Abstract
Stimulation of osteoblast differentiation from mesenchymal stem cells is a potential strategy for bone repair. Bone morphogenetic proteins (BMPs) that induce osteoblastic differentiation have been successfully used in humans to treat fractures. Here we outline a new approach to the stimulation of osteoblast differentiation using small molecules that stimulate BMP activity. We have identified the amiloride derivative phenamil as a stimulator of osteoblast differentiation and mineralization. Remarkably, phenamil acts cooperatively with BMPs to induce the expression of BMP target genes, osteogenic markers, and matrix mineralization in both mesenchymal stem cell lines and calvarial organ cultures. Transcriptional profiling of cells treated with phenamil led to the identification of tribbles homolog 3 (Trb3) as a mediator of its effects. Trb3 is induced by phenamil selectively in cells with osteoblastic potential. Both Trb3 and phenamil stabilize the expression of SMAD, the critical transcription factor in BMP signaling, by promoting the degradation of SMAD ubiquitin regulatory factor 1. Small interfering RNA-mediated knockdown of Trb3 blunts the effects of phenamil on BMP signaling and osteogenesis. Thus, phenamil induces osteogenic differentiation, at least in part, through Trb3-dependent promotion of BMP action. The synergistic use of small molecules such as phenamil along with BMPs may provide new strategies for the promotion of bone healing.
Bone is continuously remodeled throughout life by a tightly coupled process involving absorption by osteoclasts and formation by osteoblasts. Dysregulation of this coupled remodeling can lead to diseases such as osteoporosis (18, 19). The precursors of osteoblasts are pluripotent cells known as mesenchymal stem cells (MSCs) (3, 7, 36). MSCs are viewed as potential tools for therapeutic intervention in diseases related to impaired function of osteoblasts because they can be derived from bone marrow, manipulated in culture, and administered back to donor individuals (12, 13, 24, 39, 40). However, the mechanistic pathways that drive differentiation of MSCs along the osteoblast lineage are not completely understood. Therefore, elucidation of molecular mechanisms underlying osteogenesis not only is important for our understanding of bone development but also may advance strategies for bone repair. Targeting therapeutic molecules to bone in order to enhance the bone-forming activity of osteoblast precursors may aid in the treatment of bone disease.
Osteoinductive factors are required to drive the lineage-specific differentiation of MSCs into osteoblastic cells in culture. Osteoblast differentiation is influenced by multiple signaling pathways, including transforming growth factor β1, Hedgehog, Wnt, fibroblast growth factors, insulin-like growth factor 1, and bone morphogenetic proteins (BMPs) (8, 20, 25, 46, 49). Strategies employing BMPs have been successfully used with animals and humans to regenerate bones (16, 22, 27). However, the high cost and supraphysiologic doses of BMPs necessary to achieve osteoinductive activity illustrate the need for additional strategies for the stimulation of osteoblast differentiation and bone formation in vivo (23, 32, 48). Recent studies of zebrafish have validated the concept of employing small molecules to modulate BMP activity in vivo (50). Similarly, certain oxysterols have been shown to activate sonic hedgehog (SHH) and to stimulate osteoblastic differentiation and bone formation (1, 15). However, small-molecule BMP stimulators that are able to bypass the need for high doses of BMP and induce bone formation remain to be identified.
Here we show that the small molecule phenamil, a derivative of the diuretic amiloride, induces osteoblastic differentiation and mineralization of mouse MSCs. Phenamil and BMPs show additive effects on the expression of BMP target genes, osteogenic markers, and matrix mineralization in M2-10B4 (M2) MSCs as well as in calvarial organ cultures. We show that phenamil acts, at least in part, by inducing the expression of tribbles homolog 3 (Trb3), a previously identified positive regulator of BMP signaling (9, 33, 51). We further show that phenamil reduces the protein level of SMAD ubiquitin regulatory factor 1 (Smurf1) and induces expression of SMAD, the critical transcription factor in BMP signaling. These results suggest that phenamil or related small molecules may represent a novel strategy for increasing BMP activity in the clinical setting.
MATERIALS AND METHODS
Reagents.
Phenamil, amiloride, benzamil, and dimethyl amiloride hydrochloride (DMA) were purchased from Sigma. BMP2, BMP7, and SHH were from R&D Systems. Oligonucleotides were from Integrated DNA Technologies. Small interfering RNAs (siRNAs) for Trb3 and a nonspecific control were purchased from Dharmacon.
Cell culture.
The M2-10B4 (M2) mouse marrow stromal cells were purchased from ATCC and maintained as previously described. M2 cells were differentiated in RPMI containing 5% fetal bovine serum (FBS), 3 mM β-glycerophosphate, 50 μg/ml ascorbic acid, and antibiotics. Mouse embryonic stem (ES) cells were maintained in minimal essential medium with 10% FBS, 2 mM l-glutamine, 1 mM sodium pyruvate, 50 μM β-mercaptoethanol, and leukemia inhibitory factor. For osteogenesis, ES cells were directly plated at a cell density of 10,000/well in 0.1% gelatin-coated six-well plates. After 3 days, the medium was replaced with phenamil and osteogenic medium (minimal essential medium, 20% FBS, 2 mM l-glutamine, 1 mM sodium pyruvate, 50 μM β-mercaptoethanol, 50 μg/ml ascorbic acid, and 10 mM β-glycerophosphate with phenamil or dimethyl sulfoxide [DMSO]). The culture was maintained at 37°C with 5% CO2 and fed every 2 days. siRNAs for Trb3 and a nonspecific control from Dharmacon were resuspended according to the manufacturer's instructions. siRNAs were transfected using Lipofectamine RNAiMAX (Invitrogen). A concentration of 10 nM siRNAs was transfected into 80% confluent M2 cells.
Transient transfection and reporter assay.
M2 cells at 80% confluence were transfected using Lipofectamine (Invitrogen). Gli-luc for HH activity and BRE-luc for BMP activity were used (15). Transfected cells were treated for 48 h with phenamil (10 μM), BMP (100 ng/ml), and SHH (100 ng/ml), and then luciferase activity was measured using the dual-luciferase reporter 1000 assay system (Promega). Firefly luciferase activities were normalized with Renilla luciferase activity.
Mouse calvarial organ culture.
Mouse calvariae were obtained from 5-day-old neonatal CD1 mice by aseptic excision. The calvariae were cleared of soft tissue and washed in phosphate-buffered saline. These mouse calvariae (four calvariae per well) were incubated in six-well plates containing Dulbecco modified Eagle medium with 10% heat-inactivated FBS, supplemented with 1 mM sodium pyruvate, 100 U/ml penicillin, and 100 U/ml streptomycin for 2 hours before the treatment. Each calvaria was placed in a well of 24-well plates containing Dulbecco modified Eagle medium with 10% heat-inactivated FBS, 3 mM β-glycerophosphate, 50 μg/ml ascorbic acid, and antibiotics as described above. After 72 h of incubation with BMP, phenamil, or a combination, calvariae were homogenized with an Ultra-Turrax homogenizer (IKA Works, Inc., Wilmington, NC). Total RNA was extracted with the RNA isolation kit from Stratagene (La Jolla, CA) according to the manufacturer's instructions.
