Abstract
Both casein kinase 1 delta (CK1δ) and epsilon (CK1ɛ) phosphorylate core clock proteins of the mammalian circadian oscillator. To assess the roles of CK1δ and CK1ɛ in the circadian clock mechanism, we generated mice in which the genes encoding these proteins (Csnk1d and Csnk1e, respectively) could be disrupted using the Cre-loxP system. Cre-mediated excision of the floxed exon 2 from Csnk1d led to in-frame splicing and production of a deletion mutant protein (CK1δΔ2). This product is nonfunctional. Mice homozygous for the allele lacking exon 2 die in the perinatal period, so we generated mice with liver-specific disruption of CK1δ. In livers from these mice, daytime levels of nuclear PER proteins, and PER-CRY-CLOCK complexes were elevated. In vitro, the half-life of PER2 was increased by ∼20%, and the period of PER2::luciferase bioluminescence rhythms was 2 h longer than in controls. Fibroblast cultures from CK1δ-deficient embryos also had long-period rhythms. In contrast, disruption of the gene encoding CK1ɛ did not alter these circadian endpoints. These results reveal important functional differences between CK1δ and CK1ɛ: CK1δ plays an unexpectedly important role in maintaining the 24-h circadian cycle length.
Circadian rhythms are rhythms in gene expression, metabolism, physiology, and behavior that persist in constant environmental conditions with a cycle length near 24 h. In mammals, the circadian timing system is hierarchical. The primary pacemaker regulating circadian behavioral rhythms is located in the suprachiasmatic nuclei (SCN) of the hypothalamus. Most cell types express circadian clock genes and will express rhythmicity in vitro. In vivo, the SCN entrains peripheral oscillators through a complex set of physiological and hormonal rhythms (31, 32, 36).
At the molecular level, circadian oscillations are governed by a cell-autonomous negative-feedback loop in which transcription factors drive the expression of their own negative regulators, leading to oscillation between periods of transcriptional activation and repression (reviewed in references 32 and 36). The bHLH-PAS containing transcription factors CLOCK or NPAS2 form heterodimers with BMAL1. These heterodimers binds to E-box elements within regulatory regions of Period (Per1, Per2, and Per3) and Cryptochrome (Cry1 and Cry2) genes to stimulate their transcription. Approximately 12 h after transcriptional activation, PER and CRY proteins reach concentrations sufficient to form repressor complexes that inhibit the activity of the CLOCK/NPAS2:BMAL1 heterodimer, reducing the transcription of Per and Cry genes and subsequently relieving PER/CRY-mediated negative feedback. E-box-mediated expression of other transcription factors, including members of the DBP/HLF/TEF and nuclear orphan receptor families (e.g., Rev-Erbα and ROR-A), provides a mechanism for clock control of genes with diverse promoters and with gene expression peaks occurring at a variety of phases.
Posttranslational modifications of circadian clock proteins play a well-established role in the regulation of circadian cycle length. In both flies and mammals, phosphorylation of PER proteins by casein kinase 1 (CK1) proteins is thought to play a key step in determining the speed of the circadian clock (reviewed in reference 5). Mammalian cell culture studies indicate that the phosphorylation of PER proteins by CK1 epsilon (CK1ɛ) regulates their subcellular localization, likely affects their transcription repression capability, and promotes their degradation through a proteasomal pathway dependent upon the F-box proteins β-TrCP1 and β-TrCP2 (12, 29, 34, 35). Interference with β-TrCP1 activity lengthens circadian period in oscillating fibroblasts (27). The CK1 inhibitor IC261 and the proteasome inhibitors MG132 and lactacystin also lengthen period in fibroblasts (12). CRY proteins are also subjected to phosphorylation and degradation cycles that regulate circadian period. The F-box protein FBXL3 plays a key role in regulating CRY1 stability; mutations inactivating this gene increase circadian cycle length (6, 16, 30). Collectively, these studies indicate that the duration of activity of the PER/CRY repressor complex, regulated primarily by the stability of PER and CRY proteins, dictates the cycle length of the molecular oscillator (15).
Genetic studies also support an important role for casein kinase action on PER proteins in regulating circadian period. A mutation in the Syrian hamster CK1ɛ gene, tau, shortens the circadian period of behavioral rhythms. Biochemically, the tau mutation (CK1ɛtau, a T178C substitution) differentially affects the activity of the kinase protein, reducing general kinase activity while increasing activity at specific residues of the PER proteins (14, 23). The tau mutation is a gain-of-function mutation with respect to circadian substrates, resulting in decreased PER stability and a reduction in circadian period length in tau mutant hamsters and mice (14, 24). In humans, familial advanced sleep phase syndrome (FASPS) is a circadian-based sleep disorder, in which affected individuals have a short circadian period and an advanced phase of the sleep-wake cycle. One study identified a FASPS pedigree with a mutation in human PER2 (hPER2; S662G mutation); this mutation prevents a priming phosphorylation, thus preventing CK1-mediated phosphorylation (33). A second study identified a dominant mutation within the kinase domain of CK1δ in a family with FASPS (38). Modeling this mutation in mice and flies revealed alterations in period length (38).
In the circadian field, much of the attention on mammalian casein kinases has focused on CK1ɛ. The few studies examining CK1δ suggest that it plays a role similar to CK1ɛ. For example, CK1δ, like CK1ɛ, phosphorylates PER proteins, reducing their stability in vitro (7, 38), and both CK1δ and CK1ɛ are present in PER/CRY repressor complexes in vivo (21). Despite these similarities, the role of CK1δ in the molecular clockwork is not well understood. We report here results demonstrating important differences between CK1δ and CK1ɛ in the regulation of circadian cycle length, based on studies utilizing mice in which these genes have been inactivated.
MATERIALS AND METHODS
Generation of a CK1δ targeting construct.
The CK1δ targeting construct (Fig. 1B) contained 10 kb of genomic sequence including a portion of intron 1, exon 2, and a portion of intron 2. A 5′ loxP site was added to intron 1, and a floxed neomycin and thymidine kinase (Neo/TK) cassette was inserted into intron 2. (Exon and intron numbering is in reference to coding exons.) The GenBank accession for this genomic region is for the reverse strand. For simplicity, nucleotides (nt) 32310522 to 32410522 of GenBank accession no. NT_165773.1 were selected, and the reverse complement was taken. With this numbering scheme, the Csnk1d genomic region is nt 18713 to 48305, and the A of ATG in exon 1 is nt 19036.
FIG. 1.
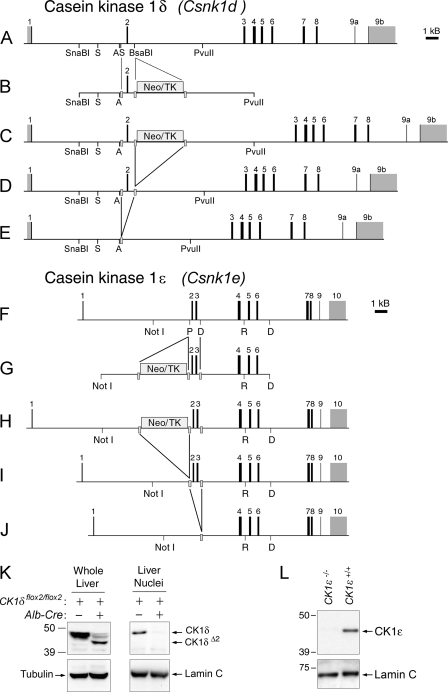
CK1δ and CK1ɛ targeting constructs and gene products. (A to E) CK1δ targeting constructs and gene products. (A) CK1δ (Csnk1d) gene. Restriction sites key to the manipulations described in text are indicated (A, AatII; S, StuI). Exons are numbered in reference to coding sequence. (B) Targeting construct. A loxP site (gray bar) was introduced into intron 1 at a StuI site, and a floxed Neo/TK cassette was inserted into intron 2 at a BsaBI site. (C) Targeted allele after homologous recombination in ES cells. (D) Targeted allele after in vitro treatment of ES cells with Cre recombinase to excise the Neo/TK cassette An ES cell clone with this allele (CK1δflox2/+) was microinjected into blastocysts to generate founder chimeras. (E) In vivo excision of exon 2 by Cre recombinase leads to the deleted allele, CK1δΔ2. (F to J) CK1ɛ targeting constructs and gene products. (F) CK1ɛ (Csnk1e) gene. Restriction sites key to the manipulations described in the text are indicated (P, PshAI; D, HinDIII; R, EcoRI). Exons are numbered in reference to coding sequence. (G) Targeting construct. A loxP sites was introduced into intron 3 at a HindIII site, and a floxed Neo/TK cassette was inserted into intron 1 at a PshAI site. (H) Targeted allele after homologous recombination in ES cells. (I) Targeted allele after in vitro treatment of ES cells with Cre recombinase to excise the Neo/TK cassette. An ES cell clone heterozygous for this allele (CK1ɛflox2-3/+) was selected for microinjection. (J) In vivo excision of exons 2 and 3 by Cre recombinase leads to the deleted allele, CK1ɛ−. (K) Western blot with an antibody to CK1δ. Samples are from an animal with floxed alleles of CK1δ (AlbCre negative, CK1δflox2/flox2) and an animal with hepatocyte-specific disruption of CK1δ (AlbCre+ CK1δflox2/flox2). In whole-cell extracts (left) a band of more rapid mobility is detected, representing CK1δΔ2 protein. In proteins extracted from liver nuclei (right), CK1δ is readily detected, while the CK1δΔ2 protein appears unable to accumulate in the nucleus. A very small amount of full-length CK1δ protein present in the AlbCre+ CK1δflox2/flox2 sample is likely due to a small population of other cell types (nonhepatocytes) in the liver. In panels K and L, antibodies to α-tubulin and lamin C were used to verify the equivalence of loading of whole-cell and nuclear extract samples, respectively. (L) Western blot of proteins extracted from liver nuclei of a control mouse (CK1ɛ+/+) and a mouse with whole-body disruption of CK1ɛ (CK1ɛ−/−), probed with an antibody to CK1ɛ.
The targeting construct was generated as follows. The SV129AB genomic DNA library was screened using a probe directed to intron 2, and three positive clones were identified. One clone was selected as the source of sequence for the construct. A 1.8-kb StuI fragment from the 5′ arm (corresponding to nt 24380 to 26257) was manipulated in several steps to add the loxP sequence from plasmid SJ36 (provided by S. N. Jones) at its 3′ end. An additional 5′ sequence was added by digesting this plasmid with EcoRV (a plasmid-derived site) and AatII and inserting a SnaBI-AatII fragment from the initial genomic clone (nt 22987 to 26165), resulting in a construct containing the 5′ arm from SnaBI (nt 22987) through StuI (nt 26257) to which the loxP site had been added at the 3′ end. Next, a 6.3-kb fragment including exon 2 and the 3′ arm was isolated by digesting the original genomic clone with StuI and PvuII (generating a fragment containing nt 26257 to 32657). This piece was cloned into the SmaI site of the plasmid containing the 5′ arm, resulting in an insert containing the genomic sequence from the SnaBI site at nt 22987 to the PvuII site at nt 32657, to which a loxP site had been added at the location of the StuI site at nt 26257. Finally, the floxed Neo/TK cassette from plasmid SJ41 (provided by S. N. Jones) was isolated, blunted with Klenow fragment, and inserted at a unique BsaBI site in intron 2 (nt 27319). Sequence analysis was used to confirm the orientation of the loxP sites and correct assembly of the construct.
