Abstract
The catalytic subunit of herpes simplex virus DNA polymerase (Pol), a member of the B family polymerases, possesses both polymerase and exonuclease activities. We previously demonstrated that a recombinant virus (YD12) containing a double mutation within conserved exonuclease motif III of the Pol was highly mutagenic and rapidly evolved to contain an additional leucine-to-phenylalanine mutation at residue 774 (L774F), which is located within the finger subdomain of the polymerase domain. We further demonstrated that the recombinant L774F virus replicated DNA with increased fidelity and that the L774F mutant Pol exhibited altered enzyme kinetics and impaired polymerase activity to extension from mismatched primer termini. In this study, we demonstrated that addition of the L774F mutation to the YD12 Pol did not restore the exonuclease deficiency. However, the polymerase activity of the YD12 Pol to extension from mismatched primer termini and on the nucleotide incorporation pattern was altered upon addition of the L774F mutation. The L774F mutation-containing YD12 Pol also supported the growth of viral progeny and replicated DNA more efficiently and more accurately than did the YD12 Pol. Together, these studies demonstrate that a herpes simplex virus Pol mutant with a highly mutagenic ability can rapidly acquire additional mutations, which may be selected for their survival and outgrowth. Furthermore, the studies demonstrate that the polymerase activity of HSV-1 Pol on primer extension is influenced by sequence context and that herpes simplex virus type 1 Pol may dissociate more frequently at G·C sites during the polymerization reaction. The implications of the findings are discussed.
Herpes simplex virus (HSV) DNA polymerase consists of the catalytic subunit of the polymerase (Pol) and the processivity factor UL42. The Pol subunit contains three well-defined activities: polymerization (replication), exonuclease proofreading (editing), and UL42 binding (5, 6, 28). The UL42 binding activity is mediated by amino acid residues located at the C terminus (5, 6). Although the UL42 binding residues are unique to certain alphaherpesvirus DNA polymerases, the sequences comprising the polymerase and exonuclease domains are conserved among the B family (or the α-like) polymerases (2-4). The exonuclease domain of the HSV type 1 (HSV-1) Pol contains conserved exonuclease I (Exo I), II, and III motifs, whereas the polymerase domain contains seven conserved regions (I to VII); conserved region IV overlaps with the Exo II motif. The Exo III motif is located within the δ region C, which is highly conserved among the B family polymerases (Fig. 1A). These conserved regions are located within the palm, the thumb, and the finger subdomains, which comprise the structural components of the polymerase domain. The crystal structure of the HSV-1 Pol subunit revealed three grooves that form the putative polymerase, exonuclease, and DNA binding sites. The putative exonuclease site is defined as a groove formed between the exonuclease domain and the tip of the thumb subdomain. The palm and thumb subdomains form a groove proposed to be the putative duplex DNA binding site for both the editing and the polymerization complexes (23). Thus, the polymerase and exonuclease domains of HSV-1 are structurally and functionally interconnected (1, 7, 16, 21, 23, 27, 28), although they are organized into two different domains.
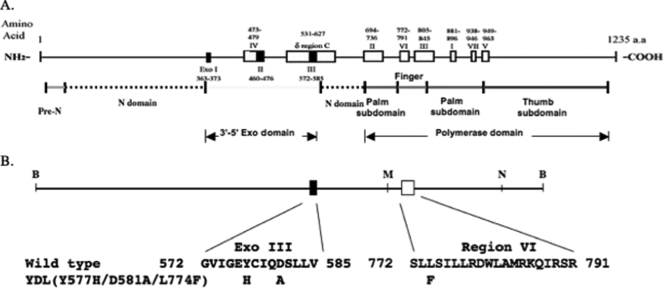
FIG. 1.
(A) Schematic diagram of the conserved regions and motifs within HSV-1 Pol. The relative locations of the conserved regions of HSV-1 Pol are shown at the top; regions I to VII and δ region C are represented by open boxes. The conserved exonuclease motifs I, II, and III are indicated with closed boxes. The functional and structural domains (determined by crystal structure analysis [23]) of the HSV-1 Pol are shown below. The N-terminal domain (N domain) is composed of two regions separated by the 3′-to-5′ exonuclease domain (23). (B) Schematic diagram of wild-type and mutant YDL Pol. The BamHI fragment of the wild-type pol from the plasmid pHC629 is shown at the top. The relative location of conserved region VI and the Exo III motif are shown below, with corresponding wild-type and mutant (YDL) amino acid sequences. B, BamHI; M, MstI; N, NotI.
The high fidelity of DNA replication is achieved by three different mechanisms: nucleotide discrimination during the polymerization reaction, editing immediately after the polymerization reaction, and postreplication repair. HSV-1 mutant Pol containing mutations within the conserved regions of the polymerase domain can result in altered enzyme kinetics and DNA replication fidelity (8, 9, 11, 12, 18, 26). Similarly, mutation of conserved Exo domain residues can lead to the loss of exonuclease activity and to altered nucleotide selection and incorporation kinetics as well as the mutator phenotype (1, 10, 13, 14, 21, 25). Our previous studies demonstrated that a mutant Pol (YD12) containing a tyrosine-to-histidine substitution at residue 577 and an aspartic acid-to-alanine substitution at residue 581 (Y577H/D581A) is exonuclease deficient (exo−) and that recombinant virus expressing the mutant Pol exhibits a mutator phenotype in vivo (14). However, this recombinant virus rapidly evolved to contain an additional leucine-to-phenylalanine substitution at residue 774 (L774F), which is located within conserved region VI of the polymerase domain (18). Interestingly, a recombinant virus containing the L774F Pol mutation exhibits increased fidelity of DNA replication (18). Our recent study also demonstrated that the mutant L774F Pol exhibits altered enzyme kinetics (26). These results led to the hypothesis that the emerged L774F mutation in the context of the YD12 Pol mutant may also affect enzyme activity, DNA replication, and fidelity.
