Abstract
The xenotropic murine leukemia virus-related virus (XMRV) has recently been detected in prostate cancer tissues and may play a role in tumorigenesis. It is currently unclear how this virus is transmitted and which factors promote its spread in the prostate. We show that amyloidogenic fragments known as semen-derived enhancer of virus infection (SEVI) originating from prostatic acid phosphatase greatly increase XMRV infections of primary prostatic epithelial and stromal cells. Hybrid simian/human immunodeficiency chimeric virus particles pseudotyped with XMRV envelope protein were used to demonstrate that the enhancing effect of SEVI, or of human semen itself, was at the level of viral attachment and entry. SEVI enhanced XMRV infectivity but did not bypass the requirement for the xenotropic and polytropic retrovirus receptor 1. Furthermore, XMRV RNA was detected in prostatic secretions of some men with prostate cancer. The fact that the precursor of SEVI is produced in abundance by the prostate indicates that XMRV replication occurs in an environment that provides a natural enhancer of viral infection, and this may play a role in the spread of this virus in the human population.
Viruses are etiologic agents of various human cancers, including cervical carcinoma (caused by human papillomavirus), Kaposi's sarcoma (caused by human herpesvirus 8), hepatocellular carcinoma (caused by hepatitis B virus and hepatitis C virus), and adult T-cell leukemia (caused by human T-cell leukemia virus type 1) (6). Genetic and epidemiologic evidence suggests that prostate cancer may also have an infectious etiology, although a causative agent has not been identified (4, 12). The gammaretrovirus xenotropic murine leukemia virus-related virus (XMRV) is a candidate human tumor virus based on its association in human prostate tumors with a reduced-activity variant of the antiviral gene, RNASEL (also known as the hereditary prostate cancer 1 gene or HPC1) (17) and because it is a member of a viral family known to cause leukemias and lymphomas in different mammalian species (8). Interferon, through its effector RNase L, potently inhibits XMRV replication (5). XMRV integration sites in human prostate cancer tissues were mapped to cancer breakpoints, common fragile sites, micro-RNA genes, and cancer-related genes (11). Many of these genes are implicated directly or indirectly in prostate cancer and metabolic pathways that affect prostate cancer, including androgen signaling. XMRV has also been observed in prostate tissue from a nonfamilial prostate cancer patient and in an individual without prostate cancer (7). The possible role of XMRV in prostatic cancer raises questions about its ability to infect the prostate and the route of viral transmission.
Recently, it has been shown that fragments of prostatic acid phosphatase (PAP), an abundant nonspecific protein phosphatase produced by the prostate (18) and secreted in semen in large quantities (about 2 mg/ml) (16), form amyloid fibrils that drastically enhance human immunodeficiency virus type 1 (HIV-1) infection (14). The fibrils of PAP248-286, termed semen-derived enhancer of virus infection (SEVI), enhanced the infectious virus titer by several orders of magnitude by capturing HIV-1 virions and promoting their attachment to target cells. The ability of SEVI to promote the interaction between virions and the cell surface is independent of the viral glycoprotein and hence is not restricted to HIV-1, although subsequent fusion between the viral and cellular membranes still required gp120, CD4, and an appropriate coreceptor (14). A recent study indicates that the positive charges on SEVI (pI = 10.21) promote infectivity by neutralizing negative-charge repulsion between HIV particles and the cell surface (15).
Because SEVI originates from the prostate (the organ from which XMRV infection was discovered [17]) and promotes viral attachment in a relatively nonspecific manner, we sought to determine its effect on XMRV infection. Here we demonstrate that XMRV infectivity is greatly enhanced by SEVI or human semen and that XMRV RNA is detectable in expressed prostatic secretions (EPS) from human tumor-bearing prostates.
MATERIALS AND METHODS
Cell culture.
DU145 and CEM cells were grown in complete RPMI containing 10% fetal bovine serum (FBS), penicillin (100 units/ml; Life Technologies), and streptomycin (100 units/ml; Life Technologies). Human foreskin fibroblasts (HFF) (a gift from P. Pellett, Wayne State University, Detroit, MI) and CaSki, HeLa M, and HEK 293T cells were cultured in complete Dulbecco modified Eagle medium containing 10% FBS, penicillin, and streptomycin. PrSC and PrEC cells were purchased from Lonza Walkersville Inc. and were grown in SCGM Bulletkit and PrEGM Bulletkit, respectively, according to the manufacturer's instructions. All cells were grown at 37°C in an atmosphere of 5% CO2.
XMRV preparations and infectivity assays.
XMRV was generated from cDNAs obtained from prostate cancer case VP62 and was subsequently transfected in LNCaP cells to produce virus stocks as previously described (5). The 50% tissue culture infective dose (TCID50) was determined as described by the ACTG Laboratory Technologist Committee (https://www.actgnetwork.org/login.aspx?ReturnUrl=%2fpub%2fdownload%2flabmanual%2f43-ALM-TCID50-Determination.pdf).
Effect of SEVI on viral infectivity.
Synthetic PAP fragments were obtained from VIRO Pharmaceuticals GmbH & Co. KG (Hannover, Germany). Peptides were dissolved in phosphate-buffered saline (PBS) and converted to amyloid fibrils by overnight agitation at 37°C at 1,400 rpm using an Eppendorf Thermomixer as described previously (14). Virus infections were performed in triplicate in 12-well or 24-well plates with 2.5 × 104 or 1.3 × 104 cells, respectively, plated 1 day before infection. Cells were at about 20% confluence at the time of infection. SEVI fibrils were mixed with XMRV, incubated at 23°C for 5 min, and further diluted with serum-free medium. SEVI-virus mixtures were added to cells, incubated for 3 h, removed, and replaced with serum-containing medium. Cells were subsequently cultured for 3 days, and infectivity was determined. To assess the effect of azidothymidine (AZT) (Sigma-Aldrich), DU145 cells were infected with 1 TCID50/1,000 cells of XMRV with or without SEVI (50 μg/ml) for 3 h, which was then replaced with complete medium. After overnight incubation, the medium was removed and replaced with fresh medium with or without AZT (10 μM). Cells were subsequently cultured, and RNAs were harvested at 0, 1, 2, 3, and 6 days postinfection.
Immunofluorescence assay.