Generation of a CK1ɛ targeting construct.
The CK1ɛ construct (Fig. 1G) consisted of 9.2 kb of genomic sequence beginning in intron 1 and ending in intron 6, into which a Neo/TK cassette flanked by loxP sites was introduced into intron 1, and a loxP site was introduced into intron 3. (Exon and intron numbering below is relative to the first coding exon, ignoring the existence of a 5′ noncoding exon.) The final targeting construct contained genomic sequence from nt 9205 to 18494 of GenBank accession NT_081921.
The targeting construct was generated as follows. The SV129AB genomic DNA library was screened using a probe directed to exon 2 of CK1ɛ. One of six positive clones was selected as the source of sequence for the construct. A 7.2-kb NotI-EcoRI fragment of the CK1ɛ genomic locus corresponding to nt 9205 to 16433 (including exons 2, 3, and 4) was subcloned. A loxP site from plasmid SJ36 was inserted into the HindIII site at nt 12814, just 3′ of exon 3. The 3′ arm was extended by ligating a 2.1-kb EcoRI-HindIII fragment (nt 16434 to 18494) to the EcoRI site at the 3′ end of construct 2. Finally, the floxed Neo/TK cassette from plasmid SJ41 was inserted into a unique PshAI site at nt 12204, just 5′ of exon 2. Sequence analysis was used to confirm the orientation of the loxP sites and to confirm correct assembly of the construct.
Generation of mice bearing targeted alleles.
Embryonic stem (ES) cell manipulations were performed by using standard procedures by the University of Massachusetts (UMass) Medical School Mouse Modeling Core facility. After transfection with targeting constructs, G418-resistant clones were isolated and Southern blot analysis of genomic DNA was used to identify ES cell clones in which homologous recombination had occurred and in which the integrity of the loxP sites was maintained (Fig. 1C and H). Selected clones (one each for CK1δ and CK1ɛ) were transfected with a Cre recombinase expression plasmid, and subclones were screened by PCR to distinguish those with the recombination between the first and third loxP sites (leading to exon plus Neo/TK cassette excision) from the desired subclones in which only the Neo/TK cassette was removed (Fig. 1D and I). Southern blot analysis with probes flanking and within the construct was used to identify subclones appropriate for microinjection. ES cells from the selected subclones were microinjected into C57BL/6J blastocysts by the UMass Medical School Mouse Modeling Core Facility. Chimeric founder mice were crossed to C57BL/6J mice, and agouti offspring were genotyped. Mice found to have the floxed allele were subsequently backcrossed to C57BL/6J.
Generation, maintenance, and genotyping of study animals.
All animal studies were reviewed and approved by the Institutional Animal Care and Use Committee at UMass Medical School. Mice used for studies had been backcrossed to the C57BL/6J background for at least 10 generations. Wild-type or littermate controls were studied simultaneously in each experiment.
To generate animals with whole-body excision of the floxed exons, animals with a floxed allele were crossed to a line expressing Cre recombinase in the testis (Protamine-Cre, C57BL/6J genetic background, generously provided by Roger Davis, UMass Medical School). Males with a floxed allele and the Protamine-Cre transgene were bred to C57BL/6J females, and offspring were assessed for deletion of the floxed region. In subsequent generations of backcrossing, the Protamine-Cre transgene was avoided.
Mice expressing Cre recombinase from the albumin promoter (AlbCre) purchased from the Jackson Laboratories [Bar Harbor, ME; B6.Cg-Tg(Alb-cre)21Mgn/J]. AlbCre+ mice were crossed to mice bearing a floxed allele of either CK1δ or CK1ɛ. Through intercrossing, lines homozygous for the floxed alleles were generated in which some animals also had the AlbCre transgene. Littermates lacking AlbCre were used as controls.
To assess the presence or absence of the Albumin-Cre transgene, a mixture consisting of two primer pairs was used. Forward (LilF, 5′-ACCTGAAGATGTTCGCGATTATCT-3′) and reverse (LilR, 5′-ACCGTCAGTACGTGAGATATCTT-3′) primers for Cre recombinase amplified an ∼370-bp band in only those samples containing the AlbCre transgene. Forward (5′-GCAAGAAGAACTAAGGAAAATTCAAGAGCAACTTCAGATGGTCCATGGTCAAGGGCTACAGTT-3′) and reverse (5′-TAGTGCCCTAGATGGCCCTGTTGG-3′) primers to the Clock gene were used to generate a ∼470-bp band from each sample, as a control for template integrity. The same two Cre primers were used to track the Protamine-Cre transgene.
Founder animals of the mPER2::LUC line on a C57BL/6J background were generously provided by Joseph Takahashi (Northwestern University). In these mice, the Per2 gene has been altered to express a fusion protein of mPER2 and firefly luciferase (41). To optimize expression levels in peripheral tissues, the mPER2::LUC line with an simian virus 40 polyadenylation signal was used (37). mPer2::luciferase genotyping methods were as described by Yoo et al (41), using a common forward primer (P2lucF 5′-CTGTGTTTACTGCGAGAGT-3′), a wild type-specific reverse primer (P2lucR1, 5′-GGGTCCATGTGATTAGAAAC-3′), and a luciferase knockin allele-specific reverse primer (P2lucR2, 5′-TAAAACCGGGAGGTAGATGAGA-3′).
For PCR genotyping at the CK1δ locus, the primer set ACR2 was used. This three-primer set consists of a forward primer located 5′ of the 5′ loxP site (A, 5′-CAGCCCTAGTTATCGTAACATCG-3′), a forward primer located just 5′ of the 3′ loxP site (C, 5′-GTAGTTTGCCTGATGAAACAGGAGC-3′), and a reverse primer (R2, 5′-GCTGAGGCCAATAAAGAGTCTGTCACATG-3′) located 3′ of the 3′ loxP site. The genotyping primer locations and products are shown in Fig. S1A and B in the supplemental material.
The genotype at the CK1ɛ locus was determined by using a set of three primers: forward primer F51 (5′-AACCTCAAATGCTGACTGGGTAGG-3′, located upstream of the 5′ loxP site); forward primer F53 (5′-GGGGCTGAGGGAGACTATAACGTG-3′), located between the 5′ and 3′ loxP sites; and reverse primer R52 (5′-TCCACCAGACCTTCCCCACTATTA-3′, located 3′ of the 3′ loxP site). The genotyping primer locations and products are shown in Fig. S1C and D in the supplemental material.
Except where otherwise noted, mice were maintained in a 12-h light/12-h dark lighting cycle with lights on at 0700. Animals for time point collections were transferred from the general housing room to a specialized facility in which the lighting conditions in each ventilated closet could be independently regulated. Within this facility, light was provided by fluorescent tubes and was ∼200 lx at the level of the cages. Dim red light from fluorescent tubes was on continuously, including periods referred to as constant darkness. Studies of protein and gene expression rhythms were conducted on the first day in constant darkness, with sampling beginning at circadian time 2 (CT02) or CT04 (e.g., 14 or 16 h after lights out).
Mouse line nomenclature.
The official symbol for the gene encoding CK1δ is Csnk1d. The Csnk1d allele in which exon 2 is flanked by loxP sites, which we refer to as CK1δflox2, is more properly called Csnk1dtm1drw (JAX stock no. 10487). The allele in which exon 2 has been excised by Cre-mediated recombination in the germ line, which we refer to as CK1δΔ2 is more properly called Csnk1dtm1.1drw.
Similarly, The official symbol for the gene encoding CK1ɛ is Csnk1e. The Csnk1e allele in which exons 2 and 3 are flanked by loxP sites, which we refer to as CK1ɛflox2-3, is more properly called Csnk1etm1drw (JAX stock no. 10489). The allele in which exons 2 and 3 has been excised by Cre-mediated recombination in the germ line, which we refer to as CK1eD2-3 ▵▵ CK1ɛ−, is more properly called Csnk1etm1.1drw.
Transfections.
All transfections were done into HEK293 cells cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum. Transfection was carried out by using Lipofectamine and Plus Reagent (Invitrogen) according to the manufacturer's protocol. Expression constructs were transfected at concentrations of 0.5 μg/35-mm plate, except for transfections assessing mPER2 stability in the presence or absence of MG132, where the amount of CK1-expressing plasmids was increased to 1.5 μg/plate.
Isolation of hepatic nuclei.
At selected times on the first cycle in constant darkness, mice were euthanized by carbon dioxide asphyxiation, and livers were collected in ice-cold saline. Nuclei were purified by sucrose gradient centrifugation as previously described (21).
Antibodies, Western blotting, and immunoprecipitation.
Antibodies for circadian proteins have been previously described and validated (21, 22). Proteins from liver nuclear extracts were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and detected with antibodies to mCLOCK (CLK-1-GP), mPER1 (PER1-1-GP), mPER2 (PER2-1-GP), mCRY1 (C1-GP), mCK1δ (CK1δ-GP), and CK1ɛ (CK1ɛ-GP) as described previously (21, 22). For most studies, a commercial anti-polymerase II (Pol II) antibody (8WG16, MMS-126R; Covance) was used to determine the equivalence of loading across lanes. Additional loading controls were monoclonal antibodies to alpha tubulin (Clone DM1A, T6199; Sigma) and lamin C (LC28.26; Developmental Studies Hybridoma Bank). Horseradish peroxidase-conjugated secondary antibodies were purchased from Santa Cruz. Signal detection was accomplished by using the Western Lightning Chemiluminescence Reagent Plus (Perkin-Elmer). Semiquantitative analysis of Western blots made use of a Fuji LAS-1000 system and ImageReady software.
Immunoprecipitation was performed as previously described (21). Liver nuclear extracts were immunoprecipitated using PER2-1-GP, and Western blots were probed with a subset of the antibodies listed above.
Protein extracts from transfected HEK293 cells were immunoprecipitated using an anti-V5 antibody (R960-25; Invitrogen), and blots were probed with antibodies to V5 (R960-25; Invitrogen) and c-Myc (A-14, sc-789; Santa Cruz). Anti-Flag M2 monoclonal antibody (F-1804) was purchased from Sigma.
Assessment of CK1δ activity.
Cotransfection experiments in HEK 293 cells were used to assess the ability of full-length casein kinase proteins to immunoprecipitate with PER protein, to promote an alteration in PER protein electrophoretic mobility, and to promote proteasome-mediated degradation of PER proteins.