MATERIALS AND METHODS
Cells and viruses.
The cell lines Vero and Pol A5 and the newly constructed cell lines, YD12-7 and YDL-8, were maintained in Dulbecco's modified Eagle's medium supplemented with 5% newborn calf serum (YDL refers to Pol with Y577H/D581A/L774F mutations). HSV-1 wild-type strain KOS was propagated on Vero cells as described previously (29). Recombinant virus HP66, a pol null mutant in which the pol open reading frame was replaced by the lacZ gene (24), was propagated in Pol A5 cells as described previously (14).
Plasmids.
The Y577H/D581A/L774F Pol mutant expression plasmid pSVK-YDL was constructed from the Y577H/D581A Pol mutant expression pSVK-YD12 (14) by replacing a 750-bp MstI-NotI fragment of pHC629 (18) (Fig. 1B). The plasmid pHC629 contains a 3.3-kbp BamHI fragment of the pol gene isolated from recombinant YD12 virus-infected cell DNA, which contains the engineered Y577A/D581A double substitutions plus the newly acquired L774F mutation (18). The same MstI-NotI DNA fragment was also used to replace the corresponding DNA fragment in pBlue-pol, the transfer vector for construction of the recombinant baculovirus (14), to obtain the pBlue-YDL plasmid. Plasmids pSVK-YDL and pBlue-YDL were sequenced to confirm the presence of mutations corresponding only to Y577A/D581A/L774F, using the primers previously described (14).
Construction of cell lines.
To construct cell lines, approximately 5 × 105 Vero cells were seeded onto 60-mm2 tissue culture plates overnight. Two micrograms of the plasmid pSVK-YD12 or pSVK-YDL was cotransfected with 0.1 μg pSV2-neo into Vero cells, using Lipofectamine 2000 according to the manufacturer's instruction. The transfected cells were then selected with 750 μg/ml G418 for 2 weeks. Single foci were isolated, amplified, and tested for the ability to support the growth of HP66 virus. One cell line each was randomly selected, and DNA was isolated, amplified by PCR, and sequenced to confirm the presence of the expected pol mutations.
Single-cycle growth curve.
The single-cycle growth curve assay was performed as described previously (20). Briefly, the 1 × 105 cells preseeded onto 12-well plates were infected with HP66 at a multiplicity of infection of 3. Infected cells were harvested at different time points after infection, and virus yields were determined by plaque assays, using Pol A5 cells.
Real-time PCR quantification of viral DNA.
The amount of viral DNA synthesized during the single-cycle growth curve assays was quantified by real-time PCR as described previously (20), using the 73-bp target sequences of the HSV-1 UL29 gene.
Real-time reverse transcription-PCR quantification of pol expression.
Total RNA was prepared from infected cells, using Trizol reagent (Invitrogen). The cDNA was synthesized using the iScript cDNA synthesis kit (Bio-Rad) and subjected to real-time PCR quantification as described previously (20), except that two pol primers, pol-1333 (5′-GAGATGCTGTTGGCCTTCAT-3′) and pol-1418 (5′-GGCCAGTCGAAGTTGATGAT-3′), were used to amplify an 86-bp target sequence of the pol.
Mutagenesis assay.
Recombinant HP66 virus was used to examine the mutation frequency of the lacZ gene propagated in cells expressing the wild-type or mutant Pol as described previously (17, 19). Briefly, cells were inoculated with 100 PFU of HP66 and incubated for 72 h. Progeny virus was harvested and analyzed by plaque assay on Pol A5 cells to identify clear or light-blue plaques (indicating mutated lacZ) after X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining. The lacZ mutation frequency was determined as the ratio of the number of mutant plaques to the total number of plaques examined. The chi-square test was used to determine the statistical significance of the mutation frequency differences.
Recombinant baculovirus, protein expression, and purification.
Recombinant baculoviruses expressing wild-type, L774F, and YD12 Pols have been described previously (14, 26). Recombinant baculovirus expressing the YDL mutant Pol was constructed as previously described (14), using the pBlue-YDL plasmid. Baculoviruses were used to infect Sf9 insect cells, and the recombinant wild-type or mutant Pols were purified as described previously (14) to homogeneity (data not shown).
Polymerase and exonuclease activity assays.
Polymerase activity was examined by measuring the Pol's ability to incorporate deoxynucleoside triphosphates (dNTPs) into a defined primer-template pair. The primer-template pairs were comprised of a 5′-32P-labeled 17-mer oligonucleotide annealed to an equal molar amount of a 34-mer oligonucleotide template. Each template contained a base allowing for formation of a matched or mismatched primer-template (p/t). The p/t (5 nM) was incubated with HSV-1 Pol (10 nM) in buffer A [20 mM Tris-HCl, 0.1 mM EDTA, 40 μg/ml bovine serum albumin, 4% (vol/vol) glycerol, 3 mM MgCl2, 5 mM dithiothreitol, 150 mM (NH4)2SO4] containing 100 μM each of four dNTPs. The reaction mixtures (10 μl each) were incubated at 37°C for 10 or 30 min for matched or mismatched p/t, respectively, and terminated with 4 μl of loading buffer. The reaction products were analyzed on 15% denaturing polyacrylamide gel containing 8 M urea.
The 3′-to-5′ exonuclease activity of the Pol was analyzed by measuring degradation of a labeled primer under the same conditions in the absence of dNTPs. The integrated band intensities were quantified using PhosphorImager analysis (Molecular Dynamics), and the relative activity was determined by comparison to wild-type Pol activity.
Drug sensitivity assays.
The standard gap-filling reactions using activated salmon sperm DNA as the substrates were performed as described previously (14) to examine the sensitivity of wild-type and mutant Pols to phosphonoacetic acid (PAA). The Klenow fragment (KF) of Escherichia coli Pol I was included as a control.
