Abstract
To understand how DEXD/H-box proteins recognize and interact with their cellular substrates, we have been studying Prp28p, a DEXD/H-box splicing factor required for switching the U1 snRNP with the U6 snRNP at the precursor mRNA (pre-mRNA) 5′ splice site. We previously demonstrated that the requirement for Prp28p can be eliminated by mutations that alter either the U1 snRNA or the U1C protein, suggesting that both are targets of Prp28p. Inspired by this finding, we designed a bypass genetic screen to specifically search for additional, novel targets of Prp28p. The screen identified Prp42p, Snu71p, and Cbp80p, all known components of commitment complexes, as well as Ynl187p, a protein of uncertain function. To examine the role of Ynl187p in splicing, we carried out extensive genetic and biochemical analysis, including chromatin immunoprecipitation. Our data suggest that Ynl187p acts in concert with U1C and Cbp80p to help stabilize the U1 snRNP-5′ splice site interaction. These findings are discussed in the context of DEXD/H-box proteins and their role in vivo as well as the potential need for more integral U1-snRNP proteins in governing the fungal 5′ splice site RNA-RNA interaction compared to the number of U1 snRNP proteins needed by metazoans.
Nuclear precursor mRNA (pre-mRNA) splicing takes place in the spliceosome, a large dynamic complex consisting of over 100 proteins and five small nuclear RNAs (snRNAs) (32, 70). During spliceosome assembly, the U1 small nuclear ribonucleoprotein particle (snRNP) first contacts the pre-mRNA 5′ splice site (5′ss), followed by binding of the U2 snRNP to the branch site and the joining of the U5-U4/U6 tri-snRNP (32, 64, 70). The step in which U1 snRNP binds to the 5′ss is arguably one of the most critical, because it probably commits pre-mRNA to the splicing pathway (38, 48, 49, 60, 74). In the budding yeast Saccharomyces cerevisiae in vitro system, two U1-snRNP-containing commitment complexes (CCs), CC1 and CC2, can be detected by native gel electrophoresis prior to the U2 snRNP's joining to form the prespliceosome (38, 60). CC1, whose formation is dependent on a functional 5′ss, appears to be a kinetic precursor to CC2, whose formation requires both a functional 5′ss and branch site and the participation of the branch-site-binding protein (BBP) and Mud2p, which are likely equivalent to SF1 and U2AF65, respectively, in the mammalian system (1-3, 75).
Accumulating evidence suggests that formation of the canonical 5- to 7-bp RNA duplex between U1 snRNA and the 5′ss region is not sufficient to cause a stable CC to form in the yeast system (59, 62, 78); protein-RNA contacts are also important. For example, Zhang and Rosbash (77) identified eight proteins, all present in CCs, that make physical contact with the pre-mRNA at or near the 5′ss. Four of these proteins, U1C, U1-70K, Snu56p, and Nam8p, are integral parts of the U1 snRNP (20), and another three, SmB, SmD1, and SmD3, belong to the seven-member ring that binds the conserved Sm site present on U1, U2, U4, and U5 snRNAs (33, 71). The remaining protein, Cbp80p, is a subunit of the nuclear cap-binding complex (CBC), which also contains a second subunit, Cbp20p (28, 39). Interestingly, despite being a non-snRNP factor, Cbp80p is known to collaborate with U1 snRNP to help form or stabilize CC1 (8, 40). Furthermore, the contact between the C-terminal tails of SmB, SmD1, and SmD3 and the pre-mRNA may contribute to stabilizing the U1 snRNP/pre-mRNA interaction (76). Finally, Du and Rosbash (11) more recently showed that U1C is capable of selecting splice-site-like sequences in which the first four nucleotides, GUAU, are identical to the first four nucleotides of the yeast splice-site consensus sequence.
Once fully assembled, the spliceosome must progress through a number of major structural and conformational changes to form the catalytic center; these include a series of highly orchestrated RNA-RNA rearrangements (53, 64, 70). Some of these are mutually exclusive; i.e., the formation of one RNA duplex requires the disruption of another. For example, the base-pairing interaction between the U1 snRNA and the 5′ss is replaced by a U6 snRNA/5′ss pairing. This exchange appears to be coupled to U4/U6 RNA unwinding (53, 64, 70). It is now known that splicing factors belonging to the ATPase II superfamily (18), which are also termed the DEXD/H-box proteins (5, 43), promote spliceosomal RNA rearrangements (64). However, the precise roles of most DEXD/H-box proteins remain unclear.
It has been nearly 2 decades since DEXD/H-box proteins were first proposed to be RNA helicases (44). Over the years, a wealth of data revealed that DEXD/H-box proteins are essential in most, if not all, RNA-related pathways, e.g., splicing, mRNA export, and ribosomal biogenesis (5, 43, 64). Their modes of action in vivo remain a mystery, however. For example, Lorsch and Herschlag (45, 46) proposed that DEXD/H-box proteins may perform functions which are distinct from RNA unwinding and include mediating large-scale RNA structural rearrangements, disrupting protein-RNA or protein-protein interactions, and functioning as fidelity sensors in RNA-RNA interactions and rearrangements. Indeed, recent data confirm that DEXD/H-box proteins can catalyze protein displacement in a manner independent of RNA duplex unwinding (30). Therefore, the essential functions of DExH/D proteins can be exerted on a wide range of RNP substrates. This “RNPase” (or ATPase for RNP remodeling) hypothesis appears especially attractive in light of the fact that RNA duplexes in vivo are rarely more than ∼10 contiguous base pairs in length and that they often require protein binding for stabilization (21, 63). To fully understand how DEXD/H-box proteins function in the cell, it is critical to identify their physiological substrates.
Inspired by our previous finding that the requirement for Prp28p, an essential DEXD/H-box splicing factor, can be bypassed by mutations that alter the YHC1 gene, which encodes U1C protein (7), we sought to exploit the bypass concept to deepen our understanding of the role of Prp28p in splicing. The underlying hypothesis is that bypass mutations define gene products that Prp28p may counteract. Here we describe the outcome of this approach and provide a detailed analysis of Ynl187p, a novel protein that probably contributes to stabilizing the U1 snRNP-5′ss interaction.
MATERIALS AND METHODS
Screen for prp28Δ bypass mutants.
All yeast procedures were done using standard protocols (22). Strain YTC585 (MATa prp28Δ::TRP1 yhc1Δ::HIS3 ura3-1 lys2::hisG ade2-1 trp1-1 his3-11 leu2-3,112 can1-100 pCA8107 [PRP28/ADE2/URA3] pYHC1005 [YHC1/LEU2]) was irradiated by UV light on YPD (1% yeast extract, 2% Bacto Peptone, 2% glucose) plates to achieve an 80 to 90% killing rate. A total of 35,000 surviving colonies were replica plated onto 5-fluoroorotic acid (5-FOA) plates and incubated at 30°C for 5 days. Five 5-FOA-resistant (Ura3-negative [Ura3−]) and red (Ade2−) colonies were chosen, grown, and transformed with pCA8032 (PRP28/URA3) and pYHC1053 (YHC1/ADE2). Following the loss of pYHC1005, all transformants remained resistant to 5-FOA, indicating that the mutations were chromosomal. Genomic DNAs isolated from bypass candidates were digested with HindIII and probed with a 1.4-kb 32P-labeled KpnI-XmnI fragment representing most of the PRP28 open reading frame (ORF). None of the five strains tested showed any residual PRP28 presence. Diploids were made by mating mutant strains to strain YTC286 (MATα prp28Δ::TRP1 ade2-1 can1-100 his3-11 leu2-3,112 trp1-1 ura3-1 pCA8032), and all diploids exhibited the 5-FOA− phenotype, indicating recessive mutations. Segregants derived from the third backcross containing chromosomal YHC1 and pCA8032 were used for subsequent analysis. To clone the wild-type alleles of the bypass mutations, bypass strains were transformed with a YCp50 (LEU2) genomic library. 5-FOA− colonies were identified upon performing replica plating onto 5-FOA plates. Recovered plasmids that reproducibly yielded the 5-FOA− phenotype were selected for DNA sequencing from both ends of their inserts. Recloning and retesting of individually PCR-amplified ORFs identified YNL187, PRP42, and SNU71 as genes responsible for the 5-FOA− phenotype. Mutant bypass alleles were also amplified by PCR from the bypass mutant strains and sequenced to determine the corresponding mutations.
Immunolocalization and precipitation of Ynl187p.