ALP assay and 45Ca incorporation assay.
Colorimetric measurement of alkaline phosphatase (ALP) activity was performed on whole-cell lysates after 3 to 4 days of treatment as previously described (15). 45Ca incorporation assay as a measure of matrix mineralization was done after 14 to 21 days of culture as previously described (28).
RNA and protein analysis.
Total RNA was isolated using TRIzol reagent (Invitrogen). Total RNA (0.5 μg) was reverse transcribed using IScribe (Bio-Rad) according to the manufacturer's instructions. Real-time quantitative PCR (SYBR green) analysis was performed on a 7900HT Fast real-time PCR system (Applied Biosystems). Expression was normalized to 36B4. The following primers were used for real-time PCR: GLI1 forward (gliF) (5′-GCTTGGATGAAGGACCTTGTG-3′) and reverse (gliR) (5′-GCTGATCCAGCCTAAGGTTCTC-3′), Patched 1 forward (patchF) (5′-TTCTGCTGCCTGTCCTCTTATC-3′) and reverse (patchR) (5′-CCTGCTGTGCTTCGTATTGC-3′), osteocalcin (OCN) forward (ocnF) (5′-TCTCTCTGACCTCACAGATGCC-3′) and reverse (ocnR) (5′-TACCTTATTGCCCTCCTGCTTG-3′), ALP forward (alpF) (5′-AAACCCAGAACACAAGCATTCC-3′) and reverse (alpR) (5′-TCCACCAGCAAGAAGAAGCC-3′), ID1 forward (id1F) (5′-GCGAGATCAGTGCCTTGG-3′) and reverse (id1R) (5′-CTCCTGAAGGGCTGGAGTC-3′), Trb1 forward (trb1F) (5′-CCTGAAGCTCAGGAAGTTCG-3′) and reverse (trb1R) (5′-CCAGGCTTTCCAGTCTAAGC-3′), Trb2 Forward (trb2F) (5′-TGGAGGGAGACCACGTTTT-3′) and reverse (trb2R) (5′-TGGAGGGAGACCACGTTTT-3′), Trb3 forward (trb3F) (5′-CTGAGGCTCCAGGACAAGA-3′) and reverse (trb3R) (5′-CCTGCAGGAAACATCAGCA-3′), OSF-2 forward (OSF2F) (5′-AAGCTGCGGCAAGACAAG-3′) and reverse (OSF2R) (5′-TCAAATCTGCAGCTTCAAGG-3′), Cyp26b1 forward (cyp26b1F) (5′-ACATCCACCGCAACAAGC-3′) and reverse (cyp26b1R) (5′-GGGCAGGTAGCTCTCAAGTG-3′), FST forward (fstF) (5′-TGGATTAGCCTATGAGGGAAAG-3′) and reverse (fstR) (5′-TGGAATCCCATAGGCATTTT-3′), SLC6a9 forward (slcF) (5′-TTCCCTTTAAGAAAGCCACCT-3′) and reverse (slcR) (5′-AGACAACAGGCCTCAAGAGC-3′), RGS forward (rgsF) (5′-CAGAAGACTTGAAATTCTGTGTGC-3′) and reverse (rgsR) (5′-TGCCTCCAAAGTGAATAAGCA-3′), EGFl9 forward (egfl9F) (5′-ATGACTGCAGCTCCCACTG-3′) and reverse (egfl9R) (5′-ATCCTCACACAGCGCTCAC-3′), osterix forward (osxF) (5′-GCTAGAGATCTGAGCCGGGTA-3′) and reverse (osxR) (5′-AAGAGAGCCTGGCAAGAGG-3′, and collagen I forward (col IF) (5′-ACCTAAGGGTACCGCTGGA-3′ and reverse (col 1R) (5′-TCCAGCTTCTCCATCTTTGC-3′). Isolation of nuclear extract and immunoblotting were performed as described previously (10). Protein extract was resuspended in NuPage LDS sample buffer (Invitrogen) and loaded on a 4 to 12% NuPage gel (Invitrogen). Smurf1 antibody (Cell signaling) was used at a 1:500 dilution, SMAD1/5/8 antibody (Santa Cruz Biotech) was at 1:1,000, and HMG-1 (BD Pharmingen) was at 1:5,000.
Microarrays.
Total RNA from M2 cells treated with phenamil for 24 h was prepared using TRIzol and further purified using RNeasy columns (Qiagen). cDNA preparation and hybridization to chips were performed by the UCLA Core Facility. Data were analyzed using Beadstring.
RESULTS
The amiloride derivative phenamil induces osteoblast differentiation and mineralization.
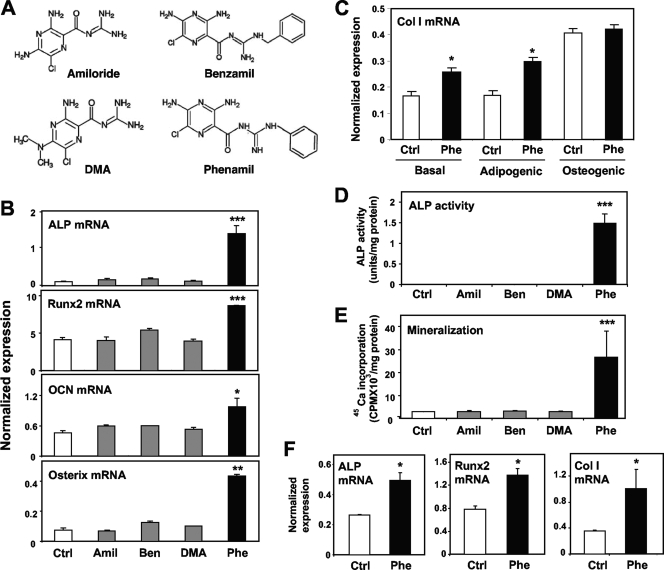
In the course of studying the effects of small molecules on gene expression in MSCs, we serendipitously discovered that the amiloride derivative phenamil promotes the expression of genes linked to osteoblastic differentiation (Fig. 1A). Treatment of M2-10B4 (M2) mouse MSCs for 6 days with 10 μM phenamil markedly stimulated expression of the genes encoding ALP, Runx2, OCN, and osterix (Fig. 1B). Similar effects of phenamil on markers of osteoblast differentiation were also observed in the C3H 10T1/2 MSC line (see Fig. S1 in the supplemental material). Collagen I is also an important marker of bone differentiation. We found that phenamil promoted collagen I expression in M2 cells in the absence of osteogenic medium (i.e., without ascorbic acid and β-glycerophosphate) (Fig. 1C). However, culture of M2 cells in osteogenic medium alone was sufficient to induce collagen I expression, and this was not further induced by phenamil.
FIG. 1.