RESULTS
We previously constructed recombinant viruses expressing Pol with amino acid substitutions in the highly conserved Exo III motif (14). The mutant Pols were defective in exonuclease activity and exhibited a mutator phenotype, which led to the emergence of heterogeneous viral progeny (14). Subsequent plaque purification experiments demonstrated that a single plaque derived from the YD12 recombinant virus had acquired an additional L774F mutation. This virus, named YDL (Y577H/D581A/L774F) acquired the L774F mutation during or prior to the second passage and rapidly became the predominant population prior to or during the fifth passage (18). This led us to characterize the effects of the L774F mutation on the Pol's phenotype and to determine that conserved region VI of the HSV-1 Pol plays a role in the polymerization reaction and regulates DNA replication fidelity (18). Because the YD12 recombinant virus rapidly evolved to acquire the L774F Pol mutation, we also hypothesized that the additional L774F mutation was beneficial for DNA replication and virus growth in the context of the YD12 Pol mutant.
YDL Pol exhibited improved viral progeny yields and more-efficient DNA synthesis compared to those exhibited by YD12 Pol.
We first compared the abilities of YDL and YD12 Pol enzymes to support virus growth. Because recombinant viruses may acquire further pol mutations over time, we constructed cell lines expressing either YD12 or YDL Pol and used these to propagate HP66, a pol null mutant. Although this approach does not prevent the development of an additional mutation(s) in the viral genome, the Pol expressed from the cell lines is not expected to contain additional mutations. The newly constructed cell lines, YD12 and YDL, which expressed YD12 Pol and YDL Pol, respectively, and were able to support the growth of HP66 mutant virus (Fig. 2A), were used in this study. The integrated mutant pol in each cell line was amplified by PCR and sequenced to confirm the presence of the expected mutations (data not shown).
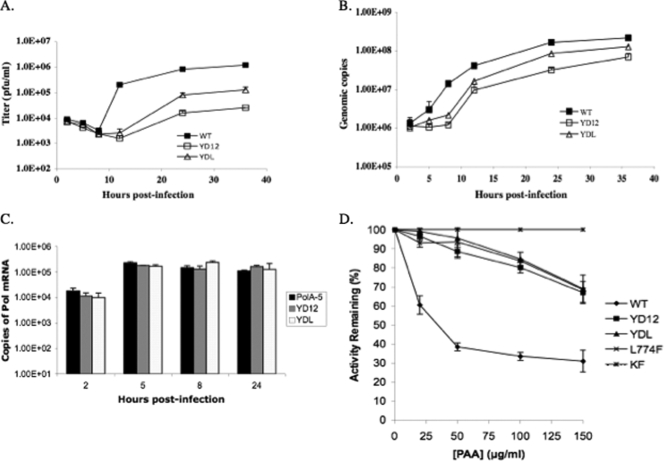
FIG. 2.
(A) Propagation of HP66 virus in Pol cell lines. A single-cycle growth assay was performed to measure the yield of HP66 propagated in the indicated cell lines. Approximately 1 × 105 cells were infected with HP66 at a multiplicity of infection of 3. The infected cells were harvested at different time points after infection, and the viral titer (PFU/ml) was measured by plaque assay on Vero cells. (B) Amounts of HP66 viral DNA synthesized in cell lines containing integrated wild-type or mutant pol. Infected cells harvested from the single-cycle growth assay were subjected to DNA isolation and purification. The relative amounts of viral DNA synthesized in the indicated cell lines were then quantified using real-time PCR amplification of a 73-bp target sequence of UL29. (C) Pol mRNA expression levels in cell lines. HP66-infected cells were harvested at 2, 5, 8, and 24 h after infection and subjected to RNA extraction and cDNA synthesis. The relative amounts of cDNA in the indicated cell lines were then quantified using real-time PCR amplification of an 86-bp target sequence of UL30. (D) PAA resistance of mutant Pols. PAA sensitivity assays were performed as described in Materials and Methods.
A single-cycle growth curve was used to examine the ability of the mutant Pol cell lines to support HP66 replication. The results from two independent experiments are shown in Fig. 2A. The Pol A5 cell line expressing wild-type Pol supported the growth of HP66 virus, which increased after 8 h and peaked at 36 h postinfection. Both the YD12 and YDL cell lines also supported the growth of HP66. However, YDL cells produced only a slightly increased viral titer between 8 and 12 h postinfection. In YD12 cells, an increase of viral titer was not detected until after 12 h postinfection. Viral progeny in both YD12 and YDL cells notably increased after 12 h and peaked at 36 h postinfection. Interestingly, YD12 cells supported HP66 replication with 50-fold lower efficiency than did Pol A5 cells, whereas YDL cells had only 10-fold lower viral replication efficiency than did Pol A5 cells. Thus, the triple-point mutant YDL Pol was fivefold more efficient in supporting viral replication than was the double-point mutant YD12 Pol.
To measure the amount of viral DNA synthesized in each cell line, aliquots of infected cells from the single-cycle growth curve assays were subjected to DNA extraction, and the amount of viral DNA was quantified using real-time PCR. Figure 2B shows the average amount of DNA synthesized in these cell lines. In Pol A5 cells, the viral DNA copy number started to increase at 2 h postinfection and peaked at 36 h postinfection. Consistent with the observed delay of virus replication in the YD12 and YDL cell lines, viral DNA synthesis in the YD12 cells become obvious only after 8 h postinfection, and only a slight increase in viral DNA was observed in the infected YDL cells between 2 and 8 h postinfection. Also, maximal viral DNA synthesis in YDL and YD12 cells was two- and threefold lower, respectively, than that in Pol A5 cells. These results demonstrated that although YD12 Pol is defective for viral growth and has a modest effect on viral DNA replication compared with the wild-type Pol, acquisition of the L774F mutation improved these parameters in comparison to those of the parent YD12 Pol.
Equal levels of expression of pol in cell lines.
