Abstract
Several respiratory viruses, including influenza virus and severe acute respiratory syndrome coronavirus (SARS-CoV), produce more severe disease in the elderly, yet the molecular mechanisms governing age-related susceptibility remain poorly studied. Advanced age was significantly associated with increased SARS-related deaths, primarily due to the onset of early- and late-stage acute respiratory distress syndrome (ARDS) and pulmonary fibrosis. Infection of aged, but not young, mice with recombinant viruses bearing spike glycoproteins derived from early human or palm civet isolates resulted in death accompanied by pathological changes associated with ARDS. In aged mice, a greater number of differentially expressed genes were observed than in young mice, whose responses were significantly delayed. Differences between lethal and nonlethal virus phenotypes in aged mice could be attributed to differences in host response kinetics rather than virus kinetics. SARS-CoV infection induced a range of interferon, cytokine, and pulmonary wound-healing genes, as well as several genes associated with the onset of ARDS. Mice that died also showed unique transcriptional profiles of immune response, apoptosis, cell cycle control, and stress. Cytokines associated with ARDS were significantly upregulated in animals experiencing lung pathology and lethal disease, while the same animals experienced downregulation of the ACE2 receptor. These data suggest that the magnitude and kinetics of a disproportionately strong host innate immune response contributed to severe respiratory stress and lethality. Although the molecular mechanisms governing ARDS pathophysiology remain unknown in aged animals, these studies reveal a strategy for dissecting the genetic pathways by which SARS-CoV infection induces changes in the host response, leading to death.
Severe acute respiratory syndrome coronavirus (SARS-CoV) is a zoonotic pathogen, likely originating from bats, civets, and raccoon dogs, that entered the human population to cause a global epidemic in 2002 and 2003. The epidemic was severe, resulting in about 8,000 human infections that often progressed to severe acute respiratory disease, pneumonia, and diarrhea, culminating in an ∼10% fatality rate (10). Advanced age (>60 years old) was significantly associated with increased SARS-related deaths due to rapidly progressive respiratory compromise (acute respiratory distress syndrome [ARDS]) (13, 14). This pulmonary damage is believed to be caused by both direct viral effects and immunopathological factors (18). Aging is associated with increased morbidity in a variety of viral infections (e.g., West Nile virus, influenza virus, SARS-CoV, and noroviruses), perhaps because the elderly respond poorly to new antigens compared to younger populations (41, 48). In addition, a typical feature of advanced age is a general increase in the production and levels in plasma of proinflammatory cytokines, resulting in an age-related increase of inflammation (48). However, the molecular mechanisms governing SARS-CoV pathogenesis are unclear because of the lack of well-defined animal models that exhibit severe pulmonary disease in vulnerable aged populations.
Mortality rates for ARDS, the most severe form of acute lung injury, approach ∼50%, and the disease affects ∼1,000,000 people/year worldwide, ranking it among the most difficult challenges in critical-care medicine (31). Early ARDS is characterized by diffuse alveolar damage (DAD), which includes a protein-rich edema, an exudative phase with hyaline membranes, and inflammation leading to surfactant dysfunction and severe hypoxia, usually occurring within the first 10 days of acute lung injury. This may progress to an organizing-phase DAD that progresses to pulmonary fibrosis, and both occurred following SARS-CoV infection. ARDS has been reported following SARS-CoV or influenza virus infection and is sporadically reported following herpesvirus, respiratory syncytial virus, adenovirus, varicella virus, and hantavirus infections, as well (9, 11, 30, 39, 42, 51); however, few well-defined ARDS animal models exist. Select SARS-CoV strains and H5N1 influenza virus are robust models for ARDS in rodents (47, 56), yet the molecular mechanisms governing ARDS pathogenesis following virus infection are still unclear, especially when placed into the context of aging. Consequently, despite intensive investigation, there are no specific clinical therapies for ARDS except supportive care.
Although the pathogenic mechanisms of SARS-CoV infection remain poorly defined, studies of a limited number of human serum (21, 24) and lung (7, 8, 20) samples have provided some insight. During SARS-CoV infection in humans, a dysregulated cytokine and inflammatory response has been proposed to be the cause of the pulmonary pathology characterized at autopsy. High levels of type I interferons and interferon-stimulated genes (ISGs), such as CXCL10, interleukin 6 (IL-6), and IL-8 genes, were seen in these patients. Some of these genes, IL-6 and IL-8 genes, as well as others, including tumor necrosis factor alpha (TNF-α) and IL-1β genes, have also been associated with ARDS (4, 37), though gene and protein expression profiles from these patient samples were likely impacted by clinical treatments (steroids and mechanical ventilation) and concurrent preexisting disease, such as chronic heart failure, diabetes, and asthma. In addition, these patient samples are more likely to represent late-stage disease. Animal models of SARS-CoV that recapitulate the ARDS outcome in humans, especially that seen in the elderly, may reveal the initial host-virus responses that contribute to severe end stage lung disease.
We recently isolated an isogenic panel of recombinant infectious-clone (ic) SARS-CoV strains bearing different spike (S) glycoproteins derived from a zoonotic SARS-CoV strain, icHC/SZ/61/03, and from strains identified during the late and early phases of the human epidemic, icUrbani and icGZ02, respectively (47, 57). In aged mice, icUrbani infection caused mild disease with transient weight loss (44), while icGZ02 and icHC/SZ/61/03 resulted in severe lung infection progressing to acute-onset DAD, ARDS, and death (47). In contrast, young mice replicated virus in the absence of clinical disease symptoms and lung pathology, regardless of the origins of the S glycoprotein. These data implicate changes in the S glycoprotein gene as major components of SARS-CoV pathogenesis. Here, we compare host responses over time in nonlethal and lethal mouse models of SARS-CoV using gene expression data to more fully understand the impacts of aging and viral-spike variation on the host response that regulates severe disease phenotypes and the onset of ARDS.
MATERIALS AND METHODS
Viruses and cells.
The generation and characterization of each of the recombinant ics (icUrbani, icGZ02, and icHC/SZ/61/03) have been described previously (47, 57). Briefly, all recombinant icSARS-CoV strains were propagated on Vero E6 cells in Eagle's minimal essential medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal calf serum (HyClone, Logan, UT), kanamycin (0.25 μg/ml), and gentamicin (0.05 μg/ml) at 37°C in a humidified CO2 incubator. All work was performed in a biological safety cabinet in a biosafety level 3 laboratory containing redundant exhaust fans. Personnel were equipped with powered air-purifying respirators with high-efficiency particulate air and organic vapor filters (3 M, St. Paul, MN), wore Tyvek suits (DuPont, Research Triangle Park, NC), and were double gloved.
Mouse infection.
Female BALB/cAnNHsd mice (young [8 weeks old] or aged [12 months old]; Harlan, Indianapolis, IN) were anesthetized with a ketamine (1.3 mg/mouse)-xylazine (0.38 mg/mouse) mixture administered intraperitoneally in a 50-μl volume. Groups of 20 mice were intranasally inoculated with phosphate-buffered saline (PBS) (mock infection) or 105 PFU of icUrbani, icGZ02, or icHC/SZ/61/03 in a 50-μl volume. The weights of the mice were monitored daily. At 3, 12, 24, 48, and 72 h postinfection (p.i.), four mice from each group were euthanized, and lung and serum samples were collected. Because the mice died by 96 h p.i. (47), mice were sacrificed through 72 h p.i. to ensure good-quality RNA for gene expression studies. For each mouse, one half of a lung was frozen at −70°C for RNA isolation. One half of a lung from each mouse was taken for RNA extraction, one quarter of a lung was assayed for viral titers by plaque assay, and one quarter was saved for immunohistochemical detection of the SARS-CoV nucleocapsid (N) protein. All mice were housed under sterile conditions in individually ventilated Sealsafe cages using the SlimLine system (Tecniplast, Exton, PA). Experimental protocols were reviewed and approved by the Institutional Animal Care and Use Committee at the University of North Carolina, Chapel Hill.
Virus titers in tissue samples.
Lung samples were weighed and homogenized in five equivalent volumes of PBS to generate a 20% solution. The solution was centrifuged at 13,000 rpm under aerosol containment in a table top centrifuge for 5 min, the clarified supernatant was serially diluted in PBS, and 200-μl volumes of the dilutions were placed onto monolayers of Vero E6 cells in six-well plates. Following 1 hour of incubation at 37°C, the cells were overlaid with 0.8% agarose-containing medium. Two days later, the plates were stained with neutral red and the plaques were counted.
Immunohistochemistry.
