Abstract
Mcl-1 is a member of the Bcl2-related protein family that is a critical mediator of cell survival. Exposure of cells to stress causes inhibition of Mcl-1 mRNA translation and rapid destruction of Mcl-1 protein by proteasomal degradation mediated by a phosphodegron created by glycogen synthase kinase 3 (GSK3) phosphorylation of Mcl-1. Here we demonstrate that prior phosphorylation of Mcl-1 by the c-Jun N-terminal protein kinase (JNK) is essential for Mcl-1 phosphorylation by GSK3. Stress-induced Mcl-1 degradation therefore requires the coordinated activity of JNK and GSK3. Together, these data establish that Mcl-1 functions as a site of signal integration between the proapoptotic activity of JNK and the prosurvival activity of the AKT pathway that inhibits GSK3.
Mcl-1 is an antiapoptotic member of the Bcl2 family. Gene knockout studies of mice demonstrate that Mcl-1 is essential for embryonic development and for the survival of hematopoietic cells (28-30). Studies of the stress response have demonstrated that Mcl-1 plays an important role in the sensitization of cells to apoptotic signals (1, 11, 25). Thus, exposure to UV radiation causes the rapid degradation of Mcl-1 and the release of proapoptotic partner proteins from Mcl-1 complexes (e.g., Bim). The mechanism of rapid Mcl-1 destruction is mediated by the combined actions of two different pathways. First, the exposure to stress causes phosphorylation of the alpha subunit of eukaryotic initiation factor 2 (eIF-2α) on the inhibitory site Ser-51 that prevents translation of Mcl-1 mRNA (1, 11, 25). Second, Mcl-1 is rapidly degraded by the ubiquitin-dependent proteasome pathway (27). Together, these pathways cause a rapid reduction in Mcl-1 expression. This loss of Mcl-1 may be a required initial response for the apoptosis of cells exposed to stress (25).
The E3 ubiquitin protein ligase Mule/ARF-BP1 contains a BH3 domain that interacts with Mcl-1 and can initiate ubiquitin-dependent degradation of Mcl-1 (39). Recent studies have demonstrated that rapid stress-induced degradation of Mcl-1 is mediated by an alternative pathway involving the E3 ubiquitin protein ligase β-TrCP, which binds a stress-induced phosphodegron created by the phosphorylation of Mcl-1 by glycogen synthase kinase 3 (GSK3) (7, 21). How the exposure to stress causes GSK3-mediated phosphorylation of Mcl-1 is unclear, but GSK3 has been shown to directly phosphorylate Mcl-1 (7, 21). Mcl-1 phosphorylation and degradation may therefore be controlled by the prosurvival AKT pathway, which can negatively regulate GSK3 (7, 21).
Mcl-1 is critically involved in the regulation of cell survival and is therefore subject to regulation by multiple mechanisms (26). Thus, Mcl-1 gene expression is regulated by many growth factors and cytokines (26), and Mcl-1 mRNA is regulated by microRNA pathways (24). The Mcl-1 protein is stabilized by binding TCTP (20) and the BH3-only protein Bim (4). In contrast, the BH3-only protein Noxa binds and destabilizes Mcl-1 (4, 36). Moreover, it is established that Mcl-1 is phosphorylated by several protein kinases on sites that may regulate Mcl-1 function. Phosphorylation of human Mcl-1 (hMcl-1) on Ser-64 (a site that is not conserved in other species) may enhance antiapoptotic activity by increasing the interaction of Mcl-1 with Bim, Noxa, and Bak (18). Phosphorylation on Ser-121 and Thr-163 may inhibit the antiapoptotic activity of hMcl-1 (15), and phosphorylation on Thr-163 may increase hMcl-1 protein stability (9). The conserved GSK3 phosphorylation site Ser-159 (and possibly Ser-155) can initiate rapid proteasomal degradation of hMcl-1 (7, 21). Together, these findings suggest that the function of Mcl-1 is very tightly regulated.
The results of previous studies have implicated the c-Jun N-terminal protein kinase (JNK) in the regulation of Mcl-1 (15, 18). The purpose of this study was to test whether Mcl-1 is a target of signal transduction by JNK. We demonstrate that a key function of JNK is to prime Mcl-1 for phosphorylation by GSK3. JNK is required for GSK3-mediated degradation of Mcl-1 in response to stress. Coordinated regulation of the stress-activated JNK pathway and the AKT-inhibited GSK3 pathway is therefore required for stress-induced Mcl-1 degradation.
MATERIALS AND METHODS
Reagents.
The proteasome inhibitor MG132, the JNK inhibitor TAT-JBD, and 1NM-PP1 were purchased from Calbiochem. Active recombinant GSK3β was obtained from Cell Signaling. The caspase inhibitor zVAD.fmk was obtained from R&D Systems. Bacterially expressed GST fusion proteins were purified by affinity chromatography using glutathione-Sepharose 4B (Pharmacia).
Mice.
Mice with disruptions of the Jnk1 or Jnk2 gene (10, 38) or a knock-in mutation in the Jnk2 gene (Jnk2MG) (16) have been described previously. B6;129X1-Mcl1tm3Sjk/J mice with a floxed Mcl-1 (Mcl-1LoxP) allele (29) and Gt(ROSA)26Sortm1(cre/Esr1)Nat/J mice that express a 4-hydroxytamoxifen-inducible Cre recombinase (2) were obtained from The Jackson Laboratory and backcrossed to the C57BL/6J strain.
Cell culture.
We have previously described wild-type, Jnk1−/− Jnk2−/− (17), and Jnk1−/− Jnk2MG/MG primary murine fibroblasts (16). We also prepared E13.5 primary murine fibroblasts from Mcl-1LoxP/LoxP Gt(ROSA)26Sor-CreEsr1 mice. COS7 cells were obtained from the American Type Culture Collection. These cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine (Invitrogen). Some cultures were treated with 4-hydroxytamoxifen (5). Transfection assays were performed using plasmid expression vectors and the Lipofectamine reagent (Invitrogen), and cells were harvested at 48 h posttransfection. Retroviral transduction of murine fibroblasts was performed using methods described previously (19).
Plasmids.
The mammalian expression vectors for human Mcl-1 (pcDNA3.1-hMcl-1 [wild type and Ser-159Ala]) were provided by Ulrich Maurer (21). We constructed the Thr-163Ala point mutation using the QuikChange mutagenesis kit (Stratagene) using pcDNA3.1-hMcl-1 (wild type) as a template and the primers T163A S (5′-CGG GTC ACT ACC CTC GGC GCC GCC GCC AGC-3′) and T163A AS (5′-GCT GGC GGC GGC GCC GAG GGT AGT GAC CCG-3′). The three hMcl1 cDNAs (wild type, Ser-159Ala, and Thr-163Ala) were also blunt-end cloned into the SalI site of the retroviral expression vector pBABE-IRES-Puro for transduction assays.