Since the effects of Pol mutation on virus and DNA replication were examined on different cell lines, it is important to examine whether the cell lines may express different level of the pol, contributing to the altered phenotypes. We performed real-time reverse transcription-PCR to quantify the relative level of the pol transcripts expressed in these cells infected with HP66. At 2, 5, 8, and 24 h postinfection, these cells expressed no significant difference of the pol transcripts, although there was a 10-fold increase in pol transcripts in these cell lines after 5 h of infection, compared with those at 2 h postinfection (Fig. 2C). As a control, the pol transcripts in KOS-infected Vero cells were 103-fold higher than the pol expressed in these cells lines infected by HP66 (data not shown). Therefore, the altered phenotypes of Pol mutants are not due to the expression level of Pol in these cells.
YDL Pol replicated viral DNA with higher fidelity than did YD12 Pol.
The YD12 mutant Pol exhibited a dramatic decrease in virus growth (a 50-fold decrease), but with only a modest effect on DNA replication (a threefold decrease). On the other hand, the YDL mutant Pol exhibited a much smaller difference in impact between the virus yields and the amounts of DNA synthesized. Interestingly, the L774F Pol appeared to improve DNA replication fidelity compared with the wild-type Pol (18). These led to the hypothesis that the L774F mutation in the context of YD12 Pol may improve DNA replication fidelity to produce a smaller amount of noninfectious virus, which in turn has less impact on virus yields and on the efficiency of DNA synthesis. To test this hypothesis, we performed the lacZ mutagenesis assay to measure the mutation frequency of the lacZ gene, which was engineered in HP66 virus, propagated in each Pol-expressing cell line. The Pol A5 cells mediated replication of the lacZ gene with a mean mutation frequency of 0.060% in two independent experiments, whereas the YD12 Pol cells exhibited an ∼10-fold higher lacZ mutation frequency than did Pol A5 cells (P < 0.0001; chi-square test) (Table 1). The YDL Pol cells replicated the lacZ gene at a fivefold higher mutation frequency than did the wild-type Pol cells and, therefore, exhibited a lower mutation frequency than did the YD12 Pol cells (P = 0.0005 and P = 0.0003, in two independent experiments). Thus, the addition of the L774F mutation in the Exo III mutant YD12 Pol background improved the fidelity of DNA replication, although the fidelity was lower than that of the wild-type Pol.
TABLE 1.
lacZ mutation frequency of Pol mutantsa
| Cell lineb | No. of white/ light-blue plaques | Total no. of plaques | Mutation frequency (%)c |
|---|---|---|---|
| Pol A5 (I) | 5 | 7,685 | 0.065 |
| Pol A5 (II) | 3 | 5,535 | 0.054 |
| YD12 (I) | 104 | 1,921 | 5.41 (P < 0.0001) |
| YD12 (II) | 83 | 1,820 | 4.56 (P < 0.0001) |
| YDL (I) | 41 | 1,709 | 2.40 (P = 0.0005) |
| YDL (II) | 37 | 1,628 | 2.27 (P = 0.0003) |
The lacZ mutagenesis assay was performed by inoculation of 100 PFU into cells. Infected cells were incubated for 72 h. Progeny viruses were harvested and subjected to plaque assay, followed by X-Gal staining. The mutation frequency was determined as the ratio of the number of clear plaques plus light-blue plaques to the total number of plaques.
Two independent experiments (I and II) of each cell line were examined for the lacZ mutagenesis assay.
The mutation frequency derived from YD12 cells was compared to that derived from Pol A5 cells. The mutation frequency derived from YDL cells was compared to that derived from YD12 cells. The P values were determined by chi-square test.
YDL triple mutant Pol was exo−.
The increased fidelity of the YDL Pol in comparison to that of the YD12 Pol may arise from altered polymerase activity and/or proofreading activity. We therefore examined whether the additional L774F mutation restored the exonuclease activity of the exo− YD12 Pol. The mutant YDL Pol was expressed and purified from baculovirus-infected Sf9 cells and examined for exonuclease activity. Although the wild-type Pol exhibited 3′-to-5′ exonuclease activity, demonstrated by degradation of a labeled single-stranded oligonucleotide primer (Fig. 3A, ladder in lane 2), both the YD12 and YDL Pols were not able to degrade the 5′-labeled DNA (Fig. 3A, lanes 3 and 4). The control exonuclease-proficient (exo+) KF of Pol I exhibited exonuclease activity (Fig. 3A, lane 5), whereas exo− KF failed to create the ladder bands (Fig. 3A, lane 6), as expected. YD12 Pol, YDL Pol, and exo− KF were also unable to degrade p/t's containing either a C·G match or C·T mismatch primer terminus (Fig. 3A, lanes 8, 9, 11, 13, 14, and 16), whereas the wild-type Pol and exo+ KF were able to degrade these primers (Fig. 3A, lanes 7, 10, 12, and 15). These results demonstrated that the YDL Pol does not exhibit exonuclease activity.
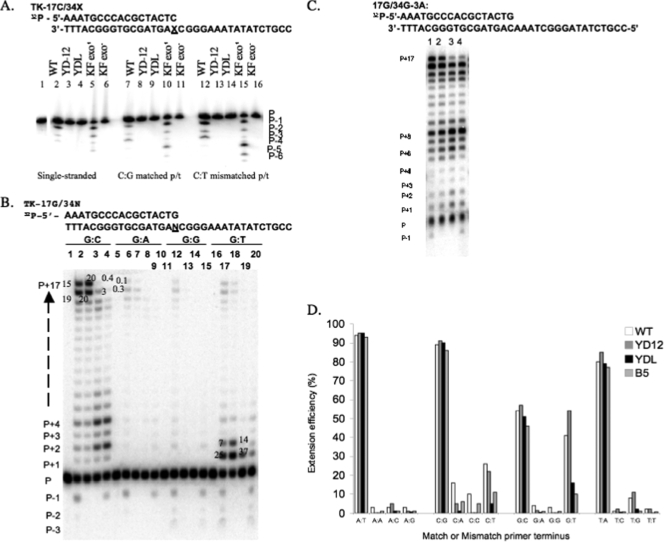
FIG. 3.