All tissues were fixed in 4% paraformaldehyde in PBS (pH 7.4) for at least 7 days prior to being submitted to the Histopathology Core Facility (University of North Carolina, Chapel Hill) for paraffin embedding. For detection of SARS-CoV antigen, serial sections were deparaffinized and rehydrated, and antigen was retrieved by incubating the sections at 100°C for 15 min with citric acid buffer, pH 6.0. Slides were cooled for 20 min on ice in the same solution. Endogenous peroxidase was blocked with 3% hydrogen peroxide. The slides were briefly washed with PBS/0.05% Tween 20 and incubated with serum from ferrets infected with SARS-CoV (1/400 in PBS/0.1% bovine serum albumin) or normal ferret serum (1/400) for 1 h at room temperature. After being washed, the sections were incubated with horseradish peroxidase-labeled goat-anti-ferret immunoglobulin G (KPL, Inc., MD) in PBS/0.1% bovine serum albumin for 1 h at room temperature. Peroxidase activity was revealed by incubating the slides in 3-amino-9-ethylcarbazole (Sigma, St. Louis, MO) for 10 min, resulting in a bright-red precipitate, followed by counterstaining with hematoxylin. Tissue sections from noninfected mice were used as negative controls. Lung sections from an experimentally SARS-CoV-inoculated ferret euthanized at 24 h p.i. were used as positive controls. Using polyclonal ferret sera directed against the SARS-CoV N protein, viral N protein was detectable as a measure of replication in the lungs of mice no earlier than 24 h p.i.
RNA isolation and genomic precharacterization.
One half of a lung from each mouse was transferred to solution D (4 M guanidine thiocyanate with 25 mM sodium citrate, 0.5% sarcosyl, and 0.1 M 2-mercaptoethanol) and homogenized, and RNA was extracted and purified as described previously (25). Using an Agilent 2100 bioanalyzer (Agilent Technologies Inc., Palo Alto, CA), total RNA samples from individual mice extracted at the time points 3, 12, 24, 48, and 72 h p.i. were characterized and judged to be pure and intact. The RNA samples were then tested by relative quantitative real-time reverse transcription (RT)-PCR (described below) for the presence of the SARS-CoV N gene and cellular transcripts Cxcl9, Cxcl10, Irf7, and B2m to determine if individual samples could be pooled to generate one representative experimental sample to be used for gene expression analysis. Most samples had similar viral and cellular transcripts; therefore, equal-mass quantities of total RNA extracted from individual samples were pooled to create an experimental gene expression sample. Mice at 12 h p.i. showed considerable variation in viral transcripts, with only one out of four young mice and three out of four aged mice with detectable viral transcripts; therefore, a different pooling strategy was used. A representative young-mouse sample of host response to virus at 12 h p.i. was created from total RNA from the one individual sample with viral transcripts, and a representative aged-mouse sample of host response to virus at 12 h p.i. was created from equal-mass quantities of total RNA from the three individual samples for aged mice with viral transcripts. In contrast, viral and cellular transcripts were equivalent at all other time points, allowing the samples to be pooled.
Oligonucleotide microarray analysis.
cRNA probes were generated from each pooled RNA sample using the Agilent Low RNA Input Fluorescent Linear Amplification kit (Agilent Technologies Inc., Palo Alto, CA). Each pooled experimental RNA sample was then cohybridized with its corresponding reference sample, i.e., young experimental mice versus young mock-infected mice and aged experimental mice versus aged mock-infected mice. In addition, one microarray experiment was performed comparing the pooled young mock-infected reference to the pooled aged mock-infected reference to directly gain insight into the effects of aging. All microarray hybridizations were performed with mouse oligonucleotide arrays (G4121B; Agilent Technologies Inc.). Slides were scanned with an Agilent DNA microarray scanner, and image analysis was performed using Agilent Feature Extractor Software (Agilent Technologies Inc.). Each microarray experiment was done with four technical replicates, two with reverse dye hybridization (28). All data were entered into a custom-designed database (Expression Array Manager) and analyzed with Resolver 6.0 (Rosetta Biosoftware, Seattle, WA) and Spotfire DecisionSite for Functional Genomics 8.1 (Tibco Spotfire, Somerville, MA). Ingenuity Pathway Analysis 6.0 (Ingenuity Systems, Redwood City, CA) was used to functionally annotate genes according to biological processes and canonical pathways. Genes were included in all analyses based on the criteria that the absolute expression level change was twofold or greater with a P value of ≤0.01, unless otherwise stated.
A gene set enrichment analysis (GSEA) (53) was used to identify sets of genes annotated by the same function in the gene ontology (GO) (1) that were differentially expressed during infection. In preparation for this analysis, the Rosetta Resolver System was used to determine the P value of the differential expression of each gene compared to the mock sample. In short, given a list of genes for a single virus/time point combination ranked in decreasing order by the −log(P value), GSEA allows the identification of gene sets that are concentrated at the top of the list (signifying significant differential expression). For every function in GO, a GSEA was applied to each virus/time point combination. A dynamic program method was used to determine the exact P value for each GO function (27). Only the significances of GO functions with a depth of at least 4 in the GO hierarchy were tested to avoid nonspecific terms. For each function, plots of the −log(P value) were generated for each virus to track the significance of the function across all time points.
Relative quantitative real-time RT-PCR.
Relative quantitative real-time RT-PCR was performed to precharacterize individual mouse samples before pooling them, to validate a number of cellular-gene expression changes detected by microarray in pooled samples, and to further characterize a subset of transcripts in an expanded time course of viral infection. Total-RNA samples were treated with DNase using a DNA-free DNase kit (Ambion, Inc. Austin, TX). cDNA was generated using RT reagents and random hexamers (Applied Biosystems, Foster City, CA). Relative quantitative real-time RT-PCR was run on the ABI 7500 PCR system, using TaqMan chemistry (Applied Biosystems, Foster City, CA). Primers and probes specific for the nucleoprotein gene of SARS-CoV have been described previously (12), and primers and probes specific for mouse cellular genes were purchased from Applied Biosystems as “gene expression assays.” Targets were run in quadruplicate following protocols supplied by Applied Biosystems (Foster City, CA).
Microarray data accession numbers.
All data described in this report, including sample information, intensity measurements, gene lists, error analysis, microarray content, and microarray hybridization conditions, will be available after publication in the public domain through Expression Array Manager at http://expression.viromics.washington.edu in accordance with proposed standards (6).
RESULTS
S glycoproteins affect clinical outcomes and viral replication in young and aged mice infected with icSARS-CoV.
As previously shown, infection of young BALB/c mice with icUrbani, icGZ02, or icHC/SZ/61/03 did not result in weight loss or other clinical signs of disease, regardless of the origin of the S glycoprotein (Fig. 1A) (47). Infection of aged BALB/c mice with icUrbani resulted in only minor weight loss of 5% with no other signs of clinical disease. In contrast, aged mice infected with either icGZ02 or icHC/SZ/61/03 exhibited significant weight loss of >10% by 72 h p.i. (Fig. 1B) and exhibited clinical symptoms, including hunching, ruffled fur, and labored breathing. As seen previously in the lungs, DAD, hyaline membrane formation, and alveolitis were noted with icGZ02 and icHC/SZ/61/03, but not after icUrbani infection (reference 47 and data not shown).
FIG. 1.
Differences in clinical outcomes did not correlate with differences in viral replication efficiencies. Weight loss (A and B) and lung titer (C and D) results are shown for 8-week-old (young) (A and C) and 12-month-old (aged) (B and D) female BALB/c mice infected with icUrbani, icGZ02, and icHC/SZ/61/03. (A and B) Body weights of infected mice were measured on a daily basis (n = 5 per group). Weight changes are expressed as the mean percent changes for infected animals relative to the initial weights at day zero. Lung tissues were harvested from infected mice at 3, 12, 24, 48, and 72 h p.i. and assayed for infectious virus as described in Materials and Methods. Tissue samples from five mice were analyzed at each time point. The error bars represent standard deviations.
In young mice, icUrbani replicated to peak titers of 8 ×107 PFU/g in lungs, while icGZ02 replicated to peak titers of 2 × 107 PFU/g and icHC/SZ/61/03 replicated to peak titers of 6 × 105 PFU/g of lung tissue under identical conditions (Fig. 1C). Viral titers peaked at 24 h p.i. for icGZ02 and icHC/SZ/61/03, after which icGZ02 titers dropped while icHC/SZ/61/03 titers remained consistent. Conversely, icUrbani replicated as efficiently as icGZ02 during the first 24 h p.i. but continued to replicate and reached peak titers at 48 h p.i. While clear differences in viral replication were observed, this did not extend to clinical differences, which were absent in young mice.
In aged mice, peak titers were 1 log unit higher for icGZ02 and 2 log units higher for icHC/SZ/61/03 than titers in young mice (Fig. 1D). Similar to that in young mice, viral replication peaked at 24 h p.i. for icGZ02 and icHC/Z/61/03 and at 48 h p.i. for icUrbani. Unique to aged mice, viral titers dropped 1 to 2 log units after reaching peak titers for all viruses. While clear differences in clinical outcomes were observed between nonlethal icUrbani and lethal icGZ02 and icHC/SZ/61/03 isolates, these differences could not be attributed to virus growth efficiencies in the lungs of infected aged mice, suggesting that intrinsic properties of the S glycoprotein contributed to differences in pathogenesis.