A bacterial expression vector (pGEX4T-1-mMcl-1) and a mammalian expression vector (pCDNA3-Flag-mMcl-1) for mouse Mcl-1 were provided by Hsin-Fang Yang-Yen (20). The Thr-144Ala, Thr-144Asp, and Ser-140Ala mutants were created using the primers T144A S (5′-GGC TCT CTG CCC AGC GCT CCG CCG CCG CCC G-3′) and T144A AS (5′-CGG GCG GCG GCG GAG CGC TGG GCA GAG AGC C-3′), T144D S (5′CGG CTC TCT GCC CTC GGA TCC GCC GCC GCC CG 3′) and T144D AS (5′CGG GCG GCG GCG GAT CCG AGG GCA GAG AGC CG 3′), S140A S (5′-CCG GGG CCG ACG GGG CCC TGC CCT CCA CGC CGC CG-3′) and S140A AS (5′-CGG CGG CGT GGA GGG CAG GGC CCC CGT CGG CCC CGG-3′). The wild-type and mutant mMcl-1 cDNAs were also blunt-end cloned into the SalI site of the retroviral expression vector pBABE-IRES-Puro for transduction assays.
Expression vectors for wild-type and K85A mutant HA-GSK3β (12) were obtained from Addgene (Addgene plasmids 14753 and 14755). The bacterial expression vector for GST-Bcl3 was provided by Alain Chariot (32).
Immunoblot analysis.
Immunoblot analysis was performed using antibodies to AKT and phospho-Ser473 AKT (Cell Signaling), GSK3β and phospho-Ser-9/21 GSK3α/β (Cell Signaling), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Santa Cruz), hMcl-1 (Pharmingen), mMcl-1 (Rockland), and α-tubulin (Covance). The pSer159 hMcl-1 antibody was provided by Ulrich Maurer (21). Immune complexes were detected using enhanced chemiluminescence with X-ray film or a Kodak 4000MM imaging station.
Mass spectroscopy.
Mcl-1 isolated by immunoprecipitation was examined after sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) by staining with Brilliant Blue G Colloidal. The Mcl-1 gel band was excised and processed for digestion with trypsin (Promega; 12.5 ng/μl in 50 mM ammonium bicarbonate, pH 8.9) or Glu-C (Sigma-Aldrich; 125 ng/μl in 100 mM ammonium bicarbonate, pH 7.8) (3). Phosphopeptides were purified using an IMAC column and eluted onto a column packed with C18 beads (23). The column was placed in-line with a tapered electrospray column packed with C18 beads on a QStar XL or QStar Elite mass spectrometer (Applied Biosystems). Peptides were eluted using a 20-min gradient (0 to 70% acetonitrile in 0.2 M acetic acid; 50 nl/min). Data were collected using the mass spectrometer in data-dependent acquisition mode to collect tandem mass spectra and examined using Mascot software (Matrix Science). Mass spectra matching phosphorylated peptides derived from Mcl-1 were examined manually to identify phosphorylation sites and fragmentation ions.
In vitro kinase assays.
Protein kinase assays using [γ-32P]ATP and protein (glutathione S-transferase [GST]-ATF2, GST-Bcl-3, GST-c-Jun, GST-Elk-1, and GST-mMcl-1) substrates were performed using standard methods with immunopurified epitope-tagged mitogen-activated protein (MAP) kinases and GSK3β isolated from transiently transfected COS7 cells (35). The phosphorylation reaction was terminated after 30 min at 30°C by the addition of Laemmli sample buffer, and the phosphorylated proteins were examined by SDS-PAGE and phosphorimager analysis.
Two-step kinase assays were performed to examine the effect of prior phosphorylation of GST-Mcl-1 by JNK on the subsequent phosphorylation of GST-mMcl-1 by GSK3β. We transiently expressed Flag-JNK1 in COS7 cells. Lysates were prepared using Triton lysis buffer (20 mM Tris [pH 7.4], 137 mM NaCl, 2 mM EDTA, 1% Triton X-100, 10% glycerol, 25 mM β-glycerophosphate, 1 mM sodium vanadate, 1 mM phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, and 10 μg/ml aprotinin). The Flag-JNK1 was immunoprecipitated using 1 μg M2 monoclonal antibody (Sigma) immobilized on 10 μl of protein G-Sepharose (Pharmacia) and washed three times with Triton lysis buffer and twice with kinase buffer (25 mM HEPES [pH 7.4], 25 mM β-glycerophosphate, 25 mM MgCl2, 2 mM dithiothreitol, and 0.1 mM sodium vanadate). Kinase assays were performed using 5 μM GST-Mcl-1 and 1 mM ATP (30 min at 30°C). Some first-step kinase assays were performed using 5 μM JNK inhibitor (TAT-JBD). The supernatant was collected, and the GST-Mcl-1 was isolated using 10 μl glutathione-Sepharose 4B (Pharmacia) at 4°C (1 h) and then washed twice with kinase buffer. The second-step protein kinase assay was then performed (30 min at 30°C) by addition of active GSK3β (500 U; Cell Signaling) and 50 μM [γ-32P]ATP (10 Ci/mmol). Phosphorylated GST-Mcl-1 was examined by SDS-PAGE, autoradiography, and phosphorimager analysis.
Apoptosis assays.
Cell death was examined by measurement of DNA fragmentation using the Cell Death Detection Elisaplus kit (Roche) according to the manufacturer's recommendations (19).
Colony assays.
Fibroblasts were exposed to UV radiation (60 J/m2) and cultured for 16 h. These cells were replated (2 × 105 cells/100-mm dish), incubated for 14 days, fixed in methanol, and stained with 0.5% crystal violet.
RESULTS
JNK is required for rapid UV-stimulated degradation of Mcl-1.