In addition, the ability of casein kinase constructs to promote the incorporation of [γ-32P]ATP into PER proteins was assessed. Constructs used to assess in vitro kinase activity encoded proteins truncated at amino acid residue 320 to eliminate the C-terminal autophosphorylation region of the protein which can inhibit activity (17). Truncated, wild-type CK1δ (CK1δ/Τ) and a truncated construct from which residues encoded by exon 2 were deleted (CK1δΔ2/T) were initially cloned into the pCMV-Tag 2B vector (Stratagene), between the BamHI and EcoRI restriction sites.
In vitro activity of truncated CK1δ and CK1δΔ2 was assessed using kinase proteins produced in two different ways. In one approach, the C-terminal truncated kinase constructs were subcloned into the pCMV-Tag 2B vector (Stratagene), which added a Flag tag to the N terminus; kinase protein was produced by transfection in HEK 293 cells, followed by immunoprecipitation using anti-Flag M2 affinity gel (F2426; Sigma). In the second approach, the CK1δ constructs were subcloned into the bacterial expression vector pET-30a (Novagen) and expressed, and kinase proteins were purified using the S tag introduced by the vector.
For kinase protein production in HEK 293 cells, Flag-tagged C-terminal truncated wild-type CK1δ (Flag-CK1δ/T) and CK1δΔ2 (Flag-CK1δΔ2/T) constructs were transfected into HEK 293 cells, and 48 h later the cells were lysed in EB buffer (20 mM HEPES [pH 7.5], 100 mM NaCl, 5% glycerol, 1 mM EDTA, 0.05% Triton X-100, 1 mM dithiothreitol, complete protease inhibitor cocktail [Roche]) and immunoprecipitated with anti-Flag M2 affinity gel for 12 h at 4°C. The beads were then washed three times with EB buffer and mixed with either V5-mPER1 or V5-mPER2 proteins produced in TNT-reticulocyte lysate (L4960; Promega) in the presence of 10 μCi of l-[35S]methionine (Perkin-Elmer). These mixtures were then incubated at 30°C for 30 min in kinase reaction buffer (30 mM HEPES [pH 7.5], 7 mM MgCl2, 50 μg of bovine serum albumin/ml, 50 μM ATP, 0.5 mM dithiothreitol) supplemented with 5 μCi of [γ-32P]ATP (Perkin-Elmer) as described by Akashi et al. (2). The phosphorylation of the mPER substrates was detected by SDS-PAGE subjected to autoradiography using two films. The film closest to the SDS-PAGE (film A) was used to visualize both the phosphorylated 35S/32P-labeled and nonphosphorylated 35S-labeled mPER substrates. The second film (film B), further from the SDS-PAGE, was used to detect exclusively the phosphorylated 32P-labeled mPER substrates, since the 35S signal is quenched by film A. Kinase reaction samples were also subjected to Western blotting and probed with anti-Flag M2 monoclonal antibody (Sigma) to detect CK1δ proteins.
For studies using bacterially expressed kinases, BL21 cells (Stratagene) were transformed with the kinase construct in the pET-30a expression vector, and expression was induced with IPTG (isopropyl-β-d-thiogalactopyranoside). Bacteria were pelleted by centrifugation and then sonicated, with several cycles of sonication and pelleting to improve extraction efficiency. The extracted supernatant was subjected to ultracentifugation, and the supernatant containing the expressed protein was incubated with S-tag bead slurry (Novagen) overnight at 4°C. Pelleted beads were washed extensively with Wash-Bind buffer and were then resuspended in 30 mM HEPES buffer. The kinase assays were performed as described above, using [35S]methionine-labeled V5-mPER1 or V5-mPER2 proteins produced in TNT-reticulocyte lysates, in the presence of [γ-32P]ATP. Detection procedures were as described above, except that CK1δ was detected using antibody CK1δ-GP (24).
Assessment of circadian behavioral rhythms and phase-shifting responses to light.
To assess locomotor activity rhythms, mice were housed individually in cages equipped with a running wheel. Running wheel revolutions were detected by a magnetic reed switch detector connected to a computer-based monitoring system. ClockLab data collection software (Actimetrics) stored activity in 1-min bins. ClockLab data analysis software was used to produce double-plotted actograms and for χ2 periodogram estimation of period and rhythm amplitude. Periodogram analysis was conducted on two consecutive 21-day epochs beginning 10 days after entry into constant dark. Three experiments were conducted, each one comparing one of the mutant genotypes to wild-type controls. The wild-type controls from the three experiments did not differ and so were pooled.
Phase-shifting responses to light were assessed in a subset of the mice used for period determination (n = 9 to 12 per genotype) after the mice had been in constant darkness for >3 months. The lights were activated in the closet in which groups of cages were kept, exposing the entire cohort of animals to light without physically disturbing them. Light pulses were 1 h in duration and occurred at 10- to 14-day intervals. Retrospective analysis allowed determination of the circadian time of the animal at the time of the start of the pulse, where CT12 is activity onset. The phase of activity onset before the pulse was assessed by using ClockLab analysis software. The phase after the pulse was determined by using ClockLab to determine activity onsets on days 4 to 10 after the pulse (days 1 to 3 were omitted to exclude transient responses) and extrapolating to the day on which the shift occurred. Phase shifts are the difference, in hours, between the phase of activity onsets before and after the light pulse. Phase shifts are binned into 2-h intervals and are plotted at the midpoint of the interval.
Assessment of hepatic gene expression by real-time PCR.
Quantitative real-time PCR was performed using TaqMan probes with an ABI SDS 7000 instrument (Applied Biosystems, Foster City, CA) as previously described (9). Each experiment included equal representation of all genotypes and time points. Each sample was assessed in duplicate. The data for each transcript were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) using the 2−ΔΔCT method. To compare relative transcript levels between genotypes, all data points were normalized to the average expression level in the control samples across the time series. The data were also expressed as the “rhythm amplitude,” calculated by setting the value at the time of lowest expression for that genotype equal to 1.0 and expressing other points within the same time series as the fold increase over this minimum.
Per2::LUC bioluminescence rhythms from liver explants.
PER2::LUC bioluminescence was recorded and analyzed as previously described (10, 40, 41). For studies involving tissue explants, mice were euthanized by carbon dioxide asphyxiation in the late afternoon, and tissues were rapidly dissected and placed in ice-cold 1× Hanks buffered saline. Three 2- to 3-mm tissue explants from the edge of a liver lobe were prepared from each liver. Each explant was cultured individually by placing it on a sterile Millicell culture plate insert (Millipore) within a well of a Hamamatsu LM-2400 luminometer. The culture medium was low bicarbonate DMEM containing glucose, 2% fetal bovine serum, and 0.1 mM luciferin (40). Bioluminescence was recorded by the luminometer for 1 min at 15-min intervals. Explants that gave acceptable bioluminescence levels (above 5,000 cps; >90% of explants prepared) were analyzed individually to determine circadian period as described by Izumo et al. (18). The data were normalized to the average luciferase activity produced over the duration of the recording period and are plotted as the difference from the centered 24-h moving average (10). Period values for all explants from each liver were averaged to give one value for each animal; sample sizes in figures represent the number of animals from which explants were prepared.
Per2::LUC bioluminescence rhythms from MEFs.
Primary mouse embryonic fibroblasts (MEFs) expressing mPER2::LUC were prepared from fetuses collected on gestational days 12 to 14. Each fetus was used to generate a separate line, whose genotype was confirmed by PCR. Serum shock to synchronize cellular oscillators (4) was accomplished by replacing maintenance media (DMEM supplemented with 10% heat-inactivated fetal bovine serum) with DMEM containing 50% horse serum for 2 h, followed by serum-free, low-bicarbonate DMEM containing 0.1 mM luciferin. Except for serum shocks, cultures were maintained in the recording apparatus without a medium change throughout the period of recording. Within each experiment, triplicate cultures were examined for each MEF cell line of a given genotype, and sample sizes reported represent the number of MEF cell lines examined in triplicate. Mutant and control cell lines of equivalent passage number were processed in parallel within each experiment. For analysis, data were normalized to the average luciferase activity produced over the duration of the recording period and are plotted as the difference from the centered 24-h moving average as previously described (10). Period was determined as described by Izumo et al. (18).
Estimation of PER2::LUC bioluminescence half-life.
Liver explants derived from animals expressing PER2::LUC were used to estimate the half-life of mPER2. Tissue explants were cultured 1 to 3 h before lights out. At the time of anticipated peak bioluminescence the following day, approximately 24 h after explant preparation, cycloheximide (80 μM) was added, and the decline in bioluminescence was recorded in the LM-2400 for 8 h. The half-life was calculated from normalized bioluminescence levels: the bioluminescence at the time of cycloheximide addition was set to 100%, the amount remaining at 8 h was set to 0%, and then the half-life was estimated by using GraphPad Prism, as described by Meng et al. (24).
RESULTS
Casein kinase gene disruption strategy.
To analyze the effects of disruption of CK1δ and CK1ɛ in vivo and to allow tissue-specific disruption of these genes, we generated lines of mice in which exons essential for kinase activity could be deleted using the Cre-loxP recombination system (see Fig. 1 and Materials and Methods). Targeted mutagenesis was used to introduce loxP sites into introns 1 and 3 of the CK1δ (Csnk1d) gene, allowing Cre-mediated excision of exon 2 (Fig. 1A to E). Similarly, loxP sites were introduced into introns 1 and 4 of the CK1ɛ (Csnk1e) gene, allowing Cre-mediated excision of exons 2 and 3 of CK1ɛ (Fig. 1F to J). In both genes, we included exon 2 in the region flanked by loxP sites (“floxed”), since exon 2 encodes a portion of the ATP-binding domain essential for kinase activity. Mutations within exon 2 (K38R or K38A) lead to inactive kinase proteins (2, 11, 14). Thus, we expected that Cre-mediated recombination of the floxed alleles would disrupt kinase activity. Reverse transcription-PCR analysis verified that the insertion of loxP sites did not alter the transcripts arising from the floxed alleles.
Targeted disruption of CK1δ.
Breeding of mice bearing the floxed CK1δ allele to mice expressing Cre recombinase in the testis (Protamine-Cre) led to germ line disruption of CK1δ. Intercrosses involving mice heterozygous for the excised allele (CK1δΔ2/+) failed to produce mice at weaning that were homozygous for the excised allele. The genotype distribution of pups at weaning (49 CK1δ+/+:62 CK1δ+/Δ2:0 CK1δΔ2/Δ2) was not consistent with the expected 1:2:1 Mendelian ratio but is consistent with a previous report (38) indicating that mice homozygous for disruption of CK1δ die during the perinatal period. Through timed pairings of heterozygous mice, we were able to recover homozygous mutant (CK1δΔ2/Δ2) mice late in gestation, albeit at a lower rate than expected (12.5% of 64 fetuses at gestational days 18 to 19). The homozygous mutant mice weighed ∼30% less than heterozygous and wild-type littermates (P < 0.05, Student t test) but were fully formed and were alive at the time of collection. The perinatal lethality of the homozygous mutant mice limited further analysis to adult mice heterozygous for the mutation and to primary MEFs. Therefore, we made use of a tissue-specific Cre recombinase “driver” line to circumvent the lethal phenotype of whole-body knockouts. In view of the considerable background regarding biochemical and molecular analysis of circadian rhythms in liver tissue, we focused our efforts on using a liver-specific Cre recombinase line, in which Cre recombinase is regulated by the albumin gene promoter specifically expressed in hepatocytes (26).