Phenamil induces osteoblastic differentiation and mineralization in MSCs. (A) Chemical structures of amiloride derivatives. (B to D) Effects of amiloride derivatives on markers of osteoblastic differentiation. Phenamil stimulated expression of osteogenic genes, ALP activity, and mineralization. (B) M2 cells were treated for 6 days with 10 μM amiloride derivatives, and expression of osteoblastic genes encoding ALP, Runx2, osteocalcin, and osterix were measured by real-time PCR. (C) M2 cells were culture under basal conditions, adipogenic conditions (insulin, dexamethasone, and IBMX), or osteogenic conditions (ascorbic acid and β-glycerolphosphate). (D) M2 cells were treated for 3 days as indicated. (E) Mineralization was determined after 3 weeks of treatment by 45Ca incorporation assay. (F) Mouse ES cells were treated for 3 days with 10 μM phenamil, and expression of the osteoblastic genes for collagen I (Col I), ALP, and Runx2 were measured by real-time PCR. *, P < 0.05; **, P < 0.005; ***, P < 0.0005. Error bars indicate standard errors of the means.
Furthermore, treatment of M2 cells for 3 days with phenamil dramatically induced ALP activity (Fig. 1D). Treatment for 3 weeks induced mineralization measured by 45Ca incorporation (Fig. 1E). Remarkably, phenamil also induced collagen I, ALP, Runx2, and other osteogenic gene products in mouse ES cells (Fig. 1F and data not shown). The amilorides are a well-characterized class of small molecules known to inhibit epithelial sodium channel (ENaC) activity (2, 5, 6, 17, 29). Due to this activity, amilorides are widely used as diuretics in humans (30, 37). To determine the correlation between the osteogenic effects of phenamil and ENaC-blocking activity, we assayed three additional amiloride derivatives that are also known to inhibit ENaC. Interestingly, only phenamil was able to promote osteogenic gene expression and mineralization (Fig. 1B, D, and E). In addition, mesenchymal M2 cells do not express the epithelial transporters ENaCα, ENaCβ, and ENaCγ (data not shown). Together, these data strongly suggest that the osteogenic effects of phenamil are not related to its previously known effects on sodium channels.
Phenamil and BMP have additive effects on osteoblast differentiation and mineralization.
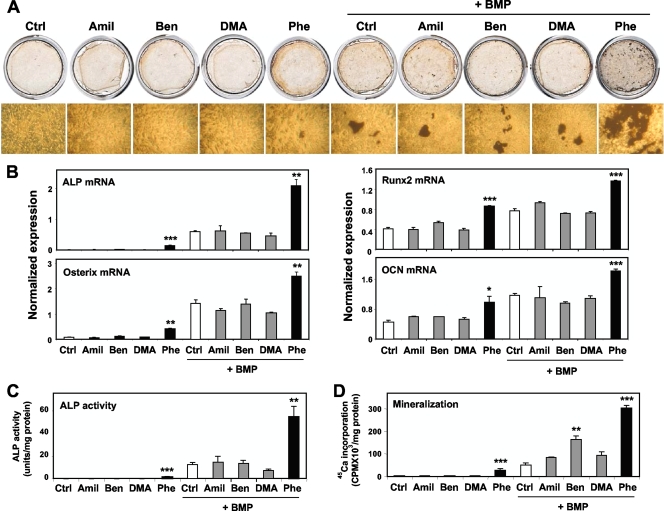
BMP is an osteogenic growth factor used in human therapeutics. To our knowledge, very few if any small molecules that enhance BMP actions in osteogenesis have been identified. To test whether phenamil would further enhance osteoblast differentiation driven by BMP signaling, we treated M2 cells with amiloride derivatives in combination with BMP7 or BMP2 (100 ng/ml). As shown in Fig. 2A, phenamil and BMP7 had synergistic effects on osteoblast differentiation as determined by Von Kossa staining. Similar results were obtained with BMP2 (data not shown). In combination with BMP, phenamil, but not other amiloride derivatives, strongly promoted ALP, Runx2, osterix, and OCN mRNA expression (Fig. 2B). Furthermore, ALP activity and 45Ca incorporation were also synergistically stimulated by phenamil and BMP7 or BMP2 (Fig. 2C and D and data not shown). These data show that phenamil works cooperatively with BMPs to promote osteoblast differentiation and mineralization.
FIG. 2.
Phenamil and BMPs work cooperatively to promote osteoblastic differentiation and mineralization in MSCs. Phenamil and BMP additively enhanced osteoblastic differentiation when combined with BMP as determined by Von Kossa staining, real-time PCR, mineralization, and ALP activity. Phenamil but not other derivatives further stimulated osteoblastic differentiation driven by BMP treatments. (A) M2-10B4 cells were treated for 4 weeks with 10 μM amiloride derivatives in the presence of BMP, and osteoblastic differentiation was determined by Von Kossa staining. (B) M2-10B4 cells were treated with amiloride derivatives with BMP7 for 6 days, and then expression of ALP, Runx2, OCN, and osterix was measured. (C) Mineralization was determined after 3 weeks of treatment by 45Ca incorporation assay. (D) M2-10B4 cells were treated with amiloride derivatives and BMP7 or BMP2 for 3 days, and ALP activity was determined. *, P < 0.05; **, P < 0.005; ***, P < 0.0005. Error bars indicate standard errors of the means.
The osteogenic effect of phenamil results from modulation of BMP signaling.
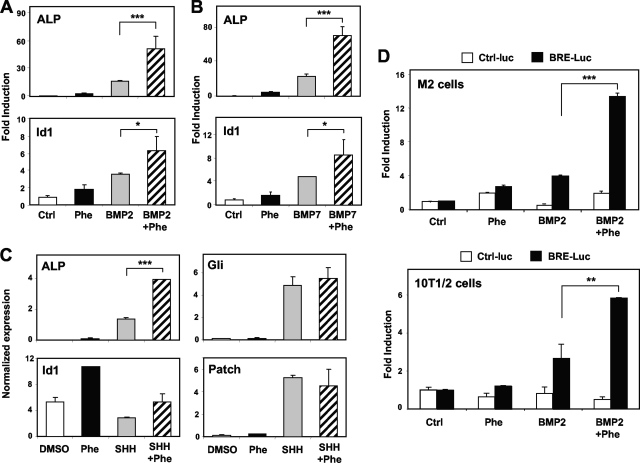
Based on the additive effects of phenamil and BMPs in osteoblastic differentiation, we postulated that phenamil might be acting to potentiate BMP signaling. To test this possibility, we examined expression of the classic BMP target gene Id1, along with other BMP responsive genes. M2 cells were treated with 100 ng/ml of BMP2 or BMP7 and 10 μM phenamil for 72 h. As shown in Fig. 3A and B, phenamil and either BMP2 or BMP7 had additive effects on Id1 and ALP mRNA.
FIG. 3.
Osteogenic effects of phenamil are mediated by BMP signaling. M2 cells were treated with BMP2, BMP7, or SHH (100 ng/ml) and 10 μM phenamil for 72 h. (A and B) Expression of ALP and Id1 mRNA was additively induced by phenamil and either BMP2 or BMP7. (C) Treatment with SHH and phenamil additively induced ALP. However, SHH failed to upregulate Id1 expression. In addition, phenamil did not affect known SHH target genes, for Gli1 and Patch. (D) M2 cells and 10T1/2 cells were transiently transfected with a BMP-specific luciferase reporter construct (BRE-Luc) and treated with phenamil (10 μM), BMP2 (100 ng/ml), or a combination of both BMP2 and phenamil for 2 days. The combination of phenamil and BMP synergistically promoted BMP-dependent reporter activity in both cell types. *, <0.05; **, P < 0.005; ***, P < 0.0005. Error bars indicate standard errors of the means.