DU145 cells were trypsinized, plated on eight-well culture slides (BD Biosciences), and subsequently cultured after viral infections. Slides were washed with PBS and then fixed with cold acetone. Immunofluorescence was performed using a rat monoclonal antibody against spleen focus-forming virus (SFFV) Gag p30 (R187; ATCC catalog no. CRL-1912) as a marker for XMRV-infected cells(3). Each well was incubated for 1 h in a humidified box at 37°C with antibody to Gag diluted 1:200 in 10% normal goat serum-5% glycine-PBS. After washing with PBS three times, secondary antibody (Molecular Probes; Alex Fluor 568-conjugated goat anti-rat) diluted in the same buffer was added and incubated for 1 h. After three PBS washes, the slides were mounted with Vectashield (Vector Laboratories) containing DAPI (4′,6′- diamidino-2-phenylindole) and an antifade reagent. Fluorescence microscopic analysis and image processing were done using a Leica DMIRB microscope with a Retiga EXi Camera and QCapture Pro software (QImaging Corporation).
Effects of semen on viral infectivity.
Semen was obtained from control men or postvasectomy men, frozen immediately, and stored at −80°C. DU145 cells were seeded at 80 to 90% confluence at the day of infection in 96-well plates. Fresh XMRV supernatants obtained from chronically infected LNCaP cells were passed through a 0.2-μm filter (Whatman, Florham Park, NJ) and diluted 1:100. Semen was thawed immediately prior to use; supplemented with 50 μg/ml gentamicin (Gibco), 100 U/ml penicillin G sodium, and 100 μg/ml streptomycin; and vortexed, and fivefold dilutions in cell culture medium were prepared. Each semen dilution was mixed with XMRV dilutions at 50, 10, 2, 0.4, and 0% of semen; incubated at 23°C for 10 min; and further diluted 1:10 with cell culture medium. The semen-virus mixtures were resuspended, added to confluent cells in triplicate, and incubated at 37°C for 3 h. (Adverse effects of semen of cell proliferation were minimized by performing experiments on confluent monolayers.) The medium was removed, replaced with fresh medium, and cultured for 1 day. Cells were trypsinized at 1 day postinfection and moved to 12-well plates for further cell culturing. Infectivity was measured by quantitative reverse transcription-PCR (qRT-PCR) at 3 days postinfection.
Preparation of PSA, PSMA, and PAP.
Prostate-specific antigen (PSA) and PAP were purchased from Sigma-Aldrich Co. and reconstituted according to the manufacturer's instructions. Prostate-specific membrane antigen (PSMA) was a gift from Warren Heston (Cleveland Clinic, Cleveland, OH).
Plasmids and experiments with recombinant viruses.
The simian immunodeficiency virus SIVmac-based retroviral vector pSIvec1ΔenvLuc was prepared by replacing the green fluorescent protein cDNA in pSIvec1ΔenvGFP (9) with the luciferase sequence using the AgeI and NotI cloning sites. The HIV-1 Rev-expressing plasmid pCMV-Rev and pSVIII-ΔKS, which contains a nonfunctional HIV-1 envelope used to determine nonspecific virus uptake, are described elsewhere (1, 9). Plasmid pTH-XMRV-coEnv contains a codon-optimized synthetic full-length version of the XMRV envelope under the control of the cytomegalovirus promoter. To generate infectious SIV/HIV (SHIV) particles capable of a single round of infection, 8 × 106 HEK-293T cells were transfected in 10-cm dishes by the calcium phosphate method with 15 μg pSIvec1ΔenvLuc, 5 μg pCMV-Rev, and 0.5 μg pTH-XMRV-coEnv or pSVIIIΔKS. At 2 days posttransfection, supernatants were harvested and centrifuged at 6,000 rpm for 10 min to remove residual cells. HIV-based retroviral particles pseudotyped with the XMRV envelope were generated the same way except that pNL4-3.Luc.R−E− was cotransfected with pTH-XMRV-coEnv. Concentrations of the virus preparations were assessed and normalized based on a Gag-specific enzyme-linked immunosorbent assay. For entry experiments, HEK 293T or CaSki target cells were sown in 96-well plates (20,000 cells/well). The next day, virus supernatants were preincubated with SEVI or semen for 15 min. In experiments with SEVI, the virus-SEVI suspension was directly added to the cells, whereas in experiments with semen, 10 μl of the virus supernatant was preincubated with the same volume of semen or fivefold dilutions thereof. These suspensions were put on cells containing 280 μl medium, resulting in a 15-fold dilution of semen. To further minimize semen-mediated cytotoxic effects (10) the entire supernatant was replaced after 2 h by fresh medium containing 50 μg/ml of gentamicin. Infection was determined after 2 days by measuring the luciferase activity in cell lysates using the luciferase assay system (Promega) according to manufacturer's instructions.
Isolation of RNA in EPS.
The prostatic fluids were collected in RNase-free microcentrifuge tubes by manually milking secretions from prostate after surgery, flash frozen, and stored at −80°C until RNA isolation. RNA isolation from 100- to 200-μl samples was performed using a MagMAX viral RNA isolation kit (Ambion, TX) with some modifications stated below. For each isolation, an EPS sample was added to 602 μl of lysis/binding solution containing 300 μl of lysis/binding solution concentrate, 2 μl of carrier RNA, and 300 μl of isopropanol. This was followed by addition of 40 μl of bead mix containing 20 μl of RNA binding beads and 20 μl of lysis/binding enhancer. The washing step was performed following the manufacturer's protocol, and elution was in 40 to 60 μl of preheated elution buffer. Generally the amount of RNA obtained was in the range of 50 to 100 ng/μl.
Depletion of XPR1 mRNA using siRNA.
DU145 cells at about 10% confluence were grown in 12-well plates with antibiotic-free RPMI medium containing 10% FBS for 24 h before small interfering RNA (siRNA) transfection. Predesigned ON-TARGETplus SMART pool siRNA reagent for XPR1 and control siRNA were purchased from Dharmacon Inc. SMART pool siRNAs against XPR1 (catalog no. L-005752-00) were 5′-ACACUAAGGUAUUGAUAGA-3′, 5′-UCGAUUACCUCUACAACUU-3′, 5′-GCUUGCCGCUGUAUUUAAA-3′, and 5′-GGCCUUUCCUCAUUUAGUU-3′, and control siRNA was ON- TARGETplus nontargeting siRNA 1 (catalog no. D-001810-01-20) against 5′-UGGUUUACAUGUCGACUAA-3′. siRNA transfection was performed in triplicate according to the manufacturer's instructions using DharmaFECT transfection reagent 1. Cells were incubated with the transfection medium for 48 h and further challenged by XMRV infection.
qRT-PCR.
