activity concomitant with the loss of ability to elongate. The requirement for P-TEFb recruitment to allow productive elongation also ensures that the CTD will be modified to activate downstream RNA processing reactions (17). In contrast, P-TEFb recruitment to the U2 gene occurs at a very early stage and is checkpoint independent (Fig. 6).
FIG. 6.
Nucleosome depletion circumvents the need for P-TEFb for efficient elongation of transcription. (A) Within the transcription units of protein-coding genes, the presence of nucleosomes causes a “road block” for pol II that becomes a checkpoint due to NELF recruitment. NELF recruitment may be due to polymerase stalling and/or the increase in nucleosomes. P-TEFb activity is required to overcome NELF/nucleosome-induced arrest. CTCF may play a role in positioning the NELF/nucleosome checkpoint. Nucleosomes are depicted as cylinders with two turns of DNA wrapped around them. The angled arrow indicates the start of transcription. (B) In the absence of nucleosomes on the U2 transcription unit, NELF is not recruited. NELF recruitment together with an increase in nucleosome occupancy causes termination of transcription. CTCF may play a role in positioning the NELF/nucleosome terminator.
The elongation checkpoint occurs later in the transcription cycle of the β-actin gene than in those of the other protein-coding genes studied, suggesting that checkpoint positioning is either quite variable or that the precise positioning plays a gene-specific role.
What turns NELF into a transcription terminator?
In the case of the U2 genes, pol II reaches the nucleosome/NELF roadblock after transcribing the short gene, and transcription is terminated. The demonstration that NELF functions as a termination factor in this case further expands the repertoire of functions performed by this complex, which has also been shown to play a role in 3′ processing of replication-activated histone mRNAs (41). Why is the block to pol II on the U2 genes not traversed? All elongation factors tested are recruited to U2 and β-actin genes, suggesting that pol II should be able to transcribe through the nucleosome block at the end of the U2 transcription unit. However, the timing of P-TEFb recruitment and Ser2 phosphorylation is not the same in expression of U2 snRNA and β-actin genes. For snRNA genes, Ser2 of the pol II CTD will already be phosphorylated when NELF is encountered, which might trigger termination rather than conversion to productive elongation. Ser7 is also phosphorylated early in transcription of snRNA genes, whereas this mark is highest at the 3′ end of the β-actin gene and other protein-coding genes (12). Ser7 phosphorylation is required to recruit the snRNA gene-specific Integrator complex to the pol II CTD (17, 18). Thus, “early” phosphorylation of Ser2 and Ser7 and Integrator recruitment could affect critical interactions between the pol II CTD and elongation/termination factors at the checkpoint, causing termination to occur. Alternatively, P-TEFb may be inactive when it reaches the end of the transcription unit and be unable to phosphorylate NELF to release the block. In this scenario, different timings of P-TEFb activity lead to different outcomes. This is an attractive possibility, since inhibition of P-TEFb activity has no effect on the U2 gene transcription profile (40).
Another notable difference is that H3K36 trimethylation is found only on the β-actin genes. In addition to CTD phosphorylation, trimethylation of H3K36 in yeast requires expression of Spt6, which has roles in elongation of transcription (11, 60). It is therefore possible that differences in H3K36 trimethylation reflect the fact that some other elongation factors are differentially recruited to the U2 and β-actin genes.
NELF recruitment and chromatin structure.
How is NELF recruited specifically where nucleosome occupancy increases? One possibility is that NELF is recruited when the polymerase is slowed down by nucleosomes. Transcriptional control of the HIV type 1 genome is known to require NELF association with the TAR RNA element at the 5′ ends of HIV transcripts to block transcription close to the site of initiation. P-TEFb- dependent release of the block is accompanied by the recruitment of histone-modifying enzymes and ATP-dependent chromatin-remodeling complexes (49), suggesting that chromatin structure also plays a role in this block to transcription of the viral genome in infected cells. In many Drosophila protein-coding genes, NELF is associated with the region immediately downstream of the transcription start site, where paused polymerase is also located (30). However, no specific RNA sequences that might play a role in recruitment have been identified (30). Chromatin structure analysis indicates that many Drosophila genes have a nucleosome in the vicinity of the transcription start site (38). In light of these and our findings, it is logical to propose that NELF/paused polymerase generally colocalize with a nucleosome on protein-coding genes in higher eukaryotes.