To localize Ynl187p, plasmids pBTE3011 (YNL187-HA/pRS315), pBTE3032 (YNL187-HA/pRS425), and pBTE3042 (YNL187/pRS425) were transformed into YTC71 (MATα ynl187Δ::kanMX4 his3Δ0 leu2Δ0 lys2Δ0 ura3Δ0) to yield strains YTC801, YTC867, and YTC1235, respectively. As a control for nuclear localization, YTC819 (MATa prp42Δ::kanMX4 his3Δ0 leu2Δ0 lys2Δ0 ura3Δ0 pPRP42018 [PRP42-HA/pRS315]) was used. Indirect immunofluorescence microscopy was performed as described previously (41). Cells harvested at early log phase were spheroplasted and incubated with antihemagglutinin (anti-HA) mouse monoclonal antibody (BAbCo) at a 1:2,000 dilution and then with Cy3-conjugated anti-mouse immunoglobulin G (Jackson ImmunoRearch Labs) at a 1:600 dilution. Images were acquired using a Nikon Microphot-FX microscope equipped with a SenSys camera (Photometrics). To detect potential association of Ynl187p with snRNAs, 20-μl aliquots of splicing extracts (protein concentration, 12.5 mg/ml) made from strains YTC826 (MATα prp42::LEU2 arg4 leu2-3,112 trp1-289 ura3-52 pBM150 [GAL1::PRP42-HA/URA3]; a gift from B. Rymond) and YTC801 (see above) were mixed with 10 μl of protein A-agarose beads (Invitrogen) prebound by rabbit anti-HA.11 polyclonal antibody (Covance) in a total volume of 50 μl of buffer D (20 mM HEPES [pH 7.9], 0.2 mM EDTA, 20% glycerol, 0.05% Nonidet P-40, 0.5 mM dithiothreitol) containing either 100 mM or 150 mM KCl. The mixtures were incubated at 4°C for 2 h on a Nutator. Beads were washed with ice-cold buffer D containing either 100 mM or 150 mM KCl. After washes, RNA was extracted and ethanol precipitated for Northern analysis as previously described (4, 47). The input control corresponded to a 1:20 dilution of the total RNA extracted from 20 μl of splicing extract. Probes for the Northern analysis were a mixture of equimolar ratios of DNA fragments corresponding to U1, U2, U4, U5, and U6 snRNAs, which were labeled with 32P by random priming.
Testing the prp28Δ-bypassing activity of various mutant alleles.
A plasmid counter-selection procedure was used for scoring mutations capable of bypassing the prp28Δ mutation. This was done by crossing a strain containing chromosomal a prp28Δ mutation complemented by a PRP28/URA3 plasmid to various strains harboring deletions of genes of interest and, when the gene in question was essential, a corresponding plasmid-borne mutant allele (e.g., smd3Δ pRS315-smd3ΔC). The resulting diploids were sporulated and haploid strains containing both chromosomal deletions (e.g., prp28Δ smd3Δ), and the necessary plasmids (e.g., pRS316-PRP28 and pRS315-smd3ΔC) were isolated. Each of these haploid strains was examined for its ability to grow on 5-FOA plates.
Synthetic lethality screen and test.
To search for mutations synthetically lethal to the ynl187Δ mutation, strain YTC1046 (MATa ynl187Δ::kanMX4 ade2 ade3 leu2 lys2 ura3 pBTE3029 [YNL187/ADE3/URA3]) was UV irradiated (see above). Out of the ∼20,000 colonies screened, one mutant was identified which exhibited the anticipated 5-FOA− and red-colony phenotypes. Further analysis revealed that the synthetic lethality phenotype was linked to the inability to lose YNL187 and to a single recessive mutation. The wild-type allele corresponding to the mutation was cloned using the same YCp50 library described above, except that in this case the acquisition of the presumed wild-type allele would result in 5-FOA+ and red- and white-sectoring phenotypes. Eight library plasmids were isolated, sequenced, and found to contain a common ORF corresponding to CBP80. The mutant cbp80-101 allele was amplified by PCR from the mutant genomic DNA and sequenced. Introduction of the wild-type CBP80 clone, but not the cbp80-101 clone, into the synthetically lethal mutant relieved the lethality. To test the genetic interaction between the ynl187Δ mutation and various U1C-[L13] alleles, an experimental design similar to the bypass test described above was used. In this case, the haploid tester strains contained ynl187Δ::kanMX4 yhc1Δ::HIS3 and pYHC1023 (pRS316-YHC1/YNL187) as well as a U1C-[L13] allele (7) on pRS315. These strains were tested for growth on the 5-FOA plates.
Genetic analysis of the U1/5′ss RNA duplex.
To examine the ability of the ynl187Δ mutation to relieve 5′ss/U1 snRNA hyperstabilization, strains YTC639 (MATa cup1::ura3 leu2 trp1 lys2 ade2 his3 ura3) and YTC813 (MATa ynl187Δ::kanMX4 cup1::ura3 leu2 trp1 lys2 ade2 his3 ura3) were separately transformed with each of the two intron-containing ACT1-CUP1 reporter plasmids (63). Plasmid bJPS28 allows 10 bp and bJPS30 6 bp to form between the endogenous U1 snRNA and the 5′ss of the ACT1-CUP1 reporter pre-mRNA. The assay was done as previously described (7) using plates containing 0.5 mM Cu2+.
Splicing and 4-thioU cross-linking assays.
Splicing extracts were prepared by the liquid-nitrogen method (65) from the isogenic strains YTC71 (ynl187Δ), YTC780 (ynl187Δ prp28Δ), and YTC771 (wild type) and used for conducting standard splicing reaction (65) and CC formation (12) experiments. The 4-thio-uridine (4-thioU) cross-linking experiments were done as described previously (77) using a 32P-labeled WT-72 transcript as a splicing substrate containing randomly incorporated 4-thioU (Tri-link Biotechnologies). WT-72 was incubated in yeast extracts for 20 min at 25°C before UV cross-linking took place. For chase experiments, a 400-fold excess of unlabeled WT-72 transcript was added at the end of the 20-min incubation. Reactions were allowed to continue for 0, 12.5, or 25 min before being subjected to UV irradiation. To examine the proteins cross-linked to WT-72, the UV-irradiated CC was first precipitated with anti-Prp40p antibody (a gift from P. Siliciano) bound to protein A-agarose beads. After washes, RNA was degraded by adding 2 μl of RNase T1/A mix (Ambion) to the beads for 10 min of incubation at 37°C. The 32P-labeled proteins were then analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and autoradiography.
ChIP assay and real-time PCR.
We followed an established chromatin immunoprecipitation (ChIP) procedure (37) to monitor cotranscriptional recruitment of U1C and Ynl187p. PCRs were done using 96-well plates, with each 20-μl reaction mixture consisting of 1 μl of IP DNA (input dilution, 1:80), 10 μl of iQ SYBR green Supermix (Bio-Rad), and a primer pair (0.2 μM each). Five sets of primer pairs (37) were used for all the experiments. The reaction conditions consisted of 95°C for 3 min, followed by 40 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s. All samples in a single PCR run were assayed in triplicate experiments. All data represent the averages of the results of at least three independent experiments, with the error bars in the figures displaying average deviations. The enrichment of U1C above the background level was initially calculated using the following formula: [2(input CT − IP CT)]U1C-TAP strain − [2(input CT − IP CT)]U1C-non-TAP strain, where CT is the threshold cycle reported by the instrument for each PCR. To account for potential variations in transcript levels in different genetic backgrounds, the initial data were further computed on the basis of per-transcript levels by using the level of 25S rRNA, a polymerase I transcript, as an internal control. The formula used for normalization was as follows: [2(RDN251 CT − ACT1 CT)]mutant/[2(RDN251 CT − ACT1 CT)]wild type, where RDN251 CT and ACT1 CT represent the threshold cycles for amplifying regions of 25S rRNA and an intron-containing region of the ACT1 transcript, respectively. For the ChIP analysis of Ynl187-HA, 2.5 μl of anti-HA High Affinity antibody (clone 3F10; Roche) was used for each IP with 20 μl of Dynabeads protein G (Invitrogen). The untagged wild-type YNL187 strain was used as the background. The enrichment of Ynl187-HA over the background level was calculated using the following formula: [2(input CT − IP CT)]Ynl187-HA strain − [2(input CT − IP CT)]Ynl187-non-HA strain, where CT represents the threshold cycle reported by the instrument for each PCR. As a basal-line control, the ChIP value obtained from a transcriptionally repressed region in the telomere of chromosome VI was assigned a numerical value of 1. This region was amplified by oligonucleotides TEL1 (CGTGTGTAGTGATCCGAACTCAGT) and TEL2 (GACCAGTCCTCATTTCCATCAATAG).
Probing U1 snRNP by native gel electrophoresis.
We followed an established protocol to monitor the electrophoretic mobility of the U1 snRNP (20, 47). Yeast splicing extracts were mixed with an equal volume of R buffer (50 mM HEPES [pH 7.5], 2 mM magnesium acetate, 20 mM EDTA, 1 μg/ml of total yeast RNA, and 1 mg/ml of heparin), incubated on ice for 10 min, and then electrophoresed on a 3% polyacrylamide (60:1 acrylamide/bis-acrylamide)-0.5% agarose composite gel. After electrophoresis, RNAs were electrotransferred to a GeneScreen membrane (NEN) for Northern analysis. The preparation of the U1 probe containing digoxigenin-11-dUTP and the subsequent hybridization procedures were done per the instructions provided by the commercial source (digoxigenin High Prime DNA labeling and detection starter kit II; Roche). After hybridization, the resulting chemiluminescence signals were imaged using a Molecular Imager Gel Doc XR system (Bio-Rad).
RESULTS
Identification of novel bypass suppressors of the prp28Δ mutation.