S glycoproteins affect the tropism of SARS-CoV replication in lungs of infected mice.
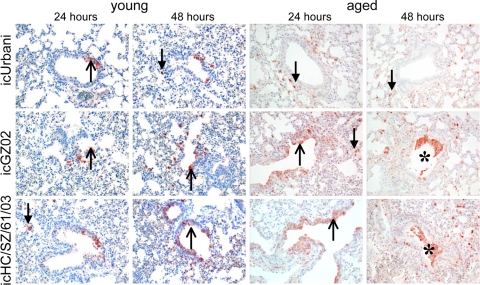
As the recombinant SARS-CoV panel was isogenic except for defined sequence variations unique to each S glycoprotein gene (47), we next determined if these spike changes altered virus tropism in the lungs of young and aged animals at early and late time points p.i. Using polyclonal ferret sera directed against the SARS-CoV N protein, unique patterns of viral N protein distribution were clearly detectable in the lungs of all infected young and aged mice by 24 h p.i. (Fig. 2). With icUrbani, a small number of scattered alveolar epithelial cells, with morphology consistent with type II pneumocytes, stained positive for SARS-CoV N protein at 24 h p.i. in both young and aged animals. Increasing numbers of alveolar epithelial cells that expressed viral N protein were observed at 48 and 72 h p.i. (Fig. 2 and data not shown). In contrast, the lungs of mice infected with icGZ02 and icHC/SZ/61/03 SARS-CoV displayed distinctly different infection patterns and cell tropism. Viral N protein expression was localized in a small number of bronchial and bronchiolar epithelial cells in the airways and scattered alveolar cells with type II pneumocyte appearance as early as 12 h p.i. (data not shown). By 24 h p.i., viral N protein expression was localized in a moderate number of bronchial and bronchiolar epithelial cells and a few scattered alveolar epithelial cells, again with morphology consistent with type II pneumocytes. By 48 h p.i. the majority of viral N protein was detected in these alveolar epithelial cells, with occasional detection in bronchial cells, bronchiolar cells, and cellular debris that was evident in the preterminal bronchioles. By 72 h p.i., viral N protein expression could be detected only in alveolar epithelial cells with type II pneumocyte appearance, with no bronchial or bronchiolar involvement and little cellular debris. These data indicate that the changes in the SARS-CoV S glycoprotein altered the tropism and pathogenesis in aged mice.
FIG. 2.
Tropism of lethal SARS-CoV is primarily for bronchial and bronchiolar cells at early stages of infection, while that of nonlethal SARS-CoV is primarily for alveolar epithelial cells. With SARS-CoV icUrbani (top row), alveolar epithelial cells of aged mice showed SARS-CoV N immunohistochemical staining, with greater numbers of alveolar epithelial cells being stained (arrows with closed arrowheads) as the infection progressed. With icGZ02 and icHC/SZ/61/03 (middle and bottom rows), SARS-CoV N staining was observed in many bronchial and bronchiolar cells (arrows with open arrowheads) and few alveolar epithelial cells at 24 h p.i. in aged mice. At 48 and 72 h p.i., the numbers of bronchial and bronchiolar cells positive for N decreased while the number of alveolar epithelial cells positive for N increased. In addition, positive staining was also observed in intraluminal debris (asterisks). Immunoperoxidase with hematoxylin counterstain was used. Magnification, ×20.
Decreased ACE2 expression correlates with lethal outcome of SARS-CoV infection.
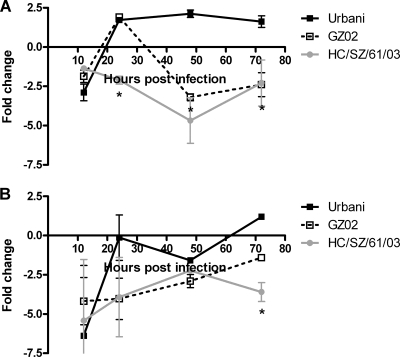
Since the SARS-CoV receptor ACE2 has been shown to be downregulated by the SARS-CoV S glycoprotein (32), causing deregulation of the rennin-angiotensin system and exaggeration of acute lung failure, we hypothesized that the expression pattern of ACE2 would be differentially altered during lethal infections compared with nonlethal infections. Several attempts to detect murine ACE2 in the lungs of infected mice by immunohistochemistry were unsuccessful. As an alternative, we used real-time RT-PCR to examine the regulation of ACE2 in infected lungs (Fig. 3). In aged mice, at 48 h p.i., ACE2 was downregulated three- and fivefold during lethal icGZ02 and icHC/SZ/61/03 infections, respectively (Fig. 3A), supporting the idea that downregulation of ACE2 leads to acute lung failure. In contrast, the nonlethal icUrbani-infected mice upregulated ACE2 twofold over mock-infected mice at 48 h (Fig. 3A), correlating with mouse survival and indicating that the icUrbani S glycoprotein is not as pathogenic as the icGZ02 or icHC/Sz/61/03 S glycoprotein. In young mice, ACE2 expression was significantly downregulated only after infection with icHC/SZ/61/03 at 72 h p.i. (Fig. 3B). These data show that early downregulation of ACE2 is associated with lethal disease and therefore that ACE2 may potentially play a protective role in SARS-CoV-induced lung injury.
FIG. 3.
Expression of ACE2 is downregulated in lethal infection of aged mice. Quantitative RT-PCR using TaqMan chemistry was performed on individual lung samples from aged (A) and young (B) animals at 12, 24, 48, and 72 h p.i. The changes over aged and young mock controls, respectively, are plotted. The error bars represent standard deviations. *, P < 0.01 (analysis of variance; Bonferroni posttest compared to icUrbani).
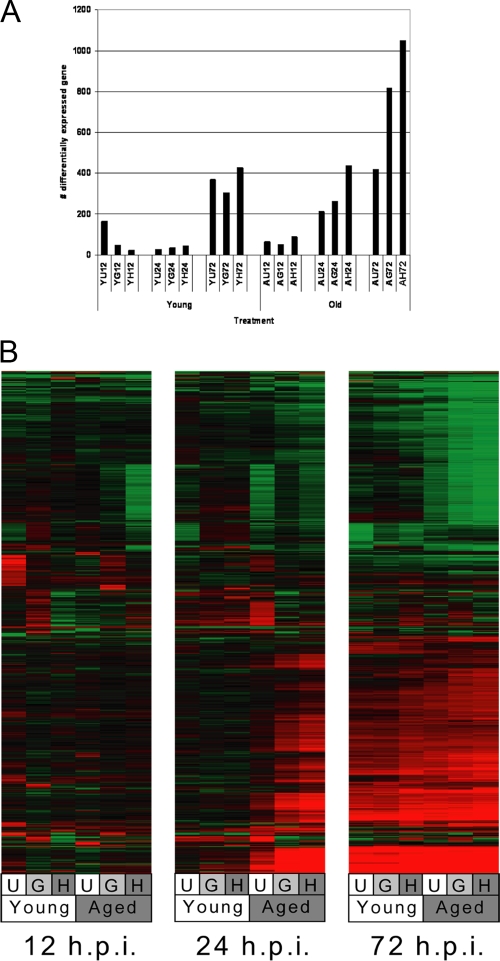
Global gene expression in lungs of aged SARS-CoV-infected mice shows an early host response to virus compared with that in young SARS-CoV-infected mice.
Pulmonary gene expression comparison in young and aged mice infected with nonlethal and lethal SARS-CoV strains provides an opportunity to identify unique expression signatures associated with lethal infection that could reveal the molecular mechanisms of SARS-CoV pathogenesis. In young mice, a late response to SARS-CoV infection was observed at 72 h p.i., with similar numbers of genes differentially expressed between mice infected with the different SARS-CoV isolates at each time point (Fig. 4A). In contrast, the host response in aged mice was characterized by an early (24-h p.i.) onset of differential gene expression that continued up to 72 h p.i. In aged mice, a greater number of genes were differentially expressed during infection with lethal SARS-CoV isolates than with the nonlethal isolate, with the lethal icHC/SZ/61/03 strain inducing greater numbers (∼200 more) of genes at 24 and 72 h p.i. than the lethal icGZ02 strain. The number of differentially regulated genes correlated with pathogenicity, as in previous results (47), showed that icUrbani does not cause lethal disease, icGZ02 causes a pathogenic disease that kills 75% of infected mice by day 4, and icHC/SZ/61/03 causes lethal disease in 100% of infected mice starting as early as day 3.