Excision of exon 2 from CK1δ leads to in-frame splicing of exon 1 to exon 3. The mutant CK1δΔ2 protein product lacks residues 26 to 62 and is detectable at lower levels, as demonstrated by Western blot analysis of whole-cell extracts from liver tissue (Fig. 1K; see below). The absence of exon 2 interferes with nuclear accumulation of CK1δΔ2 (Fig. 1K). Nevertheless, expression of the mutant CK1δΔ2 protein raises the possibility that the product from the excised allele retains functional activity. This possibility has been extensively investigated, and excluded, as discussed below.
CK1δ lacking residues encoded by exon 2 (CK1δΔ2) is a nonfunctional protein.
The presence of a protein product from the CK1δΔ2 allele raises the possibility that this protein retains functional activity. Alternatively, the CK1δΔ2 product could interfere with the function of other casein kinases, behaving as a dominant negative. To investigate these possibilities, we used a cotransfection approach to assess the activity of CK1δΔ2 using mouse PER proteins mPER1 and mPER2 as substrates. Specifically, we assessed the ability of CK1δΔ2 to coprecipitate with mPER1 and mPER2, to promote the phosphorylation of the mPER proteins (as determined by reduced electrophoretic mobility and 32P incorporation), and to promote their proteasome-mediated degradation.
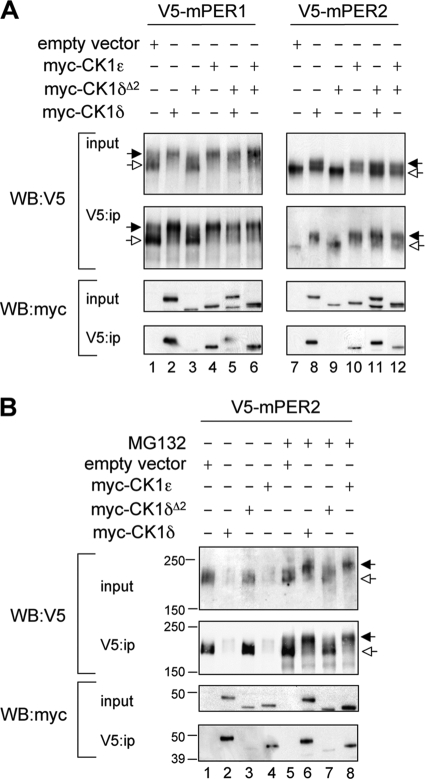
Cotransfection experiments in HEK293 cells revealed that mPER1 and mPER2 coimmunoprecipitated with CK1δ and CK1ɛ but not with CK1δΔ2 (Fig. 2). In addition, CK1δΔ2 did not interfere with the ability of CK1δ or CK1ɛ to bind to mPER substrates (Fig. 2A, lanes 5, 6, 11, and 12). Figure 2 also shows the impact of casein kinases to reduce the electrophoretic mobility of mPER substrates, which is characteristic of hyperphosphorylated PER proteins (21). The mobility of both mPER1 and mPER2 was reduced by coexpression with CK1δ or CK1ɛ, but not with CK1δΔ2. Furthermore, CK1δΔ2 did not interfere with the activity of either wild-type kinase.
FIG. 2.
CK1δΔ2 is a functional null. (A) Assessment of the functional activity of CK1δΔ2 coexpressed with mPER proteins. Myc-tagged casein kinases (CK1δ, CK1ɛ, and CK1δΔ2) and V5-tagged mPER proteins were coexpressed in HEK293 cells (input), and protein complexes were immunoprecipitated with an antibody to V5 (V5:ip). Kinase activity is reflected by an upward mobility shift of mPER proteins (indicated by the filled arrow; seen in the V5 Western blots in lanes 2, 4, 5, 6, 8, and 10, but not in lanes 1, 3, 7, and 9). Hypophosphorylated mPER proteins are indicated by the open arrows. The lower panel shows that myc-tagged CK1δ (lanes 2 and 8) and CK1ɛ (lanes 4 and 10) coprecipitate with the PER proteins, while CK1δΔ2 does not (lanes 3 and 9). Notably, expression of CK1δΔ2 does not interfere with the mobility shift or coprecipitation activities of CK1δ and CK1ɛ (lanes 5, 6, 11, and 12). The results shown are representative of four independent experiments. (B) CK1-mediated proteasomal degradation of mPER2. Cellular coexpression studies similar to those described above were conducted in HEK293 cells, except that a higher concentration of kinase plasmid was used to further promote PER protein degradation. Studies were conducted in the absence (left) or presence (right) of MG132 (10 μM). The black and white arrows indicate hyper- and hypophosphorylated mPER2, respectively. CK1δ and CK1ɛ promote degradation of mPER2 (lanes 2 and 4), while CK1δΔ2 does not (lane 3). In the presence of MG132, hyperphosphorylated mPER2 accumulates rather than being degraded. CK1δ and CK1ɛ promote accumulation of hyperphosphorylated mPER2 (lanes 5 and 7), while CK1δΔ2 does not (lane 6). The results shown are representative of three independent experiments.
Phosphorylation of mPER1 and mPER2 by CK1ɛ or CK1δ is a key step promoting their degradation by the proteasome (2, 7, 12, 27, 29). Therefore, we examined PER degradation as another assay of functional kinase activity. HEK293 cells were cotransfected with mPER2 and with a larger amount of full-length CK1δ, CK1ɛ, or CK1δΔ2, sufficient to promote PER degradation (Fig. 2B). Both CK1δ and CK1ɛ promoted hyperphosphorylation and degradation of mPER2 (Fig. 2B, lanes 2 and 4), while CK1δΔ2 did not (Fig. 2B, lane 3). In the presence of the proteasome inhibitor, MG132, a lower-mobility form of mPER2 corresponding to its hyperphosphorylated state accumulated in cells cotransfected with CK1δ or CK1ɛ but not in those cotransfected with CK1δΔ2. In cells expressing CK1δΔ2, the hypophosphorylated form of mPER2 remained at levels comparable to the empty vector control. Similar results were found with mPER1 (data not shown). These data confirm that CK1δΔ2 is not able to phosphorylate mPER1 or mPER2 and further reveals that CK1δΔ2 does not promote proteasomal degradation of mPER proteins.
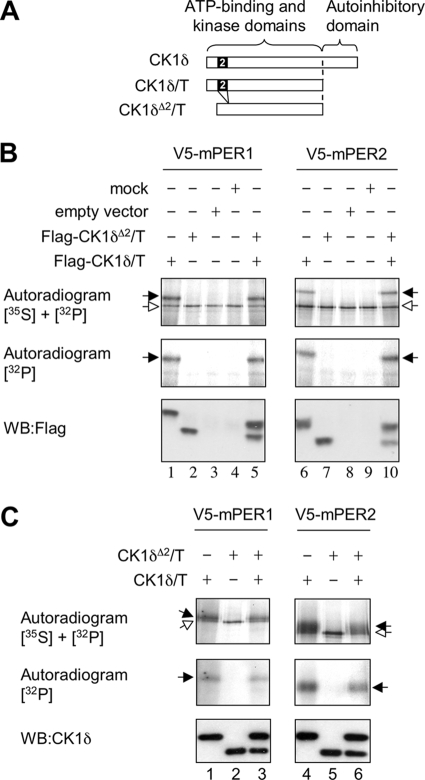
We also analyzed the kinase activity of CK1δΔ2 in vitro (Fig. 3). Constructs encoding CK1δ and CK1δΔ2 were truncated at residue 320 to remove an inhibitory autophosphorylation domain (17) (Fig. 3A). Constructs were transfected into HEK293 cells, and the kinases were immunopurified. V5-tagged mPER1 and mPER2 were generated in vitro and used in in vitro kinase assays in the presence of [γ-32P]ATP. mPER1 and mPER2 incorporated 32P when incubated with immunopurified CK1δ but not when incubated with CK1δΔ2 (Fig. 3B). Furthermore, phosphorylation of mPER1 and mPER2 by CK1δ was not obstructed by CK1δΔ2. The inability of truncated CK1δΔ2 to phosphorylate mPER substrates or to interfere with the activity of CK1δ was further confirmed by using bacterially purified, truncated kinase proteins (Fig. 3C).
FIG. 3.
Purified CK1δΔ2 lacks functional activity in vitro. (A) Schematic illustration of constructs in which the autoinhibitory C-terminal domain was removed by truncation of CK1δ at residue 320 (of 415). The CK1δΔ2 construct was truncated at the corresponding residue. (B) In vitro kinase assays reveal that truncated CK1δ phosphorylates PER proteins, whereas CK1δΔ2 does not. The phosphorylation of mPER1 and mPER2 was assessed by in vitro kinase reactions performed in the presence of [γ-32P]ATP. Flag-tagged, truncated CK1δ (CK1δ/Τ), or CK1δΔ2 (CK1δΔ2/T) were expressed in HEK293 cells and purified by using anti-Flag affinity beads. A Flag-tagged empty expression vector was used as a negative control. V5-tagged mPER1 and V5-tagged mPER2 were produced by in vitro translation in the presence of [35S]methionine and purified with anti-V5 antibody. “Mock” samples are TNT-produced PER proteins not subjected to the kinase reaction. The upper panels show an autoradiogram to detect both 35S and 32P incorporated into the V5-tagged proteins. The center panel shows a second autoradiogram revealing 32P incorporated into V5-tagged mPER proteins. (The 35S signal was quenched by placing a second film between the blot and this film.) The lower panel shows a Western blot verifying the expression of Flag-tagged kinase proteins in each sample. Phosphorylated (black arrow) and nonphosphorylated mPER proteins (white arrow) are indicated in the top panel. The truncated CK1δ produces an upward mobility shift of mPER proteins (lanes 1 and 6) and incorporation of 32P. CK1δΔ2 lacks these activities (lanes 2 and 7). These activities of CK1δ were not disrupted by coexpression of CK1δΔ2 (lanes 5 and 10). The results shown are representative of three experiments. (C) His/S-tagged, truncated CK1δ and CK1δΔ2 were expressed in bacteria and purified by immunoprecipitation. V5-tagged mPER1 and V5-tagged mPER2 were produced in rabbit reticulocyte lysates in the presence of 35S and purified with anti-V5 antibody. In vitro kinase reactions were performed in the presence of [γ-32P]ATP. The upper panels show an autoradiogram revealing both 35S and 32P incorporated into the V5-tagged mPER proteins. The center panels show autoradiograms of 32P incorporated into V5-tagged mPER proteins, with the 35S signal quenched by another film. Phosphorylated (black arrow) and nonphosphorylated (white arrow) mPER proteins are indicated in the top panel. The lower panel shows a Western blot with an antibody to CK1δ, verifying the expression of kinase proteins in each sample. The truncated CK1δ produces an upward mobility shift of mPER proteins (lanes 1,4) and the incorporation of 32P. CK1δΔ2 lacks these activities (lanes 2 and 5). These activities of CK1δ were not disrupted by coexpression of CK1δΔ2 (lanes 3 and 6). (Note that the PER input amounts were lower in lanes 3 and 6 than in lanes 1 and 4, as revealed by the lower intensity of the hypophosphorylated band of the upper autoradiogram. Notably, the ratio of signal in the two autoradiograms for each lane was roughly equal.) The results shown are representative of three experiments.