Cells were rinsed with PBS and drained thoroughly prior to RNA isolation. RNA was isolated from the cells using TRIzol (Invitrogen) according to the manufacturer's instructions. The quantity of RNA extracted was determined using a NanoDrop ND 1000 spectrophotometer (NanoDrop Technologies). qRT-PCR was performed in duplicate using the AgPath-ID one-step RT-PCR kit (Ambion Inc.).
For measuring XMRV RNA in viral growth infectivity assays, each reaction was performed in an individual tube and the mixture was made up to 25 μl containing 100 to 200 ng RNA, 12.5 μl of 2× RT-PCR buffer, 0.45 μl of 50 μM XMRV env forward and reverse primer, 0.65 μl of 10 μM env probe, human GAPDH (glyceraldehyde-3-phosphate dehydrogenase) oligonucleotides and probe as internal control (Applied Biosystems), and H2O. Conditions for the PCR were 10 min at 45°C (RT), 10 min at 95°C (RT inactivation and initial denaturation), and then 40 cycles, each consisting of 15 s at 95°C and 45 s at 58°C. Primers against XMRV env (5′-TCAGGACAAGGGTGGTTTGAG-3′ forward [Q7472F] and 5′-GGCCCATAATGGTGGATATCA-3′ reverse [Q7527R]) with probe TAM-TTAACAGGTCCCCATGGTTCACGACC-BHQ-2 (ENV-PRO) and primers against XMRV gag (5′-GGACTTTTTGGAGTGGCTTTGTT-3′ forward [Q445T] and 5′-GCGTAAAACCGAAAGCAAAAAT-3′ reverse [Q528R]) with probe FAM-ACAGAGACACTTCCCGCCCCCG-BHQ-1 (F480PRO-BHQ) were used for qRT-PCR, which bind to the p15E regions of env and gag RNAs, respectively.
For detection of XMRV RNA in EPS, qRT-PCR was done for the gp70 region of XMRV env RNA in duplicate assays with 200 ng RNA as described above, except the cycle number was increased from 40 to 55. Primers against XMRV env were 5′GGCCGAGAGAGGGCTACT-3′ forward (Q6124F) and 5′-TGATGATGATGGCTTCCAGTATGC-3′ reverse (Q6197R), and the probe was FAM-CACATCCCCATTTGCC-MGB (ENV 6159R), as a 20× master mix (Applied Biosystems) diluted to 1× in the final reaction mixtures. In vitro-transcribed 1.95-kb XMRV env RNA was used in each reaction for generation of a standard curve.
XPR1 mRNA levels were determined using primers 5′-TTCTTCATGGTGACGTTTGCA-3′ (forward) and 5′-ATGATATAAAAGACAATCCACAGGTAAAAG-3′ (reverse) and probe Cy3-TACAGCACTCACAAAGAACGAGGTCACTCG-BHQ-2.
Nested RT-PCR.
To avoid potential problems with laboratory DNA contamination, nested RT-PCR was performed with separate reagents in a separate laboratory room designated to be free of high-copy amplicon or plasmid DNA. Negative controls were included in every experiment. Tth DNA polymerase-based RT-PCR analysis was performed for XMRV RNA in EPS. The variable region of the env gp70 gene was chosen, as this region distinguishes it from polytropic and ecotropic murine leukemia viruses. For first round of PCR, oligonucleotides were with 6200R (5′-CCCATGATGATGATGGCTTCCAGTATGC-3′) and 5922F (5′-GCTAATGCTACCTCCCTCCTGG-3′), both at 20 μM. For the second round, oligonucleotides were 5942F (5′-GGGGACGATGACAGACACTTTCC-3′) and 6159R (5′-CACATCCCCATTTGCCACAGTAG-3′), both at 10 μM. Tth DNA polymerase (USB Corporation, Cleveland, OH) was used for the first-round synthesis following the manufacturer's protocol with minor modifications. RNA (200 to 300 ng) was added to a total volume of 14 μl containing 1.5 μl of 20 μM 6200R and 0.4 μl of 10 mM deoxynucleoside triphosphates on ice. RNA was denatured and annealed by placing the tubes at 70°C for 5 min followed by immediate chilling on ice for at least 2 min. After addition of 2 μl of 5× Tth reaction buffer (0.5× final concentration), 2 μl of 9 mM MnCl2 solution, 1 μl of Tth DNA polymerase, and 1 μl of RNase inhibitor (40 units), reaction mixtures were incubated on the bench top at about 25°C for 5 min, followed by incubation at 57°C for 30 min for the RT step. For first-round PCR, 80 μl of mixture containing 20 μl of 5× chelate buffer, 1.5 μl of 20 μM of 5922F, and 3 μl of 10 mM deoxynucleoside triphosphates was added. The PCR conditions were 94°C for 2 min followed by 45 cycles consisting of 94°C for 30 s, 57°C for 30 s, and 72°C for 45 s followed by a final extension of 2 min at 72°C. Three microliters of the first-round reaction product was used for the second round, consisting of 25 μl of 2× Hot Start IT Taq master mix (USB Corporation, Cleveland, OH), 1 μl each of 10 μM 5942F and 6159R oligonucleotides, and 2 μl of 25 mM MgCl2 in a total volume of 50 μl. The PCR conditions were 94°C for 2 min followed by 35 cycles consisting of 94°C for 30 s, 57°C for 30 s, and 72°C for 45 s followed by a final extension of 2 min at 72°C. The PCR products were purified using the Wizard SV PCR and gel clean-up system (Promega, WI), sequenced, and aligned with known XMRV sequences.
RESULTS
SEVI promotes XMRV infection of human prostatic and nonprostatic cell types.