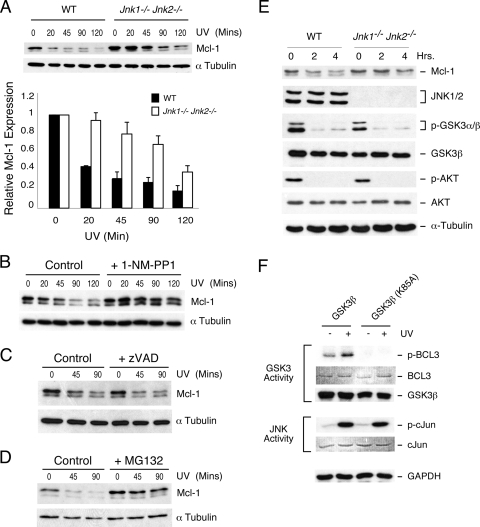
Mcl-1 is a short-lived protein that is degraded by the ubiquitin-mediated proteasome pathway (27). Indeed, the Mcl-1 protein is rapidly eliminated from cells exposed to stress, in part because of translational inhibition caused by phosphorylation of eIF-2α on the inhibitory site Ser-51 (1, 11, 25). This exposure to stress also causes activation of the JNK signal transduction pathway (6). To test whether JNK may play a role in the rapid degradation of Mcl-1 in cells exposed to stress, we examined the effect of UV radiation on Mcl-1 degradation in wild-type and Jnk1−/− Jnk2−/− fibroblasts. The JNK-deficient fibroblasts were found to exhibit reduced Mcl-1 degradation (Fig. 1A). Indeed, the half-life of Mcl-1 in wild-type fibroblasts (34 ± 6.2 min) was significantly shorter than that in JNK-deficient fibroblasts (121 ± 11 min) following exposure to UV radiation (mean ± standard deviation [SD]; n = 6; P < 0.0005). This observation suggested that JNK may contribute to Mcl-1 degradation. To confirm this finding, we employed a chemical genetic approach using fibroblasts isolated from mice with a germ line point mutation that confers sensitivity of JNK to inhibition by the drug 1NM-PP1 (16). This analysis demonstrated that JNK inhibition markedly decreased UV-stimulated degradation of Mcl-1 (Fig. 1B). We conclude the JNK activity is required for the rapid degradation of Mcl-1 caused by exposure of cells to stress.
FIG. 1.
JNK is required for stress-induced degradation of Mcl-1. (A) Wild-type (WT) and Jnk1−/− Jnk2−/− fibroblasts were exposed to UV radiation (60 J/m2). Lysates prepared from these cells at different times postirradiation were examined by immunoblot analysis by probing with antibodies to Mcl-1 and α-tubulin. The amounts of Mcl-1 and α-tubulin were quantitated using a Kodak 4000MM imaging station. The half-life of Mcl-1 in wild-type fibroblasts (34 ± 6.2 min) was significantly greater (P < 0.0005) than that in JNK-deficient fibroblasts (121 ± 11 min) (mean ± SD; n = 6). (B) Jnk1−/− Jnk2MG/MG fibroblasts were incubated without and with 10 μM 1NM-PP1 and exposed to UV radiation (60 J/m2), and the expression of Mcl-1 and α-tubulin was examined by immunoblot analysis. (C) Wild-type fibroblasts were incubated without and with the caspase inhibitor zVAD.fmk (20 μM) for 60 min and then exposed to UV radiation (60 J/m2). The amounts of Mcl-1 and α-tubulin at different times postirradiation were examined by immunoblot analysis. (D) Wild-type fibroblasts were incubated without and with the proteasome inhibitor MG132 (10 μM) for 60 min and then exposed to UV radiation (60 J/m2). The amounts of Mcl-1 and α-tubulin at different times postirradiation were examined by immunoblot analysis. (E) Wild-type and Jnk1−/− Jnk2−/− fibroblasts were transferred to serum-free medium. Lysates prepared at different times after serum withdrawal were examined by immunoblot analysis by probing with antibodies to Mcl-1, JNK1/2, phospho-Ser9/21-GSK3α/β, GSK3β, phospho-Ser473-AKT, AKT, and α-tubulin. (F) COS7 cells expressing hemagglutinin-tagged GSK3β were exposed to UV radiation (60 J/m2). Cell lysates were prepared at 40 min postirradiation and examined by immunoblot analysis using antibodies to GSK3β and GAPDH. Studies were performed using wild-type and kinase-inactive (K85A) GSK3β. JNK and GSK3β activities were measured in an immune complex kinase assay with [γ-32P]ATP using the protein substrates GST-cJun and GST-Bcl3, respectively. GST-Bcl3 and GST-cJun were stained after SDS-PAGE with Coomassie blue, and the phosphorylated proteins were detected by autoradiography.
It is established that Mcl-1 degradation can be mediated by at least two different pathways mediated by caspases (13, 22) and the proteasome (27). To test the requirement of caspases for Mcl-1 degradation, we examined the effect of treatment of cells with the broad-spectrum caspase inhibitor zVAD-fmk; no significant effect of caspase inhibition on Mcl-1 degradation was detected (Fig. 1C). In contrast, inhibition of the proteasome with the drug MG132 blocked the effect of UV radiation to decrease Mcl-1 expression (Fig. 1D). Together, these data demonstrate that UV-induced Mcl-1 degradation is mediated by the proteasome pathway.
JNK is required for Mcl-1 degradation caused by inhibition of AKT.
It is established that serum withdrawal causes degradation of Mcl-1 by an AKT/GSK3-dependent signaling pathway (7, 21). This mechanism of Mcl-1 degradation may be JNK independent. To test this hypothesis, we examined Mcl-1 expression in wild-type and Jnk1−/− Jnk2−/− fibroblasts. Serum withdrawal caused inhibition of AKT and decreased inhibitory phosphorylation of GSK3 in both wild-type and JNK-deficient fibroblasts (Fig. 1E). These changes were associated with loss of Mcl-1 protein expression in wild-type fibroblasts. However, serum withdrawal did not cause loss of Mcl-1 in JNK-deficient fibroblasts (Fig. 1E). Together, these data indicate that JNK is required for Mcl-1 degradation caused by the AKT/GSK3 pathway. This observation suggests cooperation between the JNK and AKT/GSK3 signaling pathways in the regulation of Mcl-1. This cooperative mechanism may be relevant not only to serum withdrawal but to Mcl-1 regulation by other forms of cell stress. Indeed, we found that UV radiation caused the activation of both JNK and GSK3 (Fig. 1F).
Mcl-1 is a JNK substrate.
