Abstract
GAP1IP4BP is a member of the GAP1 family of Ras GTPase-activating proteins (GAPs) that includes GAP1m, CAPRI, and RASAL. Composed of a central Ras GAP-related domain (RasGRD), surrounded by amino-terminal C2 domains and a carboxy-terminal PH/Btk domain, these proteins, with the notable exception of GAP1m, possess an unexpected arginine finger-dependent GAP activity on the Ras-related protein Rap1 (S. Kupzig, D. Deaconescu, D. Bouyoucef, S. A. Walker, Q. Liu, C. L. Polte, O. Daumke, T. Ishizaki, P. J. Lockyer, A. Wittinghofer, and P. J. Cullen, J. Biol. Chem. 281:9891-9900, 2006). Here, we have examined the mechanism through which GAP1IP4BP can function as a Rap1 GAP. We show that deletion of domains on either side of the RasGRD, while not affecting Ras GAP activity, do dramatically perturb Rap1 GAP activity. By utilizing GAP1IP4BP/GAP1m chimeras, we establish that although the C2 and PH/Btk domains are required to stabilize the RasGRD, it is this domain which contains the catalytic machinery required for Rap1 GAP activity. Finally, a key residue in Rap1-specific GAPs is a catalytic asparagine, the so-called asparagine thumb. By generating a molecular model describing the predicted Rap1-binding site in the RasGRD of GAP1IP4BP, we show that mutagenesis of individual asparagine or glutamine residues that lie in close proximity to the predicted binding site has no detectable effect on the in vivo Rap1 GAP activity of GAP1IP4BP. In contrast, we present evidence consistent with a model in which the RasGRD of GAP1IP4BP functions to stabilize the switch II region of Rap1, allowing stabilization of the transition state during GTP hydrolysis initiated by the arginine finger.
The Ras-like family of small GTPases are ubiquitously expressed, evolutionarily conserved proteins that, by undergoing conformational changes in response to the alternate binding of GDP and GTP, function as binary switches (28, 31, 35). The GDP-bound “off” state and the GTP-bound “on” state recognize distinct effector proteins, thereby allowing the regulation of a variety of downstream signaling events (28, 31, 35). While Ras is the best-known and best-studied Ras-like GTPase, Rap1 has recently attracted considerable attention (reviewed in reference 20).
Rap1 was originally identified through its ability, when overexpressed, to reverse the phenotype of K-Ras-transformed NIH 3T3 cells (19). As Ras and Rap1 have very similar effector regions, the ability of Rap1 to reverse the transformed phenotype appeared to arise through an ability to compete with K-Ras effectors. For example, Rap1 binds the Ras effector Raf1 but this does not lead to its activation (11). This is consistent with a simple model in which Rap1 functions as a Ras antagonist (6, 37). However, recent work has challenged this view. Increasing evidence points to Rap1 interacting with its own panel of effectors through which it controls cell-cell adhesion and cell-matrix interactions (reviewed in reference 20).
Like that of other GTPases, the activation of Ras and Rap1 is regulated through guanine nucleotide exchange factors, which control activation by stimulating the exchange of GDP for GTP. Inactivation is driven by GTPase-activating proteins (GAPs). These enhance the intrinsic GTPase activity of Ras and Rap1, thereby leading to GTP hydrolysis. A wide variety of guanine nucleotide exchange factors and GAPs specific for these GTPases have been identified (14). Through the arrangement of different modular domains, these proteins are regulated following the activation of cell surface receptors. This occurs either through direct association with the activated receptor or indirectly through second messengers (4, 5, 14, 41).
Mammalian proteins capable of functioning as Ras GAPs include NF1 (3, 27, 40), p120GAP (38), the semaphorin 4D receptor plexin-B1 (29), and members of the GAP1 (reviewed in reference 41) and SynGAP (DAB2IP, nGAP, and SynGAP) families (10, 18, 39). These function as Ras GAPs by supplying a catalytic arginine residue—the arginine finger—into the active site of Ras. This stabilizes the transition state of the GTPase reaction, increasing the reaction rate by more than 1,000-fold (1, 33, 34).
Rap1 GAPs include Rap GAPs I and II, the SPA-1 family (SPA-1, SPAR, SPAL, and E6TP1), and tuberin (16, 17, 26, 32). Unlike Ras, Rap1 does not possess the catalytic glutamine residue that is critical for GTP hydrolysis in Ras. This fundamental difference means that the mechanisms by which Ras and Rap1 GAPs function are distinct. Rap1 GAPs do not employ a catalytic arginine residue (8, 9); instead, they provide a catalytic asparagine—the asparagine thumb—to stimulate GTP hydrolysis (15). Here the asparagine carboxamide side chain has a function similar to that of the glutamine residue in Ras, stabilizing the position of the nucleophilic water and γ-phosphate in the transition complex (15, 36).
Given such distinct catalytic mechanisms, surprisingly, some Ras GAPs, while having no detectable sequence homology with any Rap1 GAPs, are capable of stimulating the GTPase activity of Rap1. The first protein found to display such dual activities was GAP1IP4BP (13) (also known as RASA3, GAPIII, and R-Ras GAP). This is a member of the GAP1 family, which also comprises GAP1m, CAPRI, and RASAL (2, 23-25). These proteins are characterized by a domain architecture comprising amino-terminal tandem C2 domains, a highly conserved central Ras GAP-related domain (RasGRD), and a carboxy-terminal pleckstrin homology (PH) domain that is associated with a Bruton's tyrosine kinase (Btk) motif (41). Consistent with the presence of the RasGRD, all proteins display Ras GAP activity, although each is differentially regulated following receptor stimulation (41). With the notable exception of GAP1m, all GAP1 proteins also possess efficient Rap1 GAP activity (22). Such dual specificity is not restricted solely to GAP1 proteins. Recently, C2 domain-containing SynGAP—a neuronal Ras GAP—has also been shown to display Rap1 GAP activity (21), an activity that appears to require, alongside the RasGRD, the presence of a single C2 domain (30).
Here we have examined the mechanism behind the dual Ras and Rap1 GAP activities of GAP1IP4BP. Through the generation of a series of GAP1IP4BP/GAP1m chimeras, we have established that while the C2 domains of GAP1IP4BP are required to stabilize the RasGRD, these domains do not supply catalytic residues required for Rap1 GAP activity. Rather, the Rap1 GAP catalytic machinery appears to reside solely within the RasGRD. By the site-directed mutagenesis of selected asparagine and glutamine residues within this domain—selected following the generation of a predicted molecular model of the GAP1IP4BP RasGRD-Ras(Rap1) complex—we establish that the ability of GAP1IP4BP to function as a Rap1 GAP does not occur via a mechanism that utilizes a classic asparagine thumb. Rather, we suggest that the GAP1IP4BP RasGRD functions to stabilize the switch II region of Rap1 in a manner that allows a catalytic arginine finger from GAP1IP4BP to drive the hydrolysis of GTP.
MATERIALS AND METHODS
cDNAs.
The human isoforms of the GAP1IP4BP, GAP1m, Ras, and Rap1 cDNAs were used in this study.
Generation of GAP1IP4BP deletion constructs. GAP1IP4BPΔPH/C-tail was generated by digesting pEGFP-C1-GAP1IP4BP with ApaI and religating the larger fragment, thereby deleting the PH/Btk domains and the C terminus. GAP1IP4BPΔC2, GAP1IP4BPΔGRD/PH/C-tail, and GAP1IP4BPΔC-tail were generated as previously described (7).