FIG. 4.
Global gene expression profiles show a prominent kinetic difference in gene expression, with aged mice initiating an earlier gene transcription response to viral infection than young mice. (A) Global gene expression data are the results of a comparison of gene expression in the lungs of experimental mice versus gene expression in the lungs of mock-infected mice, and genes were included if they met the criteria of an absolute change of ≥2-fold (P ≤ 0.01) in at least two experiments. Each bar represents one microarray experiment (n = 4). Y, young; A, aged; U, icUrbani; G, icGZ02; H, icHC/SZ/61/03 (12, 24, and 72 h p.i.). (B) Genes were included in the heat map if they showed an absolute change of ≥2-fold (P ≤ 0.01) in at least one experiment. These data were plotted as a heat map in which each matrix entry represents a gene expression value. Red corresponds to gene expression higher than that of the reference; green corresponds to lower gene expression. Data are presented for young and aged mice infected with either icUrbani (U), icGZ02 (G), or icHC/SZ/61/03 (H) at the indicated time points.
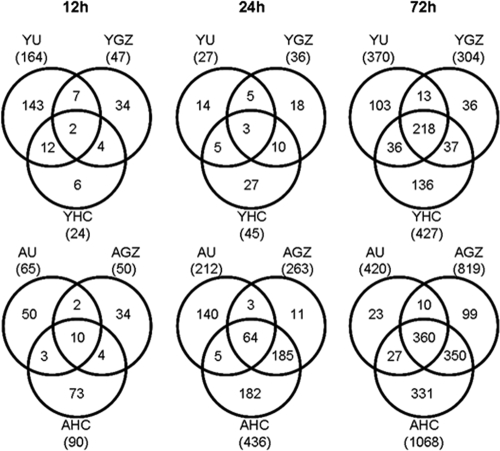
Plotting global gene expression profiles as heat maps showed few commonly regulated genes between the three different viruses at early time points in both young and aged mice, while similarities became apparent at 72 h p.i. (Fig. 4B). At this later stage of infection in aged and young mice, all three viruses induced a large number of unique genes but also showed the greatest number of common genes (Fig. 5). Notably, at 72 h p.i., aged mice displayed approximately 10-fold-greater numbers of differentially expressed genes specific to both lethal virus infections than young mice. This trend was also noted at 24 h p.i., and the gene responses specific to lethal isolates in the aged mice outnumbered the gene responses common to all three isolates in aged mice. Interestingly the number of differentially expressed genes unique to lethal infection with icHC/SZ/61/03 in aged mice was threefold higher than the number of genes unique to lethal infection with icGZ02. These data suggest that the lethal strains may produce severe disease phenotypes by different molecular mechanisms, and survival outcomes may be decided as early as 24 h p.i.
FIG. 5.
Viruses show more similar profiles of differentially expressed genes as time progresses. The Venn diagram shows the overlap of genes with ≥2-fold change (P ≤ 0.01) between icUrbani, icGZ02, and icHC/SZ/61/03 in young and aged mice over time. Data are presented for young (Y) and aged (A) mice infected with either icUrbani (U), icGZ02 (GZ), or icHC/SZ/61/03 (HC) at the indicated time points p.i. The numbers in the centers were shared by all three viruses.
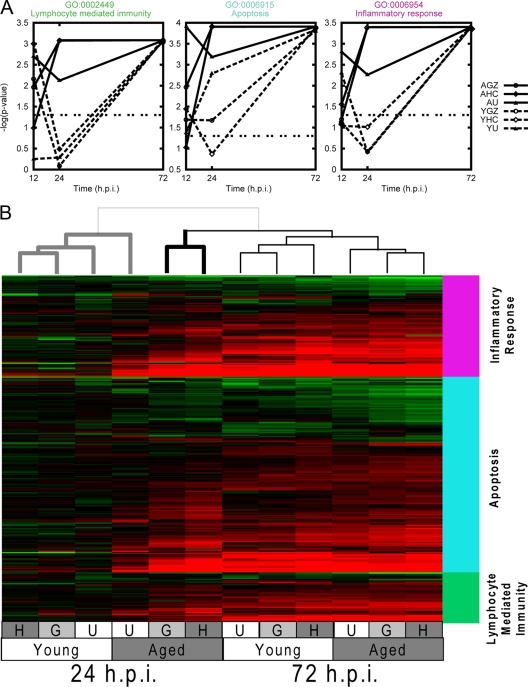
In order to better understand the biological functions associated with lung pathology and lethal disease at early stages of infection, we analyzed the microarray results using GO functional annotations. A GSEA (described in Materials and Methods) of the GO functionally annotated data was used to determine whether specific functional classes of genes were differentially regulated in response to infection. This analysis revealed significant differential expression of genes associated with immune response and apoptotic functional classes to SARS-CoV infection at 24 h p.i. (Fig. 6A). Lymphocyte-mediated immunity, apoptosis, and inflammatory response functional classes were significantly correlated, with the severity of disease with icGZ02- and icHC/SZ/61/03-infected aged animals showing the greatest statistical association of these classes. Interestingly, by 72 h p.i., these functional classes were no longer correlated with lethal disease but instead showed a common response to SARS-CoV infection. These results could also be observed when the data were plotted as a heat map: the host response to virus infection established at 24 h p.i. seem to determine the outcome for aged mice infected with lethal SARS-CoV strains, which was different from aged mice infected with a nonlethal strain and all infected young mice (Fig. 6B). Both of these graphical formats showed that the response at 24 h is an important determinant of the disease outcome due to an aberrant host response.
FIG. 6.
Functional enrichment of disease-related pathways reveals the importance of kinetics in the host response. (A) GSEA (described in Materials and Methods) was performed on the change data for all experiments using GO classes. GO categories 6954 (inflammatory response), 6915 (apoptosis), and 2449 (lymphocyte-mediated immunity) were chosen for further investigation based on their high P values. The P values for these categories were plotted as a function of time p.i. for infections of young (Y) and aged (A) animals infected with icUrbani (U), icGZ02 (G), or icHC/SZ/61/03 (H). (B) Expression data are shown for young and aged mice infected with icUrbani (U), icGZ02 (G), or icHC/SZ/61/03 (H) for 24 and 72 h p.i. Genes were selected by GSEA within the GO categories inflammatory response (pink bar), apoptosis (blue bar), and lymphocyte-mediated immunity (green bar), with the additional condition that at least one experiment met the cutoff change of >1.5-fold (P < 0.01). The experiments are clustered using the unweighted-pair group method using average linkages, with similarity determined by Euclidean distance and ordering by average value. The column dendrogram, showing the relative similarity of the experiments, is at the top. The data for aged icUrbani-infected mice at 24 h p.i. clusters more distantly with the data for young mice at 24 h p.i. (bold gray line) than the data for aged mice at 24 h p.i. Bold black line, icGZ02 and icHC/SZ/61/03 data.
Host response at 24 h after lethal infection.
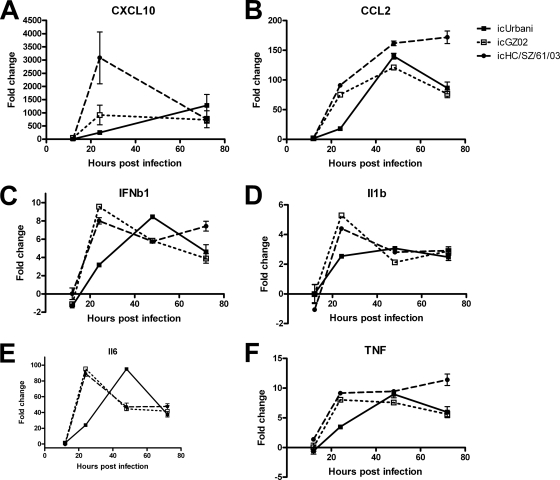
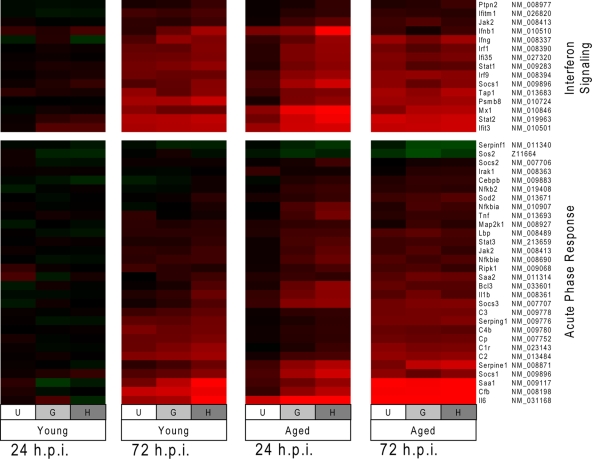
Since the host response at 24 h p.i. is clearly a determinant of the disease outcome, data from this time point were further examined to identify regulatory pathways that might provide a possible explanation for lung pathology and mortality. Ingenuity pathway analysis was used to identify the top canonical pathways induced in the 24-h p.i. data sets from the lethal models. Interferon signaling and acute-phase response were found to be the top canonical pathways induced during icGZ02 and icHC/SZ/61/03 infection. The kinetics of a subset of genes from the interferon signaling and acute-phase response pathways was confirmed by real-time RT-PCR (Fig. 7). As with the functional annotation, expression of genes in these canonical pathways was correlated with pathogenicity in the aged mice at 24 h p.i., but not in young mice (Fig. 8). The data again revealed that similarities became apparent among the different infection systems at 72 h p.i. Of particular note, the acute-phase response pathway included a number of cytokines previously identified as important in the development of ARDS, including IL-1β, IL-6, and TNF (7, 8), as well as several members of the complement cascade.