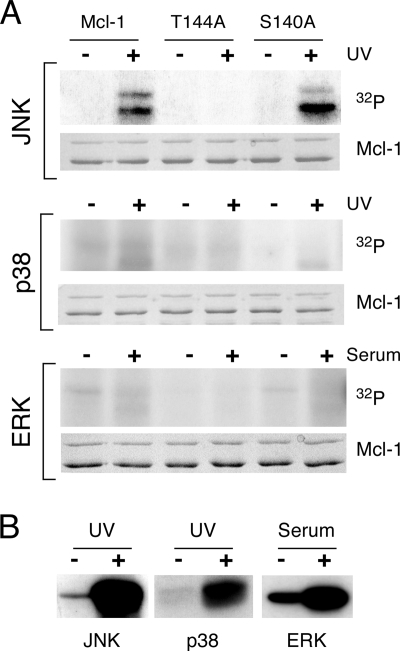
JNK phosphorylates human Mcl-1 on Ser-64, Ser-121, and Thr-163 (15, 18). We examined the phosphorylation of mouse Mcl-1 (mMcl-1) by MAP kinases in vitro (Fig. 2). mMcl-1 was phosphorylated by JNK, but little phosphorylation of mMcl-1 by extracellular signal-regulated kinase (ERK) or p38 MAP kinase was detected. Ser-64 of hMcl-1 is not conserved in the mouse (it is replaced by Glu) and is therefore not a potential mMcl-1 phosphorylation site. However, both Ser-121 and Thr-163 of hMcl-1 are conserved in mMcl-1. Mutagenesis studies demonstrated that the site corresponding to Thr-163 of hMcl-1 (Thr-144) was the major site of mMcl-1 phosphorylation by JNK, because replacement of Thr-144 with Ala strongly suppressed JNK-mediated phosphorylation of mMcl-1. Control studies demonstrated that mutation of mMcl-1 at Ser-140 caused no defect in JNK-mediated phosphorylation of mMcl-1 (Fig. 2A). Together, these data demonstrate that mMcl-1 is phosphorylated on Thr-144 by JNK.
FIG. 2.
Mcl-1 is a JNK substrate in vitro. (A) MAP kinases (JNK, p38α, and ERK2) were immunopurified from COS cells exposed to UV radiation or 10% fetal bovine serum. Protein kinase assays were performed using 50 μM [γ-32P]ATP (10 μCi/mmol) and GST-mMcl-1. The effect of replacement of Thr-144 or Ser-140 with Ala was examined. (B) Control assays were performed using JNK1, p38α MAP kinase, and ERK2 and the substrates GST-cJun, GST-ATF2, and GST-Elk-1, respectively.
JNK primes Mcl-1 for phosphorylation by GSK3.
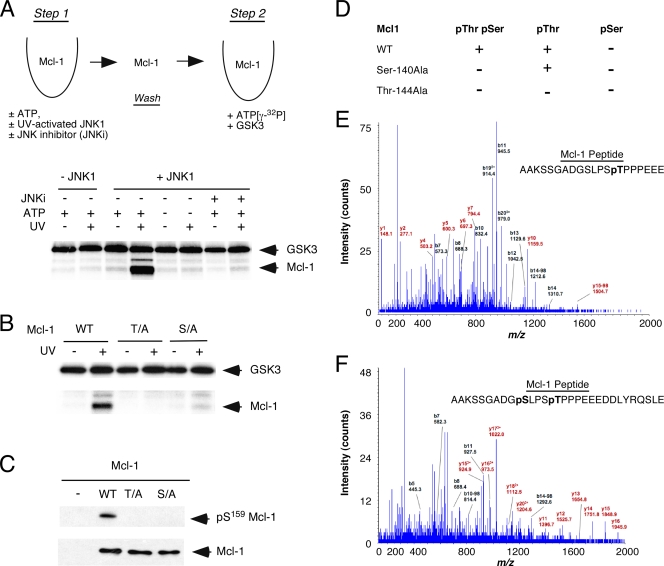
It has been reported that GSK3 can phosphorylate hMcl-1 on Ser-159 (21). This site of phosphorylation is conserved in mMcl-1 (Ser-140). However, we found that GSK3 was unable to phosphorylate mMcl-1 in an in vitro kinase assay (Fig. 3A). Priming phosphorylation of the substrate is often required prior to recognition by the GSK3 protein kinase (8). Consensus sequence analysis (Ser/Thr-X-X-X-phospho-Ser/Thr) indicated that the residue on Mcl-1 identified as the major phosphorylation site for JNK (Fig. 2) may act as the priming site for Mcl-1 phosphorylation by GSK3 (8). To test this hypothesis, we examined Mcl-1 using a two-step in vitro kinase assay (Fig. 3A). This analysis demonstrated that GSK3 phosphorylated Mcl-1 only after prior phosphorylation of Mcl-1 by JNK. Indeed, GSK3 phosphorylation of Mcl-1 was blocked by the absence of JNK, the presence of a JNK inhibitor, or the absence of ATP in the priming phosphorylation reaction (Fig. 3A). These observations suggested that JNK may serve as a kinase that primes mMcl-1 for phosphorylation by GSK3.
FIG. 3.
JNK primes Mcl-1 for phosphorylation by GSK3. (A) Upper panel, schematic illustration of the in vitro kinase assay. GST-mMcl-1 was incubated (30 min at 30°C) in step 1 without and with 1 mM ATP, JNK1, UV-activated JNK1, and/or the JNK inhibitor (JNKi) TAT-JBD (30-μl final volume). The GST-mMcl-1 was bound to glutathione-agarose beads, washed, and then incubated (30 min at 30°C) in step 2 with 500 units of GSK3β plus 50 μM [γ-32P]ATP (10 μCi/mmol) (30-μl final volume). The products of the phosphorylation reaction were examined by SDS-PAGE and autoradiography. Lower panel, the two-step in vitro kinase assay was performed using wild-type Mcl-1 as the substrate. The effect of changes in step 1 of the kinase reaction (addition of ATP, JNK, UV-activated JNK, and/or JNK inhibitor [JNKi] TAT-JBD) was examined. (B) The two-step in vitro kinase assay was performed using mMcl-1 as the substrate. The effect of replacement of the JNK phosphorylation site (Thr) or the GSK3 phosphorylation site (Ser) with Ala was examined (T/A and S/A). Step 1 of the in vitro kinase assay was performed using JNK or UV-activated JNK. WT, wild type. (C) COS cells were transiently transfected without or with an expression vector for hMcl-1. The effect of replacement of the JNK phosphorylation site (Thr) or the GSK3 phosphorylation site (Ser) with Ala was examined (T/A and S/A). Phosphorylation of hMcl-1 on the GSK3 phosphorylation site in vivo was examined by immunoblot analysis using antibodies to phosphoSer159-hMcl-1 and hMcl-1. (D) COS7 cells expressing hemagglutinin-tagged mMcl-1 were treated with the proteasome inhibitor MG132 (10 μM) and incubated (60 min). The cells were then exposed to 60 J/m2 UV radiation. Protein extracts were prepared at 40 min postirradiation. The mMcl-1 protein was isolated by immunoprecipitation and SDS-PAGE. mMcl-1 peptides obtained after digestion with trypsin or Glu-C were examined by mass spectroscopy. Peptides corresponding to the phosphodegron motif were identified. The detection of phosphorylation at a single site (pSer or pThr) or dual phosphorylation (pThr pSer) is summarized. The effect of replacement of Thr-144 and Ser-140 with Ala is shown. (+, phosphopeptide detected; −, phosphopeptide not detected). (E and F) Representative spectra of phosphopeptides obtained after digestion of wild-type mMcl-1 with Glu-C.