Generation of GAP1IP4BP and GAP1m C2 domain chimeras.
Site-directed mutagenesis was used to introduce the following unique restriction sites into GAP1IP4BP and GAP1m: an XbaI site between the C2A and C2B domains and a SpeI site at the end of the C2B domain. The restriction sites were introduced sequentially with the QuikChange XL kit (Stratagene) according to manufacturer's instructions with pEGFP-C1-GAP1IP4BP and pEGFP-C1-GAP1m as templates. The primers used were GAP1IP4BP (XbaI) fwd (5′-GGGCAAAGTGCATCTAGAGCTGCGGCTGAGCG-3′), GAP1m (XbaI) fwd (5′-GGGTAAAGTTCATCTAGAATTAAAACTGAATG-3′), GAP1IP4BP (SpeI) fwd (5′-CTCTGCGGGACCTACTAGTGAAGTCTGCGGATG-3′), and GAP1m (SpeI) fwd (5′-CTTTGAAAACTTTACTAGTAAAATCACCAGATG-3′). The reverse primers were exactly complementary to the forward primers, and the underlined sequences indicate the positions of the restriction sites. Introduction of the SpeI restriction site resulted in an amino acid change from L308 to V in GAP1IP4BP and from L334 to V in GAP1m. Following the generation of the hybrid proteins, the SpeI site was therefore removed from each protein with the QuikChange XL kit (Stratagene) and primers GAP1IP4BP/m (minus SpeI) fwd (5′-CTCTGCGGGACCTGCTGTTAAAATCACCAGATG-3′) and GAP1m/IP4BP (minus SpeI) fwd (5′-CTTTGAAAACTTTGCTGCTGAAGTCTGCGGATG-3′). Again, the reverse primers were exactly complementary to the forward primers.
Generation of GAP1IP4BP and GAP1m PH/Btk domain chimeras.
The PH/Btk domain chimeras of GAP1IP4BP and GAP1m were generated as described in reference 12.
Generation of GAP1IP4BP and GAP1m GRD chimeras.
Site-directed mutagenesis was used to introduce the following unique restriction sites into GAP1IP4BP and GAP1m: a SpeI site at the end of the C2B domain and an XbaI site after the GRD. The restriction sites were introduced sequentially with the QuikChange XL kit (Stratagene) according to the manufacturer's instructions by using pEGFP-C1-GAP1IP4BP and pEGFP-C1-GAP1m as templates. The primers used were GAP1IP4BP (SpeI) fwd (5′-CTCTGCGGGACCTACTAGTGAAGTCTGCGGATG-3′), GAP1m (SpeI) fwd (5′-CTTTGAAAACTTTACTAGTAAAATCACCAGATG-3′), GAP1IP4BP (XbaI bp1671) fwd (5′-CGGTGAAGAACTTTCTAGATTTGATTTCGTCC-3′), and GAP1m (XbaI bp1754) fwd (5′-CAGTTAAAAAGTTTCTAGATGAAATTTCATC-3′. The reverse primers were exactly complementary to the forward primers, and the underlined sequences indicate the positions of the restriction sites. Following the generation of the hybrid proteins, the SpeI site was removed from each protein with the primers described above.
Ras pulldown assays.
A glutathione S-transferase (GST) fusion with the Ras-GTP-binding domain from Raf-1 (GST-RBD) was purified from Escherichia coli BL21(DE3) cells harboring plasmid pGEX KG containing the Raf Ras-binding domain (amino acids 1 to 149). After induction of a bacterial culture (optical density at 600 nm between 0.4 and 0.6) for 3 h at 37°C with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), cells were lysed by sonication in phosphate-buffered saline (PBS) containing 1 mM EDTA, 1% (vol/vol) Triton X-100, 10 μg/ml aprotinin, and 10 μg/ml leupeptin. The lysate was clarified by centrifugation, and the resultant supernatant was stored in aliquots at −80°C. On the required day, aliquots were thawed prior to incubation with glutathione-Sepharose (Amersham Pharmacia Biotech) for 1 h at room temperature. The Sepharose beads were washed twice with PBS-1 mM EDTA-1% (vol/vol) Triton X-100 before finally being suspended as a 1:1 slurry. This was used immediately in pull-down assays. Here, dishes (100 mm) of CHO-T cells (8 × 105 cells) were transiently transfected by lipofection (GeneJuice; Novagen) with 2.5 μg H-Ras cDNA and 1 μg control vector or a vector encoding the particular GAP1 protein. The cells were serum starved for 2 h at 37°C in serum-free F-12 (Ham; Gibco) prior to the experimental procedures. Cells were lysed in 1 ml of ice-cold extraction buffer (50 mM HEPES [pH 7.5], 100 mM NaCl, 1 mM EGTA, 5 μg/ml benzamidine, 5 μg/ml aprotinin, 5 μg/ml leupeptin, 5 μg/ml pepstatin A, 5 μg/ml trypsin inhibitor, 0.5 mM phenylmethylsulfonyl fluoride, and 1 mM dithiothreitol containing 1% [vol/vol] Triton X-100 and 10 mM MgCl2). Nucleus-free supernatants were incubated with GST-RBD on glutathione-Sepharose beads at 4°C for 30 min. The beads were then collected by centrifugation and washed three times with ice-cold PBS-0.1% (vol/vol) Triton X-100-10 mM MgCl2. Ras proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and visualized by immunoblotting on nitrocellulose filters with pan-Ras antibodies (F132; Santa Cruz Biotechnology) and enhanced chemiluminescence (Amersham Pharmacia Biotech). Blots were analyzed by volume integration with ImageQuant software (Molecular Dynamics) as previously described (22).
Rap1 pulldown assays.
For the analysis of active Rap, a GST fusion with the Rap-GTP-binding domain from RalGDS (GST-RalGDS) was used (22). Dishes (100 mm) of CHO-T cells (8 × 105 cells) were transiently cotransfected by lipofection (GeneJuice; Novagen) with 2.5 μg of hemagglutinin (HA)-tagged Rap1A cDNA and 1 μg control vector or a vector encoding the particular GAP1 protein. The cells were serum starved for 2 h at 37°C in serum-free F-12 (Ham) medium at 24 h posttransfection. Cell lysis and Rap1A-GTP pulldown assays were carried out as described above. Immobilized Rap1A was detected with HA antibodies (Y-11; Santa Cruz Biotechnology). Blots were analyzed by volume integration with ImageQuant software (Molecular Dynamics) (22).
Molecular modeling.