We further tested the effect of phenamil on the expression of a BMP-specific reporter construct in transfection assays. M2 cells and 10T1/2 cells were transiently transfected with BMP specific-luciferase reporter (BRE-luc), and then treated with phenamil (10 μM), BMP2 (100 ng/ml), or both for 2 days. Both BMP2 and phenamil modestly induced reporter in both MSCs lines (Fig. 3D). However, consistent with the additive effects observed on induction of BMP target genes, BMP reporter activity strongly stimulated by cotreatment of phenamil and BMP (up to 14-fold). These observations suggest that BMP and phenamil stimulate BMP signaling through distinct but complementary mechanisms.
To establish that phenamil's effect on BMP signaling was not simply secondary to enhanced osteoblast differentiation, we tested another osteogenic growth factor, SHH. Treatment with SHH (100 ng/ml) and phenamil additively induced osteogenic markers such as ALP (Fig. 3C and data not shown). This observation is consistent with the previous findings showing that BMP and SHH pathways have additive effects on bone formation (44). However, SHH failed to upregulate Id1 expression, suggesting that Id1 expression is selectively responsive to BMP activation. Additionally, phenamil did not affect expression of the known SHH target gene products, Gli1 and Patch (Fig. 3C). Furthermore, phenamil had no effect on SHH activity as measured in luciferase-based Gli transcriptional reporter assays (data not shown). These data strongly suggest that phenamil's osteogenic activity is linked to modulation of BMP signaling.
BMP and phenamil have additive effects in ex vivo assays.
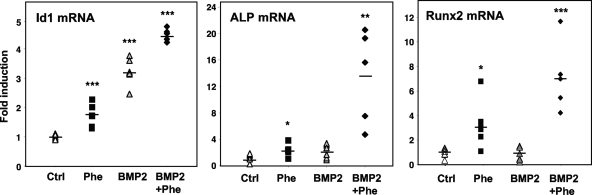
To gain additional insight into the efficacy of phenamil in promoting osteoblast differentiation, we employed a well-established organ culture assay of mouse calvariae (11, 21). Cultures were established and incubated in the presence of phenamil for 72 h. Determination of gene expression revealed that both phenamil and BMP modestly induced ALP and Runx2 expression (Fig. 4). Remarkably, however, cotreatment with phenamil and BMP robustly induced ALP and Runx2 expression. Id1 expression was induced by phenamil or BMP alone, and the combination also had an additive effect. Collagen I expression was high at baseline in these cultures and was not further induced by either phenamil or BMP (data not shown). Consistent with our data from MSCs, these results indicate that BMP and phenamil act cooperatively to promote osteogenic differentiation.
FIG. 4.
BMP2 and phenamil have additive effects in ex vivo assays of bone differentiation. Organ cultures of mouse calvariae (11, 21) were established and incubated in the presence of phenamil and/or BMP for 72 h. Phenamil and BMP additively enhanced osteoblastic differentiation. Id1 expression was also induced by phenamil or BMP alone, and the combination has an additive effect. Data are presented as mean ± standard errors of the means. Dots and bars in scatter plots represent individual organ culture (n = 5 or 6 per group) and average, respectively. *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
Identification of genes induced by phenamil.
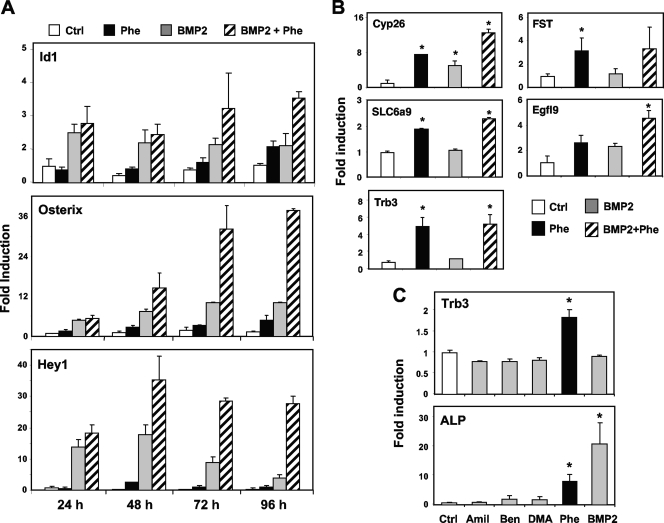
Analysis of the time course of osteoblastic differentiation revealed that phenamil's effects on Id1 expression were substantially delayed compared to those of BMP. As shown in Fig. 5A, whereas BMP induced expression of its known target Id1 within 24 h, maximal induction by phenamil required 48 h or longer. Furthermore, the additive effects of BMP and phenamil were most apparent after 48 h. Similar effects were observed on the BMP targets gene products Hey1 and osterix. These results led us to hypothesize that the effects of phenamil may be achieved through induction of a secondary mediator, rather than by direct modulation of BMP signaling. We therefore sought to identify phenamil-regulated genes that might play a role in BMP signaling and osteogenic differentiation. Our strategy was based on the hypothesis that candidate mediators of phenamil action would be induced by phenamil but not BMP. We reasoned that genes whose expression was induced by both BMP and phenamil were more likely to be primary BMP targets. Additionally, we postulated that candidate genes would be specifically induced by phenamil but not by other amiloride derivatives.
FIG. 5.
Identification of genes regulated by phenamil. (A) Time course of effects of phenamil on BMP signaling. While BMP induces Id1, Hey1, and osterix expression within 24 h, maximal induction by phenamil requires 48 h or longer. Additive effects of BMP and phenamil are also most apparent after 48 h. (B) Microarray analysis of M2 cells treated with either DMSO or phenamil for 24 h. Selected genes showing a statistically significant difference of twofold or greater between DMSO- and phenamil-treated samples are shown. Real-time PCR confirmed the differential expression of Trb3, EGFl9, FST, Cyp26b1, and SLC6a9 in response to 24 h of phenamil treatment. (C) Selective regulation of Trb3 by phenamil. Trb3 displays a pattern of phenamil-specific induction. ALP is induced by both BMP and phenamil treatments at 48 h. *, P < 0.05. Error bars indicate standard errors of the means.
To identify genes mediating phenamil effects, we profiled gene expression in M2 cells treated with DMSO or phenamil for 24 h. At this time point, Id1 stimulation by phenamil is not detectable, reducing the likelihood that primary BMP targets would be identified in our screen. Genes showing a statistically significant difference of twofold or greater between DMSO- and phenamil-treated samples are presented in Fig. S2 in the supplemental material. Real-time PCR confirmed differential expression of Trb3, EGFl9, FST, Cyp26b1, and SLC6a9 in response to 24 h of phenamil treatment (Fig. 5B). Interestingly, recent studies have identified Trb3 as a BMP receptor-interacting protein that positively affects BMP signaling (9). Among our amiloride derivatives, the ability to induce Trb3 expression was specific for phenamil (Fig. 5C). Furthermore, induction of Trb3 and Id1 expression by phenamil was relatively specific for osteogenic cells and was not observed in preadipocytes, fibroblasts, or hepatocytes (data not shown; see Fig. S3 in the supplemental material). Together, these data suggest that Trb3 may be a mediator of phenamil action in BMP signaling and ultimately in osteogenic differentiation.