It is established that some GSK3 substrates are phosphorylated in the absence of priming phosphorylation (8). Indeed, it has been reported that priming may not be required for phosphorylation of Mcl-1 by GSK3 (7). To test whether the JNK-mediated priming site (Thr-144) was required for GSK3 phosphorylation of mMcl-1 on Ser-140, we examined the effect of replacing these phosphorylation sites with Ala using the two-step in vitro kinase assay (Fig. 3B). This analysis demonstrated that GSK3 phosphorylated mMcl-1 only after prior phosphorylation by activated JNK and that this GSK3-mediated phosphorylation required both the JNK-mediated priming site (Thr-144) and the GSK3 phosphorylation site (Ser-140) on mMcl-1 (Fig. 3B). Both of these phosphorylation sites are located within the phosphodegron motif (DGpS140LPSpT144) of Mcl-1 (7, 21).
Mutational analysis of Mcl-1 phosphorylation in vivo.
In vitro studies demonstrate that phosphorylation on the JNK site acts to prime Mcl-1 for phosphorylation by GSK3 (Fig. 3A). To test the biological relevance of this finding, we examined the in vivo phosphorylation of the mMcl-1 phosphodegron motif by mass spectroscopy (Fig. 3D to F). Studies of wild-type mMcl-1 demonstrated dual phosphorylation of the phosphodegron motif (DGpS140LPSpT144) on Ser-140 plus Thr-144 (Fig. 3D). The phosphodegron motif was also phosphorylated on Thr-144 alone. In contrast, no phosphorylation of the phosphodegron motif on Ser-140 alone was detected. Together, these data indicate that Thr-144 may act as a priming site for mMcl-1 phosphorylation on Ser-140 in vivo. This conclusion is consistent with the finding that Ser-140Ala mMcl-1 was phosphorylated on Thr-144 (Fig. 3D). In contrast, no phosphorylation of the phosphodegron motif of Thr-144Ala mMcl-1 was detected (Fig. 3D).
To obtain independent evidence that the priming phosphorylation site was required for GSK3-mediated phosphorylation of Mcl-1 in vivo, we examined the phosphorylation of Mcl-1 by immunoblot analysis using an antibody to phospho-hMcl-1 that specifically recognizes the GSK3 phosphorylation site. As expected, replacement of the Ser residue that is phosphorylated by GSK3 with Ala prevented detection of Mcl-1 by immunoblot analysis (Fig. 3C). This analysis confirmed the specificity of the phospho-Mcl-1 antibody. Mutation of the JNK phosphorylation site that primes Mcl-1 for phosphorylation by GSK3 in vitro (Fig. 3A and B) also prevented the phosphorylation of Mcl-1 on the GSK3 site in vivo (Fig. 3C). Together, these data demonstrate that Mcl-1 is a conditional substrate for GSK3 that requires priming phosphorylation on the JNK site in vivo.
Phosphorylation is required for UV-stimulated Mcl-1 degradation.
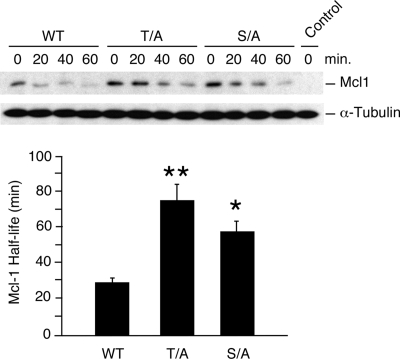
Exposure of fibroblasts to UV radiation causes rapid degradation of mMcl-1. To test whether mMcl-1 phosphorylation is required, we compared the UV-stimulated degradation of wild-type and phosphorylation-defective mMcl-1 proteins (Fig. 4). The half-life of wild-type Mcl-1 was significantly shorter than that of the phosphorylation-defective mMcl-1 proteins (Thr-144Ala and Ser-140Ala). These data confirm that both the JNK phosphorylation site (Thr-144) that primes Mcl-1 for phosphorylation by GSK3 and the GSK3 phosphorylation site Ser-140 are required for UV-stimulated Mcl-1 degradation.
FIG. 4.
Phosphorylation is required for UV-stimulated Mcl-1 degradation. Murine Mcl-1LoxP/LoxP Gt(ROSA)26Sor-CreEsr1 fibroblasts were transduced with an mMcl-1 (wild type [WT], Thr-144Ala, or Ser-140Ala) expression vector or an empty pBABE-IRES-Puro vector (control) and selected by treatment with 2 μg/ml puromycin (2 days). The endogenous Mcl-1 gene was deleted by treatment of the cells with 1 μM 4-hydroxytamoxifen (1 day). The half-life of the mMcl-1 protein was examined following exposure of the cells to UV radiation (60 J/m2). The amounts of Mcl-1 and α-tubulin were detected by autoradiography and quantitated using a Kodak 4000MM imaging station. The half-lives of wild-type and phosphorylation-defective Mcl-1 proteins (T/A and S/A) are presented. The half-lives of the mutant proteins were significantly longer than that of wild-type Mcl-1 (mean ± SD; n = 6; *, P < 0.05; **, P < 0.01).
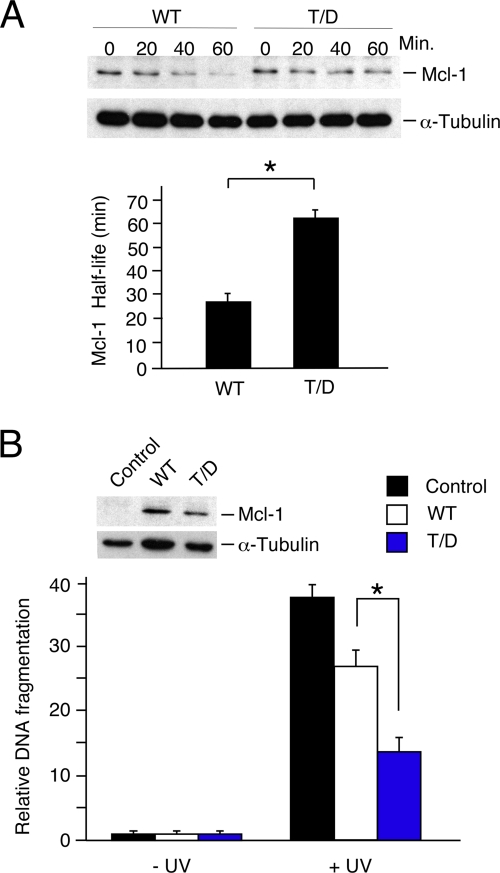
A phosphomimetic mutation (Thr-144Asp) that results in negative charge at the priming phosphorylation site also caused stabilization of mMcl-1 (Fig. 5A). This is most likely because of a requirement for phosphate at the priming site (rather than simply a negative charge) for subsequent phosphorylation of target proteins by GSK3 (33).
