Abstract
The outer capsid of the nonenveloped mammalian reovirus contains 200 trimers of the μ1 protein, each complexed with three copies of the protector protein σ3. Conformational changes in μ1 following the proteolytic removal of σ3 lead to release of the myristoylated N-terminal cleavage fragment μ1N and ultimately to membrane penetration. The μ1N fragment forms pores in red blood cell (RBC) membranes. In this report, we describe the interaction of recombinant μ1 trimers and synthetic μ1N peptides with both RBCs and liposomes. The μ1 trimer mediates hemolysis and liposome disruption under conditions that promote the μ1 conformational change, and mutations that inhibit μ1 conformational change in the context of intact virus particles also prevent liposome disruption by particle-free μ1 trimer. Autolytic cleavage to form μ1N is required for hemolysis but not for liposome disruption. Pretreatment of RBCs with proteases rescues hemolysis activity, suggesting that μ1N cleavage is not required when steric barriers are removed. Synthetic myristoylated μ1N peptide forms size-selective pores in liposomes, as measured by fluorescence dequenching of labeled dextrans of different sizes. Addition of a C-terminal solubility tag to the peptide does not affect activity, but sequence substitution V13N or L36D reduces liposome disruption. These substitutions are in regions of alternating hydrophobic residues. Their locations, the presence of an N-terminal myristoyl group, and the full activity of a C-terminally extended peptide, along with circular dichroism data that indicate prevalence of β-strand secondary structure, suggest a model in which μ1N β-hairpins assemble in the membrane to form a β-barrel pore.
Infection of a cell by a nonenveloped virus requires perforation or disruption of a cellular membrane so that the viral genome can access cytoplasmic factors. Probably the most striking process of this kind is carried out by the injection machinery of some bacteriophages, as visualized by electron microscopy more than 40 years ago (49). Among the nonenveloped animal viruses, members of several families (picornaviruses, polyomaviruses, and reoviruses) have an N-terminally myristoylated capsid protein that is believed to participate in the membrane-perturbing mechanism (15, 41, 45).
The myristoylated component of mammalian orthoreoviruses is a 76-kDa protein known as μ1 (41) (Fig. 1). It is one of eight proteins that make up the large, icosahedral capsid (20). Together with its “chaperone,” σ3, the μ1 protein associates into a threefold-symmetrical heterohexamer, μ13σ33 (31, 60), which forms a fenestrated T=13 icosahedral lattice surrounding a 70-nm core particle (20, 44). During infection, the reovirus core enters the cytoplasm and directs transcription of viral mRNA from each of the 10 double-stranded RNA genomic segments (11, 46). The core remains intact, and mRNA synthesis and 5′ capping are primarily accomplished by core proteins λ3 and λ2, respectively (18, 34, 44, 52). Translocation of the reovirus core from an extracellular milieu to the cytoplasm probably occurs in an endosome or related structure, and μ1 appears to be a principal agent of this process (1, 3, 10, 11, 19, 27, 29, 31, 33, 36, 37, 38, 42, 56).
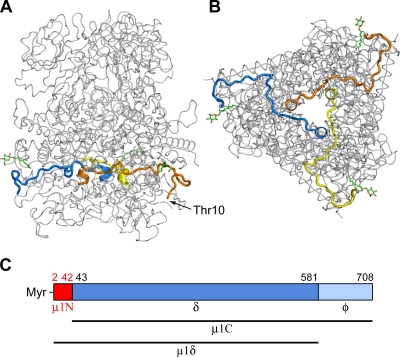
FIG. 1.
(A, B) Location of μ1N in the μ1 trimer. The three subunits of μ1 in the μ13σ33 crystal structure (31) are shown from the side (A) and bottom (B) views. The three copies of μ1N are shown as yellow, orange, and blue ribbons, and Thr10, the first visible N-terminal residue, is indicated. β-Octyl glucoside from the crystallization buffer, which occupies a hydrophobic pocket proposed ordinarily to sequester the myristoyl group (31, 60), is shown in green. (C) Diagram of the μ1 cleavage products.