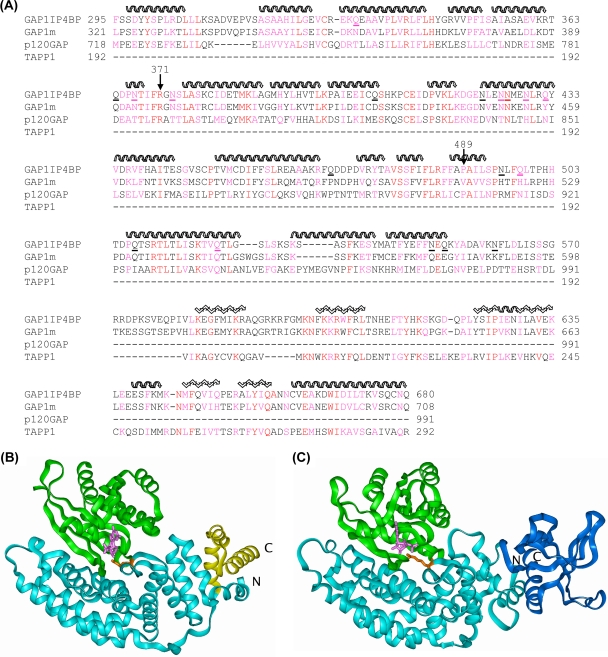
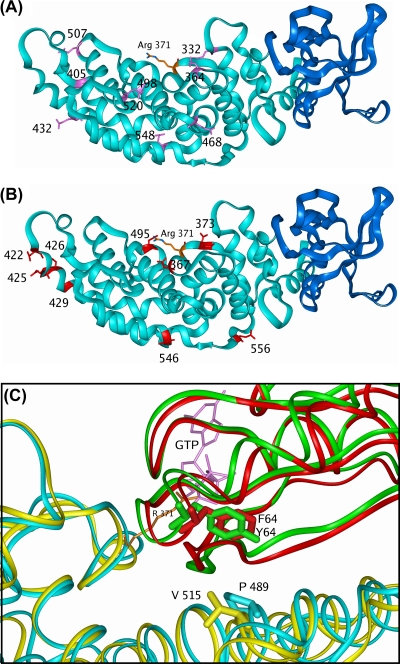
BLAST searching of the protein database with the sequence of GAP1IP4BP picks out p120GAP with a good E value (3 × 10−25) and a residue identity of 27% over the GRD (amino acids 292 to 550 of GAP1IP4BP). Beyond the GRD, the sequence matches the PH domains of Btk and TAPP1 with similar identities (30%) but with fewer gaps in the latter. Similar results are obtained with the sequence of GAP1m, as expected, since GAP1IP4BP and GAP1m have 60% sequence identity. Homology models of GAP1IP4BP and GAP1m were built according to the sequence alignment shown below (see Fig. 5A) with the templates p120GAP (1WER) and TAPP1 (1EAZ). Residue replacement, loop rebuilding (indels), and side chain repacking were performed with InsightII-2005 (Accelrys). Ras and Rap complexes of GAP1IP4BP and GAP1m were generated based on the structure of the p120GAP/RAS complex (1WQ1) and the corresponding GTPase structures (5P21 and 1C1Y, respectively). Models were relaxed by energy minimization according to the following general protocol: hydrogen atoms were added consistent with pH 7, followed by soaking with a 10-Å layer of water. Protein backbone atoms were restrained to their initial positions with a force constant of 100 kcal/Å, reducing to 0.5 kcal/Å over a total of 3,000 steps of conjugate gradient minimization with the consistence valence force field in Discover 2.98 (Accelrys).
FIG. 5.
Molecular modeling of the RasGRD of GAP1IP4BP. (A) The sequence alignment used for modeling of GAP1IP4BP and GAP1m on p120GAP (GRD 1WER) and TAPP1 (PH domain 1EAZ). The secondary structure of the templates is indicated as follows: helix, wavy overlining; sheet, zigzag overlining. Residue identities are in red, and similarities are in magenta. The asparagine and glutamine residues in GAP1IP4BP that were mutated to alanine are underlined. (B) Structure of the Ras/p120GAP complex (1WQ1). The p120GAP GRD is shown as a light blue ribbon with the residues beyond L991 shown in yellow. Ras is shown as a green ribbon, GDP is shown as pink sticks, and R789 is shown as orange sticks. (C) Model of the Ras/GAP1IP4BP complex. The GAP1IP4BP GRD (modeled on p120GAP) is shown as a cyan ribbon with the residues beyond E573 shown in dark blue and modeled as a PH domain. Ras is shown as a green ribbon, GDP is shown as pink sticks, and R371 is shown as orange sticks.
RESULTS
For GAP1IP4BP to function as an efficient Rap1 GAP, both amino- and carboxy-terminal flanking regions on either side of the RasGRD are required.
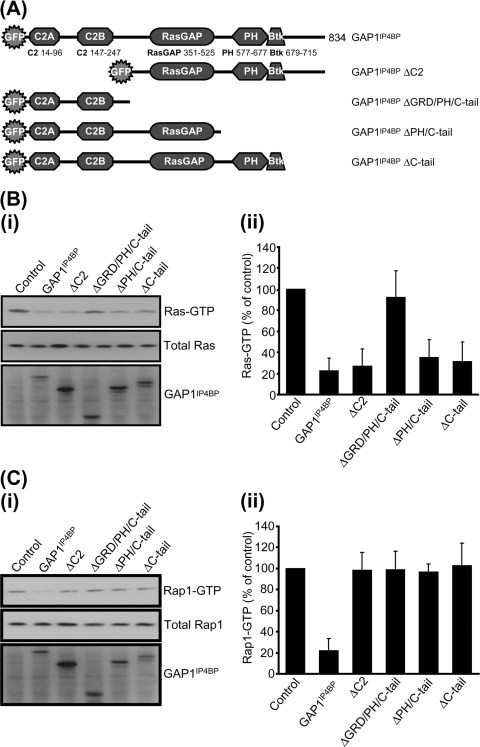
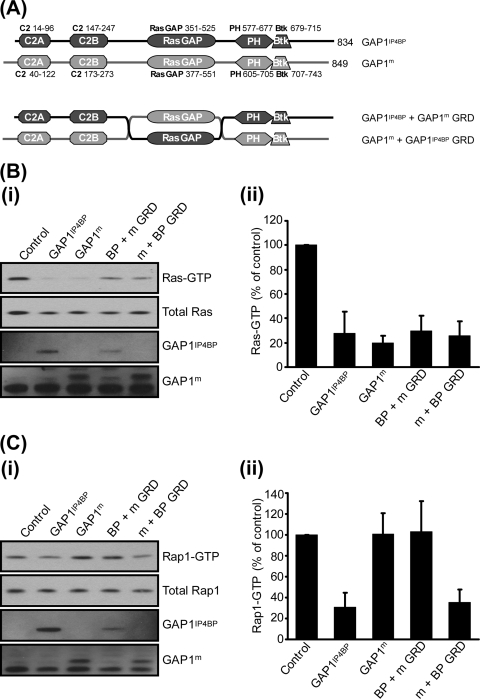
Previously, we established that while full-length GAP1IP4BP displays robust dual Ras and Rap1 GAP activities (13, 22), the isolated RasGRD loses its efficiency as a Rap GAP (22). These data led to the proposal that other regions outside of the RasGRD are necessary to support the catalytic activity of GAP1IP4BP toward Rap1 (22). To examine this in more detail, we generated and analyzed a panel of deletion mutant forms of GAP1IP4BP (Fig. 1A). In pull-down assays, deletion of either the tandem amino-terminal C2A and C2B domains or the PH/Btk domain and the carboxyl-terminal tail had no significant effect on the Ras GAP activity of GAP1IP4BP (Fig. 1B). As expected, however, removal of the RasGRD resulted in catalytically inactive mutant GAP1IP4BP. These data are entirely consistent with our previous in vitro analysis and support the conclusion that the catalytic machinery required for Ras GAP activity is located solely within the RasGRD of GAP1IP4BP (22). In contrast, while full-length GAP1IP4BP displayed robust Rap1 GAP activity, deletion of the tandem amino-terminal C2 domains or of the PH/Btk domain and the carboxyl-terminal tail resulted in mutant GAP1IP4BP proteins that were unable to display significant Rap1 GAP activity (Fig. 1C). These data establish, therefore, that both amino- and carboxy-terminal flanking regions on either side of the RasGRD are required for GAP1IP4BP to function as an efficient Rap1 GAP.