The factors required for transcription-independent clearing of nucleosomes from the human β-actin and U2 genes remain to be identified. Transcription-independent clearing of nucleosomes also occurs upon activation of the Drosophila Hsp70 gene, and promoter-specific factors are involved (47). Likewise, snRNA gene-specific promoter factors like PTF/SNAPc/PBP (26) may be responsible for clearing the way for efficient P-TEFb-independent transcription on U2 genes. It has recently been shown that NELF-dependent pol II pausing on many Drosophila protein-coding genes is required to maintain a high level of polymerase at the promoter and depletion of promoter-proximal nucleosomes (20), raising the possibility that NELF itself is somehow linked to nucleosome depletion. However, NELF knockdown does not cause a change in the profile of histones over the U2 gene (S. Egloff and S. Murphy, unpublished results), indicating that the relationship between NELF and chromatin structure is complex and possibly gene type and/or species specific.
CTCF is a key component of insulators or boundary elements that ensure that enhancers activate transcription of only legitimate target genes (55). However, it does not appear to be functioning as part of an insulator on the U2 and β-actin genes where it is located within or close to the end of a transcription unit. The proximity of CTCF to the point of NELF recruitment suggests that it participates in checkpoint structure or function. Recent findings implicate CTCF in nucleosome positioning (19), raising the possibility that it may help to nucleate and position the checkpoint.
Finally, it appears that only a small proportion of polymerases associated with the DNA template successfully negotiate the roadblock on active protein-coding genes. This early elongation checkpoint is therefore very powerful, even when transcription is not terminated, and provides a major point of potential regulation of gene expression. In vivo dynamic studies of pol II transcription have revealed that only 1% of polymerase-gene interactions lead to completion of an mRNA (15). The NELF/nucleosome roadblock detected on protein-coding genes is likely to actively participate in this “selection process.” In the most extreme cases, no pol II will be released from the block leading to poised polymerase (48) or termination of transcription, as we have seen in the U2 snRNA genes. Finding out what exactly controls the proportion of pol II that ultimately escapes into productive elongation remains an important challenge for future research.
Supplementary Material
Acknowledgments
We thank Jong-Bok Yoon and Bob Roeder for the anti-PTF antibodies, Hiroshi Handa for NELF-E cDNA and the NELF-E antibody, Zbig Dominski for the Int11 antibody, Dirk Eick for the Ser7 antibody, and Nick Proudfoot and Mathilde Calligé for critical reading of the manuscript.
This work was supported by Medical Research Council (United Kingdom) and Wellcome Trust grants to S.M. and a fellowship from King Saud University to H.A.-R.
Footnotes
Published ahead of print on 18 May 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Barski, A., S. Cuddapah, K. Cui, T. Y. Roh, D. E. Schones, Z. Wang, G. Wei, I. Chepelev, and K. Zhao. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129823-837. [DOI] [PubMed] [Google Scholar]
- 2.Belotserkovskaya, R., S. Oh, V. A. Bondarenko, G. Orphanides, V. M. Studitsky, and D. Reinberg. 2003. FACT facilitates transcription-dependent nucleosome alteration. Science 3011090-1093. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein, B. E., M. Kamal, K. Lindblad-Toh, S. Bekiranov, D. K. Bailey, D. J. Huebert, S. McMahon, E. K. Karlsson, E. J. Kulbokas III, T. R. Gingeras, S. L. Schreiber, and E. S. Lander. 2005. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120169-181. [DOI] [PubMed] [Google Scholar]