FIG. 7.
Confirmation of select gene expression results using RT-PCR. Quantitative RT-PCR for Cxcl10 (A), Ccl2 (B), IFN-β (C), IL-1β (D), IL-6 (E), and TNF (F) was performed on all separate lung samples from aged animals at 12, 24, 48, and 72 h p.i., with the exception of that for Cxcl10, which was not performed at 48 h. The data are shown as changes over mock-infected controls. The error bars represent standard deviations.
FIG. 8.
Canonical pathway regulation correlates with pathogenesis and lethality. At 24 h p.i., the top canonical pathways associated with lethal infections of aged mice (the significance criteria were >1.5-fold change with a P value of <0.01) are interferon signaling (P = 2.75 × 10−14 for HC/SZ/61/03 and P = 4.75 × 10−14 for GZ02) and acute-phase response (P = 1.62 × 10−9 for HC/SZ/61/03 and P = 5.2 × 10−9 for GZ02). These pathways are not as significantly upregulated by pathogenic but nonlethal infection (aged animals infected with Urbani, interferon signaling, P = 5.26 × 10−5, and acute-phase response, P = 1.88 × 10−2). Expression data for genes in these pathways are shown as a heat map for young and aged mice infected with icUrbani (U), icGZ02 (G), or icHC/SZ/61/03 (H) for 24 and 72 h p.i. The “NM” numbers on the right are GenBank accession numbers.
In addition, upon SARS-CoV infection in either young or aged mice, both beta interferon (IFN-β) and IFN-γ, but not IFN-α, were significantly upregulated, as previously observed in other animal models of SARS-CoV infection (3, 12). Many of the ISGs were also upregulated, including Ifit3 and Irf9 genes. The temporal regulation of many of these individual cytokine genes mirrored the kinetics of the global gene expression of host responses; strong upregulation of these genes was observed by 24 h p.i. in aged mice infected with icGZ02 and icHC/SZ/61/03, but these same responses during nonlethal icUrbani infection were delayed until 72 h p.i. These data establish the aged-mouse model as an animal model for virus-induced ARDS and suggest that interferon signaling and the acute-phase response, including the complement system, play pivotal roles in the outcome of viral infection and SARS-CoV pathogenesis.
Differences in the host responses between two lethal SARS-CoV strains.
In order to assess the host response differences between the two lethal SARS-CoV strains, which could be potential determinants of disease outcome, a gene interaction network was created, associated with early host response (24 h p.i.). The network was created by including genes that showed differential regulation that was unique in response to the lethal icGZ02 and icHC/SZ/61/03 infections (Fig. 9). The network analysis is useful to assess genes acting in concert within and between different functional processes and represents a combined interaction network generated with the top two networks identified by ingenuity pathway analysis (details are provided in the legend to Fig. 9). The interaction network shows the four broad functional classes of immune response, apoptosis, cell cycle control, and translational control/endoplasmic reticulum (ER) stress, acting in concert and shows that icGZ02 infection regulates a subset of the genes whereas icHC/SZ/61/03 infection regulates all of the genes in the network. This enhanced host response to icHC/SZ/61/03 infection at 24 h p.i. provides a possible explanation for prior mortality data (47) that showed that icGZ02 killed 75% of the animals by day 4 whereas HC/SZ/61/03 killed 100% of the animals starting as early as day 3. The enhanced 24-h p.i. host response to icHC/SZ/61/03 infection is of particular interest, because at this stage of infection, the lungs of aged mice show viral loads of icHC/SZ/61/03 (6 × 105 PFU/g) lower than the viral loads of icGZ02 (2 × 107 PFU/g) (Fig. 1), and therefore, this earlier and enhanced response may potentially represent a host immunopathologic contribution to SARS pathogenesis.
FIG. 9.
A network of genes suggests lethality in aged mice. The network was created by importing genes meeting the selection criteria (≥1.5-fold change; P ≤ 0.01 at 24 h p.i. in either the icHC/SZ/61/03-infected aged-mouse or icGZ02-infected aged-mouse experiments, but not the icUrbani-infected aged-mouse experiment) in ingenuity pathways analysis (IPA). The top two networks built by IPA were chosen and merged, keeping only “direct” IPA-curated interactions. Genes were separated into functional classes based on ingenuity functional annotations. The circles represent individual genes or protein complexes in the network and are colored according to the expression pattern observed in aged animals infected with icGZ02 spike variant virus at 24 h p.i. (A) or icHC/SZ/61/03 spike variant virus at 24 h p.i (B). The arrows represent direct biological interactions. The circles colored in shades of red represent genes upregulated in lethal infection versus mock infection. Pink indicates 1.5- to 2.0-fold change, red indicates 2.0- to 5.0-fold change, and dark red represents a >5-fold change. Likewise, light-green circles represent downregulation and indicate a 1.5- to 2.0-fold change. Gene names colored blue are differentially regulated by icHC/SZ/61/03 infection, but not icGZ02 infection, and are therefore associated with increased lethality.
DISCUSSION
Despite intensive studies, there are currently no specific therapies for ARDS, especially following virus infection, and model systems are desperately needed. While several viruses are known to cause ARDS, including SARS-CoV and influenza virus (9, 11, 30, 39, 42, 51), the molecular mechanisms of virus-induced ARDS are not well characterized due to the limited number of available animal models. The recent reconstruction of two lethal SARS-CoV infection models in mice that recapitulate the age-related pathogenic phenotype noted in humans (47) provided us with the novel opportunity to characterize host response kinetics and to more fully develop the model for characterizing the molecular mechanisms of virus-induced ARDS. The main goals of this paper were to use systems biology approaches with SARS-CoV-infected aged and young mice to (i) characterize disease progression as distinguishable gene expression profiles and (ii) validate the aged-mouse model of SARS as representative of ARDS.
In this study, both lethal and nonlethal isolates grew to high titers in the lungs of infected mice but showed different tropisms and abundances of virus-infected cells in the lungs, which were correlated with increased pathological findings. The molecular mechanisms governing tropism differences imparted by the S glycoprotein are unknown. It has been shown that mutations in the S glycoprotein are responsible for differences in receptor affinity during adaptation (34). All of the S glycoprotein mutants used in this study required ACE2 as their receptors. However, expression of ACE2 alone was not sufficient for replication of the zoonotic strains in human airway epithelium cells (47). These data suggest that the differences in tropism could reflect differential affinities for murine ACE2 receptors or for DC/L-Sign coreceptors (34). Future studies to characterize localizations and levels of ACE2 in murine lung cell types are imperative, as type I and type II human pneumocytes, as well as human bronchial epithelial cells, express ACE2 (19, 50). Differences in the tropism of target cells and disease severity have also been observed between infections with human and avian influenza virus strains (55). We speculate that like that of influenza virus, SARS-CoV tropism is a necessary factor but is not sufficient to explain differences in the severity of disease progression (55). Our observation that downregulation of ACE2 correlates with lethality agrees with earlier studies showing that SARS-CoV S glycoprotein worsens acute lung failure in mice (32). ACE2 has been suggested to regulate the host response by exaggerating acute lung failure through deregulation of the rennin-angiotensin system, pulmonary vascular tone/permeability, and epithelial cell survival (23, 31, 32, 35); therefore, future experiments evaluating SARS-CoV-induced lung pathology during modulation of RAS gene family members would be informative.