To activate μ1 for productive infection, σ3 must be removed by proteolysis (4, 5, 14, 17, 21, 38, 51). The resulting infectious subviral particle (ISVP) is relatively stable but can be triggered in vitro to undergo further rearrangement to a distinct particle known as the ISVP* (10). The ISVP→ISVP* transition involves a substantial conformational change in μ1 (to a species designated μ1*), including at least partial dissociation of the subunits of the trimer, greatly increased protease sensitivity, enhanced hydrophobicity, and reorganization of the μ1 N-terminal domains (10, 58). Elution of the attachment protein σ1 and activation of particle-associated transcriptase activity also accompany this transition (10).
Factors that promote ISVP→ISVP* conversion in vitro include increased temperature, larger monovalent cations such as K+ and Cs+, and higher particle concentration (2, 6, 7, 8, 10, 36, 37). The promoting factor in the cell is unknown, but low pH is clearly not required (23, 24, 51).
Two specific cleavages of μ1 take place during entry (Fig. 1). The proteolytic removal of σ3 that leads to the ISVP also generates a C-terminal fragment of μ1, known as φ (13 kDa) (39). This fragment remains particle associated until ISVP→ISVP* conversion (29, 39). Membrane penetration does not appear to require the production of φ, as cleavage-resistant variants of μ1 are still infectious (9, 12). An autolytic N-terminal cleavage in μ1 also occurs at some point in the virion→ISVP* transition, creating fragment μ1N (residues 2 to 42 plus the N-terminal myristoyl moiety); its complement is known as μ1C, and the large central fragment between the autolytic cleavage and the φ cleavage is known as δ (40, 41). The cleaved μ1N peptide is released from the particle during the ISVP→ISVP* transition (3, 29, 42).
ISVP→ISVP* conversion in vitro imparts hemolytic activity (10). Released μ1N binds the membranes of coincubated RBCs and is sufficient to induce the formation of small pores, 4 to 9 nm in diameter (3, 29). The μ1 mutation N42A, in which Asn42, the likely nucleophile in the autocleavage reaction, is changed to Ala, prevents autocleavage but not μ1 structural rearrangement (42, 58). Cores recoated with μ1 bearing the N42A mutation [μ1(N42A)] are defective, both for infectivity and for hemolysis (42).
These observations suggest that the release of μ1N from ISVP* is necessary for penetration and that μ1N participates directly in a process of membrane perforation, allowing the reovirus core to enter the cytoplasm. In earlier work, we found that we could induce a conformational change in particle-free μ1, using conditions similar to those that promote the ISVP→ISVP* transition (58). We now describe membrane disruption, both of RBCs and of liposomes, by μ1* produced in this way. Autolytic cleavage and release of μ1N from the rest of μ1* are not necessary for pore formation in liposomes, but synthetic, myristoylated μ1N peptide (myr-μ1N) is sufficient. Conserved characteristics of the μ1N amino acid sequence suggest possible structural mechanisms for this process.
MATERIALS AND METHODS
Expression and purification of μ13σ33 heterohexamers.
Wild-type (WT) reovirus type 1 Lang (T1L) σ3 and either WT or mutant T1L μ1 were coexpressed in insect cells from recombinant baculovirus and purified as described previously (10, 31, 58). Baculoviruses containing μ1 mutations N42A (42), A319E (11), and K459E (36) were made as described.
Viruses and recoated cores.
Spinner-adapted mouse L929 mouse fibroblasts (L cells) and Spodoptera frugiperda clone 21 and Trichoplusia ni TN-BTI-564 (High Five) insect cells (Invitrogen) were grown as described previously (1). Reovirus T1L was grown in spinner cultures of mouse L929 cells, purified according to the standard protocol (22), and stored at 4°C in virion buffer (VB) (150 mM NaCl, 20 mM MgCl2, 10 mM Tris [pH 7.5]). Recoated cores (RCs) were made without σ1 as described previously (1), using baculovirus-expressed WT T1L σ3 and either WT T1L μ1 (12) or T1L μ1(N42A) (42). Virions and recoated cores were dialyzed extensively against VB, and particle concentrations were determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Nonpurified ISVPs (or pRCs) were made by the digestion of virions (or RCs) at a concentration of 1 × 1013 particles/ml with 200 μg/ml chymotrypsin for 10 min at 32°C, followed by incubation on ice with 1 mM phenylmethylsulfonyl fluoride (PMSF) for at least 10 min.