These experiments show that the CK1δΔ2 protein is incapable of binding to either mPER1 or mPER2, and it does not promote their phosphorylation or proteasomal degradation. The CK1δΔ2 allele thus is a null allele regarding its role in the molecular clock. These results validate the CK1δΔ2 model for studying the impact of the absence of CK1δ on circadian function.
Targeted disruption of CK1ɛ.
Breeding of mice with the floxed allele of CK1ɛ to mice expressing Protamine-Cre led to deletion of genomic DNA containing exons 2 and 3. RT-PCR analysis shows that transcripts from the excised CK1ɛ allele splice exon 1 directly to exon 4, causing a frameshift mutation that introduces a premature stop codon. A protein product from the excised allele, if produced, would contain only 25 residues derived from exon 1, plus 36 residues due to out-of frame translation of exon 4, before reaching a premature stop codon. Exon 1 does not encode any known functional domain. Thus, we refer to mice homozygous for the excised allele throughout the body, generated using the Protamine-Cre line to induce germ line excision, as CK1ɛ−/− or CK1ɛ-deficient mice. Consistent with a recent report (24), CK1ɛ-deficient mice are viable and fertile. As expected, CK1ɛ-deficient mice lack CK1ɛ protein (Fig. 1L). Studies were also conducted using mice in which CK1ɛ was disrupted specifically in the liver, using the Albumin-Cre driver to excise floxed exons of CK1ɛ.
Subtle alterations in locomotor activity rhythms in CK1δΔ2/+ heterozygotes and CK1ɛ−/− mice.
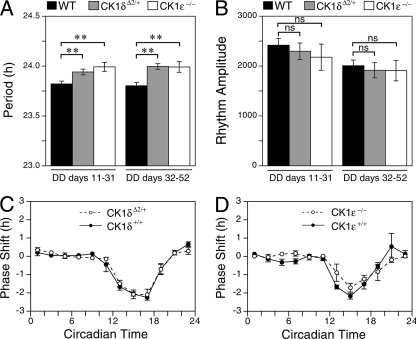
To assess whether mice with only one wild-type allele of CK1δ have alterations in circadian period, heterozygous (CK1δΔ2/+) mice and wild-type littermate controls were placed into running wheels, and locomotor activity rhythms were monitored in a light/dark cycle and then in constant darkness. CK1ɛ-deficient mice were similarly compared to wild-type controls. The period of locomotor activity rhythms and the amplitude of rhythmicity were calculated for two successive 21-day intervals, starting 10 days after entry into constant darkness. There were no differences between the wild-type control groups, so they were combined. The free-running period of CK1δΔ2/+ mice was significantly longer than controls in both epochs (Fig. 4A). The free-running period of CK1ɛ−/− mice was also significantly longer than controls (Fig. 4A). No alterations in the amplitude of rhythmicity were noted (Fig. 4B). Analysis of gene expression rhythms in the SCN revealed no differences between CK1ɛ−/− and control mice (see Fig. S2 in the supplemental material).
FIG. 4.
Locomotor activity rhythms in casein kinase mutant mice. (A) Mean period length. (B) Rhythm amplitude. Rhythm parameters were determined for two consecutive 21-day epochs in constant darkness (DD) for wild-type control mice (WT; n = 27), CK1δΔ2/+ mice, (n = 21), and CK1e−/− mice (n = 9). Asterisks indicate a significant effect of genotype within each period of study (P < 0.01, Dunnett's test). The rhythm amplitude did not differ between the genotypes (P > 0.05). (C and D) Phase-shifting responses to 1-h light pulses were similar among CK1δΔ2/+ mice, CK1e−/− mice, and wild-type control mice. Values are the mean ± the standard error of the mean (SEM) of 2 to 12 (average of 6) pulses per bin per genotype.
After more than 3 months in constant darkness, a subset of mice of each genotype were exposed to 1-h light pulses at intervals of 10 to 14 days. CK1δΔ2/+ mice, CK1ɛ-deficient mice, and wild-type mice had similar responses, with light causing phase delays during the first half of the active period (subjective night) and relatively little response at other circadian times (Fig. 4C and D).
mPER and mCRY protein rhythms are altered in CK1δ-deficient liver tissue.
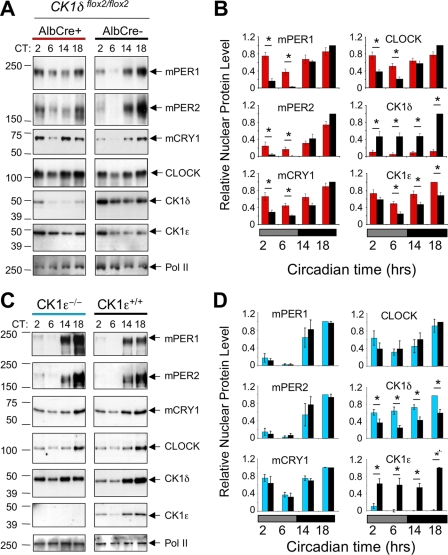
The inability of the mutant CK1δΔ2 protein to promote PER phosphorylation and proteasomal degradation suggests that mice expressing only the CK1δΔ2 variant may have altered biochemical and molecular rhythms. To circumvent the lethal phenotype of CK1δΔ2/Δ2 mice, we generated mice in which CK1δ was disrupted specifically in the liver. CK1δflox2/flox2 mice were generated which also expressed Cre recombinase from a liver-specific Albumin-Cre (AlbCre) transgene, leading to liver-specific disruption of CK1δ. CK1δflox2/flox2 littermates lacking the AlbCre transgene were used as controls. Liver tissue was collected at various times on the first day in constant darkness, nuclear fractions were isolated, and circadian proteins were analyzed by Western blotting. Figure 5A shows that peak levels of nuclear proteins were achieved during the subjective night (CT14 and CT18) in mice of both genotypes. During subjective day (CT2 to CT6), however, nuclear mPER1 and mPER2 levels were significantly higher in mice lacking CK1δ in liver, compared to littermate controls (Fig. 5A and B). This resulted in a decrease in the amplitude of PER protein oscillations. The reduced clearance of nuclear mPER proteins in CK1δ-deficient livers may be due to the inability of CK1δΔ2 to promote degradation of mPER2 as shown in Fig. 2B. Similarly, mCRY1 protein levels remained higher during subjective day in mice lacking hepatic CK1δ, while CLOCK levels were unaffected. Nuclear accumulation of mCRY proteins is tightly linked to mPER availability (21); with reduced clearance of mPER proteins, mCRY proteins are likely engaged in PER-containing protein complexes that protect them from degradation (15; see also below). Similarly, elevation of CK1ɛ in liver nuclei lacking CK1δ may reflect “compensatory” upregulation or could simply represent improved protection of one kinase in the absence of the other, because there is no competition for a limiting, stabilizing pool of mPER proteins.
FIG. 5.
Hepatic protein rhythms in CK1-deficient liver. (A) Altered protein rhythms in CK1δ-deficient liver nuclei. Liver nuclei prepared at four time points on the first day in constant darkness were probed to detect circadian proteins by Western blotting. Liver tissue was collected at the indicated circadian times (CT) from CK1δ-deficient livers (AlbCre+; CK1δflox2/flox2) and control livers (AlbCre negative; CK1δflox2/flox2). Rhythms of circadian proteins were present in CK1δ-deficient livers, but the rhythms were blunted in amplitude due to the presence of higher levels of mPER1, mPER2, and CRY1 proteins during the subjective day (CT2 and CT6). The results shown are representative of at least four experiments. (B) Quantitative analysis of hepatic protein rhythms from CK1δ-deficient livers. Relative nuclear protein levels were calculated by densitometric analysis of Western blots; data were first expressed relative to Pol II, and the maximum protein/Pol II ratio in each experiment was set to 1.0. Other values are expressed relative to this maximum. Each panel includes values from four (mCRY1) or five independent experiments generated from different tissue samples. Asterisks indicate significant difference between the genotypes at P < 0.05. (C) Unaltered protein rhythms in liver nuclei from CK1ɛ-deficient mice. Liver nuclei prepared at four circadian times on the first day in constant darkness were probed to detect circadian proteins. Rhythms of circadian proteins were present in CK1ɛ-deficient livers, and these rhythms did not differ from the rhythms in wild-type mice. The results shown are representative of three experiments, each involving different tissue samples. (D) Quantitative analysis of hepatic protein rhythms in CK1ɛ-deficient mice. Nuclear proteins from mice with whole-body disruption of CK1ɛ (CK1ɛ−/−) and wild-type controls (CK1ɛ+/+) were assessed. Three Western blots for each protein (as shown in Fig. 3C) were analyzed as described in panel B, above.
Protein expression rhythms are unaltered in CK1ɛ-deficient livers.
In parallel to the analysis of circadian protein profiles in CK1δ-deficient livers, we analyzed protein rhythms in liver tissue derived from CK1ɛ-deficient mice and wild-type controls (Fig. 5C). In contrast to the effects of liver-specific disruption of CK1δ, described above, the nuclear levels and rhythms of mPER1, mPER2, mCRY1, and CLOCK in the liver were not affected by the absence of CK1ɛ (Fig. 5C and D). The only significant difference between genotypes was a significant increase in the level of CK1δ protein in liver nuclei lacking CK1ɛ. Similar results were found when CK1ɛ was disrupted specifically in liver (see Fig. S3 in the supplemental material). These data clearly show that the circadian clock is more dependent upon CK1δ than CK1ɛ with respect to regulating clock protein rhythms in liver.
Reduced amplitude of nuclear repressor complex rhythms in CK1δ-deficient liver.