FIG. 1.
Disruption of any aspect of the domain structure perturbs the ability of GAP1IP4BP to function as a Rap GAP in vivo. (A) Schematic diagram of the GAP1IP4BP deletion constructs used in this study. (i) CHO-T cells were transiently cotransfected with 2.5 μg H-Ras (B) or 2.5 μg HA-tagged Rap1A (C) and 1 μg of the relevant GAP1IP4BP expression vector. Compared to control cells, the amount of Ras- and Rap1-GTP is significantly decreased in cells expressing wild-type GAP1IP4BP. This decrease in Ras-GTP only requires a functional GRD (cf. GAP1IP4BP with GAP1IP4BP ΔGRD/PH/C-tail; panel B). By contrast, Rap GAP activity of GAP1IP4BP requires the presence of all domains (panel C). Ras-GTP (B, part ii) and Rap1-GTP (C, part ii) levels from CHO-T cells are expressed as percentages of the pulldown level in control cells (average of six separate experiments ± the standard error of the mean).
Generation of GAP1IP4BP and GAP1m chimeras reveals that the PH/Btk domain is not required for the Rap1 GAP activity of GAP1IP4BP.
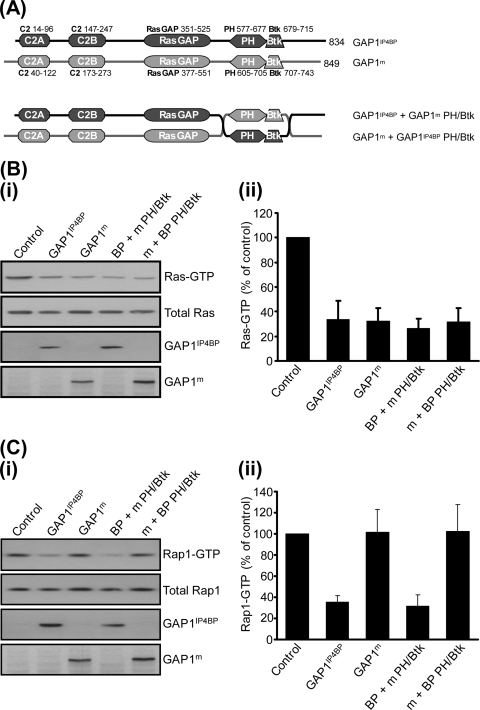
To examine the role of these flanking regions further, we took advantage of the observation that GAP1m is a specific Ras GAP that lacks the ability to function as a Rap1 GAP (22). Since GAP1IP4BP shows 60% sequence identity with GAP1m, we decided to generate full-length GAP1IP4BP-GAP1m chimeras. By utilizing such an approach, we hoped to identify the flanking domains required for the Rap1 GAP activity of GAP1IP4BP. Initially, we switched the respective PH/Btk domains between the two proteins (Fig. 2A). The chimeras that were generated—GAP1IP4BP containing the GAP1m PH/Btk domain (termed GAP1IP4BP + GAP1m PH/Btk) and GAP1m containing the GAP1IP4BP PH/Btk domain (GAP1m + GAP1IP4BP PH/Btk)—both displayed efficient GAP activity toward Ras, which was indistinguishable from the wild-type proteins (Fig. 2B). In contrast, only GAP1IP4BP containing the GAP1m PH/Btk domain displayed GAP activity toward Rap1 (Fig. 2C). No activity was detected with GAP1m containing the GAP1IP4BP PH/Btk domain (Fig. 2C). These data establish that by generating full-length GAP1IP4BP-GAP1m chimeras one can retain the Rap1 GAP activity of GAP1IP4BP and hence probe the domains required for this function. In addition, these data establish that the PH/Btk domain of GAP1IP4BP does not appear to contain the catalytic machinery and is hence dispensable for its Rap1 GAP activity.
FIG. 2.
Exchange of the PH and Btk domains between GAP1IP4BP and GAP1m has no effect on the Ras and Rap GAP activities of either hybrid protein in vivo. (A) Schematic diagram of the PH/Btk domain exchange constructs used in this study. (B, part i) CHO-T cells were transiently cotransfected with 2.5 μg Ras and 1 μg of the relevant GAP1IP4BP or GAP1m expression vector. Compared to that in control cells, the amount of Ras-GTP is significantly decreased in cells expressing wild-type GAP1IP4BP and GAP1m, as well as in cells expressing the GAP1IP4BP and GAP1m PH/Btk domain hybrid proteins. (C, part i) CHO-T cells were transiently cotransfected with 2.5 μg HA-tagged Rap1A and 1 μg of the relevant GAP1IP4BP or GAP1m expression vector. Compared to that in control cells, the amount of Rap1-GTP is only significantly decreased in cells expressing wild-type GAP1IP4BP and in cells expressing GAP1IP4BP with the PH/Btk domain from GAP1m. Wild-type GAP1m and GAP1m with the PH/Btk domain from GAP1IP4BP do not display Rap1 GAP activity. Ras-GTP (B, part ii) and Rap1-GTP (C, part ii) levels from CHO-T cells are expressed as percentages of the pulldown level in control cells (average of six separate experiments ± the standard error of the mean).
The tandem C2 domains are also not required for the Rap1 GAP activity of GAP1IP4BP.
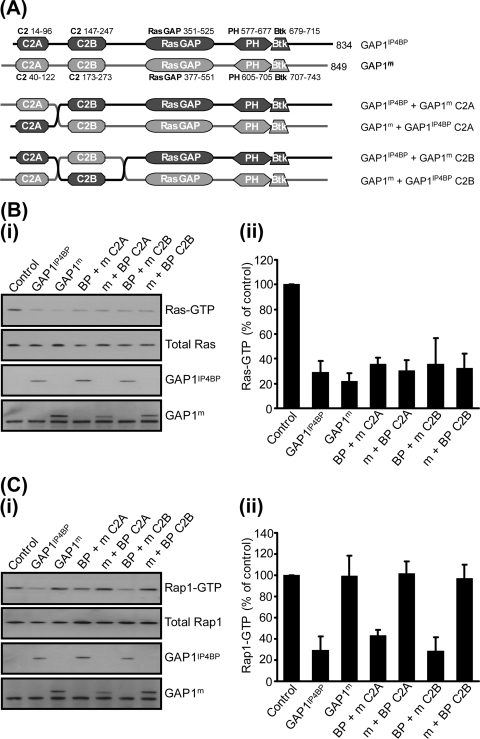
Having established that through the use of chimeras one could retain the Rap1 GAP activity of GAP1IP4BP, we next generated a series of six chimeras in which the C2A and C2B domains were swapped either individually or together (Fig. 3A; data not shown). All of these C2 domain chimeras displayed GAP activity toward Ras, which was again indistinguishable from that of the wild-type proteins (Fig. 3B). However, upon switching the C2 domains between GAP1IP4BP and GAP1m, only those chimeras retaining the GAP1IP4BP RasGRD displayed any GAP activity toward Rap1 (Fig. 3B). Taken together, therefore, these data, and those from the Btk/PH domain GAP1IP4BP-GAP1m chimeras, are consistent with a model in which the Rap1 GAP activity resides within the RasGRD of GAP1IP4BP. In such a model, the amino- and carboxy-terminal flanking regions of GAP1IP4BP are required solely to stabilize the RasGRD fold in such a way as to retain the conformation required for Rap1 GAP activity.
FIG. 3.