Synthetic peptides.
The myr-μ1N peptide, consisting of amino acids 2 to 42 of T1L μ1 and an N-terminal N-myristoyl group (myr-GNASSIVQTINVTGDGNVFKPSAETSSTAVPS LSLSPGMLN), was synthesized by standard solid-phase 9-fluorenylmethoxy carbonyl chemistry using N,N′-dicyclohexylcarbodiimide/1-hydroxybenzotriazole activation and 2.5-h acylation times. The same peptide was also prepared with RGKGR (myr-μ1N-RGKGR) added at the C terminus to enhance solubility. Myristoylation was accomplished in both cases with the anhydride in pyrimidine-4-(dimethylamino)pyridine, and the resulting peptide was cleaved and deprotected with reagent K and purified by reversed-phase high-performance liquid chromatography. The purity and primary structure were assessed by electroscopy ionization-Fourier transform ion cyclotron resonance mass spectrometry (9.4T) mass spectrometry. Peptides containing the substitutions V13N or L36D, and the C-terminal RGKGR, were also synthesized. Peptides were dissolved in dimethyl sulfoxide (DMSO), and the peptide concentration was determined in VB with 10% DMSO and 5% Triton X-100 using the BCA kit (Thermo Scientific). For experiments comparing the WT and mutant peptides, stocks in DMSO were first centrifuged for 5 min at 16,000 × g to remove aggregates, the supernatant was transferred to a fresh tube, and the peptide concentration was determined as described above. After performing liposome disruption experiments, stocks were centrifuged again, and the peptide concentration in the supernatant was within 12% of the original determination, making it unlikely that the loss of function of the mutants was due to aggregation in the stock solution.
Liposome preparation.
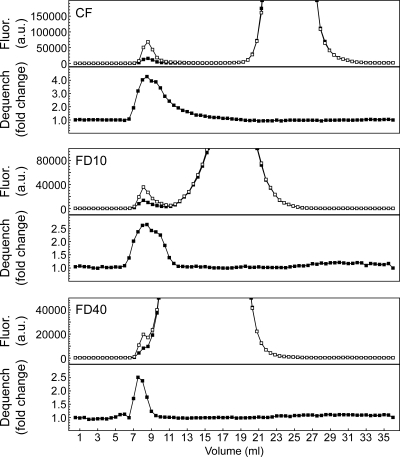
A carboxyfluorescein (CF) solution (22 mg/ml) was prepared by mixing CF with VB and adding drops of sodium hydroxide solution until the mixture became clear. The pH of the resulting solution was ∼8.0. Phosphatidylcholine (Avanti Lipids) (2 mg) dissolved in chloroform was dried under a nitrogen stream and then under vacuum for at least 2 h. The lipid film was resuspended in 200 μl VB containing CF or 100 mg/ml fluorescein isothiocyanate-conjugated dextran (FD) with molecular weights of 10,000 (FD10) or 40,000 (FD40) (Sigma). Suspensions were subjected to three freeze-thaw cycles with brief vortexing after each thaw and extruded with 35 passes through a 0.2-μm filter using a mini-extruder device (Avanti Lipids). To separate liposomes from unincorporated dye or fluorescein-conjugated dextran, the preparation was loaded onto a 24-ml Superdex S200 column (GE Healthcare) preequilibrated with VB and eluted with VB at a flow rate of 0.5 ml/min in the cold. Fractions of 0.5 ml each were collected. A 10-μl aliquot of each fraction was diluted with 90 μl VB in a 96-well plate, and fluorescence was measured using a Typhoon scanner (GE Healthcare) as described below. The identification of the peak at the void volume of the size exclusion column as liposomes containing encapsulated dye was confirmed by measuring fluorescence dequenching upon addition of 1 μl 10% Triton X-100 to each well (see Fig. 4). The two or three fractions from this peak with the largest increase in fluorescence upon addition of Triton X-100 were pooled, and stored at 4°C, and used within 24 h.