Trb3 is a mediator of the osteogenic effects of phenamil.
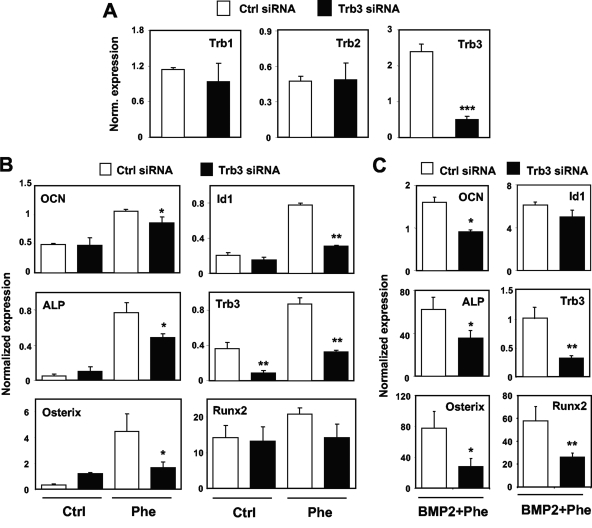
To directly test the importance of Trb3 expression in the regulation of osteogenic differentiation by phenamil, we conducted knockdown studies. Trb3 siRNAs are well validated and have been used previously by others to successfully knock down Trb3 (9). Transient transfection of M2 cells with Trb3 siRNA sequences effectively reduced mRNA levels of Trb3 but not of the related factors Trb1 and Trb2 (Fig. 6A). Remarkably, Trb3 knockdown also compromised the ability of phenamil to induce the osteoblast differentiation-related markers ALP, osterix, Runx2, and OCN in M2 cells (Fig. 6B and C). Furthermore, in accordance with the effects of phenamil in BMP signaling, knockdown of Trb3 blunted the induction of Id1 expression by phenamil or phenamil and BMP (Fig. 6B and C). Stable expression of Trb3 in 10T1/2 cell by means of a retroviral vector promoted osteogenic gene expression, in accordance with previous work, but had no effect on SHH dependent gene expression (see Fig. S4A and B in the supplemental material). Treatment with phenamil still promoted osteogenic gene expression in the presence of ectopic Trb3, but the fold induction was reduced significantly (see Fig. S4B and C in the supplemental material). These data identify Trb3 as one component of the signaling pathway by which phenamil regulates BMP signaling and osteogenic differentiation.
FIG. 6.
Trb3 mediates osteogenic effects of phenamil. (A) Transient transfection of M2 cells with Trb3 siRNA sequences effectively reduced mRNA levels of Trb3 but not of the related factors Trb1 and Trb2. (B and C) Trb3 knockdown blunts induction of ALP, OCN, and Id1 expression by phenamil in the presence or absence of BMP. *, P < 0.05; **, P < 0.005; ***, P < 0.0005. Error bars indicate standard errors of the means.
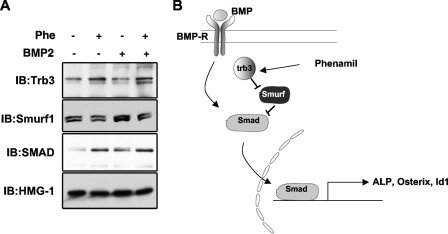
Previously, Trb3 has been shown to enhance BMP signaling by enhancing degradation of the SMAD antagonist Smurf1, resulting in stabilization of SMADs (9, 33, 51). We reasoned that if phenamil acts through Trb3, it should also affect levels of Smurf1 and SMAD. To examine this possibility, M2 cells were treated with phenamil and/or BMP for 3 days, and the expression of Smurf1 and BMP SMADs (SMAD1/5/8) was monitored by immunoblot analysis. Phenamil treatment induced Trb3 protein expression and led to a modest but highly reproducible decrease in Smurf1 expression, in both the presence and absence of BMP. Furthermore, decreased Smurf1 expression in response to phenamil was accompanied by increased SMAD expression. Taken together, our results support a model in which phenamil-induced Trb3 expression potentiates BMP signaling by negatively regulating Smurf1 and in turn increasing the abundance of BMP signal transducer SMAD1/5/8 (Fig. 7).
FIG. 7.
Phenamil potentiates BMP signaling by negatively regulating Smurf1 and increasing SMAD signaling. (A) M2 cells were treated with phenamil for 3 days, and the amounts of Trb3, Smurf1, and BMP SMADs (SMAD1/5/8) were measured by immunoblot (IB) analysis. Phenamil treatment decreased Smurf1 and increased Trb3 and SMAD levels. (B) Schematic diagram of the mechanism for regulation of the BMP signaling pathways by phenamil. Phenamil-induced Trb3 expression leads to downregulation of the BMP antagonist Smurf1, which in turn activates the BMP signaling by stabilizing the BMP signaling transducer SMAD.
DISCUSSION
Here we identify phenamil as a novel osteogenic small molecule. Phenamil induces osteogenic gene expression and mineralization in both MSC and primary calvariae organ cultures. Moreover, this effect is additive with BMP signaling in both MSCs and ex vivo assays. We also showed that phenamil mediates this response by modulating BMP signaling pathways. Mechanistic studies identified Trb3 as a phenamil-responsive gene. Inhibition of Trb3 expression with an siRNA approach blunted the induction of osteogenic markers and BMP downstream target genes by phenamil. Consistent with the known effect of Trb3 on BMP signaling (9), phenamil reduced the levels of the inhibitor Smurf1 and increased levels of the BMP signal transducer SMAD. Together, our data identify Trb3 as one component of the signaling pathway by which phenamil regulates osteoblastic gene expression and differentiation. These data also suggest potential therapeutic uses for phenamil or related compounds as BMP stimulators in bone formation.
Signaling mediated by BMP plays multiple roles in vertebrates. The dorsoventral axis is defined by BMP signaling during embryonic development, and BMPs are critical regulators of gastrulation, organogenesis, and bone formation. BMPs are also the best-characterized pharmacological osteoinductive factors (49). They induce osteoblastic differentiation in vitro and in vivo, and their potential clinical utility in bone repair is being explored actively. BMP ligands activate serine/threonine receptor kinases, leading to phosphorylation of SMAD effectors (SMAD1, SMAD5, and SMAD8). These SMADs translocate to the nucleus in complex with SMAD4, ultimately leading to induction of BMP target genes such as Id1. BMPs are also known to signal via SMAD-independent pathways. For example, BMP can signal through MAPK or protein kinase C to regulate growth in pulmonary artery muscle cells or apoptosis in osteoblast cells, respectively (34). BMP pathways are implicated in numerous diseases, including pulmonary hypertension, hereditary hemorrhagic telangiectasia syndrome, and fibrodysplasia ossificans progressiva (38, 43, 47). It is clear that complex regulatory mechanisms for BMP signaling have evolved in development and tissue homeostasis. Therefore, further understanding and fine-tuning of BMP signaling will be required to establish therapeutic tools capable of manipulating these pathways for treatment of disease.