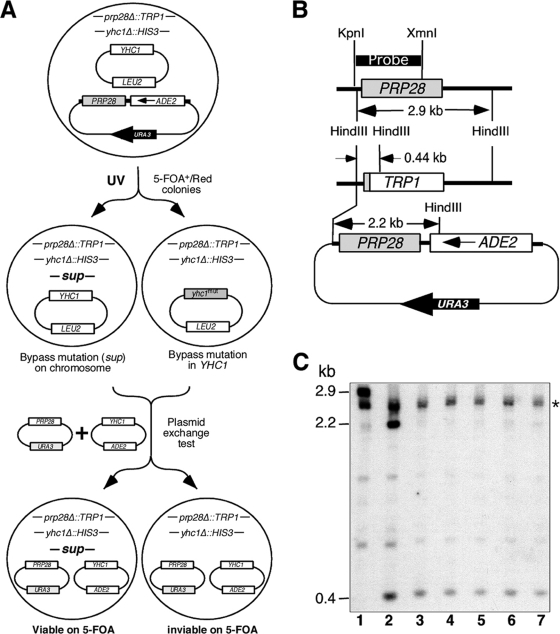
We designed a genetic screen (Fig. 1A) to purposely search for new mutations that eliminate the requirement for Prp28p. A haploid strain containing the following features was first constructed: (i) chromosomal deletions of PRP28 (prp28Δ::TRP1) and YHC1 (yhc1Δ::HIS3) and (ii) two complementing plasmids carrying PRP28 and YHC1 that were marked by URA3/ADE2 and LEU2, respectively (Fig. 1A). Because PRP28 and YHC1 are essential genes, this strain must retain both plasmids to stay alive and is thus phenotypically Ura3+/Ade2+ (5-FOA−/white colony). After UV mutagenesis, candidates able to grow on 5-FOA plates (i.e., Ura3−/5-FOA+) and form red (Ade2−) colonies were identified. These mutants were presumed to have acquired extragenic suppressors that render the URA3/ADE2/PRP28 plasmid dispensable. To exclude mutations that landed on the plasmid-borne YHC1, an ADE2-marked plasmid carrying a copy of unmutagenized YHC1 (Fig. 1A), which cannot bypass Prp28p, and a PRP28/URA3 plasmid were introduced into the mutant strains. Non-YHC1 chromosomal bypass candidates were ascertained on the basis of the ability to lose the YHC1/LEU2 plasmid and viability on 5-FOA.
FIG. 1.
Identification of YNL187 by a bypass genetic screen. (A) Strategy for isolating prp28Δ bypass suppressors. See the text for details. (B) Restriction maps of the chromosomal PRP28, prp28Δ::TRP1 (TRP1), and the plasmid (URA3/ADE2-marked)-borne PRP28. (C) PRP28 is not present in the bypass suppressor strains. DNAs isolated from bypass suppressor candidates (lanes 3 to 7) were digested with HindIII and probed for the presence of the chromosomal prp28Δ::TRP1 and the plasmid-borne PRP28. The probe hybridizes to a 2.9-kb DNA band corresponding to the chromosomal PRP28 in the wild-type strain (lane 1), which is replaced by an 0.4-kb band corresponding to prp28Δ::TRP1 and by an additional 2.2-kb band corresponding to the plasmid-borne PRP28 in the starting strain (lane 2). Only the 0.4-kb band is detected in the five bypass suppressors (lanes 3 to 7). The star indicates a band with a cross-reaction induced by the probe.
The screen uncovered 14 candidates, and 5 were validated for loss of the PRP28 plasmid by Southern blotting. DNAs isolated from these candidates were digested with HindIII and probed for the presence of the chromosomal prp28Δ::TRP1 allele and the plasmid-borne PRP28 allele. This probe (Fig. 1B) hybridized to a 2.9-kb DNA band corresponding to chromosomal PRP28 (Fig. 1C, lane 1) in a wild-type strain. As expected, in the Ade2+/Ura3+ starting strain, the 2.9-kb band was replaced by a 0.44-kb band corresponding to the prp28Δ::TRP1 allele and by an additional 2.2-kb band (Fig. 1C, lane 2) corresponding to the plasmid-borne PRP28 allele (Fig. 1B). The 2.2-kb band was not present in the Ade2−/Ura3− suppressor strains (Fig. 1C, lanes 3 to 7), indicating that the plasmid-borne PRP28 allele had been lost. It is unlikely that a portion of PRP28 not covered by the probe was still present and functional in the Ade2−/Ura3− strains. Such a truncated Prp28p would have lost the first 455 amino acids (out of a total of 588 amino acids) encompassing the conserved I to V motifs (18) essential for ATP binding, ATP hydrolysis, and RNA unwinding (43, 67). We concluded that extragenic suppressor mutations allowed bypass of Prp28p function in the Ade2−/Ura3− strains. Genetic analysis showed that, in each case, the bypass phenotype was caused by a single recessive mutation (data not shown).
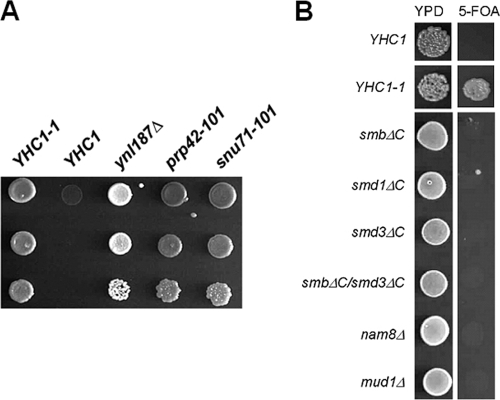
To identify genes corresponding to the bypass suppressor mutations, we screened a genomic library for clones that restore the PRP28 requirement (see Materials and Methods). This screen (Fig. 2A) uncovered three genes, PRP42 (20, 47), SNU71 (20), and YNL187W. While Prp42p and Snu71p are known and essential components of the yeast U1 snRNP, the function of Ynl187p (357 amino acids in length) is less certain and nonessential. It was, however, implicated in splicing because of its genetic interaction with Prp40p (51) and its observed two-hybrid interaction with SmB in a large-scale screen (17). More recently, Ynl187p was reported to genetically link to Nam8p and Tgs1p, a cap trimethylguanosine synthase, in two separate synthetic lethality screenings (10, 23). Computer prediction results showed that Ynl187p contains a nuclear localization signal, 146RDAVWMNQSTKKASK161, and a leucine-rich nuclear export signal (NES), 247LQHELNVLRL256. Interestingly, all three genes, PRP42, SNU71, and YNL187w, appeared to be fungus specific; their metazoan counterparts could not be clearly identified by BLAST searches (20). This may reflect differences in snRNP assembly pathways between yeast species and the metazoans (see below and Discussion).
FIG. 2.
Bypassing the prp28Δ mutation is not a general event. (A) Growth phenotypes of the suppressor strains in the background of the prp28Δ strain. Relevant genotypes of the yeast strains are shown on the top of the panel. YHC1-1 is a previously characterized dominant bypass allele. Cells were grown to saturation at 30°C, serially diluted, and spotted onto 5-FOA plates. Note that the ynl187Δ strain shown here (white in color) lacks the ade2 mutation, because it was derived from a cross between a commercial ynl187Δ strain and a laboratory prp28Δ strain. In contrast, the other strains harbor the ade2 mutation (darker in color). (B) Not all genetic perturbations of U1 snRNP can bypass the prp28Δ mutation. Relevant genotypes of the strains tested are shown at the left side of the panel. See text for detailed description.
Loss of function of Ynl187p bypasses the prp28Δ mutation.
Sequence analysis of the three mutant ynl187 alleles revealed three independent mutations: two were upstream of the coding region at −76 (C-to-T change) and −78 (C-to-G change), and the third yielded an L344P (CTA to CCA) amino acid substitution close to the Ynl187p C terminus. The upstream mutations are located within the 5′ untranslated region of the YNL187 transcript, according to a recent genome-wide analysis of the yeast transcriptional landscape (52); however, it remains to be determined whether they are part of the YNL187 promoter element. The recessive nature of all three ynl187 alleles prompted us to speculate that a loss of Ynl187p activity allows bypass of Prp28p. Indeed, a prp28Δ ynl187Δ double mutant is viable (Fig. 2A), supporting a model in which Ynl187p functions to counteract Prp28p and/or vice versa. Sequence analysis of the mutant prp42-101 allele revealed a D76Y (GAC to TAC) substitution located within the first of five tetratricopeptide repeats. This alteration may disrupt Prp42p's interaction with other splicing factors, such as Prp39p, which has six tetratricopeptide repeats (47). The snu71-101 mutation results in an L270S (TTA to TCA) substitution lying between 2 of the 19 SR-like dipeptides. It was previously speculated that Snu71p is functionally similar to human SR proteins (20).
Bypassing of the prp28Δ mutation via U1-snRNP perturbations is not a general event.
The finding that genetic alterations of U1C, U1 snRNA (7), Prp42p, Snu71p, and Ynl187p can eliminate the requirement for Prp28p raised a possibility that different perturbations of the U1 snRNP can bypass Prp28p. To test this more general hypothesis, we introduced a series of additional mutations into a prp28Δ tester strain that contained a PRP28/URA3 plasmid and assayed for their abilities to permit plasmid loss. We first examined C-terminal truncation mutations of SmB, SmD1, and SmD3, because these three proteins (i) cross-link to the 5′ss (77); (ii) may help stabilize the U1 snRNP/pre-mRNA interaction via their highly charged C-terminal tails (76); and (iii) are adjacent to U1C, according to a human U1-snRNP structure model (33). None of these three mutations alone or in a combination of SmB and SmD3 truncations permitted plasmid loss (Fig. 2B). We also tested deletion alleles of the two nonessential genes, NAM8 and MUD1. Nam8p is a U1-snRNP protein that cross-links to the 5′ss region (57), and its absence destabilizes the overall structure of U1 snRNP (20). Mud1p is the yeast counterpart of the human U1A protein, and its loss alters the conformation of the CCs (42). Again, neither the nam8Δ nor the mud1Δ mutation could bypass Prp28p (Fig. 2B). These data raise the possibility that altering the U1 snRNP alone is not sufficient to bypass Prp28p and that U1C, Prp42p, Snu71p, and Ynl187p may share certain common features that specify their unique relationship to Prp28p.