(A) Exonuclease activity of wild-type and mutant Pol enzymes. Lanes 2 to 6, a single-stranded 5′-32P-labeled 17-mer oligonucleotide (TK-17C; 5 nM) was incubated with 10 nM of the polymerase in the absence of dNTPs in a total reaction volume of 10 μl; lanes 7 to 16: the labeled 17-mer oligonucleotide was annealed to a 34-mer oligonucleotide (34X) to form C·G matched (lanes 7 to 11) and C·T mismatched (lanes 12 to 16) p/t. The underlined X residue indicates inclusion of either a G base for the matched p/t or a T base for the mismatched p/t. The p/t (5 nM) was then incubated with 10 nM of the indicated polymerase in the absence of dNTPs, and the reaction mixtures were subjected to 15% denaturing polyacrylamide gel electrophoresis analysis. The formation of a shorter ladder of bands demonstrates exonuclease activity. Lane 1, labeled 17-mer oligonucleotide alone (P); lanes 2, 7, and 12, wild-type Pol; lanes 3, 8, and 13, YD12 Pol; lanes 4, 9, and 14, YDL Pol; lanes 5, 10, and 15, exo+ KF; lanes 5, 11, and 16, exo− KF. P − 1, P − 2, etc., indicate the exonucleolytic products due to the removal of 1 base from the primer, 2 bases from the primer, etc. (B) Polymerase activity of wild-type and mutant Pol enzymes. The p/t (TK-17G/34N) was prepared by annealing a 5′-32P-labeled 17-mer oligonucleotide to a 34-mer template to form either a matched or mismatched primer terminus, as shown at the top. The underlined N residue indicates the inclusion of any of the 4 nucleotides to create either a matched or mismatched p/t; the p/t bases at these positions are indicated above the lane numbers. The p/t (5 nM) was then incubated with 10 nM of the indicated Pol enzyme in the presence of 100 μM each of four dNTPs, and reaction mixtures were subjected to 15% denaturing polyacrylamide gel electrophoresis analysis. PhosphorImager analysis was used to determined integrated band intensities. Lanes 1, 6, 11, and 16, p/t alone; lanes 2, 7, 12, and 17, wild-type Pol; lanes 3, 8, 13, and 18, YD12 Pol; lanes 4, 9, 14, and 19, YDL Pol; and lanes 5, 10, 15, and 20, L774F Pol. Relative band intensities are indicated next to specific bands (and bases) of interest. P, primer. P + 1, P + 2, etc., the products of insertion of 1 nucleotide, 2 nucleotides, etc. (C) Effect of sequence context on polymerization reactions. The sequences of p/t (TK-17G/34G-3A) used in the reactions are shown on the top. The polymerase reaction was performed as described above. The relative efficiency of primer extension was determined as described above. Relative band intensities are indicated next to specific bands (and bases) of interest. Lane 1, wild-type Pol; lane 2, YD12 Pol; lane 3, YDL Pol; lane 4, L774F Pol (B5). (D) Extension efficiency (%) of wild-type and mutant Pol enzymes. Comparison of polymerase activity results generated from p/t pairs containing all possible combinations of matched and mismatched primer termini. The p/t bases are indicated along the x axis. The extension efficiency (%) was calculated as the ratio of the sum of the extension product band intensities to unextended primer band intensity, R = ΣIP + 1/ΣIP.
Triple mutant YDL Pol exhibited no meaningful difference of polymerase activity.
The modest effects of YD12 and YDL Pols on DNA synthesis in infected cells suggest that these mutations may not significantly affect the polymerase activity. We then applied the primer extension assay to examine the ability of these Pols to incorporate dNTPs from matched primer termini (Fig. 3B). All three HSV-1 Pols were able to incorporate dNTPs efficiently for the p/t's containing A·T, T·A, and C·G matched primer termini (>80% of the primers were extended [Fig. 3D]). However, only about 50% of primers were extended by these enzymes when the p/t contained a G·C matched primer terminus (Fig. 3B and D). In these experiments, we also examined the L774F mutant Pol (B5), which had been previously demonstrated to be exo+ (26). Results demonstrated that L774F mutant Pol exhibited an extension efficiency similar to those of wild-type, YD12, and YDL Pols. Therefore, both YD12 and YDL Pols do not exhibit any meaningful difference in polymerase activity on extension from matched primer termini.
Triple mutant YDL Pol failed to polymerize the last nucleotide.
The primer extension assays revealed that the YDL Pol exhibited a nucleotide incorporation pattern distinct from those of the wild-type and YD12 Pols. For example, during the incorporation reaction from the G·C matched primer terminus, YDL Pol exhibited extension termination more frequently at positions P + 2 and P + 4 than did wild-type and YD12 Pols (Fig. 3B, compare lane 4 to lanes 2 and 3). YDL Pol was also unable to incorporate the final nucleotide into the elongating strand, whereas both wild-type and YD12 Pols incorporated the final nucleotide (Fig. 3B, compare lane 4 [0.4% incorporation of P + 17] to lanes 2 [15%] and 3 [20%]). The lack of final nucleotide incorporation was also observed for the L774F Pol (Fig. 3B, lane 5), similar to previous results (26). The inability of YDL and L774F Pols to insert the last nucleotide was also detected on p/t pairs containing the three other matched primer termini (data not shown), even though overall dNTP incorporations from matched primer termini by these Pols were similar (Fig. 3D).
Mismatch extension by YDL Pol was reduced compared to that by YD12 Pol.