Exchange of the C2A or C2B domains between GAP1IP4BP and GAP1m has no effect on the Ras and Rap GAP activities of either hybrid protein in vivo. (A) Schematic diagram of the C2A and C2B domain exchange constructs used in this study. (B, part i) CHO-T cells were transiently cotransfected with 2.5 μg Ras and 1 μg of the relevant GAP1IP4BP or GAP1m expression vector. Compared to that in control cells, the amount of Ras-GTP is significantly decreased in cells expressing wild-type GAP1IP4BP and GAP1m, as well as in cells expressing the GAP1IP4BP and GAP1m C2 domain hybrid proteins. (C, part i) CHO-T cells were transiently cotransfected with 2.5 μg HA-tagged Rap1A and 1 μg of the relevant GAP1IP4BP or GAP1m expression vector. Compared to that in control cells, the amount of Rap1-GTP is only significantly decreased in cells expressing wild-type GAP1IP4BP and in cells expressing GAP1IP4BP with the C2A or C2B domain from GAP1m. Wild-type GAP1m and the hybrid forms of GAP1m do not display Rap1 GAP activity. Ras-GTP (B, part ii) and Rap1-GTP (C, part ii) levels from CHO-T cells are expressed as percentages of the pulldown level in control cells (average of six separate experiments ± the standard error of the mean).
Switching the RasGRD of GAP1IP4BP with the corresponding domain from GAP1m confers Rap1 GAP activity upon the resultant GAP1m chimera.
If the catalytic machinery required for the Rap1 GAP activity of GAP1IP4BP is present within its RasGRD, one would predict that switching this region with the corresponding domain from GAP1m should confer upon the resultant GAP1m chimera the ability to function as a Rap1 GAP. To test this, we generated the RasGRD GAP1IP4BP-GAP1m chimeras (Fig. 4A). Consistent with the model, GAP1m containing the RasGRD from GAP1IP4BP not only possessed robust Ras GAP activity but also was able to stimulate the GTPase activity of Rap1—dual activities that were similar to those observed with wild-type GAP1IP4BP (Fig. 4B). Unfortunately, the corresponding GAP1IP4BP chimera containing the GAP1m RasGRD was not expressed as well as the wild-type protein and this made a comparative analysis of its GAP activity difficult (Fig. 4C). However, at the level of expression achieved, Ras GAP activity was observed but we could detect no significant Rap1 GAP activity. These data are entirely consistent with the catalytic machinery required for the Rap1 GAP activity of GAP1IP4BP residing solely within its RasGRD.
FIG. 4.
Exchange of the GRDs between GAP1IP4BP and GAP1m transfers the Rap1 GAP activity from GAP1IP4BP to GAP1m in vivo. (A) Schematic diagram of the GRD exchange constructs used in this study. (B, part i) CHO-T cells were transiently cotransfected with 2.5 μg Ras and 1 μg of the relevant GAP1IP4BP or GAP1m expression vector. Compared to that in control cells, the amount of Ras-GTP is significantly decreased in cells expressing wild-type GAP1IP4BP and GAP1m, as well as in cells expressing the GAP1IP4BP and GAP1m GRD hybrid proteins. (C, part i) CHO-T cells were transiently cotransfected with 2.5 μg HA-tagged Rap1A and 1 μg of the relevant GAP1IP4BP or GAP1m expression vector. Compared to that in control cells, the amount of Rap1-GTP is significantly decreased in cells expressing wild-type GAP1IP4BP and in cells expressing GAP1m with the GRD from GAP1IP4BP. Wild-type GAP1m and GAP1IP4BP with the GRD from GAP1m do not appear to have Rap1 GAP activity. Ras-GTP (B, part ii) and Rap1-GTP (C, part ii) levels from CHO-T cells are expressed as percentages of the pulldown level in control cells (average of six separate experiments ± the standard error of the mean).
Molecular modeling of the GAP1IP4BP RasGRD reveals potential residues required for its Rap1 GAP activity.
Given that the RasGRD appears to contain the machinery required for Rap1 GAP activity, we utilized the coordinates from the crystal structure of the RasGRD of p120GAP and its complex with Ras to generate a molecular model that predicted the structure of the RasGRD-Ras complex of GAP1IP4BP (Fig. 5 and Materials and Methods). This model predicted the insertion of the functionally described GAP1IP4BP arginine finger—arginine 371—into the active site of the bound Ras-GTP (22), sufficiently close for it to stabilize the transition state of the GTPase reaction (Fig. 5C). Importantly, the GAP1IP4BP RasGRD model also predicted the localization of those asparagine residues (and chemically related glutamine residues) that lie in close proximity to the Ras-GTP binding site. As Rap1-GTP is predicted to associate with p120GAP in a manner similar to that of Ras-GTP (33), the GAP1IP4BP RasGRD model suggested that asparagine residues 367, 373, and 495 line the predicted Rap1-GTP binding pocket (Fig. 6). These residues may therefore function as asparagine thumbs in stimulating the GTP hydrolysis on Rap1 (Fig. 6).
FIG. 6.
Identifying potential asparagine thumbs of GAP1IP4BP. (A) GAP1IP4BP model showing the asparagine residues (red sticks) mutated to alanine. The protein is shown as a light blue (GRD) and dark blue (PH domain) ribbon with R371 as orange sticks. (B) GAP1IP4BP model showing the glutamine residues (pink sticks) mutated to alanine. The protein is shown as a light blue (GRD) and dark blue (PH domain) ribbon with R371 as orange sticks. (C) Ribbon representations of the GAP1IP4BP (light blue)-and-Ras (green) complex overlaid on the GAP1m (yellow)-and-Rap (red) complex. Residues V515 (GAP1m) and P489 (GAP1IP4BP) are displayed as sticks to show their close proximity to GTPase residue 64 on the switch II loop (Ras, Y; Rap, F). The view is a rotation of approximately 180° on the vertical axis with respect to panels A and B. R371 in GAP1IP4BP (orange sticks) and the Ras GTP (pink sticks) are shown for reference.
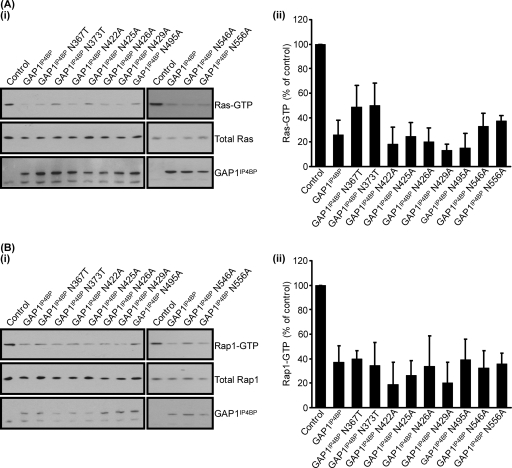
Mutagenesis of individual asparagine or glutamine residues in the proximity of the Ras-binding pocket of GAP1IP4BP has no effect on Rap1 GAP activity in vivo.
To determine whether any of the asparagines identified from the molecular model of GAP1IP4BP may constitute a catalytic asparagine thumb, we mutated each residue individually to either an alanine or, in some instances, a threonine residue (Fig. 7). The Ras GAP activity of most of these mutant proteins appeared indistinguishable from that of wild-type GAP1IP4BP (Fig. 7A). The exceptions were GAP1IP4BP(N367T) and GAP1IP4BP(N373T), where a slight reduction in Ras GAP activity was apparent (Fig. 7A). This most likely reflects the fact that these residues lie close to the catalytic arginine finger, GAP1IP4BP-R371, and hence may perturb activity through an alteration of the efficiency with which this residue is presented in these mutants. Interestingly, when assayed for in vivo Rap1 GAP activity, all of the asparagine mutant proteins appeared to display wild-type activity (Fig. 7B). Such data suggest that the ability of GAP1IP4BP to enhance the intrinsic GTPase activity of Rap1 does not require a mechanism that utilizes a classic asparagine thumb.