Ynl187p is localized in the nucleus and associated with snRNPs.
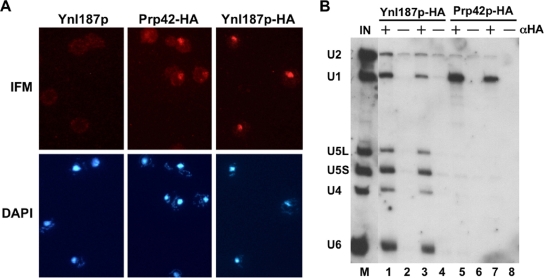
To examine the cellular localization of Ynl187p, we initially expressed a green fluorescent protein fusion of Ynl187p in the ynl187Δ strain but obtained no signal, a result which may have been due to low abundance of Ynl187p. This hypothesis was later validated in a genome-wide protein localization study (27). Our studies also found that tagging Ynl187p at its C terminus with either green fluorescent protein or TAP inactivated Ynl187p, because their expression in the ynl187Δ strain failed to restore the requirement for Prp28p (data not shown). However, an HA-tagged Ynl187p (Ynl187p-HA) proved to be functional (data not shown). Indirect immunofluorescence microscopy using an anti-HA monoclonal antibody with cells expressing Ynl187p-HA from a CEN plasmid revealed that Ynl187p is localized in the nucleus (Fig. 3A, right panels), in similarity to Prp42p (middle panels), which was previously shown to localize predominantly in the nucleus (27). Overexpression from a high-copy-number plasmid increased nuclear signal levels, with no evidence of cytoplasmic signal. Thus, these data suggest a nuclear role for Ynl187p.
FIG. 3.
Ynl187p is localized in the nucleus and associated with snRNPs. (A) Immunofluorescence microscopy (IFM) of the Ynl187p-, Ynl187p-HA-, and Prp42p-HA-expressing cells. Cells were stained with anti-HA antibody and then with Cy3-conjugated anti-mouse immunoglobulin G (top panels). Results of staining of the same cells with 4′,6-diamino-2-phenylindole (DAPI) are also shown (bottom panels). (B) Association of Ynl187p with snRNPs. Splicing extracts made from strains expressing Ynl187p-HA (lanes 1 to 4) and Prp42p-HA (lanes 5 to 8) were mixed with protein A-agarose beads alone (−) or prebound by rabbit anti-HA.11 (αHA) polyclonal antibody (+). The coprecipitated RNAs were analyzed by Northern blotting. The salt concentrations used were 100 mM NaCl (lanes 1, 2, 5, and 6) or 150 mM NaCl (lanes 3, 4, 7, and 8). The input control (IN; lane M) corresponds to a 1:20 dilution of the total RNA extracted from 20 μl of splicing extract. U1, U2, U4, U5, and U6 snRNAs are indicated.
To examine whether Ynl187p is associated with the splicing apparatus, we immunoprecipitated Ynl187p-HA with an anti-HA antibody and probed the associated snRNAs by Northern analysis. In contrast to Prp42p (47) (Fig. 3B, lanes 5 to 8), Ynl187p appears to associate with all the spliceosomal snRNPs, i.e., U1, U2, U4, U5, and U6 (Fig. 3B, lanes 1 to 4), at physiological salt concentrations. This raises the possibility that Ynl187p may have a role in snRNP assembly and/or function (see below).
Genetic analysis supports a role o Ynl187p in facilitating U1 snRNP/5′ss interaction.
We have proposed that Prp28p counteracts the stabilizing effect of U1C to promote the dissociation of the U1 snRNP from the 5′ss (7). We therefore anticipated that Ynl187p, Prp42p, and Snu71p have a role similar to that of U1C. Consistent with this notion, the newly discovered bypass suppressor strains shared a growth phenotype strikingly similar to that of the U1C bypass suppressor strain; i.e., they all failed to grow at 16°C but grew well at 30°C (data not shown). We interpreted this cold-sensitive phenotype as reflecting a strengthened U1 snRNP/5′ss interaction at low temperature, which would then lead to a requirement for Prp28p at the low temperature (7, 13).
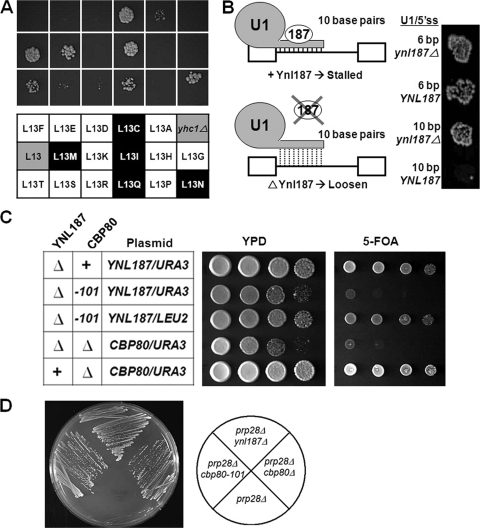
To analyze Ynl187p's functional relationship with U1C, we examined the outcome of combining ynl187Δ and U1C mutations in the presence of Prp28p. If Ynl187p were to play a role closely related to that of U1C, combining these two different bypass mutations might drastically destabilize the U1 snRNP/5′ss interaction and result in synthetic lethality. This was tested by constructing a strain containing both yhc1Δ and ynl187Δ deletions complemented by a YNL187/YHC1/URA3 plasmid. This strain was transformed with plasmids harboring each of the 17 mutant U1C-[L13] alleles that we previously analyzed (7), and the resulting strains were tested using 5-FOA to determine their ability to lose the YNL187/YHC1/URA3 plasmid. Lethality or severe growth defects on 5-FOA were observed for 10 U1C-[L13] alleles (L13A, L13D, L13E, L13F, L13G, L13H, L13K, L13P, L13R, and L13S) (Fig. 4A), all of which were previously shown to allow bypass of Prp28p (7). In contrast, cell viability was observed for the remaining seven alleles (L13C, L13I, L13L, L13M, L13N, L13Q, and L13T) (Fig. 4A), and all but L13T were previously shown to retain the requirement for Prp28p (7). The U1C-[L13] residue is located within a conserved C2H2-type zinc finger motif at a position that is typically occupied by a hydrophobic residue (36). We speculate that the 10 lethal U1C alleles may have disrupted an interaction between U1C and another spliceosomal component, thus resulting in an unstable U1 snRNP/5′ss interaction, whereas the 7 nonlethal alleles maintained this interaction (7). Overall, our data are in good agreement with a model in which Ynl187p and U1C jointly promote a stable U1 snRNP/5′ss interaction. Significantly, when tested similarly, both the prp42-101 and snu71-101 strains displayed a pattern similar to that of the ynl187Δ strain (data not shown). Taken together, these data suggest a close functional relationship among U1C, Ynl187p, Prp42p, and Snu71p at the point of U1 snRNP/5′ss interaction.
FIG. 4.
Genetic analysis suggests that Ynl187p promotes the U1 snRNP/5′ss interaction and is functionally related to Cbp80p. (A) Most mutant yhc1-[L13] alleles capable of bypassing the prp28Δ mutation are synthetically lethal with the ynl187Δ mutation. Top three rows of the panel: results of the synthetic lethality test (see text for details) scored using 5-FOA plates. The bottom three rows of the panel show the corresponding yhc1-[L13] alleles used in the test. Designations on a white background represent alleles that are known to bypass the prp28Δ mutation; designations on a gray background represent wild-type YHC1 (L13) and transformation with empty plasmid (yhc1Δ); designations on a black background represent alleles that cannot bypass the prp28Δ mutation. (B) Deletion of YNL187 suppresses hyperstabilization of the U1 snRNA/5′ss interaction. Growth at 25°C in the presence of 0.5 mM copper (Cu++) is shown for yeast containing an ACT1-CUP1 fusion reporter having a wild-type 5′ss (6 bp) or a mutated 5′ss (10 bp) that forms 10 contiguous base pairs with U1 snRNA in the presence (left, top panel) or absence (left, bottom panel) of Ynl187p. Growth at higher copper concentrations reflects greater splicing efficiency. (C) Synthetic lethality relationship between the presence of the ynl187Δ mutation and loss of function of Cbp80p. Relevant genotypes for the CBP80 alleles are indicated in the left panel. In that panel, the chromosomal wild type of each gene is indicated by “+” and deletion is indicated by “Δ.” (D) Alteration of Cbp80p can also bypass the prp28Δ mutation. Two alleles of CBP80 in which cbp80-101 carries a point mutation were tested.
A typical U1/5′ss RNA duplex consists of 6 bp, and extension of this duplex to 10 bp is detrimental to splicing and inhibitory to cell growth (63). Our model predicts that loss of Ynl187p might suppress the growth defect caused by hyperstabilizing the U1/5′ss RNA duplex (Fig. 4B). This was tested by constructing a ynl187Δ tester strain that allowed the 10-bp U1/5′ss RNA duplex to form (63). This strain was transformed with either an empty plasmid or a plasmid containing YNL187, and splicing of a CUP1 reporter gene capable of forming a 6- or 10-bp RNA duplex was monitored. Consistent with our hypothesis, loss of Ynl187p suppressed the effect of U1/5′ss hyperstabilization (Fig. 4B).