FIG. 5.
Effect of a phosphomimetic mutation at the JNK phosphorylation site on Mcl-1 protein half-life and apoptotic activity. (A) Murine Mcl-1LoxP/LoxP Gt(ROSA)26Sor-CreEsr1 fibroblasts were transduced with an mMcl-1 (wild type [WT] or Thr-144Asp) expression vector or an empty pBABE-IRES-Puro vector (control) and selected by treatment with 2 μg/ml puromycin (2 days). The endogenous Mcl-1 gene was deleted by treatment of the cells with 1 μM 4-hydroxytamoxifen (1 day). The half-life of the mMcl-1 protein was examined following exposure of the cells to UV radiation (60 J/m2). The amounts of Mcl-1 and α-tubulin were detected by autoradiography and quantitated using a Kodak 4000MM imaging station. The half-life of Thr-144Asp (T/D) mMcl-1 was significantly greater than that of wild-type Mcl-1 (mean ± SD; n = 5; *, P < 0.001). (B) Murine fibroblasts were transduced with an mMcl-1 (WT or Thr-144Asp) expression vector or an empty retroviral vector (control). Immunoblot analysis indicated the expression of the wild-type and mutated mMcl-1 proteins (inset). Cell death was measured by DNA fragmentation 16 h after UV treatment (60 J/m2) and is presented as the mean ± SD (n = 5). The antiapoptotic activity of Thr-144Asp mMcl-1 was significantly greater than that of wild-type (WT) Mcl-1 (*, P < 0.0005).
UV-stimulated cell death is suppressed by defects in Mcl-1 phosphorylation.
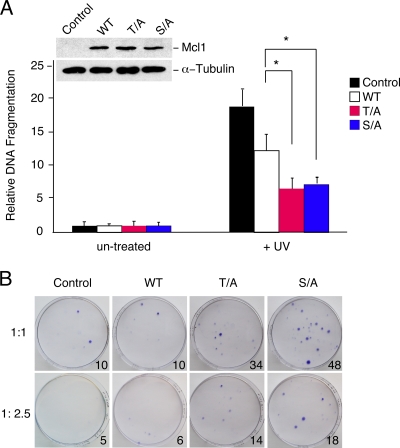
Exposure of cells to stress causes phosphorylation of eIF-2α on Ser-51 and inhibition of Mcl-1 mRNA translation (1, 11, 25). The subsequent degradation of Mcl-1 contributes to the sensitization of cells to apoptosis (25). This degradation is mediated by the E3 ubiquitin protein ligase β-TrCP, which recognizes GSK3-phosphorylated Mcl-1 (7). If JNK phosphorylation of Mcl-1 on the priming site is required for GSK3 phosphorylation, mutation of Mcl-1 at either the JNK or GSK3 phosphorylation site would be predicted to suppress apoptosis by increasing Mcl-1 function. To test this hypothesis, we examined the effect of mutating these Mcl-1 phosphorylation sites on UV-stimulated apoptosis. We found that mutation of either the JNK or the GSK3 phosphorylation site on Mcl-1 (by replacement with Ala) caused similar changes in UV-stimulated cell death (Fig. 6A). These phosphorylation-defective Mcl-1 proteins caused reduced UV-stimulated apoptotic DNA fragmentation compared with that caused by wild-type Mcl-1 (Fig. 5B and 6A). Similarly, the phosphorylation-defective Mcl-1 proteins caused increased cell survival after exposure to UV radiation compared with wild-type Mcl-1 in a colony formation assay (Fig. 6B). Together, these data confirm that both the JNK and GSK3 phosphorylation sites are required for downregulation of Mcl-1 antiapoptotic activity and are consistent with the conclusion that JNK is required for the rapid GSK3-mediated phosphorylation and degradation of Mcl-1.
FIG. 6.
Phosphorylation-defective Mcl-1 proteins suppress UV-stimulated apoptosis. (A) Murine Mcl-1LoxP/LoxP Gt(ROSA)26Sor-CreEsr1 fibroblasts were transduced with an hMcl-1 (wild type [WT], Thr163Ala, or Ser159Ala) expression vector or an empty pBABE-IRES-Puro vector (control) and selected by treatment with 2 μg/ml puromycin (2 days). The endogenous Mcl-1 gene was deleted by treatment of the cells with 1 μM 4-hydroxytamoxifen (1 day). Immunoblot analysis indicated the expression of the wild-type and mutated hMcl-1 proteins (inset). Cell death was measured by DNA fragmentation 16 h after UV treatment (60 J/m2) and is presented as the mean ± SD (n = 4). The effects of wild-type and phosphorylation-defective Mcl-1 proteins (T/A and S/A) on apoptosis are presented. Statistically significant differences are indicated (*, P < 0.05). (B) Cell survival was examined by replating the cells at 16 h postirradiation in 100-mm dishes. The cells were cultured for 14 days, and colony formation was detected by staining the dishes with crystal violet. The effects of wild-type and phosphorylation-defective Mcl-1 (T/A and S/A) proteins on apoptosis are presented. A representative image of the culture dishes is illustrated. The mean number of colonies in four separate experiments is indicated (inset, lower right side of each subpanel).
DISCUSSION
The JNKs can mediate proapoptotic signal transduction (6). The mechanism of JNK-induced apoptosis is unclear, but JNK-deficient fibroblasts exhibit defects in mitochondrial outer membrane permeabilization and the release of cytochrome c (31). JNK substrates that are implicated in apoptosis include members of the Bcl2-related protein family (34). Thus, mice with a germ line mutation in the Bim gene at the phosphorylation site Thr-112 (replacement with Ala) exhibit defects in Bim-dependent apoptosis (14). However, BimEL phosphorylation by JNK on Thr-112 is not sufficient, by itself, to fully account for JNK-induced apoptosis (14). These data suggest that BimEL may cooperate with another JNK substrate to mediate JNK-stimulated apoptosis. One candidate substrate is Mcl-1 (34). Indeed, previous studies have established that the rapid reduction of Mcl-1 expression detected in cells exposed to stress may be critical for stress-induced apoptosis (25). The mechanism of stress-induced reduction in Mcl-1 expression is mediated by phosphorylation of eIF-2α on the inhibitory site Ser-51 that prevents translation of Mcl-1 mRNA (1, 11, 25) and rapid degradation of Mcl-1 protein mediated by the ubiquitin-dependent proteasome pathway (27).