- 4.Bird, G., D. A. Zorio, and D. L. Bentley. 2004. RNA polymerase II carboxy-terminal domain phosphorylation is required for cotranscriptional pre-mRNA splicing and 3′-end formation. Mol. Cell. Biol. 248963-8969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bondarenko, V. A., L. M. Steele, A. Ujvari, D. A. Gaykalova, O. I. Kulaeva, Y. S. Polikanov, D. S. Luse, and V. M. Studitsky. 2006. Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Mol. Cell 24469-479. [DOI] [PubMed] [Google Scholar]
- 6.Boyd, D. C., I. H. Greger, and S. Murphy. 2000. In vivo footprinting studies suggest a role for chromatin in transcription of the human 7SK gene. Gene 24733-44. [DOI] [PubMed] [Google Scholar]
- 7.Brès, V., S. M. Yoh, and K. A. Jones. 2008. The multi-tasking P-TEFb complex. Curr. Opin. Cell Biol. 20334-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, S. A., A. N. Imbalzano, and R. E. Kingston. 1996. Activator-dependent regulation of transcriptional pausing on nucleosomal templates. Genes Dev. 101479-1490. [DOI] [PubMed] [Google Scholar]
- 9.Brown, S. A., and R. E. Kingston. 1997. Disruption of downstream chromatin directed by a transcriptional activator. Genes Dev. 113116-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carey, M., B. Li, and J. L. Workman. 2006. RSC exploits histone acetylation to abrogate the nucleosomal block to RNA polymerase II elongation. Mol. Cell 24481-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carrozza, M. J., B. Li, L. Florens, T. Suganuma, S. K. Swanson, K. K. Lee, W. J. Shia, S. Anderson, J. Yates, M. P. Washburn, and J. L. Workman. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123581-592. [DOI] [PubMed] [Google Scholar]
- 12.Chapman, R. D., M. Heidemann, T. K. Albert, R. Mailhammer, A. Flatley, M. Meisterernst, E. Kremmer, and D. Eick. 2007. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science 3181780-1782. [DOI] [PubMed] [Google Scholar]
- 13.Chodosh, L. A., A. Fire, M. Samuels, and P. A. Sharp. 1989. 5,6-Dichloro-1-β-d-ribofuranosylbenzimidazole inhibits transcription elongation by RNA polymerase II in vitro. J. Biol. Chem. 2642250-2257. [PubMed] [Google Scholar]
- 14.Cuello, P., D. C. Boyd, M. J. Dye, N. J. Proudfoot, and S. Murphy. 1999. Transcription of the human U2 snRNA genes continues beyond the 3′ box in vivo. EMBO J. 182867-2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Darzacq, X., Y. Shav-Tal, V. de Turris, Y. Brody, S. M. Shenoy, R. D. Phair, and R. H. Singer. 2007. In vivo dynamics of RNA polymerase II transcription. Nat. Struct. Mol. Biol. 14796-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dominski, Z., X. C. Yang, M. Purdy, E. J. Wagner, and W. F. Marzluff. 2005. A CPSF-73 homologue is required for cell cycle progression but not cell growth and interacts with a protein having features of CPSF-100. Mol. Cell. Biol. 251489-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Egloff, S., and S. Murphy. 2008. Cracking the RNA polymerase II CTD code. Trends Genet. 24280-288. [DOI] [PubMed] [Google Scholar]
- 18.Egloff, S., D. O'Reilly, R. D. Chapman, A. Taylor, K. Tanzhaus, L. Pitts, D. Eick, and S. Murphy. 2007. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science 3181777-1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu, Y., M. Sinha, C. L. Peterson, and Z. Weng. 2008. The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet. 4e1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilchrist, D. A., S. Nechaev, C. Lee, S. K. Ghosh, J. B. Collins, L. Li, D. S. Gilmour, and K. Adelman. 2008. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev. 221921-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilmour, D. S. 2009. Promoter proximal pausing on genes in metazoans. Chromosoma 1181-10. [DOI] [PubMed] [Google Scholar]
- 22.Glover-Cutter, K., S. Kim, J. Espinosa, and D. L. Bentley. 2008. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat. Struct. Mol. Biol. 1571-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gomes, N. P., G. Bjerke, B. Llorente, S. A. Szostek, B. M. Emerson, and J. M. Espinosa. 2006. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 20601-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gromak, N., S. West, and N. J. Proudfoot. 2006. Pause sites promote transcriptional termination of mammalian RNA polymerase II. Mol. Cell. Biol. 263986-3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guermah, M., V. B. Palhan, A. J. Tackett, B. T. Chait, and R. G. Roeder. 2006. Synergistic functions of SII and p300 in productive activator-dependent transcription of chromatin templates. Cell 125275-286. [DOI] [PubMed] [Google Scholar]