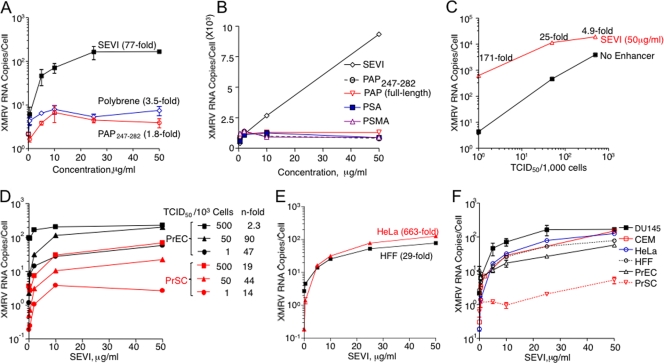
To investigate effects of SEVI on infectivity, XMRV was incubated with SEVI for 5 min prior to infecting DU145 cells at 1 TCID50 per 103 cells. XMRV RNA levels in the cells were measured after 3 days by qRT-PCR. Treatment with SEVI resulted in an impressive 77-fold enhancement of infection, as measured by XMRV env RNA copy numbers, compared to cells infected by untreated virus (Fig. 1A). In contrast, PAP247-282, which lacks the C-terminal sequence LIMY (and does not efficiently form amyloid fibrils [14]), as well as Polybrene, a cationic polymer commonly used to enhance retroviral infections, promoted XMRV infection only inefficiently (Fig. 1A). Full-length PAP and other prostate-derived proteins, including PSA and PSMA, did not affect viral infectivity (Fig. 1B). Next, we examined whether the magnitude of SEVI-mediated amplification of XMRV infection was dependent on the size of the viral inoculum. We found that enhancement of infection was greatest at the lowest dose of virus (171-fold at 1 TCID50/103 cells) and decreased to about 5-fold at 500 TCID50s/103 cells (Fig. 1C).
FIG. 1.
SEVI enhances XMRV infections of human prostatic and nonprostatic cells. Effects of SEVI on XMRV RNA yields in DU145 cells (A to C), PrEC and PrSC cells (D), HFF and HeLa M cells (E), and all of these cell types (F) are shown. For comparison, PAP247-282 and polybrene were used in panel A. Effects of SEVI, PAP247-282, full-length PAP, PSA, and PSMA on XMRV infectivity are as indicated in panel B. Infections were with 1 TCID50/103 cells of XMRV for 3 days unless otherwise indicated. XMRV env RNA levels were determined by qRT-PCR. Data are average values ± standard deviations obtained from triplicate experiments (except for panel D).
In a series of experiments we determined whether the effect of SEVI is cell type specific and also observed in relevant primary cells. We found that SEVI efficiently enhanced XMRV infection in all cell types examined (Fig. 1D, E, and F). SEVI increased viral RNA yields up to 90-fold and 44-fold, respectively, in primary prostatic epithelial (PrEC) and stromal (PrSC) cells, which are the most likely targets for XMRV replication, spread, and potential oncogenic effects in vivo (Fig. 1D). Infections of cervical carcinoma cells (HeLa M) and HFF were enhanced by SEVI by up to 663-fold and 29-fold, respectively (Fig. 1E). Comparison of the effect of SEVI at the same multiplicity of infection (1 TCID50 per 103 cells) showed that SEVI greatly enhanced XMRV infectivity on all six cell types examined in this study (Fig. 1F).
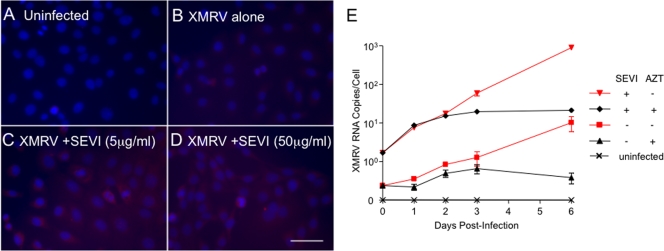
To determine whether SEVI increases the proportion of cells that become infected, indirect immunofluorescence assays (IFA) were performed. DU145 cells were infected with XMRV at 1 TCID50/103 cells in the absence or presence of SEVI. Infected cells were visualized at 3 days postinfection by IFA using a monoclonal antibody to SFFV Gag that is cross-reactive with XMRV Gag(3, 17). XMRV infections in the absence of SEVI resulted in infection of about 7.6% ± 1.8% of the cells as determined by counting brightly staining cells and all cells (Fig. 2B). In contrast, in the presence of SEVI (at either 5 or 50 μg/ml), nearly all cells (96.2% ± 1.0% and 97.6% ± 2.8%, respectively) were infected (Fig. 2C and D). To determine whether SEVI was promoting an early step in the virus replication cycle, the reverse transcription inhibitor AZT was added at 1 day postinfection. Whereas SEVI resulted in increased levels of cell-associated XMRV RNA prior to treatment with AZT, by 3 days increases in XMRV RNA levels ceased in the AZT-treated cells in either the presence or absence of SEVI (Fig. 2E). These data are consistent with enhancement of viral attachment by SEVI.
FIG. 2.
SEVI increases the fraction of cells that become infected with XMRV. (A to D) Enhancement of infectivity by SEVI was determined by IFA with a monoclonal antibody against SFFV Gag (red). DU145 cells were either uninfected or infected with 1 TCID50/103 of XMRV with 0, 5, or 50 μg/ml of SEVI for 3 days. Nuclei were counterstained with DAPI. Bar, 50 μm. (E) Effects of AZT (10 μM added at 1 day postinfection) on XMRV RNA levels in DU145 cells were determined in the absence or presence of SEVI (50 μg/ml).
SEVI potently increases the infectious titer of XMRV.
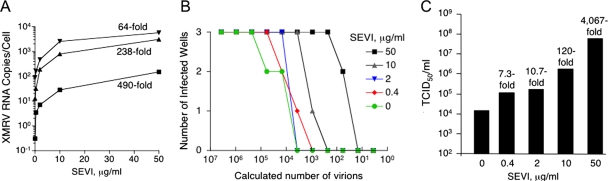
The effects of SEVI determined by standard infectivity assays may underestimate its real potency in promoting XMRV infection because some cells could be infected by multiple XMRV particles (especially at higher multiplicities of infection). Therefore, limiting-dilution assays were done with T-lymphoblastoid CEM cells, which were chosen because they grow in suspension culture, allowing dilutions of cells to be conveniently performed (14). The rationale for using suspension cells is that they can be kept in culture over 7 days until replication from single infectious units can be detected, whereas the adherent cells rapidly become too dense. In standard infectivity assays with 1 TCID50/103 cells, SEVI at 50 μg/ml enhanced XMRV RNA yields by 490-fold (Fig. 2A). At higher levels of inoculum, i.e., 50 and 500 TCID50s/103 cells, SEVI increased viral RNA yields by smaller amounts, i.e., 238- and 64-fold, respectively. In contrast, limiting-dilution analysis (see Materials and Methods) revealed a 4,067-fold enhancement in the TCID50 of XMRV with 50 μg/ml of SEVI (Fig. 3B and C). Calculation of the numbers of virions by qRT-PCR indicated that about 20 virions per culture well (seeded with 1,000 cells) were sufficient for productive XMRV infection at the highest concentration of SEVI (50 μg/ml), whereas about 80,000 virions were required to infect cells in the absence of SEVI (Fig. 3B).
FIG. 3.