The third line of evidence came from an open-ended genetic screening for mutation(s) that (when in combination with the ynl187Δ mutation) caused lethality (see Materials and Methods). This search identified a single recessive mutation located within CBP80, which encodes the large subunit of the nuclear CBC (28, 39). CBC is known to facilitate U1 snRNP binding to the cap-proximal 5′ss and to associate with pre-mRNA throughout the splicing cycle (8, 40). Sequence analysis of the mutant cbp80-101 allele revealed a C-to-T substitution at codon 382 and an A insertion in codon 381 (GAT to GAAT), the latter of which creates a reading frameshift mutation that results in a truncated gene product 393 amino acids in length (Cbp80p has 861 amino acids). These data therefore suggest a loss-of-function model for the observed synthetic lethality. Indeed, a cbp80Δ ynl187Δ double mutant strain was inviable (Fig. 4C). This finding prompted us to test whether the cbp80Δ mutation was also able to bypass the requirement for Prp28p, which proved to be the case (Fig. 4D).
Ynl187p promotes the formation of stable CCs.
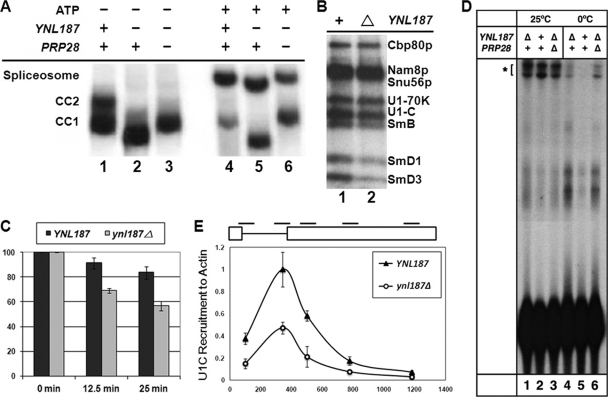
Our genetic data thus far paint a highly consistent picture of the role of Ynl187p in promoting the U1 snRNP/5′ss interaction. To probe this issue further, we first examined the formation of the CCs, which represent the initial binding of U1 snRNP to the pre-mRNA and can be accumulated by leaving ATP out of in vitro splicing reactions. Radioactively labeled pre-mRNA was incubated with wild-type, ynl187Δ, and ynl187Δ prp28Δ splicing extracts to form CCs, which were resolved by native-gel electrophoresis. A complex with increased mobility compared to that of CC1 and CC2 (Fig. 5A, compare lanes 1 and 2) was detected in the ynl187Δ reaction. This phenotype is reminiscent of the faster-migrating CCs that are seen when U1A/Mud1p (42) or Mud2p (1) is lost, but it is opposite to the result of loss of Cbp20p/Mud13p, which causes a decrease of CC mobility (8). We interpreted the observed faster-migrating phenotype as reflecting a change of conformation or composition of CCs in the absence of Ynl187p. Intriguingly, simultaneous loss of Ynl187p and Prp28p yielded a band comigrating with CC1 (lane 3). Likewise, when assayed in the presence of ATP to convert CCs to spliceosome, loss of Ynl187p yielded a spliceosome-like complex that migrated faster than the wild-type spliceosome (lanes 4 and 5), and this phenotype of faster migration was restored by a concurrent loss of Prp28p (lane 6). One possible explanation for these data (lanes 2 and 5) is that, in the absence of the opposing Ynl187p, Prp28p may alter the conformation of splicing complexes.
FIG. 5.
Biochemical analysis confirms a role of Ynl187p in promoting U1 snRNP/5′ss interaction. (A) Altered electrophoretic mobilities of splicing complexes formed in the absence of Ynl187p. Results represent CCs (CC1 and CC2) or a spliceosome formed in the absence (−) or presence (+) of ATP, respectively. (B) Altered protein-RNA contacts in the absence (Δ) of Ynl187p as detected by 4-thioU cross-linking assays. Identities of the cross-linked proteins are indicated. (C) Reduced stability of the RNA-protein complexes formed in the absence of Ynl187p. CC formation and chase assays were done as described in Materials and Methods. Complex stability was measured by use of a liquid scintillation counter after IP with an antibody against Prp40p. The value obtained after a 20-min incubation without unlabeled RNA was arbitrarily set to 100. Error bars representing the results of three experiments are shown. (D) Stability of the U1 snRNA-5′ss base pairing at two different temperatures. 32P-labeled transcripts were incubated with splicing extracts containing (+) or lacking (Δ) either or both of Ynl187p and Prp28p in standard CC reactions for 20 min at either 25°C (lanes 1 to 3) or 0°C (lanes 4 to 6). The psoralen-cross-linked species are indicated (*). (E) The ynl187Δ mutation severely reduces U1 snRNP's chromatin association. Five pairs of oligonucleotides were used to amplify different regions (short lines at the top of the top panel) of the ACT1 gene containing two exons (boxes, top panel) and an intron (connecting thin line, top panel). All data were normalized to the signal of the second oligonucleotide pair from experiments using the wild-type (YNL187) strain. The x axis of the bottom panel represents the distance in base pairs from the ATG start codon.
To inspect RNA-protein contacts within CCs, we employed a 4-thioU-labeled pre-mRNA substrate in splicing reactions and UV cross-linking (77). In the absence of Ynl187p, the levels of the cross-linked SmB, SmD1, and SmD3 were reduced (Fig. 5B, lane 2), which is consistent with the occurrence of a conformational change or structural alteration. This result fits well with the previously reported two-hybrid interaction between Ynl187p and SmB (17) and the known role of SmB, SmD1, and SmD3 in promoting the U1 snRNP/pre-mRNA interaction (76). To assess the stability of CCs, a 400-fold excess of cold pre-mRNA was added for chase experiments after an initial incubation of the hot pre-mRNA with the splicing extract (Fig. 5C). During the chase, the amount of pre-mRNA associated with the U1 snRNP was determined by IP using an anti-Prp40p antibody. The wild-type complexes were stable, with >80% recoverable counts after a 25-min chase experiment (Fig. 5C). However, in the absence of Ynl187p, the recoverable counts dropped markedly to ∼60% after a 25-min chase experiment. This indicates that the CC is indeed less stable in the absence of Ynl187p.
Recent studies of the U1C-[L13S] bypass mutation (13) showed that it decreases stable base pairing between U1 snRNA and the pre-mRNA substrate at room temperature but permits base pairing at 0°C. We used the same psoralen cross-linking assay (11, 13) to examine the effect of Ynl187p loss. At room temperature, we observed a modest reduction of cross-linking with the ynl187Δ (∼81% remaining) and ynl187Δ prp28Δ (∼78%) strains, suggesting some contribution of Ynl187p to in vitro CC formation under these standard conditions (Fig. 5D, lanes 1 to 3). At 0°C, the overall cross-linking efficiency was reduced as expected, but cross-linking was dramatically increased in ynl187Δ (5.6×) and ynl187Δ prp28Δ (8.8×) extracts compared to the wild-type extract (1×) (Fig. 5D, lanes 4 to 6). This result is highly similar to the recently reported U1C-[L13S] data (13) (see Discussion).
Because transcription and splicing are linked in vivo, we wondered whether loss of Ynl187p would also affect the association of U1 snRNP with chromatin (19, 37). To this end, we adapted a modified ChIP protocol (37) designed to measure chromatin association of the U1C protein, which represents U1 snRNP, with ACT1, an endogenous intron-containing gene. TAP-tagged U1C and formaldehyde-cross-linked chromatin were immunoprecipitated, and DNA was analyzed by quantitative real-time PCR using five sets of primers directed to different regions of the ACT1 gene (Fig. 5E).
U1C levels peaked close to the end of the intron and then decreased substantially at the beginning of exon 2 and continued to decrease toward the end of exon 2, which was a result essentially identical to what had been previously reported for U1 snRNP recruitment (19, 37). Importantly, the ynl187Δ mutation reduced U1C association to ∼50% of the wild-type level (Fig. 5E). This reduction of U1C association in the presence of the ynl187Δ mutation was unlikely to have been caused by a decrease in transcription, because there was no detectable reduction in the levels of the ACT1 transcript (data not shown). Taken together, these biochemical data validated our genetic prediction that Ynl187p has a role in promoting the stable interaction between U1 snRNP and the 5′ss.
Ynl187p is unlikely to play a significant role in U1 snRNP assembly.
An alternative explanation of the data is based on the observed synthetic lethality relationship between the ynl187Δ mutation and the prp40-nes1 allele, which encodes an altered form of Prp40p with a deletion of one of its two putative NESs (51). The observations that NESs in Prp40p are functional and that Ynl187p also contains a putative NES have led to the speculation that Prp40p and Ynl187p work in concert to export U1 snRNA to the cytoplasm (51). This suggests that Ynl187p functions in some aspect of U1 snRNP assembly.
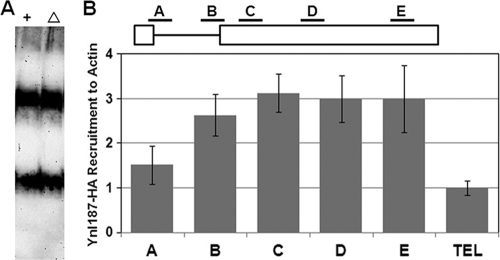
To evaluate this possibility, we first used native gel electrophoresis coupled with Northern analysis to examine U1 snRNP mobility in the presence or absence of Ynl187p. Two major forms of U1 snRNP were detected (Fig. 6A), a result that is consistent with previous findings (20, 47). However, no Ynl187p-dependent alterations were visible; there were no changes in abundance or in mobility (compare lanes 1 and 2). This is in sharp contrast to the effects of a loss of Prp42p, Nam8p/Mud15p, or U1A/Mud1p (20, 47): there were significant alterations of gel migration patterns (20, 47) or loss of the fully assembled U1 snRNP (top band, Fig. 6A) (20, 47).