The ability of the exo− YDL Pol to replicate less mutations than YD12 Pol suggested that the L774F mutation in the background of YD12 Pol may have altered polymerase activity to extend from mismatched primer termini to achieve the higher fidelity. This hypothesis is based on previous studies demonstrating that the HSV-1 Pol exhibits poor extension from mismatched primer termini even though it possesses exonuclease proofreading activity (1, 25, 26) and that the L774F mutant Pol exhibits poor extension from mismatched primer termini compared with that exhibited by the wild-type Pol (1, 25, 26). In this study, we examined whether the L774F mutation in the YD12 Pol background altered extension efficiency from any of the 12 mismatched primer termini. Similar to earlier studies (1, 25, 26) the wild-type Pol and the exo− YD12 Pol exhibited dramatically reduced extension from mismatched primer termini in a mismatch-dependent manner (Fig. 3D). The wild-type and YD12 Pols exhibited extension from a G·T mismatched primer terminus with an efficiency similar to that from a G·C matched primer terminus. Interestingly, both the L774F and YDL Pols had significantly reduced G·T mismatch extension efficiencies compared to the wild-type or YD12 Pols. Furthermore, extension of the G·T mismatched p/t by L774F and YDL Pol was limited to the first few nucleotides, whereas the wild-type and YD12 Pols exhibited final nucleotide incorporation, although considerable termination sites at P + 1 and P + 2 were observed (Fig. 3D, compare lanes 19 and 20 with lanes 17 and 18). Similar trends were also observed for other mismatched p/t extension reactions; both L774F and YDL Pols had limited extension efficiencies, extending only a few bases, whereas the wild-type and YD12 Pols displayed final nucleotide incorporation, albeit with reduced efficiency compared to extension from matched primer termini. Figure 3D summarizes the relative mismatched extension activities of these four Pols and demonstrates that the L774F mutation further impairs the polymerase activity of YD12 Pol in mismatch extension reactions.
Recombinant Pols with L774F mutation are resistant to PAA.
The recombinant YD12 Pol exhibited resistance to PAA (20). We thought to examine whether the L774F mutant Pol and the L774F mutation in the context of YD12 Pol also exhibit altered drug sensitivity. Using activated salmon sperm DNA as substrates in standard gap-filling reactions, we demonstrated that while the polymerase activity of wild-type Pol was inhibited by PAA, with an effective dose inhibiting 50% of activity at ∼35 μg/ml, all three Pol mutants (YD12, YDL, and L774F Pol) were resistant to PAA, with an effective dose inhibiting 50% of activity at >150 μg/ml. The KF of E. coli Pol I also was included in these experiments to demonstrate its resistance to PAA. Therefore, mutant Pols containing the L774F mutation were resistant to PAA (Fig. 2D), suggesting that the L774F mutation in the context of the wild-type or YD12 mutant Pol may alter the enzyme conformation.
Sequence context effect on Pol's activity.
The primer extension assays further revealed interesting nucleotide incorporation patterns by these Pols; all Pols, including the wild-type Pol, exhibited several bands with stronger intensity that mimic the locations at which the polymerase may dissociate from the p/t. For example, during the extension from the p/t containing a G·C matched primer terminus, the Pol terminated most frequently at the penultimate position of the template, and it also had more frequent pauses at the positions surrounding the G and C template bases (i.e., P + 2, P + 4) than at the other positions (Fig. 3B). Interestingly, the KF Pol I did not polymerize the reaction with a similar pattern of pauses (data not shown). Furthermore, the unique pattern of pauses mediated by HSV-1 Pol was observed in the reactions using p/t containing all four matched primer termini. These observations suggest that HSV-1 Pol may dissociate from the p/t much more easily when it encounters the incorporations of dGMP or dCMP than when it encounters those of dAMP or dTMP. To test this hypothesis, we performed primer extension assays using a different template strand 34G-3A, which contains the same nucleotide contents of the 34G template, but the CGGGAAAT bases immediately after the base complementary to the primer terminus were replaced with the AAATCGGG sequence. As expected, these Pols polymerized the reactions with a different termination pattern; the stronger pauses were found at positions surrounding to the G·C and C·G base pairs, which were located at positions P + 6, P + 7, and P + 8 (Fig. 3C), as well as those near the end of the p/t. Furthermore, these Pols exhibited higher efficiencies (∼85%, compared with 50% of the activity with the p/t of the 34G template) of nucleotide incorporations from this p/t. Although both p/t's contain the same primer terminus, it appears that the extension initiating from the incorporation of dTMP is more efficient than that starting with the insertion of dGMP. This suggests that the incorporation of dGMP is the limiting factor of the polymerization reaction, when the reaction is initiated from the primer terminus of dGMP:C. However, we cannot rule out the possibility that the Pol may stall at the positions encountering the G·C and/or C·G base pair. Further studies will be necessary to address whether the Pol fails to extend or dissociates from p/t during the incorporation of dGMP and/or dCMP.
DISCUSSION
Implications of this study on the structure and activity of HSV-1 Pol.
Although the L774 residue of HSV-1 Pol does not directly interact with incoming dNTPs based on the structural study (7), the L774F mutation may indirectly affect the conformation change of the polymerase active site upon its interaction with dNTPs and/or during the catalytic step. The data demonstrating that the L774F mutation impairs the activity of both wild-type and YD12 Pols to extend from mismatched primer termini in a mismatch-dependent manner (Fig. 3C) and that these mutant Pols are less sensitive to PAA (Fig. 2D) support this notion. Perhaps, the effect of the L774F mutation on the conformation of the polymerase active site is more restricted to binding and/or catalysis of certain bases from mismatched primer termini. Examination of presteady kinetics and structural studies of these Pols will be necessary to address these questions.