SEVI lowers the threshold for XMRV infection. (A) Effects of SEVI on XMRV RNA yields in CEM cells (1 [▪], 50 [▴], or 500 [▾] TCID50s 103 cells) at 3 days postinfection. (B) CEM cells were infected in triplicate with serial fourfold dilutions of XMRV supernatant in the presence of the indicated concentrations of SEVI. Indicated on the y axis is the number of cultures that became productively infected at 21 days postinfection. (C) Effect of SEVI on XMRV yield (TCID50) calculated based on the infectivity data from panel (B).
SEVI and semen enhance infectivity at the level of virion attachment and fusion.
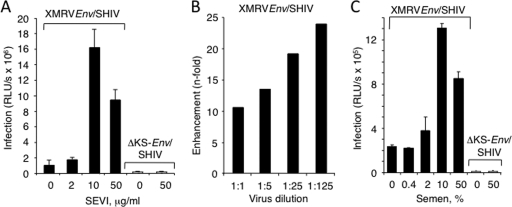
To establish that the SEVI effect was occurring at the level of viral attachment and entry, experiments were performed with infectious SHIV-derived virus particles pseudotyped with the XMRV envelope protein and expressing luciferase (XMRVenv/SHIV). To monitor nonspecific viral uptake, we used virus pseudotyped with a mutant HIV-1 env (ΔKS) that expresses a nonfunctional HIV-1 envelope protein (1, 9). The pseudotyped SHIVs are capable of only a single round of infection and thus provide an accurate and quantitative measure of infection. Virus was preincubated with SEVI for 15 min prior to dilution and added to human embryonic kidney HEK293T cells. We found that SEVI efficiently enhanced XMRVenv/SHIV infection in a dose-dependent manner, whereas HIV-1 ΔKSenv/SHIV remained noninfectious (Fig. 4A). Consistent with the results of the replication assays, the magnitude of enhancement was inversely correlated to the viral dose (Fig. 4B). These data showed that SEVI enhanced the efficiency of virus entry mediated by the XMRV envelope protein.
FIG. 4.
SEVI and semen enhance virus entry by the XMRV envelope protein. (A) Enhancement of the specific entry of an XMRV env-pseudotyped lentivirus (XMRVenv/SHIV) into HEK-293T cells by SEVI. (B) Infectivities of undiluted or serial fivefold dilutions of XMRVenv/SHIV determined with and without addition of SEVI (10 μg/ml). (C) Enhancement of XMRVenv/SHIV infectivity by semen. Virus supernatants (as indicated) were preincubated with the indicated percentage of semen for 15 min and further diluted 1:15 prior to infecting cells. Data are average values ± standard deviations obtained from triplicate experiments at 2 days postinfection.
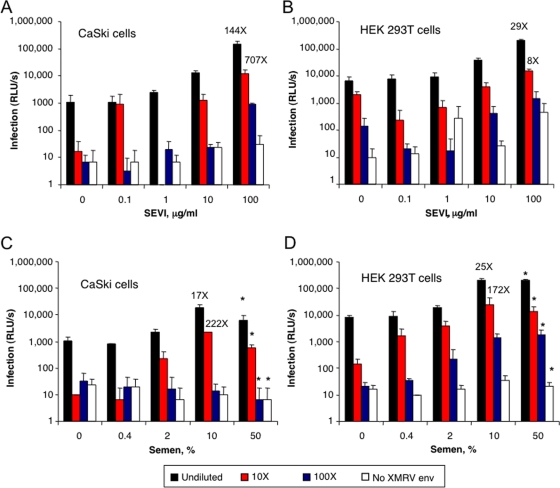
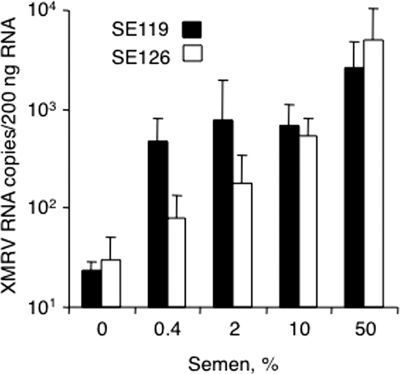
To examine the hypothesis that XMRV could be sexually transmitted, we sought to determine if semen itself could enhance its infectivity. Preincubation with 10% freshly liquefied semen enhanced XMRVenv/SHIV infection by >5-fold, whereas higher levels of semen caused a lower level of infection due to its cytotoxic effects (Fig. 4C and data not shown). Furthermore, similar experiments were performed with HIV-derived virus particles pseudotyped with the XMRV envelope protein on CaSki (epidermoid cervical carcinoma) and HEK 293T cells (Fig. 5). In these experiments, SEVI enhanced XMRVenv/HIV infectivity up to 707- and 29-fold on CaSki and 293T cells, respectively (Fig. 5A and B). In addition, there was up to 222- and 172-fold enhancement of XMRVenv/HIV infectivity by semen on CaSki and 293T cells, respectively (Fig. 5C and D). To determine whether semen also enhances infectivity of XMRV itself, semen samples from normal individuals postvasectomy (SE119 and SE126) were premixed with XMRV prior to infection of DU145 cells. Our results demonstrated that semen enhanced XMRV infection in a concentration-dependent manner up to 169-fold (Fig. 6).
FIG. 5.
SEVI and semen enhance infectivity of HIV-derived particles pseudotyped with XMRV Env. Undiluted and serial 10-fold dilutions of HIV particles encoding luciferase that were pseudotyped with XMRV Env were treated with the indicated concentrations of SEVI or semen and used to infect CaSki (A and C) or HEK 293T (B and D) cells. Nonpseudotyped particles served as the control (No XMRV Env). Infection rates were determined at 3 days postinfection by luciferase assay. Shown are average values and standard deviations derived from triplicate infections. *, slightly cytotoxic. RLU/s, relative light units per second. Numbers above bars indicate fold enhancement rates relative to untreated virus.
FIG. 6.
Effect of human semen samples (SE126 and SE119) on XMRV infectivity of DU145 cells determined by measuring XMRV env RNA levels at 3 days postinfection. Semen-XMRV mixtures were diluted 10-fold prior to infecting cells. Data are average values ± standard deviations obtained from triplicate experiments.