FIG. 6.
Ynl187p is unlikely to play a significant role in U1 snRNP assembly. (A) Deletion of YNL187 does not affect U1 snRNP mobility on a native gel. Splicing extracts made from wild-type (+) and ynl187 deletion (Δ) strains were electrophoresed on a native polyacrylamide (3%)-agarose (0.5%) gel. The RNAs were transferred to a membrane and probed by a digoxigenin-labeled U1 probe. No apparent differences were detected for the two major forms of the U1 snRNP. (B) Ynl187p is associated with the entire ACT1 gene. Ynl187-HA was immunoprecipitated by anti-HA antibody, and the associated chromatin was amplified by five oligonucleotide pairs (A to E) (see also Fig. 5E). All data were normalized against the value obtained from measurement of a transcriptionally repressed region in the chromosome VI telomere (TEL).
If Ynl187p has a direct role in splicing, it may be closely associated with chromatin. We therefore examined the chromatin association of HA-tagged Ynl187p with the ACT1 gene. Like U1 snRNP (Fig. 5E), Ynl187p is significantly associated with ACT1 by the end of its intron (primer set B) (Fig. 6B). Unlike U1 snRNP, however, Yn1187p remains present at a high level throughout exon 2 (primer sets C, D, and E) (Fig. 6B). Values are much higher than those seen with the negative control, which measured the association of Ynl187p with a transcriptionally repressed telomeric region. The chromatin-associated behavior of Ynl187p is also distinct from the reported pattern of the U2 snRNP, which first peaks with primer set C and then drops to a low level with primer sets D and E (37). Notably, the sustained association of Ynl187p throughout the entire ACT1 gene may provide a possible explanation for its apparent association with all five spliceosomal snRNPs (Fig. 3B). Taken together, with all the genetic and biochemical data presented above, the ChIP results indicate that Ynl187p plays a direct role in U1 snRNP's stable association with splicing complexes rather than in U1 snRNP assembly (see Discussion).
DISCUSSION
Although mammalian U1 snRNP is dispensable for in vitro splicing under conditions of excess SR proteins (9, 68), the base pairing between U1 snRNA and 5′ss alone is generally important for splicing under normal conditions (59, 62, 78). In the yeast system, identification of eight proteins cross-linked to the 5′ss region (57, 77) led to a proposal that these proteins may complement the energetic contribution from the formation of the 5- to 7-bp RNA duplex between pre-mRNA and U1 snRNA (77). Supporting evidence for this notion is available for U1C (7, 11, 13, 24), SmB, SmD1, SmD3 (76), and Cbp80p (15, 39, 40). To make the essential U1/U6 switch at the 5′ss during spliceosome assembly, it follows that it might be far more important to reverse protein-RNA contacts than to unwind the half-turn of RNA duplex, which can disassociate without extensive unwinding in the absence of protein cofactors (77). This hypothesis was validated in part by our previous finding that Prp28p, an otherwise essential DEXD/H-box splicing factor required for the switch of U6 for U1 at the 5′ss (63), can be eliminated by altering the U1C protein (7). Thus, U1C is likely to be one of the potential targets of Prp28p.
In light of the dense protein contacts at the 5′ss, it seems reasonable that Prp28p may need to counteract other proteins as well. We approached this issue by exploiting a bypass strategy to search for mutations that render Prp28p dispensable. This strategy identified four new potential targets of Prp28p, i.e., Prp42p, Snu71p, Cbp80p, and Ynl187p. Interestingly, three of the five total targets are components of U1 snRNP: U1C, Prp42p, and Snu71p (20). The remaining two, Cbp80p and Ynl187p, are functionally related to U1 snRNP. Although it is plausible that another class of proteins might be identified by this approach, the striking bias of our findings argues that the sole function of Prp28p is to promote the destabilization of the U1 snRNP/5′ss interaction.
Detailed mapping analysis placed the contacts of SmD1 and SmD3 within the 5′ exon, SmB at the +6 position of the intron, and Nam8p at a more distal location (between +18 and +30 and between +40 and +46) (77). Furthermore, the highly charged C-terminal tails of SmB, SmD1, and SmD3 were shown to directly contact pre-mRNA and have been suggested to function at least in part to stabilize the U1 snRNP/pre-mRNA interaction (76). It is therefore surprising that the removal of the C-terminal tail of the SmB, SmD1, or SmD3 or elimination of Nam8p failed to bypass Prp28p (Fig. 2B). Considering that U1C was also most extensively cross-linked to the intron +6 position (77) and that it can select the splice-site-like GUAU sequence in vitro (11), we anticipated that C-terminal truncation of SmB would also bypass Prp28p, assuming that the key determining factor lies in the contact at the 5′ss (e.g., at the +6 position). If so, one would predict that Prp42p and Snu71p might also come close to or contact the same region. This line of reasoning, however, is complicated by the fact that the removal of Cbp80p, which cross-links to the cap-proximal location (77), can also bypass Prp28p (Fig. 4D). Detailed structural information concerning wild-type and mutant yeast U1 snRNP, in similarity to the now-available human U1 snRNP crystal structure information (56), will be needed to fully explain these data.
It seems unlikely that Prp28p makes simultaneous contacts with U1C, Prp42p, Snu71p, and Cbp80p to “remodel” the U1 snRNP. A more plausible scenario would be that Prp28p initially contacts one or two of these proteins, thereby activating its intrinsic ATPase activity and causing major conformational changes of the 5′ss-bound U1 snRNP. Alternatively, Prp28p may track the single-stranded region of the pre-mRNA to capture nucleotides that are normally part of the U1/5′ss RNA duplex, which is almost certainly covered by proteins. It may be through this progressive “prying” motion that the propensity of the protein(s) to dissociate from the RNA is markedly increased and a large-scale RNP remodeling is ultimately achieved. However, this “RNA-contact” model (29), proposed on the basis of studying several in vitro model systems (6, 30, 31, 35, 72, 73), remains to be rigorously tested in vivo. Regardless of which model turns out to be correct, it is essential to determine when and where Prp28p touches the RNP proper, because proteins are indispensable to the U1 snRNP/5′ss interaction (77). Cross-linking experiments designed to capture the contact points of Prp28p in a splicing-dependent manner may help to resolve such issues concerning these and other competing models.
Ynl187p is unique among potential targets of Prp28p. Like Cbp80p, it is not a U1 snRNP-specific protein, although it was initially suspected to be involved in splicing, owing to its interaction with SmB as uncovered in a large-scale two-hybrid screening (17). Unlike Cbp80p, however, Ynl187p was not cross-linked to the pre-mRNA or otherwise identified as a CC component (77). So, what role could it play in splicing? In this study, we demonstrated that a loss of Ynl187p consistently compromises the U1 snRNP/5′ss interaction both in vivo and in vitro. First, the ynl187Δ mutation (i) abolishes the need for Prp28p (Fig. 2A); (ii) rescues the splicing defect caused by extending U1 snRNA/5′ss base pairing (Fig. 4B); and (iii) causes cell death when combined with U1C-[L13] bypass alleles (Fig. 4A) or with the cbp80Δ mutation (Fig. 4C). Second, the presence of a ynl187Δ mutation results in increased mobility of CCs and spliceosomes (Fig. 5A), diminished CC1 stability (Fig. 5C), decreased contacts between SmB, SmD1, and SmD3 and the 5′ss region (Fig. 5B), and prominent reduction of the cotranscriptional recruitment of U1 snRNP to the ACT1 transcript (Fig. 5E). Third, ChIP analysis also showed that Ynl187p is closely associated with the intron-containing ACT1 gene throughout its entire length (Fig. 6B). Taken together, these data suggest a working model in which Ynl187p is cotranscriptionally recruited to and remains present in the nascent mRNP throughout the splicing process. Perhaps Ynl187p, despite its low abundance in the cell, functions as an “adhesive” molecule to help in holding stably together the overall structures of CCs and the spliceosome. In this scenario, Ynl187p would be expected to associate with splicing complexes at various stages. Although this idea is speculative, it might explain why Ynl187p is found to be associated with U1, U2, U4, U5, and U6 snRNPs (Fig. 3B).
It has been proposed that Prp40p and Ynl187p work in concert and participate in U1 snRNP biogenesis to export U1 snRNA to the cytoplasm (51). If true, snRNP biogenesis in yeast may have a cytoplasmic phase akin to that seen with metazoans, in which 5′-cap trimethylation, 3′-end trimming, and Sm ring assembly take place in the cytoplasm (71). Recent data add some support to this possibility. For example, yeast snRNAs were found to cycle between the nucleus and the cytoplasm in a heterokaryon assay (55), and tRNAs (61, 66) and several small RNAs do display unexpectedly dynamic shuttling (25). However, the cycling theory may need modifications for the budding yeast system. First, the homolog of PHAX (54), an snRNA export adaptor, has not been detected in the yeast genome. Second, S. cerevisiae does not encode homologs of SMN (14) and snurportin-1 (26), which are required for Sm ring assembly in the cytoplasm and for reimporting the partially matured snRNPs, respectively. Third, the yeast enzymatic machines for 5′-cap trimethylation (50) and 3′-end trimming (69) seem to function strictly in the nucleus. Fourth, Ynl187p is apparently present exclusively in the nucleus in the steady state (Fig. 3A) and is recruited to the entire ACT1 gene (Fig. 6B), and its absence does not detectably impact U1 snRNP assembly (Fig. 6A). Thus, even if some minor aspects of U1 snRNP assembly require Ynl187p, assembly seems most likely to take place largely if not exclusively within the confines of the nucleus. In this scenario, Ynl187p may substitute for the functions of SMN but strictly in the nucleus. Although highly speculative, this idea may also offer an explanation of why Ynl187p does not seem to have a mammalian counterpart.