Amiloride, a member of a group of compounds called pyrazinecarboxamides, is a well-characterized diuretic. ENaCs and the Na+/H+ exchanger have been identified as putative molecular targets for the renal effects of amiloride and its derivatives (2, 5, 6, 17). Introduction of hydrophobic (or hydrophilic) groups on the 5-amino moiety of amiloride enhances inhibitory activity against the Na+/H+ exchanger, while addition of hydrophobic substitutes on the terminal nitrogen of the guanidine moiety of amiloride increases activity against ENaCs (29, 31). Importantly, these sodium channels do not appear to be the targets for the osteogenic effects of phenamil. Amiloride, benzamil, and DMA do not share the ability to promote osteoblastic differentiation and enhance BMP signaling, even at concentrations of as high as 100 μM. Phenamil and benzamil are also known to inhibit diamine oxidase. However, diamine oxidase, along with ENaCα, -β, and -γ, is not detectable in M2 cells by real-time PCR (data not shown). Although the precise protein targets remains to be identified, our experiments indicate that phenamil exerts its effects, at least in part, through induction of Trb3 and modulation of BMP signaling in MSCs. Identification of the direct molecular target of phenamil's effects will be an interesting avenue for future studies. The mechanism whereby phenamil induces Trb3 expression also remains to be determined. We also cannot exclude the contribution of other pathways to phenamil's effects on MSCs.
Trb3 is a pseudokinase whose expression can be regulated by stress and nutrients. It has been reported to play roles in cell cycle control, transcriptional regulation, lipid synthesis, insulin signal transduction, and BMP signaling. All members of the tribbles family (Trb1 to -3) bind to the CDC25 homolog string and promote its degradation (42). Trb3 has also been suggested to interact with many proteins, including CHOP and ATF4, to regulate their transcriptional activity (35). Trb3 is induced under fasting conditions to inhibit lipid synthesis by degrading acetyl coenzyme A, a rate-limiting enzyme in fatty acid synthesis (41). Trb3 has also been reported to suppress insulin signaling by inhibiting the activity of the protein kinase AKT (14, 26). Of particular relevance for our work is the previous observation that Trb3 acts to promote BMP signaling and osteogenesis (9). Upon BMP stimulation, Trb3 dissociates from BMPRII-TD and triggers degradation of Smurf1, leading to stabilization of SMADs and potentiation of the SMAD pathway. Similarly, our data showed that phenamil also increases SMAD and downregulates the antagonist Smurf1.
The induction of Trb3 expression in osteogenic cells by phenamil is consistent with the delayed effects of this small molecule on BMP signaling and osteogenic gene expression. Osteoblast and adipocyte differentiations are often reciprocally regulated. It is therefore interesting to note that Trb3 has been shown to suppress adipocyte differentiation by inhibiting PPARγ and C/EBPβ transcriptional activities (4, 45). Expression of Trb3, however, is not regulated by phenamil in preadipocytes (see Fig. S1 in the supplemental material).
Our studies imply that deciphering the mechanisms of action of bioactive small molecules could bring new insight into MSC biology. In particular, further dissection of the molecular mechanisms underlying the additive effects of phenamil and BMP in lineage-specific differentiation could advance our current understanding of osteogenesis. MSCs are also being contemplated for use in clinical applications associated with bone loss (24, 40). A better understanding of osteogenic inductive pathways in vivo will be required to advance MSCs as tools to promote bone regeneration. BMP2 has been successfully used as an osteoinductive agent to promote fracture healing in humans (16, 22, 27). However, the clinical use of BMPs is currently limited by cost and difficulties in delivery of concentrated recombinant protein (23, 32, 48). Hence, complementary and/or more cost-effective strategies for clinical modulation of bone formation are needed. One potential approach would be to employ BMP stimulators. Identification of small molecules that enhance BMP signaling could open the door to new therapeutic strategies. Our studies suggest that phenamil is one such candidate molecule. Phenamil induces Trb3 and perhaps other molecular mediators that render osteogenic precursors more sensitive to BMP stimulation. This and similarly acting small molecules may reduce the doses of BMPs required for effective therapeutic intervention. In addition, the discovery that pharmacological modulation of Trb3 expression can enhance osteoblast differentiation provides a second avenue for potential therapeutic development.
Supplementary Material
Acknowledgments
We thank members of the Tontonoz laboratory for fruitful discussions.
P.T. is an Investigator of the Howard Hughes Medical Institute. This work was also supported by NIH grant HL090553 to P.T. and S.G.Y.
We have nothing to disclose.
Footnotes
Published ahead of print on 11 May 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Aghaloo, T. L., C. M. Amantea, C. M. Cowan, J. A. Richardson, B. M. Wu, F. Parhami, and S. Tetradis. 2007. Oxysterols enhance osteoblast differentiation in vitro and bone healing in vivo. J. Orthop. Res. 251488-1497. [DOI] [PubMed] [Google Scholar]
- 2.Benos, D. J. 1982. Amiloride: a molecular probe of sodium transport in tissues and cells. Am. J. Physiol. 242C131-C145. [DOI] [PubMed] [Google Scholar]
- 3.Beresford, J. N. 1989. Osteogenic stem cells and the stromal system of bone and marrow. Clin. Orthop. Relat. Res. 1989270-280. [PubMed] [Google Scholar]
- 4.Bezy, O., C. Vernochet, S. Gesta, S. R. Farmer, and C. R. Kahn. 2007. TRB3 blocks adipocyte differentiation through the inhibition of C/EBPβ transcriptional activity. Mol. Cell. Biol. 276818-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blazer-Yost, B. L., and S. I. Helman. 1997. The amiloride-sensitive epithelial Na+ channel: binding sites and channel densities. Am. J. Physiol. 272C761-C769. [DOI] [PubMed] [Google Scholar]