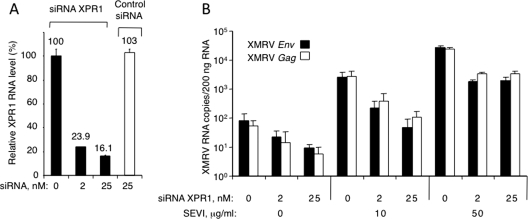
In a prior study (4), we showed that infection of hamster cells by XMRV was obtained only after expression of the putative receptor XPR1 (2, 5). Accordingly, depletion of XPR1 mRNA with siRNA in DU145 cells (Fig. 7A) resulted in a reduction in viral RNA yield of about 1 order of magnitude in both the absence and presence of SEVI (Fig. 7B). Control siRNA failed to inhibit XMRV infections in the presence of SEVI (data not shown). These results suggest that XMRV does not bypass the requirement for the cellular receptor XPR1.
FIG. 7.
SEVI does not bypass the requirement of XPR1 for infection. (A) Downregulation of XPR1 RNA by siRNA and lack of effect of control siRNA in DU145 cells at 5 days posttransfection. (B) Two days after XPR1 siRNA transfection, cells were infected with XMRV premixed with SEVI. RNA levels were measured by qRT-PCR at 3 days postinfection. Data are average values ± standard deviations obtained from triplicate experiments.
XMRV RNA isolated and sequenced from EPS.
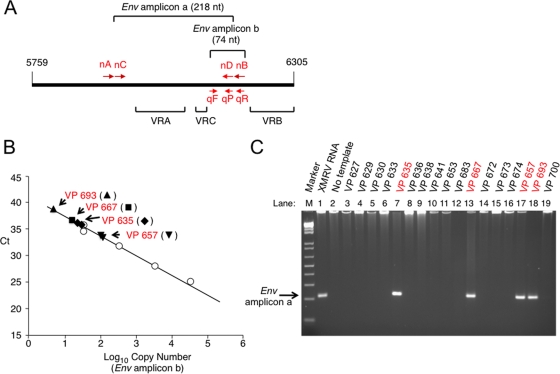
To investigate whether XMRV may interact with SEVI during naturally occurring infections in humans, we obtained EPS by manually milking prostates after radical prostatectomy of prostate cancer patients. XMRV env RNA was detected in EPS by qRT-PCR and verified by nested RT-PCR in EPS from 4 of 32 unselected prostate cancer patients (Fig. 8). The XMRV-positive patients were unremarkable with regard to age, clinical parameters, and geographical locations (Table 1). Five of these patients were homozygous QQ for RNase L, one of whom (VP667) was XMRV positive. Of the remaining 27 patients, who had at least one R variant, three were XMRV positive. To confirm that the amplified PCR products (amplicon “a” in Fig. 8A) were derived from XMRV, they were sequenced, and all were identical to previously reported strains of XMRV (VP35, VP42, and VP62) (data not shown) (17). These finding suggest that XMRV may be present in semen, where SEVI could interact with the virus and potentially promote infectivity and viral transmission.
FIG. 8.
Detection of XMRV RNA in prostatic secretions of prostate cancer patients. (A) Locations of qRT-PCR primers and probe (qF, qR, and qP for forward primer, reverse primer, and probe, respectively) and two rounds of nested PCR primers (nA and nB for first round; nC and nD for second round) in the variable regions of the XMRV gp70 env gene. qRT-PCR amplifies a region of 74 nucleotides (nt) between variable C and B regions (VRC and VRB); second-round nested RT-PCR amplifies a region of 218 nt including both the variable A region (VRA) and VRC. (B) qRT-PCR results for RNA isolated from EPS or standard control 1.95-kb env RNA (circles). Ct, cycle threshold. (C) Nested RT-PCR for env amplicon “a” of total RNA isolated from EPS. As a control, XMRV env RNA was used (lane 1). The case (VP) numbers are indicated, and the XMRV-positive cases are shown in red.
TABLE 1.
Age, clinical parameters, geographical locations, RNASEL genotypes, and presence of XMRV sequences for prostate cancer patientsa
| Patient (VP no.) | Age (yr) | Clinical stage | PSA (ng/ml) | Tumor grade | Pathological stage | Region | RNASEL | XMRV |
|---|---|---|---|---|---|---|---|---|
| 626 | 67 | T2a | 4.4 | 9 (5 + 4) | Extraprostatic extension | Northeastern OH | RQ (GA) | Negative |
| 627 | 49 | T1c | 4.1 | 6 (3 + 3) | Organ confined | Northeastern OH | RR (GG) | Negative |
| 629 | 64 | T1c | 4.1 | 7 (3 + 4) | Organ confined | Northeastern OH | RQ (GA) | Negative |
| 630 | 68 | T1c | 8.6 | 7 (3 + 4) | Organ confined | Northeastern OH | RR (GG) | Negative |
| 633 | 74 | T1c | 4.3 | 8 (4 + 4) | Extraprostatic extension | Southern FL | RQ (GA) | Negative |
| 635 | 72 | T1c | 18 | 9 (4 + 5) | Extraprostatic extension | Southern FL | RR (GG) | Positive |
| 636 | 57 | T1c | 4.4 | 7 (3 + 4) | Extraprostatic extension | Northeastern OH | RQ (GA) | Negative |
| 638 | 60 | T1c | 5.8 | 6 (3 + 3) | Organ confined | Southwestern OH | RR (GG) | Negative |
| 641 | 60 | T1c | 4.5 | 6 (3 + 3) | Extraprostatic extension | Northeastern OH | RQ (GA) | Negative |
| 647 | 60 | T1c | 2.8 | 7 (4 + 3) | Organ confined | Northeastern OH | QQ (AA) | Negative |
| 653 | 63 | T1c | 6.8 | 8 (4 + 4) | Extraprostatic extension | Northeastern OH | RR (GG) | Negative |
| 657 | 58 | T1c | 4.6 | 7 (3 + 4) | Organ confined | Northeastern OH | RR (GG) | Positive |
| 658 | 58 | NA | 0.4 | 6 (3 + 3) | Organ confined | Northeastern OH | RR (GG) | Negative |
| 663 | 65 | T1c | 6.8 | 8 (4 + 4) | Organ confined | Northeastern OH | QQ (AA) | Negative |
| 667 | 69 | T1c | 1.9 | 7 (3 + 4) | Extraprostatic extension | Southwestern OH | QQ (AA) | Positive |
| 672 | 50 | T1c | 10.6 | 7 (3 + 4) | Extraprostatic extension | Northeastern OH | RR (GG) | Negative |
| 673 | 70 | T1c | 11 | 8 (4 + 4) | Extraprostatic extension | SC | RQ (GA) | Negative |
| 674 | 52 | T1c | 4.6 | 6 (3 + 3) | Organ confined | Northeastern OH | RQ (GA) | Negative |
| 677 | 71 | T1c | 7.2 | 8 (4 + 4) | Extraprostatic extension | Southwestern OH | RR (GG) | Negative |
| 683 | 48 | T1c | 5.3 | 7 (3 + 4) | Organ confined | Northeastern OH | QQ (AA) | Negative |
| 693 | 56 | T1c | 4.2 | 6 (3 + 3) | Organ confined | Southwestern OH | RR (GG) | Positive |
| 695 | 65 | T1c | 8.7 | 7 (3 + 4) | Organ confined | Northeastern OH | RR (GG) | Negative |
| 700 | 70 | T1c | 6.5 | 7 (3 + 4) | Extraprostatic extension | Southwestern OH | RR (GG) | Negative |
| 702 | 49 | T2a | 11.5 | 9 (4 + 5) | Extraprostatic extension | Northeastern OH | RR (GG) | Negative |
| 723 | 60 | T1c | 44.3 | 9 (5 + 4) | Extraprostatic extension & nodal metastasis | SC | RQ (GA) | Negative |
| 728 | 62 | T1c | 5.6 | 6 (3 + 3) | Organ confined | Northeastern OH | RR (GG) | Negative |