FIG. 7.
Mutation of asparagine residues within the GRD of GAP1IP4BP has no effect on Ras and Rap1 GAP activities in vivo. (i) CHO-T cells were transiently cotransfected with 2.5 μg H-Ras (A) or 2.5 μg HA-tagged Rap1A (B) and 1 μg of the relevant GAP1IP4BP expression vector. Compared to that in control cells, the amount of Ras- and Rap1-GTP is significantly decreased in cells expressing wild-type GAP1IP4BP, as well as in the cell lines expressing the asparagine point mutations. Ras-GTP (A, part ii) and Rap1-GTP (B, part ii) levels from CHO-T cells are expressed as percentages of the pulldown level in control cells (average of four separate experiments ± the standard error of the mean).
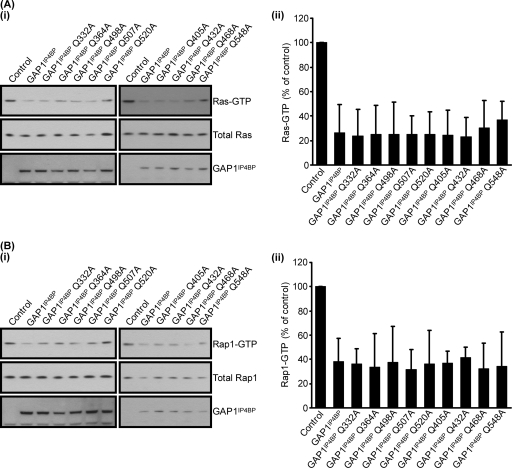
To further this analysis, we also mutated all of the glutamine residues that line the predicted GAP1IP4BP Rap1-binding pocket (Fig. 8). The Ras and Rap1 GAP activities of these mutant proteins were again indistinguishable from that of wild-type GAP1IP4BP (Fig. 8A and B, respectively). Taken together, therefore, these data are consistent with the notion that neither the asparagine nor the glutamine residues that line the predicted Rap1 binding pocket of GAP1IP4BP are required for this protein to function as a Rap1 GAP.
FIG. 8.
Mutation of glutamine residues within the GRD of GAP1IP4BP also has no effect on Ras and Rap1 GAP activities in vivo. (i) CHO-T cells were transiently cotransfected with 2.5 μg H-Ras (A) or 2.5 μg HA-tagged Rap1A (B) and 1 μg of the relevant GAP1IP4BP expression vector. Compared to that in control cells, the amount of Ras- and Rap1-GTP is significantly decreased in cells expressing wild-type GAP1IP4BP, as well as in the cell lines expressing the glutamine point mutations. Ras-GTP (A, part ii) and Rap1-GTP (B, part ii) levels from CHO-T cells are expressed as percentages of the pulldown level in control cells (average of four separate experiments ± the standard error of the mean).
Perturbing the α6c helix of GAP1IP4BP leads to a mutant protein that, while retaining Ras GAP activity, has reduced Rap1 GAP activity.
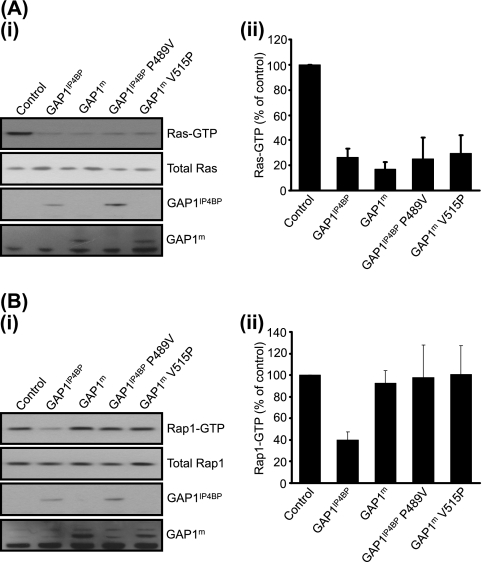
It is clear that mutation of arginine 371 of GAP1IP4BP—the catalytic arginine finger—inhibits the GAP activity against not only Ras but also Rap1 (22). Although the mechanistic details of the GAP activity must be different, given the lack of a corresponding glutamine in Rap1, the mechanism of Rap1 GAP activity may therefore be a variation upon that already described for Ras (33). With this in mind, we examined the predicted interaction surfaces between Ras and Rap1 and the RasGRD of GAP1IP4BP, focusing in particular on the α6c helix, which in the p120GAP-Ras complex forms a hydrophobic interface which stabilizes the important switch II region of Ras (33). On comparing the molecular models of the α6c helixes of GAP1IP4BP and GAP1m (Fig. 6C), the major difference was that proline 489 in GAP1IP4BP was replaced with valine 515 in GAP1m. Interestingly, when proline 489 was mutated into a valine, GAP1IP4BP(P489V) maintained its Ras GAP activity but had a clearly detectable decrease in its Rap1 GAP activity (Fig. 9). Such data are consistent with the hypothesis that the ability of GAP1IP4BP to stabilize the switch II region of Rap1 is an important component of its Rap1 GAP activity, in that it may allow an unidentified Rap1 residue to stabilize the transition state during GTP hydrolysis. However, proline 489 cannot be the only stabilizing feature of the Rap1 switch II region since the GAP1m(V515P) mutant protein did not display Rap1 GAP activity (Fig. 9).
FIG. 9.
Mutation of proline 489 to valine largely abolishes Rap1 GAP activity in GAP1IP4BP but has no effect on Ras GAP activity in vivo. (A, part i) CHO-T cells were transiently cotransfected with 2.5 μg H-Ras and 1 μg of the GAP1IP4BP, GAP1m, GAP1IP4BP (P489V), and GAP1m (V515P) expression vectors. Compared to that in control cells, the amount of Ras-GTP is significantly decreased in cells expressing wild-type GAP1IP4BP and GAP1m, as well as in cells expressing GAP1IP4BP (P489V) and GAP1m (V515P). (B, part i) CHO-T cells were transiently cotransfected with 2.5 μg Rap1 and 1 μg of the GAP1IP4BP, GAP1m, GAP1IP4BP (P489V), and GAP1m (V515P) expression vectors. Compared to that in control cells, the amount of Rap1-GTP is only significantly decreased in cells expressing wild-type GAP1IP4BP. Wild-type GAP1m, GAP1IP4BP (P489V), and GAP1m (V515P) do not appear to have Rap1 GAP activity. Ras-GTP (A, part ii) and Rap1-GTP (B, part ii) levels from CHO-T cells are expressed as percentages of the pulldown level in control cells (average of four separate experiments ± the standard error of the mean).
DISCUSSION
Here we have examined the mechanism by which the GAP1 family member GAP1IP4BP is able to function as dual Ras and Rap1 GAPs. Through the generation of a series of chimeras with GAP1m—a close relative of GAP1IP4BP that, while possessing Ras GAP activity, displays no Rap1 GAP activity (22)—we have established that the ability of GAP1IP4BP to function as a Rap1 GAP is not a consequence of either its C2 domains or its PH/Btk domain. These domains are, however, required in order to stabilize the RasGRD fold in such a way as to retain the conformation required for Rap1 GAP activity. Thus, although the isolated GAP1IP4BP RasGRD retains robust Ras GAP activity, the removal of any one of the flanking domains results in a mutant protein lacking Rap1 GAP activity (22).