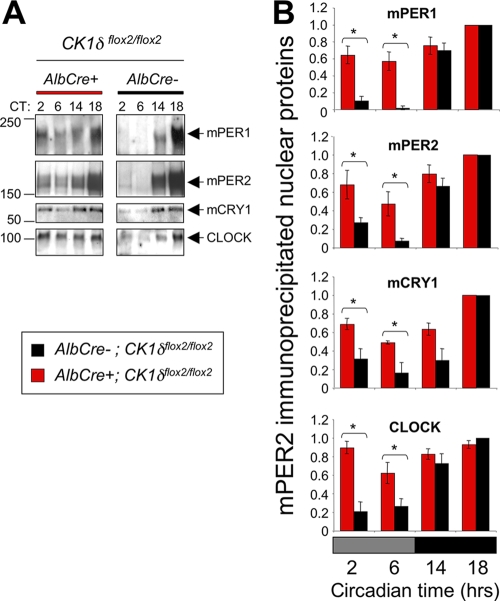
The elevated PER and CRY levels present in CK1δ-deficient liver nuclei during subjective day are likely present in circadian repressor complexes. To directly assess this, protein complexes were isolated from liver nuclear extracts using an antibody to mPER2, as previously described (21). At night, coprecipitating protein levels were comparable between mice with liver-specific disruption of CK1δ and littermates lacking the AlbCre transgene (Fig. 6). During the subjective day, however, the levels of mPER1, mCRY1, and CLOCK that coimmunoprecipitated with mPER2 were elevated in mice lacking CK1δ in the liver, relative to controls (Fig. 6). This increase in repressor complex levels would be expected to reduce CLOCK:BMAL1-mediated transcription during the subjective day.
FIG. 6.
Nuclear repressor complex levels are increased during subjective day in CK1δ-deficient liver tissues. (A) Nuclear protein extracts were purified from CK1δ-deficient (AlbCre+; CK1δflox2/flox2) and control (AlbCre−; CK1δflox2/flox2) livers collected at the indicated circadian times, and protein complexes were isolated by immunoprecipitation of mPER2. Then, mPER1, mPER2, mCRY1, and CLOCK were detected by Western blotting. Representative of three independent immunoprecipitations. (B) Quantification of three replicate immunoprecipitation experiments. Asterisks indicate significant differences between the genotypes at P < 0.05.
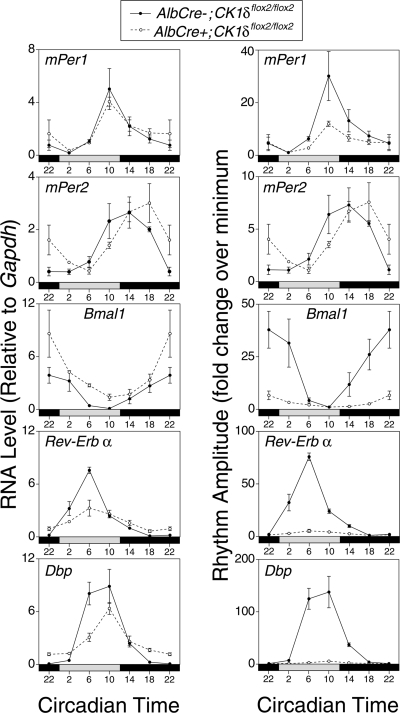
CK1δ-deficient livers have gene-specific alterations in transcript rhythms.
To determine whether the alterations in PER/CRY nuclear protein rhythms and in the abundance of nuclear repressor complexes had consequences on circadian gene expression, we investigated selected genes in CK1δ-deficient and control livers using real-time PCR (Fig. 7). Expression levels and relative rhythm amplitudes for mPer1 and mPer2 mRNAs were relatively unaffected by the absence of CK1δ. There were significant alterations in Bmal1, Rev-Erbα, and Dbp mRNA profiles, however. For both Rev-Erbα and Dbp, there was a reduction in transcript levels at the time of peak levels and an elevation in the levels at the time of nadir (Fig. 7, left), leading to a drastic reduction in the amplitude of oscillation (Fig. 7, right). The greatly reduced amplitude of Dbp and Rev-Erbα transcript rhythms likely reflects the impact of reduced rhythmicity of the PER/CRY repressor complex. The elevated level of expression of Bmal1 in mutant tissue is expected due to derepression, caused by reduced expression of the repressor, Rev-Erbα (20).
FIG. 7.
Transcript rhythms in CK1δ-deficient liver. Liver tissues were collected at six times on the first day in constant darkness, and RNA levels were assessed by real-time PCR. Panels on the left show RNA levels expressed relative to GAPDH. Panels on the right show the data from the same samples, plotted as the RNA amplitude (where each value is expressed relative to the nadir value during the circadian cycle for each genotype). Expression of the data as rhythm amplitude reveals a dramatic impact on the amplitude of the output genes Rev-Erbα and Dbp. Elevation of Bmal1 is expected due to the reduced levels of Rev-Erbα. Each value represents the mean (± the SEM) of three tissue samples assessed in duplicate, except the AlbCre+ group at CT2, where n = 2. An independent tissue collection and real-time PCR analysis of mPer1, mPer2, Rev-Erbα, and Dbp gave similar results.
The gene-specific transcriptional response in CK1δ-deficient livers is consistent with results from other mutant mice which indicate that behavioral rhythmicity emanating from the SCN can drive hepatic Per transcript and protein rhythms, even when the liver is devoid of autonomous circadian clock function (9, 10, 20). The rhythmicity of output genes appears more strongly dependent on E-box-mediated transcription and thus is more vulnerable to disruption. Thus, in vivo assessment of hepatic mPer rhythmicity in mutant mice likely underestimates the impact of a molecular lesion. Studies described below support the conclusion that the impact of tissue-specific CK1δ gene disruption is underestimated due to systemic-driven oscillation.
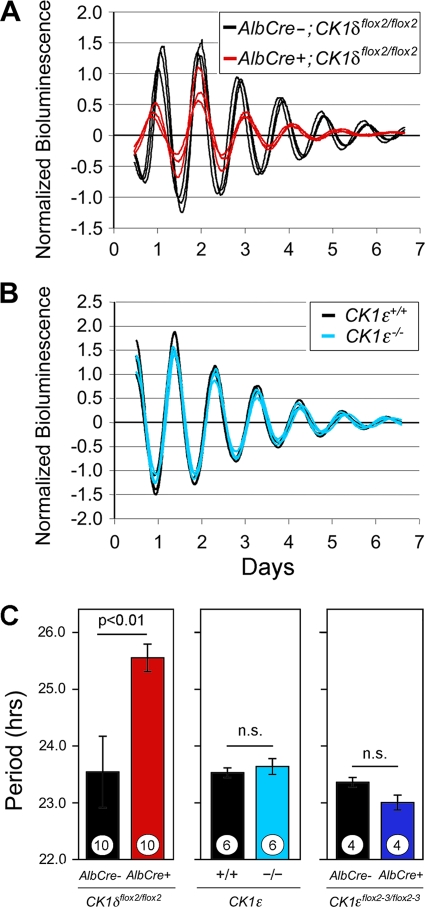
Ex vivo analysis reveals that CK1δ deficiency significantly lengthens the circadian period, while CK1ɛ deficiency does not.
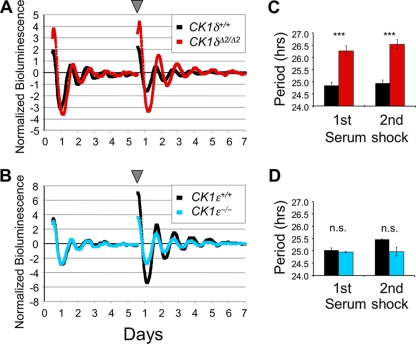
As noted above, in vivo assessment of molecular rhythms in the liver can be complicated by the hierarchical nature of the circadian timing system. To more clearly determine the consequences of CK1δ/ɛ gene disruption on the hepatic molecular oscillator, we bred our mutant mice with mice bearing a modified mPer2 gene in which luciferase was fused in-frame with mPER2 (37, 41). Liver explants were prepared from adult mice lacking CK1δ in liver (AlbCre+; CK1δflox2/flox2; Per2luc/+), and controls lacking AlbCre. mPER2::LUC bioluminescence was monitored by using photomultiplier tubes as previously described (10). The circadian period of mPER2::LUC bioluminescence rhythms in CK1δ-deficient liver tissue was ∼2 h longer than in controls (Fig. 8A and C). In contrast, there was no significant difference in the period length of liver tissues derived from CK1ɛ-deficient mice compared to the controls (Fig. 8B and C). Similarly, the period of mPER2::LUC bioluminescence rhythms in liver explants was not altered in mice with liver-specific disruption of CK1ɛ (Fig. 8C).
FIG. 8.
mPER2::LUC bioluminescence rhythms in liver explants from kinase-mutant mice. (A) Bioluminescence recordings from CK1δ-deficient liver explants (AlbCre+; CK1δflox2/flox2; Per2luc/+) and from control mice (AlbCre−; CK1δflox2/flox2; Per2luc/+). (B) Bioluminescence recordings from liver explants of CK1ɛ-deficient mice (CK1ɛ−/−; Per2luc/+) and control mice (CK1ɛ+/+; Per2luc/+). (A and B) Each panel shows three representative profiles from a single experiment. Day 0 is the day of explant preparation. The data were normalized to the average bioluminescence level over the duration of the experiment and are plotted as the difference from the centered 24-h moving average. The reduced amplitude and duration of rhythmicity in CK1δ-deficient liver explants seen in this example was not seen consistently in other experiments. (C) Mean ± SEM values for circadian cycle length (period) of the mPER2::LUC bioluminescence rhythms recorded from kinase-deficient liver explants. Values plotted within each panel represent experiments conducted simultaneously and thus are directly comparable. Significance levels are indicated within each panel. Sample sizes within each bar represent the number of animals from which explants were studied.
The ability to breed CK1δΔ2/+ heterozygotes and recover CK1δΔ2/Δ2 embryos allowed us to obtain primary MEFs lacking CK1δ while expressing PER2::LUC. Figure 9A shows the bioluminescence profiles of wild-type and homozygous mutant (CK1δΔ2/Δ2; mPer2luc/+) MEFs after synchronization by “serum shock” with 50% horse serum. The period length calculated from the bioluminescence profile was significantly longer in CK1δΔ2/Δ2 MEFs than in control MEFs (Fig. 9C). In contrast, there was no difference in the period of the bioluminescence rhythm between MEFs derived from CK1ɛ-deficient mice and wild-type controls (Fig. 9B and D). These studies complement and confirm the results from liver tissue in which the kinase genes were disrupted. Both experimental models show that CK1δ deficiency significantly lengthens circadian period in vitro, while CK1ɛ deficiency has no detectable effect on the period of bioluminescence rhythms (Fig. 8 and 9). These findings emphasize the key role played by CK1δ in regulating circadian period.
FIG. 9.
mPER2::LUC bioluminescence rhythms in kinase-deficient primary MEFs. (A and B) Representative mPER2::LUC bioluminescence rhythms recorded from MEF cultures of different genotypes. On days 0 and 5 (arrowhead), a serum shock was administered to synchronize the cells. Bioluminescence recordings from CK1δ-deficient (CK1δΔ2/Δ2) and control (CK1δ+/+) MEFs (A) and CK1ɛ-deficient mice (CK1ɛ−/−) and control (wild-type, CK1ɛ+/+) MEFs (B). (C and D) Mean circadian period of mPER2::LUC bioluminescence rhythms recorded from kinase-deficient MEFs. (C) The period of CK1δ-deficient MEFs was significantly longer than in controls (***, P < 0.001 [Student t test]). (D) The period of CK1ɛ-deficient MEFs did not differ from controls (n.s., P > 0.05). Values are the means ± the SEM of three independent experiments in each panel.