- 6.Bobkov, Y. V., and B. W. Ache. 2007. Block by amiloride derivatives of odor-evoked discharge in lobster olfactory receptor neurons through action on a presumptive TRP channel. Chem. Senses 32149-159. [DOI] [PubMed] [Google Scholar]
- 7.Caplan, A. I. 1991. Mesenchymal stem cells. J. Orthop. Res. 9641-650. [DOI] [PubMed] [Google Scholar]
- 8.Centrella, M., M. C. Horowitz, J. M. Wozney, and T. L. McCarthy. 1994. Transforming growth factor-beta gene family members and bone. Endocrinol. Rev. 1527-39. [DOI] [PubMed] [Google Scholar]
- 9.Chan, M. C., P. H. Nguyen, B. N. Davis, N. Ohoka, H. Hayashi, K. Du, G. Lagna, and A. Hata. 2007. A novel regulatory mechanism of the bone morphogenetic protein (BMP) signaling pathway involving the carboxyl-terminal tail domain of BMP type II receptor. Mol. Cell. Biol. 275776-5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chao, L. C., S. J. Bensinger, C. J. Villanueva, K. Wroblewski, and P. Tontonoz. 2008. Inhibition of adipocyte differentiation by Nur77, Nurr1, and Nor1. Mol. Endocrinol. 222596-2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, T. L., M. A. Hirst, and D. Feldman. 1979. A receptor-like binding macromolecule for 1 alpha, 25-dihydroxycholecalciferol in cultured mouse bone cells. J. Biol. Chem. 2547491-7494. [PubMed] [Google Scholar]
- 12.Dezawa, M., H. Ishikawa, Y. Itokazu, T. Yoshihara, M. Hoshino, S. Takeda, C. Ide, and Y. Nabeshima. 2005. Bone marrow stromal cells generate muscle cells and repair muscle degeneration. Science 309314-317. [DOI] [PubMed] [Google Scholar]
- 13.Dezawa, M., H. Kanno, M. Hoshino, H. Cho, N. Matsumoto, Y. Itokazu, N. Tajima, H. Yamada, H. Sawada, H. Ishikawa, T. Mimura, M. Kitada, Y. Suzuki, and C. Ide. 2004. Specific induction of neuronal cells from bone marrow stromal cells and application for autologous transplantation. J. Clin. Investig. 1131701-1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du, K., S. Herzig, R. N. Kulkarni, and M. Montminy. 2003. TRB3: a tribbles homolog that inhibits Akt/PKB activation by insulin in liver. Science 3001574-1577. [DOI] [PubMed] [Google Scholar]
- 15.Dwyer, J. R., N. Sever, M. Carlson, S. F. Nelson, P. A. Beachy, and F. Parhami. 2007. Oxysterols are novel activators of the hedgehog signaling pathway in pluripotent mesenchymal cells. J. Biol. Chem. 2828959-8968. [DOI] [PubMed] [Google Scholar]
- 16.Fleet, J. C., K. Cashman, K. Cox, and V. Rosen. 1996. The effects of aging on the bone inductive activity of recombinant human bone morphogenetic protein-2. Endocrinology 1374605-4610. [DOI] [PubMed] [Google Scholar]
- 17.Frelin, C., P. Barbry, P. Vigne, O. Chassande, E. J. Cragoe, Jr., and M. Lazdunski. 1988. Amiloride and its analogs as tools to inhibit Na+ transport via the Na+ channel, the Na+/H+ antiport and the Na+/Ca2+ exchanger. Biochimie 701285-1290. [DOI] [PubMed] [Google Scholar]
- 18.Fulfaro, F., A. Casuccio, C. Ticozzi, and C. Ripamonti. 1998. The role of bisphosphonates in the treatment of painful metastatic bone disease: a review of phase III trials. Pain 78157-169. [DOI] [PubMed] [Google Scholar]
- 19.Gangji, V., and T. Appelboom. 1999. Analgesic effect of intravenous pamidronate on chronic back pain due to osteoporotic vertebral fractures. Clin. Rheumatol. 18266-267. [DOI] [PubMed] [Google Scholar]
- 20.Giannobile, W. V., R. A. Hernandez, R. D. Finkelman, S. Ryan, C. P. Kiritsy, M. D'Andrea, and S. E. Lynch. 1996. Comparative effects of platelet-derived growth factor-BB and insulin-like growth factor-I, individually and in combination, on periodontal regeneration in Macaca fascicularis. J. Periodontal Res. 31301-312. [DOI] [PubMed] [Google Scholar]
- 21.Gong, Y., R. B. Slee, N. Fukai, G. Rawadi, S. Roman-Roman, A. M. Reginato, H. Wang, T. Cundy, F. H. Glorieux, D. Lev, M. Zacharin, K. Oexle, J. Marcelino, W. Suwairi, S. Heeger, G. Sabatakos, S. Apte, W. N. Adkins, J. Allgrove, M. Arslan-Kirchner, J. A. Batch, P. Beighton, G. C. Black, R. G. Boles, L. M. Boon, C. Borrone, H. G. Brunner, G. F. Carle, B. Dallapiccola, A. De Paepe, B. Floege, M. L. Halfhide, B. Hall, R. C. Hennekam, T. Hirose, A. Jans, H. Juppner, C. A. Kim, K. Keppler-Noreuil, A. Kohlschuetter, D. LaCombe, M. Lambert, E. Lemyre, T. Letteboer, L. Peltonen, R. S. Ramesar, M. Romanengo, H. Somer, E. Steichen-Gersdorf, B. Steinmann, B. Sullivan, A. Superti-Furga, W. Swoboda, M. J. van den Boogaard, W. Van Hul, M. Vikkula, M. Votruba, B. Zabel, T. Garcia, R. Baron, B. R. Olsen, and M. L. Warman. 2001. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell 107513-523. [DOI] [PubMed] [Google Scholar]
- 22.Govender, P. V., Y. R. Rampersaud, L. Rickards, and M. G. Fehlings. 2002. Use of osteogenic protein-1 in spinal fusion: literature review and preliminary results in a prospective series of high-risk cases. Neurosurg. Focus 13e4. [DOI] [PubMed] [Google Scholar]
- 23.Govender, S., C. Csimma, H. K. Genant, A. Valentin-Opran, Y. Amit, R. Arbel, H. Aro, D. Atar, M. Bishay, M. G. Borner, P. Chiron, P. Choong, J. Cinats, B. Courtenay, R. Feibel, B. Geulette, C. Gravel, N. Haas, M. Raschke, E. Hammacher, D. van der Velde, P. Hardy, M. Holt, C. Josten, R. L. Ketterl, B. Lindeque, G. Lob, H. Mathevon, G. McCoy, D. Marsh, R. Miller, E. Munting, S. Oevre, L. Nordsletten, A. Patel, A. Pohl, W. Rennie, P. Reynders, P. M. Rommens, J. Rondia, W. C. Rossouw, P. J. Daneel, S. Ruff, A. Ruter, S. Santavirta, T. A. Schildhauer, C. Gekle, R. Schnettler, D. Segal, H. Seiler, R. B. Snowdowne, J. Stapert, G. Taglang, R. Verdonk, L. Vogels, A. Weckbach, A. Wentzensen, and T. Wisniewski. 2002. Recombinant human bone morphogenetic protein-2 for treatment of open tibial fractures: a prospective, controlled, randomized study of four hundred and fifty patients. J. Bone Joint Surg. Am. 84A2123-2134. [DOI] [PubMed] [Google Scholar]
- 24.Horwitz, E. M., P. L. Gordon, W. K. Koo, J. C. Marx, M. D. Neel, R. Y. McNall, L. Muul, and T. Hofmann. 2002. Isolated allogeneic bone marrow-derived mesenchymal cells engraft and stimulate growth in children with osteogenesis imperfecta: implications for cell therapy of bone. Proc. Natl. Acad. Sci. USA 998932-8937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu, H., M. J. Hilton, X. Tu, K. Yu, D. M. Ornitz, and F. Long. 2005. Sequential roles of Hedgehog and Wnt signaling in osteoblast development. Development 13249-60. [DOI] [PubMed] [Google Scholar]