The observed asymmetry in the incorporation ability of G·C and C·G and the extension ability from G·C and C·G primer termini mediated by these Pols (Fig. 3D) may suggest that HSV-1 Pol has difficulty in accommodating guanine in the polymerase active site. However, the observed asymmetry in this study may also be affected by the composition of duplex DNA present in the polymerase active site; it is possible that sequence context of p/t may affect the efficiencies of nucleotide incorporations and extensions. Indeed, evidence that HSV-1 Pol exhibited different extension efficiency levels on two p/t's containing different sequence contexts (Fig. 2, compare panels C and B) and that HSV-1 Pol does not exhibit asymmetry in the incorporation ability of G and C on a different p/t used in a different study (25) supports this notion. Furthermore, the observations that HSV-1 Pol exhibited stronger terminations surrounding the G and C bases on different p/t's also suggest that HSV-1 Pol may dissociate from G·C sites more frequently (Fig. 3B and C). Along this line, it is well documented that sequence context can affect the fidelity of DNA replication. Given that the HSV genome is G·C rich, it can be expected that HSV-1 Pol will frequently dissociate from p/t's during the polymerization reactions. It will be interesting to explore whether this property will lead to more mutations and/or produce more substrates for subsequent recombination/repair events. Therefore, the in vitro studies to examine the mutation spectra will be helpful to delineate the underlined mechanisms. It is also noteworthy that this study examines only the Pol catalytic subunit in the absence of the processivity factor. Further in vitro studies are necessary to define whether the processivity factor has any effect on the pausing pattern and/or the extension efficiency. Furthermore, recombinant viruses may be constructed to harbor the appropriate mutagenesis target gene for future studies to examine the mutation spectra mediated by these Pols. These future studies may correlate the mutagenic outcomes to the impairment of Pol's activities and reveal possible mechanisms affecting DNA replication fidelity.
Evolution, selection, and survival of the HSV-1 mutant.
A DNA polymerase with intrinsic exonucleolytic proofreading can have 10- to 100-fold higher DNA replication fidelity than a polymerase that lacks proofreading activity (22). The HSV-1 YD12 exo− Pol is highly mutagenic (14, 15). Thus, the emergence of additional mutations, including those in pol, forming heterogeneous populations of progeny viruses is not surprising (13; our unpublished data). More importantly, the rapid emergence of the L774F mutation during the second viral passage and its outgrowing the parental YD12 mutant prior to the fifth passage (18) suggest that the L774F mutation in the background of the highly mutagenic YD12 Pol is beneficial for viral DNA replication and, hence, the growth of the virus. Indeed, addition of the L774F mutation to the exo− YD12 Pol allowed the virus to replicate more viral DNA and, hence, progeny virus sooner after infection, continuing through peak infection, albeit with less efficiency than the wild-type Pol (Fig. 2). It is reasonable to assume that the improved DNA replication and virus growth of YDL Pol are attributable to the L774F mutation, since the L774F Pol also replicates DNA with increased fidelity compared to the wild-type Pol (18). This, however, raises an interesting question of why HSV-1 does not evolve to contain L774F Pol. It is possible that the impairment of polymerase activity of L774F Pol, including the reduced ability to extend the last nucleotide incorporation (18) (Fig. 3B), may be disadvantageous to DNA replication, such as the synthesis of more breaks and damages in DNA, and to virus growth and evolution as well. It will be interesting to examine whether a recombinant virus expressing the L774F mutant Pol replicates with an altered amount and quality of DNA, compared to those expressed by the wild-type virus. Nevertheless, the study demonstrated that the rapid emergence of genetic diversity of HSV-1 due to the exonuclease deficiency of mutant Pol can be an ideal model for examining how mutant viruses evolve and respond to selective pressure. Furthermore, the mutant YD12 virus and another exo− mutant Y7 virus (14) can be used to isolate a variety of mutant viruses for further characterization of the gene of interest.
Acknowledgments
This work is supported by grant AI56359 from National Institutes of Health (to C.B.C.H.).
Footnotes
Published ahead of print on 6 May 2009.
REFERENCES
- 1.Baker, R. O., and J. D. Hall. 1998. Impaired mismatch extension by a herpes simplex DNA polymerase mutant with an editing nuclease defect. J. Biol. Chem. 27324075-47082. [DOI] [PubMed] [Google Scholar]
- 2.Blanco, L., and M. Salas. 1996. Relating structure to function in phi29 DNA polymerase. J. Biol. Chem. 2718509-8512. [DOI] [PubMed] [Google Scholar]
- 3.Burgers, P. M., E. V. Koonin, E. Bruford, L. Blanco, K. C. Burtis, M. F. Christman, W. C. Copeland, E. C. Friedberg, F. Hanaoka, D. C. Hinkle, C. W. Lawrence, M. Nakanishi, H. Ohmori, L. Prakash, S. Prakash, C. A. Reynaud, A. Sugino, T. Todo, Z. Wang, J. C. Weill, and R. Woodgate. 2001. Eukaryotic DNA polymerases: proposal for a revised nomenclature. J. Biol. Chem. 27643487-43490. [DOI] [PubMed] [Google Scholar]
- 4.Coen, D. M. 1991. The implications of resistance to antiviral agents for herpesvirus drug targets and drug therapy. Antivir. Res. 15287-300. [DOI] [PubMed] [Google Scholar]
- 5.Digard, P., W. R. Bebrin, K. Weisshart, and D. M. Coen. 1993. The extreme C terminus of herpes simplex virus DNA polymerase is crucial for functional interaction with processivity factor UL42 and for viral replication. J. Virol. 67398-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Digard, P., and D. M. Coen. 1990. A novel functional domain of an alpha-like DNA polymerase. The binding site on the herpes simplex virus polymerase for the viral UL42 protein. J. Biol. Chem. 26517393-17396. [PubMed] [Google Scholar]