| 730 | 70 | T1c | 14.3 | 8 (4 + 4) | Extraprostatic extension | Northeastern OH | RQ (GA) | Negative |
| 731 | 60 | T1c | 12.3 | 7 (4 + 3) | Organ confined | Northeastern OH | RQ (GA) | Negative |
| 739 | 65 | T1c | 5.1 | 7 (3 + 4) | Organ confined | Northeastern OH | RQ (GA) | Negative |
| 740 | 59 | T2a | 11.8 | 7 (4 + 3) | Organ confined | Northeastern OH | RQ (GA) | Negative |
| 742 | 63 | T2a | 7.1 | 6 (3 + 3) | Organ confined | Western PA | QQ (AA) | Negative |
| 744 | 61 | T1c | 4.4 | 7 (3 + 4) | Organ confined | Northeastern OH | RR (GG) | Negative |
Data for XMRV-positive patients are in boldface.
DISCUSSION
Potent enhancement of XMRV infectivity mediated by SEVI or semen.
Our data demonstrate that XMRV infection was dramatically enhanced by amyloid SEVI fibrils in human cell types of both prostatic and nonprostatic origin. Indeed, limiting-dilution assays showed that addition of SEVI to XMRV potently increased the TCID50. Furthermore, SEVI efficiently enhanced XMRV infectivity in a concentration-dependent manner and within the physiological range of amyloidogenic PAP248-286 in semen (∼35 μg/ml), a concentration exceeding that required to enhance XMRV infection (≥2 μg/ml) (14). In agreement with previous findings on HIV-1 (14), the effect of SEVI on XMRV infection was most pronounced at low viral doses, resembling the situation in vivo in the infected prostate (17). Results with SHIVs and HIV-derived particles pseudotyped with the XMRV Env clearly showed that the effect of SEVI on infectivity was at the level of viral attachment and subsequent entry into the cell. These results are consistent with previous findings showing that SEVI drastically enhanced HIV-1 binding to cells by capturing virions and promoting their physical interaction and fusion with target cells (14). The cationic nature of SEVI (pI = 10.2) is crucial for enhancing infectivity by neutralizing anionic interactions between virions and the cell (15). Semen also greatly enhanced the infectivity of XMRV, presumably due to the presence of endogenous SEVI, confirming results obtained with another retrovirus, HIV-1 (14), although other components affecting infectivity may be present in semen.
Previously we reported that XMRV infection of hamster cells required expression of human XPR1 (5). Here we have validated XPR1 as a receptor of XMRV by showing that reducing XPR1 levels with siRNA decreased infectivity. The magnitude of the SEVI effect was depressed by siRNA against XPR1, suggesting that SEVI does not bypass the requirement for this receptor. Similarly, the SEVI effect on HIV-1 infection does not bypass the requirement for its receptor and coreceptor (14).
Host restriction of XMRV replication by RNase L.
Previously, the presence of XMRV in prostatic tissues from prostate cancer cases was strongly associated with a homozygous reduced-activity variant of RNase L (QQ at residue 462) (17). In that study, 8 of 20 QQ patients (40%) were XMRV positive, compared with only 1 of 66 RQ and RR patients (1.5%). In the current smaller study in which we used EPS from prostate cancer patients, 1 of 5 QQ patients was XMRV positive (20%), compared with 3 of 27 patients with at least one R variant (11%) (Table 1). Including the cases we reported previously, we have analyzed prostate cancer tissue and/or EPS from a total of 152 men with prostate cancer. To date, 5 of 73 RR patients (7%), 1 of 35 RQ patients (3%), and 18 of 44 QQ patients (41%) were XMRV positive. Therefore, the correlation between the QQ genotype and the presence of XMRV remains high (P < 0.0001), although RR and RQ patients can in some instances also be XMRV positive at lower frequencies (7, 17). It is unknown whether such patients have other mutations in RNASEL or in the OAS genes responsible for synthesis of the activator (2′,5′-oligoadenylate) of RNase L.
Also, prior experiments using in situ hybridization and immunohistochemistry detected XMRV in prostatic stromal cells but not in epithelial cells (17). Interestingly, our current findings indicate that XMRV replicates to about 10-fold-higher levels in PrEC (epithelial) cells than in PrSC (stromal) cells. Differences in viral RNA yields between the PrEC and PrSC cells have not been associated with RNase L (both cell types express similar levels of RNase L protein as determined by immunoblotting, and both are heterozygous RQ for RNase L residue 462) (data not shown). Therefore, it is likely that host factors other than RNase L account for the higher viral yields obtained in PrEC cells relative to the PrSC cells.
The SEVI effect and presence of XMRV in EPS.
The presence of XMRV in EPS and the ability of both SEVI and human semen to lower the viral threshold required for productive infection of a variety of cell types derived from the genitourinary tract (prostate epithelial and stromal cells, HFF, and HeLa cells derived from human cervix) suggests sexual transmission of XMRV. Many epidemiologic studies suggest a higher risk of prostate cancer in men with a history of prostatitis or sexually transmitted infections, including syphilis, gonorrhea, human herpesvirus 8 infection, and Trichomonas vaginalis infection [reviewed in reference 13]. If a link between the virus and cancer can be made, detection of XMRV could provide an early indicator of prostate cancer development or disease aggressiveness in some patients. We show that detection of XMRV RNA in prostatic secretions by RT-PCR is a means for determining if men have been infected with this virus. Prostatic secretions that might be used in assays for XMRV RNA could be obtained from men, even without overt cancer, during digital rectal exam. Our findings suggest that SEVI may be critical for the spread of XMRV in the human prostate and hence possibly in prostate cancer.