The overall “hairpinlike” structure of the RasGRD from p120GAP and NF1 ensures that the amino- and carboxy-terminal regions of the RasGRD actually lie in close proximity to one another (34). For the GAP1 family, assuming a conservation of this basic structure, a conclusion supported by our modeling, the tandem C2 and PH/Btk domains would therefore also lie in close proximity to one another. Loss of either domain may lead to a subtle destabilization of the structure of the RasGRD, leading to the observed loss of GAP activity toward Rap1 but not Ras (22). That subtle destabilization of the RasGRD can have pronounced effects upon the GAP activity toward Rap1 is further highlighted by the GAP1IP4BP(P489V) mutant protein.
The ability to transfer the Rap1 GAP activity from GAP1IP4BP to GAP1m through exchange of the RasGRDs clearly establishes that it is the RasGRD from GAP1IP4BP that contains the necessary residues for Rap1 GAP activity. Precisely how the RasGRD of GAP1IP4BP is able to increase the intrinsic GTPases activity of Rap1 remains to be determined. While mutagenesis of the conserved catalytic arginine finger inhibits both the Ras and Rap1 GAP activities (22), mutagenesis of all of the asparagine and glutamine residues within the GRD, including those that may function as potential catalytic asparagine thumbs, has no major effect upon the Rap1 GAP activity of GAP1IP4BP. These data argue for a novel mode of Rap1 GAP activity for members of the GAP1 family that nevertheless requires the presence of an arginine finger. A similar importance of the presence of the RasGRD arginine finger has recently been described for the Rap1 GAP activity of SynGAP (30).
In contrast to the situation with SynGAP, where it has been suggested that the C2 domain may supply catalytic asparagine residues important in its Rap1 GAP activity (30), our studies find no role for the tandem C2 domains in the Rap1 GAP activity of GAP1IP4BP. In contrast, we prefer a model in which all of the required catalytic residues from GAP1IP4BP are housed within the RasGRD. We have presented evidence from deletion mutant proteins and the GAP1IP4BP(P489V) site-directed mutant protein which suggests that, unlike the Ras GAP activity, the GAP activity on Rap1 is sensitive to subtle changes in the organization of the RasGRD. It is our view, therefore, that the RasGRD of GAP1IP4BP is organized to stabilize the conformation of Rap1, and in particular its switch II region, such that residues within Rap1 stabilize the transition state during GTP hydrolysis initiated by the arginine finger. The nature of these residues and the exact mechanism through which they regulate Rap1 GAP activity await the elucidation of the crystal structure of the GAP1IP4BP RasGRD-Rap1 complex.
Acknowledgments
This work was funded by the Wellcome Trust.
We thank Hans Bos for provision of constructs for expression of GST-RalGDS and Rap1.
Footnotes
Published ahead of print on 11 May 2009.
REFERENCES
- 1.Ahmadian, M. R., P. Stege, K. Scheffzek, and A. Wittinghofer. 1997. Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nat. Struct. Biol. 4686-689. [DOI] [PubMed] [Google Scholar]
- 2.Allen, M., S. Chu, S. Brill, C. Stotler, and A. Buckler. 1998. Restricted tissue expression pattern of a novel human rasGAP-related gene and its murine ortholog. Gene 21817-25. [DOI] [PubMed] [Google Scholar]
- 3.Ballester, R., D. Marchuk, M. Boguski, A. Saulino, R. Letcher, M. Wigler, and F. Collins. 1990. The NF1 locus encodes a protein functionally related to mammalian GAP and yeast IRA proteins. Cell 63851-859. [DOI] [PubMed] [Google Scholar]
- 4.Bernards, A. 2003. GAPs galore! A survey of putative Ras superfamily GTPase activating proteins in man and Drosophila. Biochim. Biophys. Acta 160347-82. [DOI] [PubMed] [Google Scholar]
- 5.Bernards, A., and J. Settleman. 2004. GAP control: regulating the regulators of small GTPases. Trends Cell Biol. 14377-385. [DOI] [PubMed] [Google Scholar]
- 6.Bos, J. L., B. Franke, L. M'Rabet, K. Reedquist, and F. Zwartkruis. 1997. In search of a function for the Ras-like GTPase Rap1. FEBS Lett. 41059-62. [DOI] [PubMed] [Google Scholar]
- 7.Bottomley, J. R., J. S. Reynolds, P. J. Lockyer, and P. J. Cullen. 1998. Structural and functional analysis of the putative inositol 1,3,4,5-tetrakisphosphate receptors GAP1IP4BP and GAP1m. Biochem. Biophys. Res. Commun. 250143-149. [DOI] [PubMed] [Google Scholar]
- 8.Brinkmann, T., O. Daumke, U. Herbrand, D. Kuhlmann, P. Stege, M. R. Ahmadian, and A. Wittinghofer. 2002. Rap-specific GTPase activating protein follows an alternative mechanism. J. Biol. Chem. 27712525-12531. [DOI] [PubMed] [Google Scholar]
- 9.Chakrabarti, P. P., Y. Suveyzdis, A. Wittinghofer, and K. Gerwert. 2004. Fourier transform infrared spectroscopy on the Rap·RapGAP reaction, GTPase activation without an arginine finger. J. Biol. Chem. 27946226-46233. [DOI] [PubMed] [Google Scholar]
- 10.Chen, H. J., M. Rojas-Soto, A. Oguni, and M. B. Kennedy. 1998. A synaptic Ras-GTPase activating protein (p135 SynGAP) inhibited by CaM kinase II. Neuron 20895-904. [DOI] [PubMed] [Google Scholar]
- 11.Cook, S. J., B. Rubinfeld, I. Albert, and F. McCormick. 1993. RapV12 antagonizes Ras-dependent activation of ERK1 and ERK2 by LPA and EGF in Rat-1 fibroblasts. EMBO J. 123475-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cozier, G. E., P. J. Lockyer, J. S. Reynolds, S. Kupzig, J. R. Bottomley, T. H. Millard, G. Banting, and P. J. Cullen. 2000. GAP1IP4BP contains a novel group I pleckstrin homology domain that directs constitutive plasma membrane association. J. Biol. Chem. 27528261-28268. [DOI] [PubMed] [Google Scholar]
- 13.Cullen, P. J., J. J. Hsuan, O. Truong, A. J. Letcher, T. R. Jackson, A. P. Dawson, and R. F. Irvine. 1995. Identification of a specific Ins(1,3,4,5)P4-binding protein as a member of the GAP1 family. Nature 376527-530. [DOI] [PubMed] [Google Scholar]
- 14.Cullen, P. J., and P. J. Lockyer. 2002. Integration of calcium and Ras signalling. Nat. Rev. Mol. Cell Biol. 3339-348. [DOI] [PubMed] [Google Scholar]
- 15.Daumke, O., M. Weyand, P. P. Chakrabarti, I. R. Vetter, and A. Wittinghofer. 2004. The GTPase-activating protein Rap1GAP uses a catalytic asparagine. Nature 429197-201. [DOI] [PubMed] [Google Scholar]