FIG. 4.
Liposomes loaded with CF, FD10, or FD40 were separated from unincorporated dye by gel filtration. Top panels, fluorescence (Fluor.) of each column fraction before (filled squares) and after (unfilled squares) addition of Triton X-100. Bottom panels, increase in fluorescence upon addition of detergent (Dequench). a.u., arbitrary units.
For some experiments with CF-loaded liposomes only, liposomes were separated from unincorporated dye using a 4-ml Sephadex G25 PD10 column (GE Healthcare) preequilibrated and eluted with VB. The orange liposome band eluting ahead of the excess free dye was collected in ∼1 ml. Liposomes for experiments with μ13σ33 heterohexamers were prepared in the same way, except that 2.5 mg phosphatidylcholine was used, the lipid suspension was subjected to five freeze-thaw cycles, and the suspension was extruded with 21 passes through a 0.1-μm filter. For some experiments with μ13σ33, liposomes were made from a mixture of phosphatidylcholine, phosphatidylethanolamine, sphingomyelin, and cholesterol (molar ratio, 1:1:1:1).
Hemolysis assays.
Citrated bovine calf RBCs or chicken RBCs in Alsever's solution (Colorado Serum) were washed immediately before use in cold phosphate-buffered saline (PBS) (137 mM NaCl, 8 mM Na2HPO4, 1.5 mM KH2PO4, 2.7 mM KCl [pH 7.5]) supplemented with 2 mM MgCl2 (PBS-Mg) and incubated at a concentration of ∼3% (vol/vol) with either 1 mg/ml purified protein in buffer containing 6.7 mM Tris-Cl (pH 8.5), 0.7 mM MgCl2, 100 mM NaCl, and 400 mM CsCl, or with 10 μg/ml myr-μ1N-RGKGR peptide in VB with 5% DMSO. To make protease-treated RBCs, cells were incubated at a concentration of ∼10% (vol/vol) in VB containing 0.5 mg/ml proteinase K, 0.5 mg/ml trypsin, and 1.5 mM CaCl for 90 min at 37°C with gentle agitation and then chilled on ice and treated with 2 mM PMSF (from a 100 mM stock in ethanol) for 15 min. Cells were pelleted and resuspended in cold VB with 2 mM PMSF, incubated on ice for 15 min more, and then washed twice with cold VB. Mock-treated RBCs were made the same way but without proteases. Fresh RBCs (less than ∼1 week old) were required for a reasonable yield following this treatment. Protease- and mock-treated RBCs were used in hemolysis reactions containing 6 × 1012 pRCs/ml and 375 mM CsCl in VB. The RBC concentration is as indicated in Results. Following incubation for 30 min at 37°C, the unlysed cells were pelleted by centrifugation at 380 × g for 2 to 5 min, and the extent of the hemoglobin release into the supernatant was measured by its absorbance at 405 or 415 nm, relative to controls containing only the buffer and RBCs (0%) or additionally containing 0.1% Triton X-100 (100%).
Liposome disruption assays.
For kinetic experiments, fluorescence was monitored in a spectrofluorimeter (FluoroMax2; ISA, Inc.) using slit widths of 1.5 nm, an excitation wavelength of 491 or 496 nm, and an emission wavelength of 514 or 515 nm. The experiments were performed at 37°C in a stirred cuvette, and readings were taken every minute with an integration time of 2 s, closing the shutter between readings to minimize bleaching. For the liposome disruption assays with free μ1 protein, 1 μl CF-loaded liposomes was added to a cuvette containing 2.5 ml prewarmed VB. WT or mutant μ13σ33 (3 mg/ml, 40 μl) was added to the reaction 5 min after the addition of liposomes. After another 10 min, 3 μl chymotrypsin (5 mg/ml) was added to remove σ3. Protease digestion was quenched after 15 min by addition of 0.1 ml of 0.2 M PMSF, followed immediately by addition of 0.5 ml of 2 M CsCl, and the reaction was allowed to proceed for at least 25 additional minutes. At the end of the experiment, the level of complete dequenching was estimated by addition of 25 μl of 10% Triton X-100. For the liposome disruption assays with synthetic peptides, 100 μl liposomes loaded with CF, FD10, or FD40 was added to 2.4 ml prewarmed VB. After 5 min, 131.2 μl of 20× peptide stock in DMSO was added, for final peptide concentrations of 2, 5, or 10 μg/ml and 5% DMSO. After 20 additional min, the level of complete dequenching was estimated by adding Triton X-100 to 0.1% (26.6 μl of 10% stock).