- 26.Jawdekar, G. W., and R. W. Henry. 2008. Transcriptional regulation of human small nuclear RNA genes. Biochim. Biophys. Acta 1779295-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kireeva, M. L., B. Hancock, G. H. Cremona, W. Walter, V. M. Studitsky, and M. Kashlev. 2005. Nature of the nucleosomal barrier to RNA polymerase II. Mol. Cell 1897-108. [DOI] [PubMed] [Google Scholar]
- 28.Kouzarides, T. 2007. Chromatin modifications and their function. Cell 128693-705. [DOI] [PubMed] [Google Scholar]
- 29.Kristjuhan, A., and J. Q. Svejstrup. 2004. Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo. EMBO J. 234243-4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, C., X. Li, A. Hechmer, M. Eisen, M. D. Biggin, B. J. Venters, C. Jiang, J. Li, B. F. Pugh, and D. S. Gilmour. 2008. NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila. Mol. Cell. Biol. 283290-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, C. K., Y. Shibata, B. Rao, B. D. Strahl, and J. D. Lieb. 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 36900-905. [DOI] [PubMed] [Google Scholar]
- 32.Li, B., M. Carey, and J. L. Workman. 2007. The role of chromatin during transcription. Cell 128707-719. [DOI] [PubMed] [Google Scholar]
- 33.Li, B., M. Gogol, M. Carey, S. G. Pattenden, C. Seidel, and J. L. Workman. 2007. Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Genes Dev. 211422-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, M., H. P. Phatnani, Z. Guan, H. Sage, A. L. Greenleaf, and P. Zhou. 2005. Solution structure of the Set2-Rpb1 interacting domain of human Set2 and its interaction with the hyperphosphorylated C-terminal domain of Rpb1. Proc. Natl. Acad. Sci. USA 10217636-17641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mandal, S. S., C. Chu, T. Wada, H. Handa, A. J. Shatkin, and D. Reinberg. 2004. Functional interactions of RNA-capping enzyme with factors that positively and negatively regulate promoter escape by RNA polymerase II. Proc. Natl. Acad. Sci. USA 1017572-7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mangin, M., M. Ares, Jr., and A. M. Weiner. 1986. Human U2 small nuclear RNA genes contain an upstream enhancer. EMBO J. 5987-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marshall, N. F., and D. H. Price. 1995. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J. Biol. Chem. 27012335-12338. [DOI] [PubMed] [Google Scholar]
- 38.Mavrich, T. N., C. Jiang, I. P. Ioshikhes, X. Li, B. J. Venters, S. J. Zanton, L. P. Tomsho, J. Qi, R. L. Glaser, S. C. Schuster, D. S. Gilmour, I. Albert, and B. F. Pugh. 2008. Nucleosome organization in the Drosophila genome. Nature 453358-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Medlin, J., A. Scurry, A. Taylor, F. Zhang, B. M. Peterlin, and S. Murphy. 2005. P-TEFb is not an essential elongation factor for the intronless human U2 snRNA and histone H2b genes. EMBO J. 244154-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Medlin, J. E., P. Uguen, A. Taylor, D. L. Bentley, and S. Murphy. 2003. The C-terminal domain of pol II and a DRB-sensitive kinase are required for 3′ processing of U2 snRNA. EMBO J. 22925-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Narita, T., T. M. Yung, J. Yamamoto, Y. Tsuboi, H. Tanabe, K. Tanaka, Y. Yamaguchi, and H. Handa. 2007. NELF interacts with CBC and participates in 3′ end processing of replication-dependent histone mRNAs. Mol. Cell 26349-365. [DOI] [PubMed] [Google Scholar]
- 42.Ni, Z., B. E. Schwartz, J. Werner, J. R. Suarez, and J. T. Lis. 2004. Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol. Cell 1355-65. [DOI] [PubMed] [Google Scholar]
- 43.Orphanides, G., G. LeRoy, C. H. Chang, D. S. Luse, and D. Reinberg. 1998. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell 92105-116. [DOI] [PubMed] [Google Scholar]