Considering that all three viral isolates replicated to similar titers in the lungs of aged animals, specific changes in the S glycoprotein seem to have triggered an aberrant host response. Similar effects of changes in virulence have been described for variants of the influenza virus hemagglutinin and neuraminidase, which also show differences in pathogenicity, linked with upregulation of genes associated with apoptosis and tissue injury (25). SARS-CoV infection in aged mice promotes rapid and fatal disease by influencing, not the kinetics of viral replication, but rather the host response kinetics. Aged mice showed an early host response dominated by the induction of inflammatory cytokines and an acute-phase response at 24 h p.i. The differences in host gene expression, as well as cell tropism and peak viral replication, at early time points in aged mice infected with lethal SARS-CoV isolates indicate that the first 24 h p.i. are critical to the clinical outcome. This is in agreement with the observation that p.i. treatment of SARS-CoV-infected mice with a neutralizing monoclonal antibody did not protect them from death in the absence of virus replication at later time points (46). Disease outcome was most affected by lymphocyte-mediated immunity, inflammatory response, and apoptosis, as well as pathways of interferon signaling and acute-phase response. In contrast, at 72 h p.i., common patterns of regulation occurred regardless of the disease outcome. Interestingly, an enhanced host response to icHC/SZ/61/03 infection compared with icGZ02 infection was observed that provides a possible explanation for the enhanced lethality of icHC/SZ/61/03 previously observed (47). This enhanced host response was indicated by a greater number of differentially expressed genes, larger absolute change of these genes, and a more extensive network of genes acting in concert during icHC/SZ/61/03 infection. Because the more lethal virus showed a lower viral load yet a greater host response, these results suggest a potential immunopathologic contribution to disease by the early response of the host. This phenomenon is similar to responses seen in several animal models of viral pathogenesis (3, 12), including hemagglutinin/neuraminidase variants of influenza virus (25, 26), and could be described as a more inclusive example of the phenomenon previously described as a “cytokine storm” (21).
This “cytokine storm” observed in SARS-CoV-infected mice paralleled that in SARS patients, including Cxcl-10, IL-1β, IL-6, TNF, and IFN-γ (21). The kinetics of these cytokine and interferon responses in mice is similar to that in human cases of ARDS, including SARS-CoV-induced cases. Cytokines are significantly increased at the onset of ARDS in nonsurvivors compared to survivors, with persistent elevation over time predicting a poor outcome (8, 37, 54). In addition, SARS-CoV-infected patients with poor outcome show deviated ISG and immunoglobulin gene expression early in infection and persistent chemokine levels (8). Our data clearly showed that key inflammatory mediators, including IL-1β, IL-6, and TNF, were upregulated, with all responses being much more robust early during infection with the lethal isolates, in agreement with previous clinical studies (8, 54). In support of this study, IL-1 receptor-deficient mice survived infection with a lethal mouse-adapted SARS-CoV strain (49), suggesting a role for IL-1β in SARS-CoV inflammation-induced pathogenesis. These inflammatory mediators have been shown to play an important role in the exudative phase of ARDS pathophysiology (43). Interferon has also been suggested to play a role in ARDS pathophysiology, but what that role is has been less clear. Here, mouse interferon responses were increased early in lethal infection with a delayed response after nonlethal SARS-CoV infection. Previously, in vitro data have suggested that SARS-CoV infection blocks the interferon response through several mechanisms (15, 16, 29), while in vivo data from humans, nonhuman primates, and mice have suggested that early SARS-CoV infection results in robust type I and II interferon responses, as well as increased ISG expression (3, 8, 12). Studies of nonhuman primates at the cellular level bridge these results to show that cells infected with SARS-CoV cannot produce interferon but are capable of activating uninfected neighboring cells to produce an interferon response (12).
In addition to uncovering interferon signaling and components of an aberrant host response that contribute to lethality, mouse data also revealed differential gene expression in the complement cascade, as well as a novel regulatory network that could distinguish two lethal phenotypes. Complement activation has been shown to play a role in ARDS (22, 58), as well as in the pathogenesis of other viruses, including arboviruses, dengue virus, and respiratory syncytial virus (2, 38, 40). Studies regarding the involvement of the complement system in SARS-CoV pathogenesis are currently ongoing in our laboratory. Interestingly, gene expression analysis revealed a regulatory network made up of cell cycle, apoptosis, ER stress, and immune response genes working in concert to affect the progression of pathogenesis in aged animals. Both lethal phenotypes are associated with a strong regulation of immune pathways, including NF-κB signaling and upregulation of IFN-β and translational-control/ER stress pathways, including DNAJC3 (p58IPK). p58IPK was previously shown to be a virulence factor during influenza infection (17) and could play a similar role during virus-induced ARDS. During viral infection, translational-control and cell cycle genes are expected to inhibit viral replication, and immune and apoptotic pathways kill infected cells. However, if this response occurs early in infected cells with strong upregulation of these genes, uninfected tissues can also suffer the effects of this uncontrolled immune response, similar to that seen in macaque studies (12). Moreover, upregulation of apoptotic genes primes the cells for death rather than survival in a manner consistent with the two lethal phenotypes. In animals infected with icGZ02, a small number of apoptotic genes are upregulated at 24 h p.i., and this pattern is associated with the death of 75% of the mice by day 4. In contrast, infection with icHC/SZ/61/03 causes upregulation of more apoptotic genes at 24 h p.i. and is associated with a 100% mortality rate by the end of day 3. Additional cell cycle regulation observed in icHC/SZ/61/03-infected mice appears to be a result of increased lung damage. Klf6 expression was uniquely upregulated during lethal infection and has been linked to transforming growth factor beta-related repair of tissue damage (5). Although this repair mechanism is activated by 24 h p.i., it is unable to counteract the inflammatory damage already under way. As a result, we describe the icHC/SZ/61/03 host response as showing “enhanced lethality” due to the additional components of regulation identified in the lethality-associated network.
Although several other animal models of SARS-CoV infection have been described (12, 33, 36, 44, 45, 52), these models rarely showed lethality. The results varied, ranging from asymptomatic virus replication to pulmonary pathology with visible clinical symptoms. The mouse models of virus-induced ARDS used in this study lead to clinical symptoms in mice and more accurately recapitulate the age-related pathogenic phenotype seen in human populations, suggesting this system is a relevant model to study the acute phase of SARS-CoV pathogenesis and virus-induced ARDS. The time course of our study was focused on the exudative phase of ARDS, characterized by DAD and hyaline membrane formation (47), with a lethal outcome 3 to 4 days p.i.; therefore, mechanisms of fibrosis and tissue repair could not be determined. However, since several cytokines involved in fibrosis are induced in our model, we predict that the survival of lethally infected aged mice could be extended to the fibrotic phase of ARDS by using sublethal infection. How this time course extension would affect host response kinetics is of great interest. Future studies, using knockout mice and depletion studies, will investigate the roles of ARDS-associated cytokines, transforming growth factor beta-related repair mechanisms, complement cascades, cell cycle regulation, and caspase-directed apoptosis during acute virus infection with the goal of promoting normal tissue repair and inhibiting lethal disease in order to determine direct contributions to respiratory death. These studies, including comparative work with young and aged mice with low- and highly pathogenic influenza, may provide considerable insight into the effects of aging on early and/or aberrant host response.
In conclusion, the genomics data presented in this study have established two new models of virus-induced ARDS. These data provide several target cytokines that are associated with the development of ARDS and have identified several genes and pathways that potentially play roles in SARS-CoV pathogenesis. These genes and pathways will be valuable as candidates for future studies of the mechanism of SARS-CoV lethality and as potential targets for treatment. In addition, these new models of virus-induced ARDS will be invaluable for comparative analysis with other virus-induced ARDS models and may provide insight into common mechanisms of ARDS pathophysiology leading to death.
Acknowledgments
We thank Marcus J. Korth for critically reading the manuscript.
This work was supported by NIH NIAID grants P01-AI059443 and AI059136 to R.B. and NIH NHLBI grant R01-HL080621 to M.G.K.
Footnotes
Published ahead of print on 6 May 2009.