Two other relevant U1 snRNP proteins, Prp42p and Snu71p, also appear to lack mammalian counterparts. Perhaps fungi need these additional proteins to adjust the stability of U1 snRNP and/or U1 snRNP/5′ss interactions, owing to the wider temperature range they may experience compared to metazoan homeotherms. This may explain why mammalian U1 snRNA/5′ss complementarity appears to exert a more dominant effect on 5′ss selection with a more limited contribution by U1 snRNP protein factors (16, 34, 58). Finally, this also offers a possible explanation for the enhanced importance of DEXD/H-box proteins, namely, to cope with this broader range of temperatures and thermal stabilities as well as with the larger number of protein cofactors that contribute to the U1 snRNP-pre-mRNA RNA-RNA base-pairing interaction.
Acknowledgments
We thank R. Pangel and S. Hamzehzadeh for assistance with the project; B. Rymond and P. Siliciano for yeast strains and antibodies; A. Hopper and E. S. Chang for help with microscopy and graphics, respectively; and V. Gopalan, P. Herman, and A. Simcox for insightful discussions.
This study was supported by the National Science Foundation (T.-H.C.), the National Science Council (97-2311-B-001-014-MY3 to T.-H.C.), the National Institutes of Health (GM23549 to M.R.), and an Academia Sinica postdoctoral fellowship (to L.T.).
Footnotes
Published ahead of print on 18 May 2009.
REFERENCES
- 1.Abovich, N., X. C. Liao, and M. Rosbash. 1994. The yeast MUD2 protein: an interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Genes Dev. 8834-854. [DOI] [PubMed] [Google Scholar]
- 2.Abovich, N., and M. Rosbash. 1997. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell 89403-412. [DOI] [PubMed] [Google Scholar]
- 3.Arning, S., P. Gruter, G. Bilbe, and A. Kramer. 1996. Mammalian splicing factor SF1 is encoded by variant cDNAs and binds to RNA. RNA 2794-810. [PMC free article] [PubMed] [Google Scholar]
- 4.Blanton, S., A. Srinivasan, and B. C. Rymond. 1992. PRP38 encodes a yeast protein required for pre-mRNA splicing and maintenance of stable U6 small nuclear RNA levels. Mol. Cell. Biol. 123939-3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bleichert, F., and S. J. Baserga. 2007. The long unwinding road of RNA helicases. Mol. Cell 27339-352. [DOI] [PubMed] [Google Scholar]
- 6.Bowers, H. A., P. A. Maroney, M. E. Fairman, B. Kastner, R. Luhrmann, T. W. Nilsen, and E. Jankowsky. 2006. Discriminatory RNP remodeling by the DEAD-box protein DED1. RNA 12903-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, J. Y., L. Stands, J. P. Staley, R. R. Jackups, L. J. Latus, and T.-H. Chang. 2001. Specific alterations of U1-C protein or U1 small nuclear RNA can eliminate the requirement of Prp28p, an essential DEAD box splicing factor. Mol. Cell 7227-232. [DOI] [PubMed] [Google Scholar]
- 8.Colot, H. V., F. Stutz, and M. Rosbash. 1996. The yeast splicing factor Mud13p is a commitment complex component and corresponds to CBP20, the small subunit of the nuclear cap-binding complex. Genes Dev. 101699-1708. [DOI] [PubMed] [Google Scholar]
- 9.Crispino, J. D., B. J. Blencowe, and P. A. Sharp. 1994. Complementation by SR proteins of pre-mRNA splicing reactions depleted of U1 snRNP. Science 2651866-1868. [DOI] [PubMed] [Google Scholar]
- 10.Decourty, L., C. Saveanu, K. Zemam, F. Hantraye, E. Frachon, J. C. Rousselle, M. Fromont-Racine, and A. Jacquier. 2008. Linking functionally related genes by sensitive and quantitative characterization of genetic interaction profiles. Proc. Natl. Acad. Sci. USA 1055821-5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du, H., and M. Rosbash. 2002. The U1 snRNP protein U1C recognizes the 5′ splice site in the absence of base pairing. Nature 41986-90. [DOI] [PubMed] [Google Scholar]
- 12.Du, H., and M. Rosbash. 2001. Yeast U1 snRNP-pre-mRNA complex formation without U1snRNA-pre-mRNA base pairing. RNA 7133-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du, H., D. F. Tardiff, M. J. Moore, and M. Rosbash. 2004. Effects of the U1C L13 mutation and temperature regulation of yeast commitment complex formation. Proc. Natl. Acad. Sci. USA 10114841-14846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer, U., Q. Liu, and G. Dreyfuss. 1997. The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell 901023-1029. [DOI] [PubMed] [Google Scholar]
- 15.Fortes, P., J. Kufel, M. Fornerod, M. Polycarpou-Schwarz, D. Lafontaine, D. Tollervey, and I. W. Mattaj. 1999. Genetic and physical interactions involving the yeast nuclear cap-binding complex. Mol. Cell. Biol. 196543-6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freund, M., M. J. Hicks, C. Konermann, M. Otte, K. J. Hertel, and H. Schaal. 2005. Extended base pair complementarity between U1 snRNA and the 5′ splice site does not inhibit splicing in higher eukaryotes, but rather increases 5′ splice site recognition. Nucleic Acids Res. 335112-5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fromont-Racine, M., A. E. Mayes, A. Brunet-Simon, J. C. Rain, A. Colley, I. Dix, L. Decourty, N. Joly, F. Ricard, J. D. Beggs, and P. Legrain. 2000. Genome-wide protein interaction screens reveal functional networks involving Sm-like proteins. Yeast 1795-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorbalenya, A. E., and E. V. Koonin. 1993. Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 3419-429. [Google Scholar]
- 19.Görnemann, J., K. M. Kotovic, K. Hujer, and K. M. Neugebauer. 2005. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol. Cell 1953-63. [DOI] [PubMed] [Google Scholar]
- 20.Gottschalk, A., J. Tang, O. Puig, J. Salgado, G. Neubauer, H. V. Colot, M. Mann, B. Seraphin, M. Rosbash, R. Luhrmann, and P. Fabrizio. 1998. A comprehensive biochemical and genetic analysis of the yeast U1 snRNP reveals five novel proteins. RNA 4374-393. [PMC free article] [PubMed] [Google Scholar]
- 21.Gutell, R. R., N. Larsen, and C. R. Woese. 1994. Lessons from an evolving rRNA: 16S and 23S rRNA structures from a comparative perspective. Microbiol. Mol. Biol. Rev. 5810-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guthrie, C., and G. R. Fink. 1991. Methods in enzymology, vol. 194. Guide to yeast genetics and molecular biology. Academic Press, San Diego, CA. [PubMed]
- 23.Hausmann, S., S. Zheng, M. Costanzo, R. L. Brost, D. Garcin, C. Boone, S. Shuman, and B. Schwer. 2008. Genetic and biochemical analysis of yeast and human cap trimethylguanosine synthase: functional overlap of 2,2,7-trimethylguanosine caps, small nuclear ribonucleoprotein components, pre-mRNA splicing factors, and RNA decay pathways. J. Biol. Chem. 28331706-31718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heinrichs, V., M. Bach, G. Winkelmann, and R. Luhrmann. 1990. U1-specific protein C needed for efficient complex formation of U1 snRNP with 5′ splice site. Science 24769-72. [DOI] [PubMed] [Google Scholar]
- 25.Hopper, A. K. 2006. Cellular dynamics of small RNAs. Crit. Rev. Biochem. Mol. Biol. 413-19. [DOI] [PubMed] [Google Scholar]
- 26.Huber, J., U. Cronshagen, M. Kadokura, C. Marshallsay, T. Wada, M. Sekine, and R. Luhrmann. 1998. Snurportin1, an m3G-cap-specific nuclear import receptor with a novel domain structure. EMBO J. 174114-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson, J. S. Weissman, and E. K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature 425686-691. [DOI] [PubMed] [Google Scholar]
- 28.Izaurralde, E., J. Lewis, C. McGuigan, M. Jankowska, E. Darzynkiewicz, and I. W. Mattaj. 1994. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell 78657-668. [DOI] [PubMed] [Google Scholar]
- 29.Jankowsky, E., and H. Bowers. 2006. Remodeling of ribonucleoprotein complexes with DExH/D RNA helicases. Nucleic Acids Res. 344181-4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jankowsky, E., C. H. Gross, S. Shuman, and A. M. Pyle. 2001. Active disruption of an RNA-protein interaction by a DExH/D RNA helicase. Science 291121-125. [DOI] [PubMed] [Google Scholar]
- 31.Jankowsky, E., C. H. Gross, S. Shuman, and A. M. Pyle. 2000. The DExH protein NPH-II is a processive and directional motor for unwinding RNA. Nature 403447-451. [DOI] [PubMed] [Google Scholar]
- 32.Jurica, M. S., and M. J. Moore. 2003. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell 125-14. [DOI] [PubMed] [Google Scholar]
- 33.Kambach, C., S. Walke, R. Young, J. M. Avis, E. de la Fortelle, V. A. Raker, R. Luhrmann, J. Li, and K. Nagai. 1999. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell 96375-387. [DOI] [PubMed] [Google Scholar]