The E3 ubiquitin protein ligases that mediate degradation of Mcl-1 include Mule/ARF-BP1 (39) and β-TrCP (7). Stress-induced degradation of Mcl-1 appears to be mediated by a phosphodegron (DGpSLPSpT) that interacts with β-TrCP (7). Phosphorylation of this phosphodegron motif is mediated by GSK3 (7, 21). This conclusion is supported by studies of small interfering RNA-mediated knockdown of GSK3 expression (21) and by studies of fibroblasts isolated from GSK3 knockout mice (7). GSK3 is therefore a negative regulator of Mcl-1 expression. Significantly, the prosurvival protein kinase AKT is a negative regulator of GSK3. Consequently, prosurvival signaling by AKT can increase Mcl-1 expression by inhibiting GSK3-mediated Mcl-1 degradation (7, 21).
Priming phosphorylation is required for recognition of Mcl-1 as a substrate by GSK3. Thus, in vitro assays demonstrate that GSK3 does not phosphorylate Mcl-1 (Fig. 3A). However, Thr-144 within the phosphodegron motif DGpS140LPSpT144 can serve to prime Mcl-1 for phosphorylation by GSK3 on Ser-140 (Fig. 3). Our studies of JNK-deficient cells demonstrate that JNK represents the major protein kinase activity that mediates phosphorylation of Mcl-1 on the priming site (Fig. 1). However, it should be noted that our analysis was restricted to the response of murine fibroblasts to serum starvation and UV radiation. It is possible that other protein kinases may contribute to the priming phosphorylation of Mcl-1 in other cell types or in response to other stimuli.
Dual phosphorylation of the Mcl-1 phosphodegron in response to stress in fibroblasts requires activation of both JNK and GSK3. JNK can be activated by a canonical stress-induced signaling pathway (6). In contrast, the major control of GSK3 activity is mediated by negative regulation, for example, by inhibitory phosphorylation by mediated AKT (8). Efficient dual phosphorylation of the Mcl-1 phosphodegron motif may therefore be mediated by the coordinated actions of activated JNK and inhibited AKT signaling pathways. This may provide a molecular explanation for previous findings that the proapoptotic activity of JNK is suppressed by activation of the prosurvival AKT signaling pathway (37). Together, these considerations implicate Mcl-1 as a site of signal integration between proapoptotic (JNK) and prosurvival (e.g., AKT) protein kinase signaling pathways.
Acknowledgments
We thank Alain Chariot (Université de Liége), Ulrich Maurer (Universität Freiburg), and Hsin-Fang Yang-Yen (Academia Sinica) for providing essential reagents; Tammy Barrett for expert technical assistance; and Kathy Gemme for administrative assistance.
These studies were supported by a grant from the National Institutes of Health. S.M.C. is supported by a graduate research fellowship from the National Science Foundation. R.J.D. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Published ahead of print on 11 May 2009.
REFERENCES
- 1.Adams, K. W., and G. M. Cooper. 2007. Rapid turnover of Mcl-1 couples translation to cell survival and apoptosis. J. Biol. Chem. 2826192-6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Badea, T. C., Y. Wang, and J. Nathans. 2003. A noninvasive genetic/pharmacologic strategy for visualizing cell morphology and clonal relationships in the mouse. J. Neurosci. 232314-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brar, G. A., B. M. Kiburz, Y. Zhang, J. E. Kim, F. White, and A. Amon. 2006. Rec8 phosphorylation and recombination promote the step-wise loss of cohesins in meiosis. Nature 441532-536. [DOI] [PubMed] [Google Scholar]
- 4.Czabotar, P. E., E. F. Lee, M. F. van Delft, C. L. Day, B. J. Smith, D. C. Huang, W. D. Fairlie, M. G. Hinds, and P. M. Colman. 2007. Structural insights into the degradation of Mcl-1 induced by BH3 domains. Proc. Natl. Acad. Sci. USA 1046217-6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Das, M., F. Jiang, H. K. Sluss, C. Zhang, K. M. Shokat, R. A. Flavell, and R. J. Davis. 2007. Suppression of p53-dependent senescence by the JNK signal transduction pathway. Proc. Natl. Acad. Sci. USA 10415759-15764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis, R. J. 2000. Signal transduction by the JNK group of MAP kinases. Cell 103239-252. [DOI] [PubMed] [Google Scholar]
- 7.Ding, Q., X. He, J. M. Hsu, W. Xia, C. T. Chen, L. Y. Li, D. F. Lee, J. C. Liu, Q. Zhong, X. Wang, and M. C. Hung. 2007. Degradation of Mcl-1 by beta-TrCP mediates glycogen synthase kinase 3-induced tumor suppression and chemosensitization. Mol. Cell. Biol. 274006-4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doble, B. W., and J. R. Woodgett. 2003. GSK-3: tricks of the trade for a multi-tasking kinase. J. Cell Sci. 1161175-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Domina, A. M., J. A. Vrana, M. A. Gregory, S. R. Hann, and R. W. Craig. 2004. MCL1 is phosphorylated in the PEST region and stabilized upon ERK activation in viable cells, and at additional sites with cytotoxic okadaic acid or taxol. Oncogene 235301-5315. [DOI] [PubMed] [Google Scholar]
- 10.Dong, C., D. D. Yang, M. Wysk, A. J. Whitmarsh, R. J. Davis, and R. A. Flavell. 1998. Defective T cell differentiation in the absence of Jnk1. Science 2822092-2095. [DOI] [PubMed] [Google Scholar]
- 11.Fritsch, R. M., G. Schneider, D. Saur, M. Scheibel, and R. M. Schmid. 2007. Translational repression of MCL-1 couples stress-induced eIF2 alpha phosphorylation to mitochondrial apoptosis initiation. J. Biol. Chem. 28222551-22562. [DOI] [PubMed] [Google Scholar]
- 12.He, X., J. P. Saint-Jeannet, J. R. Woodgett, H. E. Varmus, and I. B. Dawid. 1995. Glycogen synthase kinase-3 and dorsoventral patterning in Xenopus embryos. Nature 374617-622. [DOI] [PubMed] [Google Scholar]
- 13.Herrant, M., A. Jacquel, S. Marchetti, N. Belhacene, P. Colosetti, F. Luciano, and P. Auberger. 2004. Cleavage of Mcl-1 by caspases impaired its ability to counteract Bim-induced apoptosis. Oncogene 237863-7873. [DOI] [PubMed] [Google Scholar]