REFERENCES
- 1.Ashburner, M., C. A. Ball, J. A. Blake, D. Botstein, H. Butler, J. M. Cherry, A. P. Davis, K. Dolinski, S. S. Dwight, J. T. Eppig, M. A. Harris, D. P. Hill, L. Issel-Tarver, A. Kasarskis, S. Lewis, J. C. Matese, J. E. Richardson, M. Ringwald, G. M. Rubin, G. Sherlock, et al. 2000. Gene ontology: tool for the unification of biology. Nat. Genet. 2525-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avirutnan, P., N. Punyadee, S. Noisakran, C. Komoltri, S. Thiemmeca, K. Auethavornanan, A. Jairungsri, R. Kanlaya, N. Tangthawornchaikul, C. Puttikhunt, S. N. Pattanakitsakul, P. T. Yenchitsomanus, J. Mongkolsapaya, W. Kasinrerk, N. Sittisombut, M. Husmann, M. Blettner, S. Vasanawathana, S. Bhakdi, and P. Malasit. 2006. Vascular leakage in severe dengue virus infections: a potential role for the nonstructural viral protein NS1 and complement. J. Infect. Dis. 1931078-1088. [DOI] [PubMed] [Google Scholar]
- 3.Baas, T., A. Roberts, T. H. Teal, L. Vogel, J. Chen, T. M. Tumpey, M. G. Katze, and K. Subbarao. 2008. Genomic analysis reveals age-dependent innate immune responses to severe acute respiratory syndrome coronavirus. J. Virol. 829465-9476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhatia, M., and S. Moochhala. 2004. Role of inflammatory mediators in the pathophysiology of acute respiratory distress syndrome. J. Pathol. 202145-156. [DOI] [PubMed] [Google Scholar]
- 5.Botella, L. M., T. Sanchez-Elsner, F. Sanz-Rodriguez, S. Kojima, J. Shimada, M. Guerrero-Esteo, M. P. Cooreman, V. Ratziu, C. Langa, C. P. Vary, J. R. Ramirez, S. Friedman, and C. Bernabeu. 2002. Transcriptional activation of endoglin and transforming growth factor-beta signaling components by cooperative interaction between Sp1 and KLF6: their potential role in the response to vascular injury. Blood 1004001-4010. [DOI] [PubMed] [Google Scholar]
- 6.Brazma, A., P. Hingamp, J. Quackenbush, G. Sherlock, P. Spellman, C. Stoeckert, J. Aach, W. Ansorge, C. A. Ball, H. C. Causton, T. Gaasterland, P. Glenisson, F. C. Holstege, I. F. Kim, V. Markowitz, J. C. Matese, H. Parkinson, A. Robinson, U. Sarkans, S. Schulze-Kremer, J. Stewart, R. Taylor, J. Vilo, and M. Vingron. 2001. Minimum information about a microarray experiment (MIAME)—toward standards for microarray data. Nat. Genet. 29365-371. [DOI] [PubMed] [Google Scholar]
- 7.Cameron, M. J., J. F. Bermejo-Martin, A. Danesh, M. P. Muller, and D. J. Kelvin. 2008. Human immunopathogenesis of severe acute respiratory syndrome (SARS). Virus Res. 13313-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cameron, M. J., L. Ran, L. Xu, A. Danesh, J. F. Bermejo-Martin, C. M. Cameron, M. P. Muller, W. L. Gold, S. E. Richardson, S. M. Poutanen, B. M. Willey, M. E. DeVries, Y. Fang, C. Seneviratne, S. E. Bosinger, D. Persad, P. Wilkinson, L. D. Greller, R. Somogyi, A. Humar, S. Keshavjee, M. Louie, M. B. Loeb, J. Brunton, A. J. McGeer, and D. J. Kelvin. 2007. Interferon-mediated immunopathological events are associated with atypical innate and adaptive immune responses in patients with severe acute respiratory syndrome. J. Virol. 818692-8706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caramello, P., F. Canta, L. Bonino, C. Moiraghi, F. Navone, F. Lipani, R. Balbiano, A. M. Caputo, and V. Gai. 2002. Puumala virus pulmonary syndrome in a Romanian immigrant. J. Travel Med. 9326-329. [DOI] [PubMed] [Google Scholar]
- 10.Chan-Yeung, M., and R. H. Xu. 2003. SARS: epidemiology. Respirology 8(Suppl.)S9-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chou, D. W., C. H. Lee, C. W. Chen, H. Y. Chang, and T. R. Hsiue. 1999. Varicella pneumonia complicated by acute respiratory distress syndrome in an adult. J. Formos. Med. Assoc. 98778-782. [PubMed] [Google Scholar]
- 12.de Lang, A., T. Baas, T. Teal, L. M. Leijten, B. Rain, A. D. Osterhaus, B. L. Haagmans, and M. G. Katze. 2007. Functional genomics highlights differential induction of antiviral pathways in the lungs of SARS-CoV-infected macaques. PLoS Pathog. 3e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding, Y., H. Wang, H. Shen, Z. Li, J. Geng, H. Han, J. Cai, X. Li, W. Kang, D. Weng, Y. Lu, D. Wu, L. He, and K. Yao. 2003. The clinical pathology of severe acute respiratory syndrome (SARS): a report from China. J. Pathol. 200282-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franks, T. J., P. Y. Chong, P. Chui, J. R. Galvin, R. M. Lourens, A. H. Reid, E. Selbs, C. P. McEvoy, C. D. Hayden, J. Fukuoka, J. K. Taubenberger, and W. D. Travis. 2003. Lung pathology of severe acute respiratory syndrome (SARS): a study of 8 autopsy cases from Singapore. Hum. Pathol. 34743-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frieman, M., M. Heise, and R. Baric. 2008. SARS coronavirus and innate immunity. Virus Res. 133101-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frieman, M., B. Yount, M. Heise, S. A. Kopecky-Bromberg, P. Palese, and R. S. Baric. 2007. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J. Virol. 819812-9824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodman, A. G., J. A. Smith, S. Balachandran, O. Perwitasari, S. C. Proll, M. J. Thomas, M. J. Korth, G. N. Barber, L. A. Schiff, and M. G. Katze. 2007. The cellular protein P58IPK regulates influenza virus mRNA translation and replication through a PKR-mediated mechanism. J. Virol. 812221-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu, J., and C. Korteweg. 2007. Pathology and pathogenesis of severe acute respiratory syndrome. Am. J. Pathol. 1701136-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamming, I., W. Timens, M. L. Bulthuis, A. T. Lely, G. J. Navis, and H. van Goor. 2004. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 203631-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He, L., Y. Ding, Q. Zhang, X. Che, Y. He, H. Shen, H. Wang, Z. Li, L. Zhao, J. Geng, Y. Deng, L. Yang, J. Li, J. Cai, L. Qiu, K. Wen, X. Xu, and S. Jiang. 2006. Expression of elevated levels of pro-inflammatory cytokines in SARS-CoV-infected ACE2+ cells in SARS patients: relation to the acute lung injury and pathogenesis of SARS. J. Pathol. 210288-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang, K. J., I. J. Su, M. Theron, Y. C. Wu, S. K. Lai, C. C. Liu, and H. Y. Lei. 2005. An interferon-gamma-related cytokine storm in SARS patients. J. Med. Virol. 75185-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huber-Lang, M., J. V. Sarma, F. S. Zetoune, D. Rittirsch, T. A. Neff, S. R. McGuire, J. D. Lambris, R. L. Warner, M. A. Flierl, L. M. Hoesel, F. Gebhard, J. G. Younger, S. M. Drouin, R. A. Wetsel, and P. A. Ward. 2006. Generation of C5a in the absence of C3: a new complement activation pathway. Nat. Med. 12682-687. [DOI] [PubMed] [Google Scholar]
- 23.Imai, Y., K. Kuba, S. Rao, Y. Huan, F. Guo, B. Guan, P. Yang, R. Sarao, T. Wada, H. Leong-Poi, M. A. Crackower, A. Fukamizu, C. C. Hui, L. Hein, S. Uhlig, A. S. Slutsky, C. Jiang, and J. M. Penninger. 2005. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature 436112-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang, Y., J. Xu, C. Zhou, Z. Wu, S. Zhong, J. Liu, W. Luo, T. Chen, Q. Qin, and P. Deng. 2005. Characterization of cytokine/chemokine profiles of severe acute respiratory syndrome. Am. J. Respir. Crit. Care Med. 171850-857. [DOI] [PubMed] [Google Scholar]
- 25.Kash, J. C., C. F. Basler, A. Garcia-Sastre, V. Carter, R. Billharz, D. E. Swayne, R. M. Przygodzki, J. K. Taubenberger, M. G. Katze, and T. M. Tumpey. 2004. Global host immune response: pathogenesis and transcriptional profiling of type A influenza viruses expressing the hemagglutinin and neuraminidase genes from the 1918 pandemic virus. J. Virol. 789499-9511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kash, J. C., T. M. Tumpey, S. C. Proll, V. Carter, O. Perwitasari, M. J. Thomas, C. F. Basler, P. Palese, J. K. Taubenberger, A. Garcia-Sastre, D. E. Swayne, and M. G. Katze. 2006. Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature 443578-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keller, A., C. Backes, and H. P. Lenhof. 2007. Computation of significance scores of unweighted Gene Set Enrichment Analyses. BMC Bioinformatics 8290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kerr, M. K., and G. A. Churchill. 2001. Statistical design and the analysis of gene expression microarray data. Genet. Res. 77123-128. [DOI] [PubMed] [Google Scholar]