- 34.Kammler, S., C. Leurs, M. Freund, J. Krummheuer, K. Seidel, T. O. Tange, M. K. Lund, J. Kjems, A. Scheid, and H. Schaal. 2001. The sequence complementarity between HIV-1 5′ splice site SD4 and U1 snRNA determines the steady-state level of an unstable env pre-mRNA. RNA 7421-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kawaoka, J., E. Jankowsky, and A. M. Pyle. 2004. Backbone tracking by the SF2 helicase NPH-II. Nat. Struct. Mol. Biol. 11526-530. [DOI] [PubMed] [Google Scholar]
- 36.Klug, A., and J. W. Schwabe. 1995. Protein motifs 5. Zinc fingers. FASEB J. 9597-604. [PubMed] [Google Scholar]
- 37.Lacadie, S. A., and M. Rosbash. 2005. Cotranscriptional spliceosome assembly dynamics and the role of U1 snRNA:5′ss base pairing in yeast. Mol. Cell 1965-75. [DOI] [PubMed] [Google Scholar]
- 38.Legrain, P., B. Seraphin, and M. Rosbash. 1988. Early commitment of yeast pre-mRNA to the spliceosome pathway. Mol. Cell. Biol. 83755-3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lewis, J. D., D. Gorlich, and I. W. Mattaj. 1996. A yeast cap binding protein complex (yCBC) acts at an early step in pre-mRNA splicing. Nucleic Acids Res. 243332-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lewis, J. D., E. Izaurralde, A. Jarmolowski, C. McGuigan, and I. W. Mattaj. 1996. A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5′ splice site. Genes Dev. 101683-1698. [DOI] [PubMed] [Google Scholar]
- 41.Li, J. M., A. K. Hopper, and N. C. Martin. 1989. N2,N2-dimethylguanosine-specific tRNA methyltransferase contains both nuclear and mitochondrial targeting signals in Saccharomyces cerevisiae. J. Cell Biol. 1091411-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liao, X. C., J. Tang, and M. Rosbash. 1993. An enhancer screen identifies a gene that encodes the yeast U1 snRNP A protein: implications for snRNP protein function in pre-mRNA splicing. Genes Dev. 7419-428. [DOI] [PubMed] [Google Scholar]
- 43.Linder, P. 2006. Dead-box proteins: a family affair—active and passive players in RNP-remodeling. Nucleic Acids Res. 344168-4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Linder, P., P. F. Lasko, M. Ashburner, P. Leroy, P. J. Nielsen, K. Nishi, J. Schnier, and P. P. Slonimski. 1989. Birth of the D-E-A-D box. Nature 337121-122. [DOI] [PubMed] [Google Scholar]
- 45.Lorsch, J. R., and D. Herschlag. 1998. The DEAD box protein eIF4A. 1. A minimal kinetic and thermodynamic framework reveals coupled binding of RNA and nucleotide. Biochemistry 372180-2193. [DOI] [PubMed] [Google Scholar]
- 46.Lorsch, J. R., and D. Herschlag. 1998. The DEAD box protein eIF4A. 2. A cycle of nucleotide and RNA-dependent conformational changes. Biochemistry 372194-2206. [DOI] [PubMed] [Google Scholar]
- 47.McLean, M. R., and B. C. Rymond. 1998. Yeast pre-mRNA splicing requires a pair of U1 snRNP-associated tetratricopeptide repeat proteins. Mol. Cell. Biol. 18353-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Michaud, S., and R. Reed. 1993. A functional association between the 5′ and 3′ splice site is established in the earliest prespliceosome complex (E) in mammals. Genes Dev. 71008-1020. [DOI] [PubMed] [Google Scholar]
- 49.Michaud, S., and R. Reed. 1991. An ATP-independent complex commits pre-mRNA to the mammalian spliceosome assembly pathway. Genes Dev. 52534-2546. [DOI] [PubMed] [Google Scholar]
- 50.Mouaikel, J., C. Verheggen, E. Bertrand, J. Tazi, and R. Bordonne. 2002. Hypermethylation of the cap structure of both yeast snRNAs and snoRNAs requires a conserved methyltransferase that is localized to the nucleolus. Mol. Cell 9891-901. [DOI] [PubMed] [Google Scholar]
- 51.Murphy, M. W., B. L. Olson, and P. G. Siliciano. 2004. The yeast splicing factor Prp40p contains functional leucine-rich nuclear export signals that are essential for splicing. Genetics 16653-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nagalakshmi, U., Z. Wang, K. Waern, C. Shou, D. Raha, M. Gerstein, and M. Snyder. 2008. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 3201344-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nilsen, T. W. 1994. RNA-RNA interactions in the spliceosome: unraveling the ties that bind. Cell 781-4. [DOI] [PubMed] [Google Scholar]
- 54.Ohno, M., A. Segref, A. Bachi, M. Wilm, and I. W. Mattaj. 2000. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell 101187-198. [DOI] [PubMed] [Google Scholar]
- 55.Olson, B. L., and P. G. Siliciano. 2003. A diverse set of nuclear RNAs transfer between nuclei of yeast heterokaryons. Yeast 20893-903. [DOI] [PubMed] [Google Scholar]
- 56.Pomeranz Krummel, D. A., C. Oubridge, A. K. Leung, J. Li, and K. Nagai. 2009. Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature 458475-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Puig, O., A. Gottschalk, P. Fabrizio, and B. Seraphin. 1999. Interaction of the U1 snRNP with nonconserved intronic sequences affects 5′ splice site selection. Genes Dev. 13569-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roca, X., R. Sachidanandam, and A. R. Krainer. 2005. Determinants of the inherent strength of human 5′ splice sites. RNA 11683-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Séraphin, B., L. Kretzner, and M. Rosbash. 1988. A U1 snRNA:pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5′ cleavage site. EMBO J. 72533-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seraphin, B., and M. Rosbash. 1989. Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosome assembly and splicing. Cell 59349-358. [DOI] [PubMed] [Google Scholar]
- 61.Shaheen, H. H., and A. K. Hopper. 2005. Retrograde movement of tRNAs from the cytoplasm to the nucleus in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 10211290-11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Siliciano, P. G., and C. Guthrie. 1988. 5′ Splice site selection in yeast: genetic alterations in base-pairing with U1 reveal additional requirement. Genes Dev. 21258-1267. [DOI] [PubMed] [Google Scholar]
- 63.Staley, J. P., and C. Guthrie. 1999. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol. Cell 355-64. [DOI] [PubMed] [Google Scholar]
- 64.Staley, J. P., and C. Guthrie. 1998. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92315-326. [DOI] [PubMed] [Google Scholar]
- 65.Stevens, S. W., and J. Abelson. 2002. Yeast pre-mRNA splicing: methods, mechanisms, and machinery. Methods Enzymol. 351200-220. [DOI] [PubMed] [Google Scholar]
- 66.Takano, A., T. Endo, and T. Yoshihisa. 2005. tRNA actively shuttles between the nucleus and cytosol in yeast. Science 309140-142. [DOI] [PubMed] [Google Scholar]
- 67.Tanner, N. K., and P. Linder. 2001. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell 8251-262. [DOI] [PubMed] [Google Scholar]
- 68.Tarn, W.-Y., and J. A. Steitz. 1994. SR proteins can compensate for the loss of U1 snRNP functions in vitro. Genes Dev. 82704-2717. [DOI] [PubMed] [Google Scholar]
- 69.van Hoof, A., P. Lennertz, and R. Parker. 2000. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol. 20441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wahl, M. C., C. L. Will, and R. Luhrmann. 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136701-718. [DOI] [PubMed] [Google Scholar]
- 71.Will, C. L., and R. Luhrmann. 2001. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 13290-301. [DOI] [PubMed] [Google Scholar]
- 72.Yang, Q., M. Del Campo, A. M. Lambowitz, and E. Jankowsky. 2007. DEAD-box proteins unwind duplexes by local strand separation. Mol. Cell 28253-263. [DOI] [PubMed] [Google Scholar]
- 73.Yang, Q., and E. Jankowsky. 2006. The DEAD-box protein Ded1 unwinds RNA duplexes by a mode distinct from translocating helicases. Nat. Struct. Mol. Biol. 13981-986. [DOI] [PubMed] [Google Scholar]
- 74.Yu, Y., P. A. Maroney, J. A. Denker, X. H. Zhang, O. Dybkov, R. Luhrmann, E. Jankowsky, L. A. Chasin, and T. W. Nilsen. 2008. Dynamic regulation of alternative splicing by silencers that modulate 5′ splice site competition. Cell 1351224-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zamore, P. D., J. G. Patton, and M. R. Green. 1992. Cloning and domain structure of the mammalian splicing factor U2AF. Nature 355609-614. [DOI] [PubMed] [Google Scholar]
- 76.Zhang, D., N. Abovich, and M. Rosbash. 2001. A biochemical function for the Sm complex. Mol. Cell 7319-329. [DOI] [PubMed] [Google Scholar]
- 77.Zhang, D., and M. Rosbash. 1999. Identification of eight proteins that cross-link to pre-mRNA in the yeast commitment complex. Genes Dev. 13581-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhuang, Y., and A. M. Weiner. 1986. A compensatory base change in U1 snRNA suppresses a 5′ splice site mutation. Cell 46827-835. [DOI] [PubMed] [Google Scholar]