Per2::luciferase stability is increased in the absence of CK1δ but is unaffected by disruption of CK1ɛ.
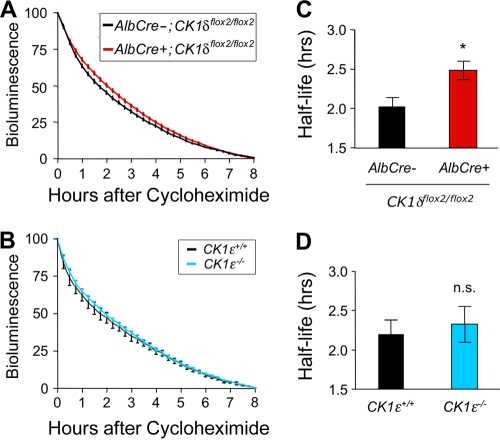
Despite the high degree of sequence similarity between CK1δ and CK1ɛ and their similar activities in most assays, the results presented above indicate that the disruption of these genes has markedly different effects on circadian clock function. The period of the circadian oscillation is lengthened significantly in liver and fibroblasts after disruption of CK1δ but not after disruption of CK1ɛ. Furthermore, nuclear mPER1, mPER2, and mCRY1 protein rhythms are blunted in the CK1δ-deficient liver, a finding suggestive of reduced clearance of the proteins, whereas these rhythms are normal in CK1ɛ-deficient livers. These results suggest that CK1δ plays a unique role in regulating PER protein turnover. To test this hypothesis, we adopted a strategy for assessing mPER2 stability described by Meng et al. (24). Cycloheximide was used to block new protein synthesis in liver explants, and the decline in mPER2::LUC bioluminescence was used to estimate mPER2 protein degradation. The half-life of mPER2::LUC bioluminescence was significantly longer in CK1δ-deficient liver explants, relative to controls (Fig. 10A and B). In contrast, the half-life of mPER2::LUC bioluminescence in liver explants from CK1ɛ-deficient mice did not differ from control (Fig. 10C and D). The longer half-life of mPER2 in tissue lacking CK1δ is likely due to the inability of CK1δΔ2 to promote the proteasome-dependent degradation of mPER proteins (Fig. 2B). Nevertheless, mPER2::LUC is still degraded in tissues lacking either CK1δ or CK1ɛ, indicating functional redundancy between CK1δ and CK1ɛ, or the presence of CK1δ/ɛ-independent pathways for degradation of mPER2.
FIG. 10.
mPER2::LUC bioluminescence half-life is increased by disruption of CK1δ but is unaffected by the absence of CK1ɛ. (A and B) Liver explants were treated with cycloheximide (80 μg/ml) at the time of peak bioluminescence on the first day in vitro, and the decline in bioluminescence was monitored for 8 h. The data were normalized to the peak level of mPER2::LUC bioluminescence and the minimum level after 8 h of recording. Values are plotted as means ± the SEM. (A) Average bioluminescence profiles from explants from CK1δ-deficient liver (AlbCre+; CK1δflox2/flox2; mPer2Luc/+) (n = 12) and control liver (AlbCre−; CK1δflox2/flox2; mPer2Luc/+) (n = 7) tissues. (B) Average bioluminescence profiles from liver explants from CK1ɛ-deficient livers and wild-type (CK1ɛ+/+; mPer2Luc/+) controls (n = 8 per genotype). (C) The calculated bioluminescence half-life was significantly (*, P < 0.025) increased in CK1δ-deficient livers, relative to controls lacking Cre recombinase. (D) mPER2::LUC bioluminescence half-life was unaltered in CK1ɛ-deficient livers.
DISCUSSION
CK1δ and CK1ɛ are generally viewed as having redundant function with respect to regulation of circadian protein stability and circadian cycle length. Although these two kinases do have overlapping functions, our data reveal an unexpectedly important role for CK1δ in regulation of circadian period length. In the absence of functional CK1δ, the dynamics of circadian protein rhythms were altered in vivo, and the output gene rhythm amplitude was dramatically reduced. mPER2 bioluminescence half-life was increased in vitro. These alterations likely cause the increase in period length observed in CK1δ-deficient fibroblasts and CK1δ-deficient liver explants in vitro. In contrast, the absence of CK1ɛ did not have marked effects on molecular oscillations in vitro or in vivo, a finding in agreement with recently published results (24). One potential explanation for this difference is that, despite their high degree of sequence identity, CK1δ and CK1ɛ may phosphorylate overlapping but distinct populations of residues on PER protein substrates.
Analysis of locomotor activity rhythms indicates that the absence of CK1ɛ has a small but significant effect, increasing circadian period, a finding consistent with results reported by Meng et al. (24). Our results show that the presence of only one functional allele of CK1δ also produced a small but significant increase in the circadian period. This is in contrast to the results of Xu et al. (38, 39), which showed no period alteration in mice heterozygous for an apparent null allele of CK1δ. These authors did report a significant effect of heterozygosity at CK1δ in the presence of mutant alleles of mPER2, however (39). The phase-response curve to light is unaltered in CK1ɛ-deficient mice, a finding that does not support the hypothesis that CK1ɛ regulates phase-shifting responses to light (1).
A more prominent circadian phenotype might be expected in a system lacking both CK1δ and CK1ɛ. Attempts to generate liver tissue or MEFs lacking both CK1δ and CK1ɛ have been unsuccessful, however. We have also been unsuccessful in obtaining simultaneous knockdown of both CK1δ and CK1ɛ in cultured cells transfected with small interfering RNAs, even though the small interfering RNAs used were effective individually (data not shown). The involvement of CK1δ/ɛ in Wnt signaling and cell cycle regulation may preclude simultaneous disruption of both genes in liver and cell lines (19). Pharmacological approaches resulting in transient disruption of kinase activity may be more fruitful for simultaneously disrupting CK1δ and CK1ɛ activity than standard (noninducible) genetic methods. Indeed, treatment with PF-670462 (a CK1δ/ɛ inhibitor with 1.8-fold selectivity for CK1ɛ over CK1δ) caused large delay shifts of rat locomotor activity rhythms (3), and cell culture studies indicate that inhibition of CK1δ/ɛ activity slows the circadian oscillator (12). The relative lack of effect of CK1ɛ disruption on circadian period suggests that the primary molecular target of these inhibitors within the circadian system is CK1δ.
Recent studies in flies suggest that vertebrate CK1δ is better able to interact with fly clock proteins than is vertebrate CK1ɛ and that CK1δ interacts with these fly components in a manner more functionally equivalent to Drosophila DOUBLETIME (13, 28). Thus, CK1δ may possess more conserved function within the circadian clockwork than CK1ɛ (13).
Our studies have emphasized the PER proteins as substrates for CK1δ and CK1ɛ. mCRY1, mCRY2, and BMAL1 can also be phosphorylated by CK1ɛ (11), and these proteins could represent additional sites of differential actions of CK1δ and CK1ɛ. BMAL1, like PER1 and PER2, appears to be a direct substrate for CK1ɛ-mediated phosphorylation, while mCRY1, mCRY2, and mPER3 require the presence of mPER1 or mPER2 as scaffolding proteins (11, 22). Other proteins within the circadian repressor complex may be phosphorylated by CK1 proteins in a complex-dependent fashion.
There is a strong link between the stability of PER proteins and circadian period (5). PER proteins have the most dynamic rhythmicity and appear to be rate-limiting for the formation and nuclear accumulation of repressor protein complexes (21). Overexpression of Per1 increases period length (25). Mutations of CK1ɛ in hamsters and mice and of CK1δ in humans alter PER dynamics and affect the circadian period (23, 24, 38). These mutations are characterized by a general loss of kinase activity and a shortened circadian period. The tau mutant CK1ɛ is now recognized as a gain-of-function mutation, with the mutant protein hyperactive with respect to specific residues of PER, reducing PER half-life and circadian cycle length (14, 24). The CK1δT44A human FASPS mutation may similarly be a gain-of-function mutation (14). This interpretation would be consistent with the dominant inheritance pattern within the affected family, the short-period phenotype of affected individuals, and the lack of a significant period-shortening phenotype in mice with only one functional copy of CK1δ (38; the present study), which would appear to rule out haploinsufficiency as a mechanism for the decreased period in humans with the CK1δT44A allele. Furthermore, transgenic expression of human CK1δT44A in mice results in a gene dosage-related decrease in the circadian period (38). Studies of the kinase activity of the CK1δT44A mutant protein are inconsistent; Xu et al. (38) reported a large (∼80%) reduction in kinase activity toward PER proteins, while Gallego et al. (14) show that the CK1δT44A protein can readily phosphorylate PER proteins coexpressed in NIH 3T3 cells. It is likely that there are important functional differences between global phosphorylation of PER substrates and the specific phosphorylation events that affect PER protein localization, activity, stability, and degradation. Identification of the specific residues within PER proteins that are phosphorylated by wild-type CK1δ and CK1ɛ and by the mutant casein kinase proteins that alter circadian period (CK1δT44A, CK1ɛT178C) will be key to understanding the detailed molecular mechanisms regulating circadian cycle length (8, 34).
Our results indicate that the absence of CK1δ reduces PER protein turnover, and this likely causes the increase in period length observed in CK1δ-deficient fibroblasts and CK1δ-deficient liver explants in vitro. In parallel experiments, the absence of CK1ɛ did not affect protein rhythms, PER stability, or the oscillator period. These results reveal an important functional difference between CK1δ and CK1ɛ. Our results show that CK1δ plays an important and previously underappreciated role in maintaining the accuracy of the mammalian circadian clock.
Supplementary Material
Acknowledgments
We thank Stephen N. Jones and the staff of the Transgenic Animal Modeling facility for assistance in generating the targeted mice and Haley Conde for assessing lethality in CK1δΔ2/Δ2 mice. The UMMS Transgenic Animal Modeling facility is supported in part by Diabetes and Endocrinology Research Center grant DK32520.
This study was supported by grants from the National Institute of Neurological Diseases and Stroke (R01 NS047141 to S.M.R. and R21 NS051458 and R01 NS056125 to D.R.W.). E.N. was supported by a long-term fellowship from the Human Frontier Science Program, R.D. was supported in part by DFG grant 525/2-1, and J.P.D. was supported in part by NIH NRSA F32 GM074277.
The content of this publication is solely the responsibility of the authors and does not necessarily represent the official views of the funding institutes or the National Institutes of Health.