- 29.Kopecky-Bromberg, S. A., L. Martinez-Sobrido, M. Frieman, R. A. Baric, and P. Palese. 2007. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 81548-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ku, A. S., and L. T. Chan. 1999. The first case of H5N1 avian influenza infection in a human with complications of adult respiratory distress syndrome and Reye's syndrome. J. Paediatr. Child Health 35207-209. [DOI] [PubMed] [Google Scholar]
- 31.Kuba, K., Y. Imai, and J. M. Penninger. 2006. Angiotensin-converting enzyme 2 in lung diseases. Curr. Opin. Pharmacol. 6271-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuba, K., Y. Imai, S. Rao, H. Gao, F. Guo, B. Guan, Y. Huan, P. Yang, Y. Zhang, W. Deng, L. Bao, B. Zhang, G. Liu, Z. Wang, M. Chappell, Y. Liu, D. Zheng, A. Leibbrandt, T. Wada, A. S. Slutsky, D. Liu, C. Qin, C. Jiang, and J. M. Penninger. 2005. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 11875-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuiken, T., R. A. Fouchier, M. Schutten, G. F. Rimmelzwaan, G. van Amerongen, D. van Riel, J. D. Laman, T. de Jong, G. van Doornum, W. Lim, A. E. Ling, P. K. Chan, J. S. Tam, M. C. Zambon, R. Gopal, C. Drosten, S. van der Werf, N. Escriou, J. C. Manuguerra, K. Stohr, J. S. Peiris, and A. D. Osterhaus. 2003. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet 362263-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, W., C. Zhang, J. Sui, J. H. Kuhn, M. J. Moore, S. Luo, S. K. Wong, I. C. Huang, K. Xu, N. Vasilieva, A. Murakami, Y. He, W. A. Marasco, Y. Guan, H. Choe, and M. Farzan. 2005. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 241634-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marshall, R. P., S. Webb, G. J. Bellingan, H. E. Montgomery, B. Chaudhari, R. J. McAnulty, S. E. Humphries, M. R. Hill, and G. J. Laurent. 2002. Angiotensin converting enzyme insertion/deletion polymorphism is associated with susceptibility and outcome in acute respiratory distress syndrome. Am. J. Respir. Crit. Care Med. 166646-650. [DOI] [PubMed] [Google Scholar]
- 36.Martina, B. E., B. L. Haagmans, T. Kuiken, R. A. Fouchier, G. F. Rimmelzwaan, G. Van Amerongen, J. S. Peiris, W. Lim, and A. D. Osterhaus. 2003. Virology: SARS virus infection of cats and ferrets. Nature 425915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meduri, G. U., S. Headley, G. Kohler, F. Stentz, E. Tolley, R. Umberger, and K. Leeper. 1995. Persistent elevation of inflammatory cytokines predicts a poor outcome in ARDS. Plasma IL-1 beta and IL-6 levels are consistent and efficient predictors of outcome over time. Chest 1071062-1073. [DOI] [PubMed] [Google Scholar]
- 38.Melendi, G. A., S. J. Hoffman, R. A. Karron, P. M. Irusta, F. R. Laham, A. Humbles, B. Schofield, C. H. Pan, R. Rabold, B. Thumar, A. Thumar, N. P. Gerard, W. Mitzner, S. R. Barnum, C. Gerard, S. R. Kleeberger, and F. P. Polack. 2007. C5 modulates airway hyperreactivity and pulmonary eosinophilia during enhanced respiratory syncytial virus disease by decreasing C3a receptor expression. J. Virol. 81991-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Merk, J., F. X. Schmid, M. Fleck, S. Schwarz, C. Lehane, S. Boehm, B. Salzberger, and D. E. Birnbaum. 2005. Fatal pulmonary failure attributable to viral pneumonia with human herpes virus 6 (HHV6) in a young immunocompetent woman. J. Intensive Care Med. 20302-306. [DOI] [PubMed] [Google Scholar]
- 40.Morrison, T. E., and M. T. Heise. 2008. The host complement system and arbovirus pathogenesis. Curr. Drug Targets 9165-172. [DOI] [PubMed] [Google Scholar]
- 41.Murasko, D. M., and J. Jiang. 2005. Response of aged mice to primary virus infections. Immunol. Rev. 205285-296. [DOI] [PubMed] [Google Scholar]
- 42.Peiris, J. S., C. M. Chu, V. C. Cheng, K. S. Chan, I. F. Hung, L. L. Poon, K. I. Law, B. S. Tang, T. Y. Hon, C. S. Chan, K. H. Chan, J. S. Ng, B. J. Zheng, W. L. Ng, R. W. Lai, Y. Guan, and K. Y. Yuen. 2003. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet 3611767-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Puneet, P., S. Moochhala, and M. Bhatia. 2005. Chemokines in acute respiratory distress syndrome. Am. J. Physiol. Lung Cell. Mol. Physiol. 288L3-L15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roberts, A., C. Paddock, L. Vogel, E. Butler, S. Zaki, and K. Subbarao. 2005. Aged BALB/c mice as a model for increased severity of severe acute respiratory syndrome in elderly humans. J. Virol. 795833-5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roberts, A., L. Vogel, J. Guarner, N. Hayes, B. Murphy, S. Zaki, and K. Subbarao. 2005. Severe acute respiratory syndrome coronavirus infection of golden Syrian hamsters. J. Virol. 79503-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rockx, B., D. Corti, E. Donaldson, T. Sheahan, K. Stadler, A. Lanzavecchia, and R. Baric. 2008. Structural basis for potent cross-neutralizing human monoclonal antibody protection against lethal human and zoonotic severe acute respiratory syndrome coronavirus challenge. J. Virol. 823220-3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rockx, B., T. Sheahan, E. Donaldson, J. Harkema, A. Sims, M. Heise, R. Pickles, M. Cameron, D. Kelvin, and R. Baric. 2007. Synthetic reconstruction of zoonotic and early human severe acute respiratory syndrome coronavirus isolates that produce fatal disease in aged mice. J. Virol. 817410-7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salvioli, S., M. Capri, S. Valensin, P. Tieri, D. Monti, E. Ottaviani, and C. Franceschi. 2006. Inflamm-aging, cytokines and aging: state of the art, new hypotheses on the role of mitochondria and new perspectives from systems biology. Curr. Pharm. Des. 123161-3171. [DOI] [PubMed] [Google Scholar]
- 49.Sheahan, T., T. E. Morrison, W. Funkhouser, S. Uematsu, S. Akira, R. S. Baric, and M. T. Heise. 2008. MyD88 is required for protection from lethal infection with a mouse-adapted SARS-CoV. PLoS Pathog. 4e1000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sims, A. C., R. S. Baric, B. Yount, S. E. Burkett, P. L. Collins, and R. J. Pickles. 2005. Severe acute respiratory syndrome coronavirus infection of human ciliated airway epithelia: role of ciliated cells in viral spread in the conducting airways of the lungs. J. Virol. 7915511-15524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spelman, D. W., and P. A. Stanley. 1983. Respiratory syncytial virus pneumonitis in adults. Med. J. Aust. 1430-431. [DOI] [PubMed] [Google Scholar]
- 52.Subbarao, K., J. McAuliffe, L. Vogel, G. Fahle, S. Fischer, K. Tatti, M. Packard, W. J. Shieh, S. Zaki, and B. Murphy. 2004. Prior infection and passive transfer of neutralizing antibody prevent replication of severe acute respiratory syndrome coronavirus in the respiratory tract of mice. J. Virol. 783572-3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Subramanian, A., P. Tamayo, V. K. Mootha, S. Mukherjee, B. L. Ebert, M. A. Gillette, A. Paulovich, S. L. Pomeroy, T. R. Golub, E. S. Lander, and J. P. Mesirov. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 10215545-15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang, N. L., P. K. Chan, C. K. Wong, K. F. To, A. K. Wu, Y. M. Sung, D. S. Hui, J. J. Sung, and C. W. Lam. 2005. Early enhanced expression of interferon-inducible protein-10 (CXCL-10) and other chemokines predicts adverse outcome in severe acute respiratory syndrome. Clin. Chem. 512333-2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Riel, D., V. J. Munster, E. de Wit, G. F. Rimmelzwaan, R. A. Fouchier, A. D. Osterhaus, and T. Kuiken. 2007. Human and avian influenza viruses target different cells in the lower respiratory tract of humans and other mammals. Am. J. Pathol. 1711215-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu, T., J. Qiao, L. Zhao, G. Wang, G. He, K. Li, Y. Tian, M. Gao, J. Wang, H. Wang, and C. Dong. 2006. Acute respiratory distress syndrome induced by avian influenza A (H5N1) virus in mice. Am. J. Respir. Crit. Care Med. 1741011-1017. [DOI] [PubMed] [Google Scholar]
- 57.Yount, B., K. M. Curtis, E. A. Fritz, L. E. Hensley, P. B. Jahrling, E. Prentice, M. R. Denison, T. W. Geisbert, and R. S. Baric. 2003. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA 10012995-13000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zilow, G., J. A. Sturm, U. Rother, and M. Kirschfink. 1990. Complement activation and the prognostic value of C3a in patients at risk of adult respiratory distress syndrome. Clin. Exp. Immunol. 79151-157. [DOI] [PMC free article] [PubMed] [Google Scholar]