- 26.Iynedjian, P. B. 2005. Lack of evidence for a role of TRB3/NIPK as an inhibitor of PKB-mediated insulin signalling in primary hepatocytes. Biochem. J. 386113-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson, E. E., and M. R. Urist. 2000. Human bone morphogenetic protein allografting for reconstruction of femoral nonunion. Clin. Orthop. Relat. Res. 200061-74. [DOI] [PubMed] [Google Scholar]
- 28.Kha, H. T., B. Basseri, D. Shouhed, J. Richardson, S. Tetradis, T. J. Hahn, and F. Parhami. 2004. Oxysterols regulate differentiation of mesenchymal stem cells: pro-bone and anti-fat. J. Bone Miner. Res. 19830-840. [DOI] [PubMed] [Google Scholar]
- 29.Kleyman, T. R., and E. J. Cragoe, Jr. 1988. Amiloride and its analogs as tools in the study of ion transport. J. Membr. Biol. 1051-21. [DOI] [PubMed] [Google Scholar]
- 30.Lant, A. F., A. J. Smith, and G. M. Wilson. 1969. Clinical evaluation of amiloride, a potassium-sparing diuretic. Clin. Pharmacol. Ther. 1050-63. [DOI] [PubMed] [Google Scholar]
- 31.Masereel, B., L. Pochet, and D. Laeckmann. 2003. An overview of inhibitors of Na(+)/H(+) exchanger. Eur. J. Med. Chem. 38547-554. [DOI] [PubMed] [Google Scholar]
- 32.Minamide, A., M. Yoshida, M. Kawakami, S. Yamasaki, H. Kojima, H. Hashizume, and S. D. Boden. 2005. The use of cultured bone marrow cells in type I collagen gel and porous hydroxyapatite for posterolateral lumbar spine fusion. Spine 301134-1138. [DOI] [PubMed] [Google Scholar]
- 33.Murakami, G., T. Watabe, K. Takaoka, K. Miyazono, and T. Imamura. 2003. Cooperative inhibition of bone morphogenetic protein signaling by Smurf1 and inhibitory Smads. Mol. Biol. Cell 142809-2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nohe, A., E. Keating, P. Knaus, and N. O. Petersen. 2004. Signal transduction of bone morphogenetic protein receptors. Cell Signal. 16291-299. [DOI] [PubMed] [Google Scholar]
- 35.Ohoka, N., S. Yoshii, T. Hattori, K. Onozaki, and H. Hayashi. 2005. TRB3, a novel ER stress-inducible gene, is induced via ATF4-CHOP pathway and is involved in cell death. EMBO J. 241243-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Owen, M. 1988. Marrow stromal stem cells. J. Cell Sci. Suppl. 1063-76. [DOI] [PubMed] [Google Scholar]
- 37.Padilla, M. C., M. J. Armas-Hernandez, R. H. Hernandez, Z. H. Israili, and M. Valasco. 2007. Update of diuretics in the treatment of hypertension. Am. J. Ther. 14154-160. [DOI] [PubMed] [Google Scholar]
- 38.Papanikolaou, G., M. E. Samuels, E. H. Ludwig, M. L. MacDonald, P. L. Franchini, M. P. Dube, L. Andres, J. MacFarlane, N. Sakellaropoulos, M. Politou, E. Nemeth, J. Thompson, J. K. Risler, C. Zaborowska, R. Babakaiff, C. C. Radomski, T. D. Pape, O. Davidas, J. Christakis, P. Brissot, G. Lockitch, T. Ganz, M. R. Hayden, and Y. P. Goldberg. 2004. Mutations in HFE2 cause iron overload in chromosome 1q-linked juvenile hemochromatosis. Nat. Genet. 3677-82. [DOI] [PubMed] [Google Scholar]
- 39.Pittenger, M. F., and B. J. Martin. 2004. Mesenchymal stem cells and their potential as cardiac therapeutics. Circ. Res. 959-20. [DOI] [PubMed] [Google Scholar]
- 40.Prockop, D. J. 1997. Marrow stromal cells as stem cells for nonhematopoietic tissues. Science 27671-74. [DOI] [PubMed] [Google Scholar]
- 41.Qi, L., J. E. Heredia, J. Y. Altarejos, R. Screaton, N. Goebel, S. Niessen, I. X. Macleod, C. W. Liew, R. N. Kulkarni, J. Bain, C. Newgard, M. Nelson, R. M. Evans, J. Yates, and M. Montminy. 2006. TRB3 links the E3 ubiquitin ligase COP1 to lipid metabolism. Science 3121763-1766. [DOI] [PubMed] [Google Scholar]
- 42.Rorth, P., K. Szabo, and G. Texido. 2000. The level of C/EBP protein is critical for cell migration during Drosophila oogenesis and is tightly controlled by regulated degradation. Mol. Cell 623-30. [DOI] [PubMed] [Google Scholar]
- 43.Shore, E. M., M. Xu, G. J. Feldman, D. A. Fenstermacher, T. J. Cho, I. H. Choi, J. M. Connor, P. Delai, D. L. Glaser, M. LeMerrer, R. Morhart, J. G. Rogers, R. Smith, J. T. Triffitt, J. A. Urtizberea, M. Zasloff, M. A. Brown, and F. S. Kaplan. 2006. A recurrent mutation in the BMP type I receptor ACVR1 causes inherited and sporadic fibrodysplasia ossificans progressiva. Nat. Genet. 38525-527. [DOI] [PubMed] [Google Scholar]
- 44.Spinella-Jaegle, S., G. Rawadi, S. Kawai, S. Gallea, C. Faucheu, P. Mollat, B. Courtois, B. Bergaud, V. Ramez, A. M. Blanchet, G. Adelmant, R. Baron, and S. Roman-Roman. 2001. Sonic hedgehog increases the commitment of pluripotent mesenchymal cells into the osteoblastic lineage and abolishes adipocytic differentiation. J. Cell Sci. 1142085-2094. [DOI] [PubMed] [Google Scholar]
- 45.Takahashi, Y., N. Ohoka, H. Hayashi, and R. Sato. 2008. TRB3 suppresses adipocyte differentiation by negatively regulating PPARgamma transcriptional activity. J. Lipid Res. 49880-892. [DOI] [PubMed] [Google Scholar]
- 46.van der Horst, G., H. Farih-Sips, C. W. Lowik, and M. Karperien. 2003. Hedgehog stimulates only osteoblastic differentiation of undifferentiated KS483 cells. Bone 33899-910. [DOI] [PubMed] [Google Scholar]
- 47.Waite, K. A., and C. Eng. 2003. From developmental disorder to heritable cancer: it's all in the BMP/TGF-beta family. Nat. Rev. Genet. 4763-773. [DOI] [PubMed] [Google Scholar]
- 48.Winn, S. R., Y. Hu, C. Sfeir, and J. O. Hollinger. 2000. Gene therapy approaches for modulating bone regeneration. Adv. Drug Deliv. Rev. 42121-138. [DOI] [PubMed] [Google Scholar]
- 49.Yamaguchi, A., T. Komori, and T. Suda. 2000. Regulation of osteoblast differentiation mediated by bone morphogenetic proteins, hedgehogs, and Cbfa1. Endocrinol. Rev. 21393-411. [DOI] [PubMed] [Google Scholar]
- 50.Yu, P. B., C. C. Hong, C. Sachidanandan, J. L. Babitt, D. Y. Deng, S. A. Hoyng, H. Y. Lin, K. D. Bloch, and R. T. Peterson. 2008. Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat. Chem. Biol. 433-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu, H., P. Kavsak, S. Abdollah, J. L. Wrana, and G. H. Thomsen. 1999. A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature 400687-693. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.