- 44.Palangat, M., D. B. Renner, D. H. Price, and R. Landick. 2005. A negative elongation factor for human RNA polymerase II inhibits the anti-arrest transcript-cleavage factor TFIIS. Proc. Natl. Acad. Sci. USA 10215036-15041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pavelitz, T., A. D. Bailey, C. P. Elco, and A. M. Weiner. 2008. Human U2 snRNA genes exhibit a persistently open transcriptional state and promoter disassembly at metaphase. Mol. Cell. Biol. 283573-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peterlin, B. M., and D. H. Price. 2006. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 23297-305. [DOI] [PubMed] [Google Scholar]
- 47.Petesch, S. J., and J. T. Lis. 2008. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell 13474-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Price, D. H. 2008. Poised polymerases: on your mark… get set… go! Mol. Cell 307-10. [DOI] [PubMed] [Google Scholar]
- 49.Quivy, V., S. De Walque, and C. Van Lint. 2007. Chromatin-associated regulation of HIV-1 transcription: implications for the development of therapeutic strategies. Subcell. Biochem. 41371-396. [PubMed] [Google Scholar]
- 50.Reinberg, D., and R. J. Sims III. 2006. de FACTo nucleosome dynamics. J. Biol. Chem. 28123297-23301. [DOI] [PubMed] [Google Scholar]
- 51.Schwabish, M. A., and K. Struhl. 2004. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 2410111-10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shilatifard, A. 2006. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem. 75243-269. [DOI] [PubMed] [Google Scholar]
- 53.Strahl, B. D., P. A. Grant, S. D. Briggs, Z. W. Sun, J. R. Bone, J. A. Caldwell, S. Mollah, R. G. Cook, J. Shabanowitz, D. F. Hunt, and C. D. Allis. 2002. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol. Cell. Biol. 221298-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wada, T., G. Orphanides, J. Hasegawa, D. K. Kim, D. Shima, Y. Yamaguchi, A. Fukuda, K. Hisatake, S. Oh, D. Reinberg, and H. Handa. 2000. FACT relieves DSIF/NELF-mediated inhibition of transcriptional elongation and reveals functional differences between P-TEFb and TFIIH. Mol. Cell 51067-1072. [DOI] [PubMed] [Google Scholar]
- 55.Wallace, J. A., and G. Felsenfeld. 2007. We gather together: insulators and genome organization. Curr. Opin. Genet. Dev. 17400-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Westin, G., J. Zabielski, K. Hammarstrom, H. J. Monstein, C. Bark, and U. Pettersson. 1984. Clustered genes for human U2 RNA. Proc. Natl. Acad. Sci. USA 813811-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wind, M., and D. Reines. 2000. Transcription elongation factor SII. Bioessays 22327-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamaguchi, Y., T. Takagi, T. Wada, K. Yano, A. Furuya, S. Sugimoto, J. Hasegawa, and H. Handa. 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 9741-51. [DOI] [PubMed] [Google Scholar]
- 59.Yoon, J. B., S. Murphy, L. Bai, Z. Wang, and R. G. Roeder. 1995. Proximal sequence element-binding transcription factor (PTF) is a multisubunit complex required for transcription of both RNA polymerase II- and RNA polymerase III-dependent small nuclear RNA genes. Mol. Cell. Biol. 152019-2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Youdell, M. L., K. O. Kizer, E. Kisseleva-Romanova, S. M. Fuchs, E. Duro, B. D. Strahl, and J. Mellor. 2008. Roles for Ctk1 and Spt6 in regulating the different methylation states of histone H3 lysine 36. Mol. Cell. Biol. 284915-4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yuo, C. Y., M. Ares, Jr., and A. M. Weiner. 1985. Sequences required for 3′ end formation of human U2 small nuclear RNA. Cell 42193-202. [DOI] [PubMed] [Google Scholar]
- 62.Zhou, Q., and J. H. Yik. 2006. The yin and yang of P-TEFb regulation: implications for human immunodeficiency virus gene expression and global control of cell growth and differentiation. Microbiol. Mol. Biol. Rev. 70646-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.