- 7.Gibbs, J. S., K. Weisshart, P. Digard, A. deBruynKops, D. M. Knipe, and D. M. Coen. 1991. Polymerization activity of an α-like DNA polymerase requires a conserved 3′-5′ exonuclease active site. Mol. Cell. Biol. 114786-4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hall, J. D., D. M. Coen, B. L. Fisher, M. Weisslitz, S. Randall, R. E. Almy, P. T. Gelep, and P. A. Schaffer. 1984. Generation of genetic diversity in herpes simplex virus: an antimutator phenotype maps to the DNA polymerase locus. Virology 13226-37. [DOI] [PubMed] [Google Scholar]
- 9.Hall, J. D., P. A. Furman, M. H. St. Clair, and C. W. Knopf. 1985. Reduced in vivo mutagenesis by mutant herpes simplex DNA polymerase involves improved nucleotide selection. Proc. Natl. Acad. Sci. USA 823889-3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hall, J. D., K. L. Orth, K. L. Sander, B. M. Swihart, and R. A. Senese. 1995. Mutations within conserved motifs in the 3′-5′ exonuclease domain of herpes simplex virus DNA polymerase. J. Gen. Virol. 762999-3008. [DOI] [PubMed] [Google Scholar]
- 11.Huang, L., K. K. Ishii, H. Zuccola, A. M. Gehring, C. B. C. Hwang, J. Hogle, and D. M. Coen. 1999. The enzymological basis for resistance of herpesvirus DNA polymerase mutants to acyclovir: relationship to the structure of alpha-like DNA polymerases. Proc. Natl. Acad. Sci. USA 96447-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hwang, C. B., and H. J. Chen. 1995. An altered spectrum of herpes simplex virus mutations mediated by an antimutator DNA polymerase. Gene 152191-193. [DOI] [PubMed] [Google Scholar]
- 13.Hwang, Y. T., and C. B. C. Hwang. 2003. Exonuclease-deficient polymerase mutant of herpes simplex virus type 1 induces altered spectra of mutations. J. Virol. 772946-2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hwang, Y. T., B. Y. Liu, D. M. Coen, and C. B. Hwang. 1997. Effects of mutations in the Exo III motif of the herpes simplex virus DNA polymerase gene on enzyme activities, viral replication, and replication fidelity. J. Virol. 717791-7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hwang, Y. T., B. Y. Liu, C. Y. Hong, E. J. Shillitoe, and C. B. Hwang. 1999. Effects of exonuclease activity and nucleotide selectivity of the herpes simplex virus DNA polymerase on the fidelity of DNA replication in vivo. J. Virol. 735326-5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hwang, Y. T., J. F. Smith, L. Gao, and C. B. Hwang. 1998. Mutations in the Exo III motif of the herpes simplex virus DNA polymerase gene can confer altered drug sensitivities. Virology 246298-305. [DOI] [PubMed] [Google Scholar]
- 17.Hwang, Y. T., Y. A. Wang, Q. Lu, and C. B. Hwang. 2003. Thymidine kinase of herpes simplex virus type 1 strain KOS lacks mutator activity. Virology 305388-396. [DOI] [PubMed] [Google Scholar]
- 18.Hwang, Y. T., H. J. Zuccola, Q. Lu, and C. B. Hwang. 2004. A point mutation within conserved region VI of herpes simplex virus type 1 DNA polymerase confers altered drug sensitivity and enhances replication fidelity. J. Virol. 78650-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang, C., Y. T. Hwang, and C. B.-C. Hwang. 2006. Herpes simplex virus type 1 recombinants without the oriL sequence replicate DNA with increased fidelity. Virology 347277-285. [DOI] [PubMed] [Google Scholar]
- 20.Jiang, C., Y. T. Hwang, J. C. W. Randell, D. M. Coen, and C. B. C. Hwang. 2007. Mutations that decrease DNA binding of the processivity factor of the herpes simplex DNA polymerase reduce viral yield, alter the kinetics of viral DNA replication, and decrease fidelity of DNA replication. J. Virol. 813495-3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhn, F. J. P., and C. W. Knopf. 1996. Herpes simplex virus type 1 DNA polymerase. Mutational analysis of the 3′-5′ exonuclease domain. J. Biol. Chem. 27129245-29254. [DOI] [PubMed] [Google Scholar]
- 22.Kunkel, T. A. 1988. Exonucleolytic proofreading. Cell 53837-840. [DOI] [PubMed] [Google Scholar]
- 23.Liu, S., J. D. Knafels, J. S. Chang, G. A. Waszak, E. T. Baldwin, M. R. Deibel, Jr., D. R. Thomsen, F. L. Homa, P. A. Wells, M. C. Tory, R. A. Poorman, H. Gao, X. Qiu, and A. P. Seddon. 2006. Crystal structure of the herpes simplex virus 1 DNA polymerase. J. Biol. Chem. 28118193-18200. [DOI] [PubMed] [Google Scholar]
- 24.Marcy, A. I., D. R. Yager, and D. M. Coen. 1990. Isolation and characterization of herpes simplex virus mutants containing engineered mutations at the DNA polymerase locus. J. Virol. 642208-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song, L., M. Chaudhuri, C. W. Knopf, and D. S. Parris. 2004. Contribution of the 3′- to 5′-exonuclease activity of herpes simplex virus type 1 DNA polymerase to the fidelity of DNA synthesis. J. Biol. Chem. 27918535-18543. [DOI] [PubMed] [Google Scholar]
- 26.Tian, W., Y. T. Hwang, and C. B. C. Hwang. 2008. The enhanced DNA replication fidelity of a mutant herpes simplex virus type 1 DNA polymerase is mediated by an improved nucleotide selectivity and reduced ability of mismatch extension ability. J. Virol. 828937-8941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang, Y. S., S. Woodward, and J. D. Hall. 1992. Use of suppressor analysis to identify DNA polymerase mutations in herpes simplex virus which affect deoxynucleoside triphosphate substrate specificity. J. Virol. 661814-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weisshart, K., A. A. Kuo, C. B. Hwang, K. Kumura, and D. M. Coen. 1994. Structural and functional organization of herpes simplex virus DNA polymerase investigated by limited proteolysis. J. Biol. Chem. 26922788-22796. [PubMed] [Google Scholar]