Footnotes
Published ahead of print on 4 May 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Agostino, P. V., S. A. Plano, and D. A. Golombek. 2008. 2008. Circadian and pharmacological regulation of casein kinase I in the hamster suprachiasmatic nucleus. J. Genet. 87467-471. [DOI] [PubMed] [Google Scholar]
- 2.Akashi, M., Y. Tsuchiya, T. Yoshino, and E. Nishida. 2002. Control of intracellular dynamics of mammalian period proteins by casein kinase I ɛ (CKIɛ) and CKIδ in cultured cells. Mol. Cell. Biol. 221693-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Badura, L., T. Swanson, W. Adamowicz, J. Adams, J. Cianfrogna, K. Fisher, J. Holland, R. Kleiman, F. Nelson, L. Reynolds, K. St Germain, E. Schaeffer, B. Tate, and J. Sprouse. 2007. An inhibitor of casein kinase I epsilon induces phase delays in circadian rhythms under free-running and entrained conditions. J. Pharmacol. Exp. Ther. 322730-738. [DOI] [PubMed] [Google Scholar]
- 4.Balsalobre, A., F. Damiola, and U. Schibler. 1998. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93929-937. [DOI] [PubMed] [Google Scholar]
- 5.Blau, J. 2008. PERspectives on PER phosphorylation. Genes Dev. 221737-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Busino, L., F. Bassermann, A. Maiolica, C. Lee, P. M. Nolan, S. I. Godinho, G. F. Draetta, and M. Pagano. 2007. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science 316900-904. [DOI] [PubMed] [Google Scholar]
- 7.Camacho, F., M. Cilio, Y. Guo, D. M. Virshup, K. Patel, O. Khorkova, S. Styren, B. Morse, Z. Yao, and G. A. Keesler. 2001. Human casein kinase Iδ phosphorylation of human circadian clock proteins period 1 and 2. FEBS Lett. 489159-165. [DOI] [PubMed] [Google Scholar]
- 8.Chiu, J. C., J. T. Vanselow, A. Kramer, and I. Edery. 2008. The phospho-occupancy of an atypical SLIMB-binding site on PERIOD that is phosphorylated by DOUBLETIME controls the pace of the clock. Genes Dev. 221758-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeBruyne, J. P., E. Noton, C. M. Lambert, E. S. Maywood, D. R. Weaver, and S. M. Reppert. 2006. A clock shock: mouse CLOCK is not required for circadian oscillator function. Neuron 50465-477. [DOI] [PubMed] [Google Scholar]
- 10.DeBruyne, J. P., D. R. Weaver, and S. M. Reppert. 2007. Peripheral circadian oscillators require CLOCK. Curr. Biol. 17R538-539. [DOI] [PubMed] [Google Scholar]
- 11.Eide, E. J., E. L. Vielhaber, W. A. Hinz, and D. M. Virshup. 2002. The circadian regulatory proteins BMAL1 and cryptochromes are substrates of casein kinase I epsilon. J. Biol. Chem. 27717248-17254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eide, E. J., M. F. Woolf, H. Kang, P. Woolf, W. Hurst, F. Camacho, E. L. Vielhaber, A. Giovanni, and D. M. Virshup. 2005. Control of mammalian circadian rhythm by CKIɛ-regulated proteasome-mediated PER2 degradation. Mol. Cell. Biol. 252795-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan, J. Y., F. Preuss, M. J. Muskus, E. S. Bjes, and J. L. Price. 2009. Drosophila and vertebrate casein kinase Iδ exhibits evolutionary conservation of circadian function. Genetics 181139-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallego, M., E. J. Eide, M. F. Woolf, D. M. Virshup, and D. B. Forger. 2006. An opposite role for tau in circadian rhythms revealed by mathematical modeling. Proc. Natl. Acad. Sci. USA 10310618-10623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gatfield, D., and U. Schibler. 2007. Proteasome keeps the circadian clock ticking. Science 3161135-1136. [DOI] [PubMed] [Google Scholar]
- 16.Godinho, S. I., E. S. Maywood, L. Shaw, V. Tucci, A. R. Barnard, L. Busino, M. Pagano, R. Kendall, M. M. Quwailid, M. R. Romero, J. O'Neill, J. E. Chesham, D. Brooker, Z. Lalanne, M. H. Hastings, and P. M. Nolan. 2007. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science 316897-900. [DOI] [PubMed] [Google Scholar]
- 17.Graves, P. R., and P. J. Roach. 1995. Role of COOH-terminal phosphorylation in the regulation of casein kinase I delta. J. Biol. Chem. 27021689-21694. [DOI] [PubMed] [Google Scholar]
- 18.Izumo, M., C. H. Johnson, and S. Yamazaki. 2003. Circadian gene expression in mammalian fibroblasts revealed by real-time luminescence reporting: temperature compensation and damping. Proc. Natl. Acad. Sci. USA 10016089-16094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knippschild, U., A. Gocht, S. Wolff, N. Huber, J. Lohlker, and M. Stoter. 2005. The casein kinase 1 family: participation in multiple cellular processes in eukaryotes. Cell Signal. 15675-689. [DOI] [PubMed] [Google Scholar]
- 20.Kornmann, B., O. Schaad, H. Bujard, J. S. Takahashi, and U. Schibler. 2007. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 5e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, C., J. P. Etchegaray, F. R. Cagampang, A. S. Loudon, and S. M. Reppert. 2001. Posttranslational mechanisms regulate the mammalian circadian clock. Cell 107855-867. [DOI] [PubMed] [Google Scholar]
- 22.Lee, C., D. R. Weaver, and S. M. Reppert. 2004. Direct association between mouse PERIOD and CKIɛ is critical for a functioning circadian clock. Mol. Cell. Biol. 24584-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lowrey, P. L., K. Shimomura, M. P. Antoch, S. Yamazaki, P. D. Zemenides, M. R. Ralph, M. Menaker, and J. S. Takahashi. 2000. Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science 288483-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meng, Q. J., L. Logunova, E. S. Maywood, M. Gallego, J. Lebiecki, T. M. Brown, M. Sladek, A. S. Semikhodskii, N. R. Glossop, H. D. Piggins, J. E. Chesham, D. A. Bechtold, S. H. Yoo, J. S. Takahashi, D. M. Virschup, R. P. Boot-Handford, M. H. Hastings, and A. S. Loudon. 2008. Setting clock speed in mammals: the CK1 epsilon tau mutation in mice accelerates circadian pacemakers by selectively destabilizing PERIOD proteins. Neuron 5878-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Numano, R., S. Yamazaki, N. Umeda, T. Samura, M. Sujino, R. Takahashi, M. Ueda, A. Mori, K. Yamada, Y. Sakaki, S.-I. T. Inouye, M. Menaker, and H. Tei. 2006. Constitutive expression of the period 1 gene impairs behavioral and molecular circadian rhythms. Proc. Natl. Acad. Sci. USA 1033716-3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Postic, C., M. Shiota, K. D. Niswender, T. L. Jetton, Y. Chen, J. M. Moates, K. D. Shelton, J. Lindner, A. D. Cherrington, and M. A. Magnuson. 1999. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knockouts using Cre recombinase. J. Biol. Chem. 274305-315. [DOI] [PubMed] [Google Scholar]
- 27.Reischl, S., K. Vanselow, P. O. Westermark, N. Thierfelder, B. Maier, H. Herzel, and A. Kramer. 2007. Beta-TrCP1-mediated degradation of PERIOD2 is essential for circadian dynamics. J. Biol. Rhythms 22375-386. [DOI] [PubMed] [Google Scholar]
- 28.Sekine, T., T. Yamaguchi, K. Hamano, M. W. Young, M. Shimoda, and L. Saez. 2008. Casein kinase I epsilon does not rescue double-time function in Drosophila despite evolutionarily conserved roles in the circadian clock. J. Biol. Rhythms 233-15. [DOI] [PubMed] [Google Scholar]
- 29.Shirogane, T., J. Jin, X. L. Ang, and J. W. Harper. 2005. SCFbeta-TRCP controls clock-dependent transcription via casein kinase 1-dependent degradation of the mammalian period-1 (Per1) protein. J. Biol. Chem. 28026863-26872. [DOI] [PubMed] [Google Scholar]
- 30.Siepka, S. M., S. H. Yoo, J. Park, W. Song, V. Kumar, Y. Hu, C. Lee, and J. S. Takahashi. 2007. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of Cryptochrome and Period gene expression. Cell 1291011-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stratmann, M., and U. Schibler. 2006. Properties, entrainment, and physiological functions of mammalian peripheral oscillators. J. Biol. Rhythms 21494-506. [DOI] [PubMed] [Google Scholar]
- 32.Takahashi, J. S., H.K Hong, C. H. Ko, and E. L. McDearmon. 2008. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat. Rev. Genet. 9764-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toh, K. L., C. R. Jones, Y. He, E. J. Eide, W. A. Hinz, D. M. Virshup, L. J. Ptacek, and Y. H. Fu. 2001. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science 2911040-1043. [DOI] [PubMed] [Google Scholar]
- 34.Vanselow, K., J. T. Vanselow, P. O. Westermark, S. Reischl, B. Maier, T. Korte, A. Herrmann, H. Herzel, A. Schlosser, and A. Kramer. 2006. Differential effects of PER2 phosphorylation: molecular basis for the human familial advanced sleep phase syndrome (FASPS). Genes Dev. 202660-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vielhaber, E., E. Eide, A. Rivers, Z. H. Gao, and D. M. Virshup. 2000. Nuclear entry of the circadian regulator mPER1 is controlled by mammalian casein kinase I epsilon. Mol. Cell. Biol. 204888-4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weaver, D. R., and S. M. Reppert. 2008. Circadian timekeeping, p. 931-957. In L. R. Squire, D. Berg, F. E. Bloom, S. du Lacv, A. Ghosh, and N. C. Spitzer (ed.), Fundamental neuroscience, 3rd ed. Academic Press, Inc., New York, NY.
- 37.Welsh, D. K., S. H. Yoo, A. C. Liu, J. S. Takahashi, and S. A. Kay. 2004. Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr. Biol. 142289-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu, Y., Q. S. Padiath, R. E. Shapiro, C. R. Jones, S. C. Wu, N. Saigoh, K. Saigoh, L. J. Ptacek, and Y. H. Fu. 2005. Functional consequences of a CKIδ mutation causing familial advanced sleep phase syndrome. Nature 434640-644. [DOI] [PubMed] [Google Scholar]
- 39.Xu, Y., K. L. Toh, C. R. Jones, J. Y. Shin, Y. H. Fu, and L. J. Ptacek. 2007. Modeling of a human circadian mutation yields insights into clock regulation by PER2. Cell 12859-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamazaki, S., and J. S. Takahashi. 2005. Real-time luminescence reporting of circadian gene expression in mammals. Methods Enzymol. 393288-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoo, S. H., S. Yamazaki, P. L. Lowrey, K. Shimomura, C. H. Ko, E. D. Buhr, S. M. Siepka, H. K. Hong, W. J. Oh, O. J. Yoo, M. Menaker, and J. S. Takahashi. 2004. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. USA 1015339-5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.