- 14.Hubner, A., T. Barrett, R. A. Flavell, and R. J. Davis. 2008. Multisite phosphorylation regulates Bim stability and apoptotic activity. Mol. Cell 30415-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inoshita, S., K. Takeda, T. Hatai, Y. Terada, M. Sano, J. Hata, A. Umezawa, and H. Ichijo. 2002. Phosphorylation and inactivation of myeloid cell leukemia 1 by JNK in response to oxidative stress. J. Biol. Chem. 27743730-43734. [DOI] [PubMed] [Google Scholar]
- 16.Jaeschke, A., M. Karasarides, J. J. Ventura, A. Ehrhardt, C. Zhang, R. A. Flavell, K. M. Shokat, and R. J. Davis. 2006. JNK2 is a positive regulator of the cJun transcription factor. Mol. Cell 23899-911. [DOI] [PubMed] [Google Scholar]
- 17.Kennedy, N. J., H. K. Sluss, S. N. Jones, D. Bar-Sagi, R. A. Flavell, and R. J. Davis. 2003. Suppression of Ras-stimulated transformation by the JNK signal transduction pathway. Genes Dev. 17629-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi, S., S. H. Lee, X. W. Meng, J. L. Mott, S. F. Bronk, N. W. Werneburg, R. W. Craig, S. H. Kaufmann, and G. J. Gores. 2007. Serine 64 phosphorylation enhances the antiapoptotic function of Mcl-1. J. Biol. Chem. 28218407-18417. [DOI] [PubMed] [Google Scholar]
- 19.Lamb, J. A., J. J. Ventura, P. Hess, R. A. Flavell, and R. J. Davis. 2003. JunD mediates survival signaling by the JNK signal transduction pathway. Mol. Cell 111479-1489. [DOI] [PubMed] [Google Scholar]
- 20.Liu, H., H. W. Peng, Y. S. Cheng, H. S. Yuan, and H. F. Yang-Yen. 2005. Stabilization and enhancement of the antiapoptotic activity of mcl-1 by TCTP. Mol. Cell. Biol. 253117-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maurer, U., C. Charvet, A. S. Wagman, E. Dejardin, and D. R. Green. 2006. Glycogen synthase kinase-3 regulates mitochondrial outer membrane permeabilization and apoptosis by destabilization of MCL-1. Mol. Cell 21749-760. [DOI] [PubMed] [Google Scholar]
- 22.Michels, J., J. W. O'Neill, C. L. Dallman, A. Mouzakiti, F. Habens, M. Brimmell, K. Y. Zhang, R. W. Craig, E. G. Marcusson, P. W. Johnson, and G. Packham. 2004. Mcl-1 is required for Akata6 B-lymphoma cell survival and is converted to a cell death molecule by efficient caspase-mediated cleavage. Oncogene 234818-4827. [DOI] [PubMed] [Google Scholar]
- 23.Moser, K., and F. M. White. 2006. Phosphoproteomic analysis of rat liver by high capacity IMAC and LC-MS/MS. J. Proteome Res. 598-104. [DOI] [PubMed] [Google Scholar]
- 24.Mott, J. L., S. Kobayashi, S. F. Bronk, and G. J. Gores. 2007. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene 266133-6140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nijhawan, D., M. Fang, E. Traer, Q. Zhong, W. Gao, F. Du, and X. Wang. 2003. Elimination of Mcl-1 is required for the initiation of apoptosis following ultraviolet irradiation. Genes Dev. 171475-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Opferman, J. T. 2007. Life and death during hematopoietic differentiation. Curr. Opin. Immunol. 19497-502. [DOI] [PubMed] [Google Scholar]
- 27.Opferman, J. T. 2006. Unraveling MCL-1 degradation. Cell Death Differ. 131260-1262. [DOI] [PubMed] [Google Scholar]
- 28.Opferman, J. T., H. Iwasaki, C. C. Ong, H. Suh, S. Mizuno, K. Akashi, and S. J. Korsmeyer. 2005. Obligate role of anti-apoptotic MCL-1 in the survival of hematopoietic stem cells. Science 3071101-1104. [DOI] [PubMed] [Google Scholar]
- 29.Opferman, J. T., A. Letai, C. Beard, M. D. Sorcinelli, C. C. Ong, and S. J. Korsmeyer. 2003. Development and maintenance of B and T lymphocytes requires antiapoptotic MCL-1. Nature 426671-676. [DOI] [PubMed] [Google Scholar]
- 30.Rinkenberger, J. L., S. Horning, B. Klocke, K. Roth, and S. J. Korsmeyer. 2000. Mcl-1 deficiency results in peri-implantation embryonic lethality. Genes Dev. 1423-27. [PMC free article] [PubMed] [Google Scholar]
- 31.Tournier, C., P. Hess, D. D. Yang, J. Xu, T. K. Turner, A. Nimnual, D. Bar-Sagi, S. N. Jones, R. A. Flavell, and R. J. Davis. 2000. Requirement of JNK for stress-induced activation of the cytochrome c-mediated death pathway. Science 288870-874. [DOI] [PubMed] [Google Scholar]
- 32.Viatour, P., E. Dejardin, M. Warnier, F. Lair, E. Claudio, F. Bureau, J. C. Marine, M. P. Merville, U. Maurer, D. Green, J. Piette, U. Siebenlist, V. Bours, and A. Chariot. 2004. GSK3-mediated BCL-3 phosphorylation modulates its degradation and its oncogenicity. Mol. Cell 1635-45. [DOI] [PubMed] [Google Scholar]
- 33.Weston, C. R., and R. J. Davis. 2001. Signal transduction: signaling specificity—a complex affair. Science 2922439-2440. [DOI] [PubMed] [Google Scholar]
- 34.Weston, C. R., and R. J. Davis. 2007. The JNK signal transduction pathway. Curr. Opin. Cell Biol. 19142-149. [DOI] [PubMed] [Google Scholar]
- 35.Whitmarsh, A. J., and R. J. Davis. 2001. Analyzing JNK and p38 mitogen-activated protein kinase activity. Methods Enzymol. 332319-336. [DOI] [PubMed] [Google Scholar]
- 36.Willis, S. N., L. Chen, G. Dewson, A. Wei, E. Naik, J. I. Fletcher, J. M. Adams, and D. C. Huang. 2005. Proapoptotic Bak is sequestered by Mcl-1 and Bcl-xL, but not Bcl-2, until displaced by BH3-only proteins. Genes Dev. 191294-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xia, Z., M. Dickens, J. Raingeaud, R. J. Davis, and M. E. Greenberg. 1995. Opposing effects of ERK and JNK-p38 MAP kinases on apoptosis. Science 2701326-1331. [DOI] [PubMed] [Google Scholar]
- 38.Yang, T., K. M. Kozopas, and R. W. Craig. 1995. The intracellular distribution and pattern of expression of Mcl-1 overlap with, but are not identical to, those of Bcl-2. J. Cell Biol. 1281173-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhong, Q., W. Gao, F. Du, and X. Wang. 2005. Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell 1211085-1095. [DOI] [PubMed] [Google Scholar]