- 16.Gao, Q., S. Srinivasan, S. N. Boyer, D. E. Wazer, and V. Band. 1999. The E6 oncoproteins of high-risk papillomaviruses bind to a novel putative GAP protein, E6TP1, and target it for degradation. Mol. Cell. Biol. 19733-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hattori, M., N. Tsukamoto, M. S. Nue-e-Kamal, B. Rubinfeld, K. Iwai, H. Kubota, H. Maruta, and N. Minato. 1995. Molecular cloning of a novel mitogen-inducible nuclear protein with a Ran GTPase-activating domain that affects cell cycle progression. Mol. Cell. Biol. 15552-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, J. H., D. Liao, L. F. Lau, and R. L. Huganir. 1998. SynGAP: a synaptic RasGAP that associates with the PSD-95/SAP90 protein family. Neuron 20683-691. [DOI] [PubMed] [Google Scholar]
- 19.Kitayama, H., Y. Sugimoto, T. Matsuzaki, Y. Ikawa, and M. Noda. 1989. A ras-related gene with transformation suppressor activity. Cell 5677-84. [DOI] [PubMed] [Google Scholar]
- 20.Kooistra, M. R. H., N. Dubé, and J. L. Bos. 2007. Rap1: a key regulator in cell-cell junction formation. J. Cell Sci. 12017-22. [DOI] [PubMed] [Google Scholar]
- 21.Krapivinsky, G., I. Medina, I. Krapivinsky, S. Gapon, and D. E. Clapham. 2004. SynGAP-MUPP1-CaMKII synaptic complexes regulate p38 MAP kinase activity and NMDA receptor-dependent synaptic AMPA receptor potentiation. Neuron 43563-574. [DOI] [PubMed] [Google Scholar]
- 22.Kupzig, S., D. Deaconescu, D. Bouyoucef, S. A. Walker, Q. Liu, C. L. Polte, O. Daumke, T. Ishizaki, P. J. Lockyer, A. Wittinghofer, and P. J. Cullen. 2006. GAP1 family members constitute bifunctional Ras and Rap GTPase-activating proteins. J. Biol. Chem. 2819891-9900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lockyer, P. J., S. Kupzig, and P. J. Cullen. 2001. CAPRI regulates Ca2+-dependent inactivation of the Ras-MAPK pathway. Curr. Biol. 11981-986. [DOI] [PubMed] [Google Scholar]
- 24.Lockyer, P. J., S. Wennstrom, S. Kupzig, K. Venkateswarlu, J. Downward, and P. J. Cullen. 1999. Identification of the ras GTPase-activating protein GAP1m as a phosphatidylinositol-3,4,5-trisphosphate-binding protein in vivo. Curr. Biol. 9265-268. [DOI] [PubMed] [Google Scholar]
- 25.Maekawa, M., S. A. Iwamatsu, T. Morishita, K. Yokota, Y. Imai, S. Kohsaka, S. Nakamura, and S. Hattori. 1994. A novel mammalian Ras GTPase-activating protein which has phospholipid-binding and Btk homology regions. Mol. Cell. Biol. 146879-6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manning, B. D., and L. C. Cantley. 2003. Rheb fills a GAP between TSC and TOR. Trends Biochem. Sci. 28573-576. [DOI] [PubMed] [Google Scholar]
- 27.Martin, G. A., D. Viskochil, G. Bollag, P. C. McCabe, W. J. Crosier, H. Haubruck, L. Conroy, R. Clark, P. O'Connell, R. M. Cawthon, M. A. Innis, and F. McCormick. 1990. The GAP-related domain of the neurofibromatosis type 1 gene product interacts with ras p21. Cell 63843-849. [DOI] [PubMed] [Google Scholar]
- 28.Mitin, N., K. L. Rossman, and C. J. Der. 2005. Signaling interplay in Ras superfamily function. Curr. Biol. 15R563-R574. [DOI] [PubMed] [Google Scholar]
- 29.Oinuma, I., Y. Ishikawa, H. Katoh, and M. Negishi. 2004. The semaphorin 4D receptor plexin-B1 is a GTPase activating protein for R-Ras. Science 305862-865. [DOI] [PubMed] [Google Scholar]
- 30.Pena, V., M. Hothorn, A. Eberth, N. Kaschau, A. Parret, L. Gremer, F. Bonneau, M. R. Ahmadian, and K. Scheffzek. 2008. The C2 domain of SynGAP is essential for stimulation of the Rap GTPase reaction. EMBO Rep. 9350-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rajalingam, K., R. Schreck, U. R. Rapp, and S. Albert. 2007. Ras oncogenes and their downstream targets. Biochim. Biophys. Acta 17731177-1195. [DOI] [PubMed] [Google Scholar]
- 32.Rubinfeld, B., S. Munemitsu, R. Clark, L. Conroy, K. Watt, W. J. Crosier, F. McCormick, and P. Polakis. 1991. Molecular cloning of a GTPase activating protein specific for the Krev-1 protein p21rap1. Cell 651033-1042. [DOI] [PubMed] [Google Scholar]
- 33.Scheffzek, K., M. R. Ahmadian, W. Kabsch, L. Wiesmuller, A. Lautwein, F. Schmitz, and A. Wittinghofer. 1997. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science 277333-338. [DOI] [PubMed] [Google Scholar]
- 34.Scheffzek, K., A. Lautwein, W. Kabsch, M. R. Ahmadian, and A. Wittinghofer. 1996. Crystal structure of the GTPase-activating domain of human p120GAP and implications for the interaction with Ras. Nature 384591-596. [DOI] [PubMed] [Google Scholar]
- 35.Schubbert, S., K. Shannon, and G. Bollag. 2007. Hyperactive Ras in developmental disorders and cancer. Nat. Rev. Cancer 7295-308. [DOI] [PubMed] [Google Scholar]
- 36.Scrima, A., C. Thomas, D. Deaconescu, and A. Wittinghofer. 2008. The Rap-RapGAP complex: GTP hydrolysis without catalytic glutamine and arginine residues. EMBO J. 271145-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stork, P. J. 2003. Does Rap1 deserve a bad Rap? Trends Biochem. Sci. 28267-275. [DOI] [PubMed] [Google Scholar]
- 38.Vogel, U. S., R. A. Dixon, M. D. Schaber, R. E. Diehl, M. S. Marshall, E. M. Scolnick, I. S. Sigal, and J. B. Gibbs. 1988. Cloning of bovine GAP and its interaction with oncogenic ras p21. Nature 33590-93. [DOI] [PubMed] [Google Scholar]
- 39.Wang, Z., C. P. Tseng, R. C. Pong, H. Chen, J. D. McConnell, N. Navone, and J. T. Hsieh. 2002. The mechanism of growth-inhibitory effect of DOC-2/DAB2 in prostate cancer. Characterization of a novel GTPase-activating protein associated with N-terminal domain of DOC-2/DAB2. J. Biol. Chem. 27712622-12631. [DOI] [PubMed] [Google Scholar]
- 40.Xu, G. F., B. Lin, K. Tanaka, D. Dunn, D. Wood, R. Gesteland, R. White, R. Weiss, and F. Tamanoi. 1990. The catalytic domain of the neurofibromatosis type 1 gene product stimulates ras GTPase and complements ira mutants of S. cerevisiae. Cell 63835-841. [DOI] [PubMed] [Google Scholar]
- 41.Yarwood, S., D. Bouyoucef-Cherchalli, P. J. Cullen, and S. Kupzig. 2006. The GAP1 family of GTPase-activating proteins: spatial and temporal regulators of small GTPase signalling. Biochem. Soc. Trans. 34846-850. [DOI] [PubMed] [Google Scholar]