Acknowledgments
We thank Beihua Dong (Cleveland, OH) for preparation of XMRV stocks, Warren Heston (Cleveland, OH) for PSMA, Philip Pellett (Detroit, MI) for HFF, and ViroPharmaceuticals for peptide synthesis and support.
These studies were supported by grant CA104943 from the NCI/NIH, grant W81XWH-07-1-338 from the U.S. Department of Defense Prostate Cancer Research Program, the V Foundation for Cancer Research, the Prostate Cancer Foundation, the Charlotte Geyer Foundation, the Maltz Family Foundation, the Mal and Lea Bank Chair, and the Wilhelm-Sander Foundation and the Deutsche Forschungsgemeinschaft.
R.H.S. and E.A.K. have the potential to receive royalties from Abbott Laboratories.
Footnotes
Published ahead of print on 29 April 2009.
REFERENCES
- 1.Bannert, N., D. Schenten, S. Craig, and J. Sodroski. 2000. The level of CD4 expression limits infection of primary rhesus monkey macrophages by a T-tropic simian immunodeficiency virus and macrophagetropic human immunodeficiency viruses. J. Virol. 7410984-10993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Battini, J. L., J. E. Rasko, and A. D. Miller. 1999. A human cell-surface receptor for xenotropic and polytropic murine leukemia viruses: possible role in G protein-coupled signal transduction. Proc. Natl. Acad. Sci. USA 961385-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chesebro, B., W. Britt, L. Evans, K. Wehrly, J. Nishio, and M. Cloyd. 1983. Characterization of monoclonal antibodies reactive with murine leukemia viruses: use in analysis of strains of friend MCF and Friend ecotropic murine leukemia virus. Virology 127134-148. [DOI] [PubMed] [Google Scholar]
- 4.De Marzo, A. M., E. A. Platz, S. Sutcliffe, J. Xu, H. Gronberg, C. G. Drake, Y. Nakai, W. B. Isaacs, and W. G. Nelson. 2007. Inflammation in prostate carcinogenesis. Nat. Rev. Cancer 7256-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong, B., S. Kim, S. Hong, J. Das Gupta, K. Malathi, E. A. Klein, D. Ganem, J. L. Derisi, S. A. Chow, and R. H. Silverman. 2007. An infectious retrovirus susceptible to an IFN antiviral pathway from human prostate tumors. Proc. Natl. Acad. Sci. USA 1041655-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fan, H. 2007. A new human retrovirus associated with prostate cancer. Proc. Natl. Acad. Sci. USA 1041449-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fischer, N., O. Hellwinkel, C. Schulz, F. K. Chun, H. Huland, M. Aepfelbacher, and T. Schlomm. 2008. Prevalence of human gammaretrovirus XMRV in sporadic prostate cancer. J. Clin. Virol. 43277-283. [DOI] [PubMed] [Google Scholar]
- 8.Goff, S. 2007. Retroviridae: the retroviruses and their replication, p. 1999-2069. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 9.Hofmann, W., D. Schubert, J. LaBonte, L. Munson, S. Gibson, J. Scammell, P. Ferrigno, and J. Sodroski. 1999. Species-specific, postentry barriers to primate immunodeficiency virus infection. J. Virol. 7310020-10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kiessling, A. A. 2005. Isolation of human immunodeficiency virus type 1 from semen and vaginal fluids. Methods Mol. Biol. 30471-86. [DOI] [PubMed] [Google Scholar]
- 11.Kim, S., N. Kim, B. Dong, D. Boren, S. A. Lee, J. Das Gupta, C. Gaughan, E. A. Klein, C. Lee, R. H. Silverman, and S. A. Chow. 2008. Integration site preference of xenotropic murine leukemia virus-related virus, a new human retrovirus associated with prostate cancer. J. Virol. 829964-9977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klein, E. A., G. Casey, and R. Silverman. 2006. Genetic susceptibility and oxidative stress in prostate cancer: integrated model with implications for prevention. Urology 681145-1151. [DOI] [PubMed] [Google Scholar]
- 13.Klein, E. A., E. A. Platz, and I. M. Thompson. 2006. Epidemiology, etiology, and prevention of prostate cancer, p. 2854-2873. In A. J. Wein, L. R. Kavoussi, A. C. Novick, A. W. Partin, and C. A. Peters, (ed.), Campbell-Walsh urology, 9th ed. Saunders, Philadelphia, PA.
- 14.Munch, J., E. Rucker, L. Standker, K. Adermann, C. Goffinet, M. Schindler, S. Wildum, R. Chinnadurai, D. Rajan, A. Specht, G. Gimenez-Gallego, P. C. Sanchez, D. M. Fowler, A. Koulov, J. W. Kelly, W. Mothes, J. C. Grivel, L. Margolis, O. T. Keppler, W. G. Forssmann, and F. Kirchhoff. 2007. Semen-derived amyloid fibrils drastically enhance HIV infection. Cell 1311059-1071. [DOI] [PubMed] [Google Scholar]
- 15.Roan, N. R., J. Munch, N. Arhel, W. Mothes, J. Neidleman, A. Kobayashi, K. Smith-McCune, F. Kirchhoff, and W. C. Greene. 2009. The cationic properties of SEVI underlie its ability to enhance human immunodeficiency virus infection. J. Virol. 8373-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ronnberg, L., P. Vihko, E. Sajanti, and R. Vihko. 1981. Clomiphene citrate administration to normogonadotropic subfertile men: blood hormone changes and activation of acid phosphatase in seminal fluid. Int. J. Androl. 4372-378. [DOI] [PubMed] [Google Scholar]
- 17.Urisman, A., R. J. Molinaro, N. Fischer, S. J. Plummer, G. Casey, E. A. Klein, K. Malathi, C. Magi-Galluzzi, R. R. Tubbs, D. Ganem, R. H. Silverman, and J. L. Derisi. 2006. Identification of a novel gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathog. 2e25. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Yeh, L. C., A. J. Lee, N. E. Lee, K. W. Lam, and J. C. Lee. 1987. Molecular cloning of cDNA for human prostatic acid phosphatase. Gene 60191-196. [DOI] [PubMed] [Google Scholar]