Endpoint experiments with pRCs were done in 20-μl reactions containing 20% liposome preparation, 3 × 1012 pRCs/ml, and 400 mM CsCl in VB. Endpoint experiments with peptides were done in 40-μl reactions containing 5% liposome preparation, 5% DMSO, and the indicated amount of peptide in VB. The reactions were incubated on ice or at 37°C for 20 min, and complete dequenching controls were lysed with 0.1% Triton X-100 at the end of the experiment. Fluorescence was measured in a clear 96-well plate with a Typhoon scanner (GE Healthcare). The samples were diluted with VB so that wells contained ∼2% liposome preparation in 100 μl, and the plate was scanned using the 532-nm laser and 526-nm SP emission filter, with the focal plane 3 mm above the platen. No cross-contamination of the signal between wells was observed. The fluorescence intensity was quantified using ImageQuant (GE Healthcare).
For detection using both the spectrofluorimeter and the Typhoon, samples were within the linear range of the instrument with the settings used, as determined by measurement of standard curves with the same settings.
Trypan blue uptake experiments.
Spinner-adapted L cells were grown as described previously (1). L cells were pelleted immediately before use by centrifugation at 380 × g for 4 min and washed three times in PBS-Mg. Thirty microliter reaction mixtures consisting of VB, L cells at a concentration of ∼10% (vol/vol), and 5% DMSO with or without 10 μg/ml myr-μ1N-RGKGR peptide were incubated at 37°C for 15 min. Immediately after the reaction, 350 μl of L-cell growth medium was added to each tube, and the cells were pelleted at 380 × g for 1 min and resuspended in 100 μl of fresh L-cell growth medium. Three-microliter aliquots of L-cell suspensions were mixed with 10 μl of a 0.2% solution of trypan blue (Cellgro) in equal parts PBS and L-cell growth medium. Between 600 and 2,000 blue and white cells were counted per sample using a hemocytometer.
Assays with released components.
The isolation of components released from virus particles was essentially as described previously (29). Briefly, pRCs (5 × 1012 particles/ml in 20 μl) were converted by incubation in VB with 400 mM CsCl at 37°C for 14 min and then centrifuged at 16,000 × g for 10 min at 4°C to pellet particles, while no-spin controls were kept on ice. Supernatants (18 μl) was added to tubes containing 6 μl CF-loaded liposomes. For the no-spin controls, 6.7 μl liposomes was added directly to the 20-μl conversion sample. After incubation at 37°C for 2 min, fluorescence was measured in a 96-well plate as described above. Siliconized low-retention tubes were used for all steps.
Trypsin sensitivity assay.
The trypsin sensitivity of μ1 was assayed by incubation with 100 μg/ml trypsin (Sigma) on ice for 45 min. The reactions were stopped by the addition of Laemmli sample buffer and boiling. Following separation on a 10% acrylamide gel by SDS-PAGE, the μ1 bands were detected either by Coomassie blue staining or by transfer to nitrocellulose and Western blotting with T1L-virion-specific serum (55) (1:2,000 dilution) followed by rabbit-specific donkey immunoglobulin G conjugated to horseradish peroxidase (Jackson Immunoresearch) (1:10,000 dilution). Antibody binding was detected with Western Lightning chemiluminescence reagents (PerkinElmer) and exposure to film.
Density gradient centrifugation.
ISVPs or pRCs (3 × 1012 particles/ml) were converted by incubation in VB with 400 mM CsCl for 30 min at 37°C. The converted samples (55 μl) were layered over 950-μl CsCl step gradients (ρ = 1.25 g/ml and 1.45 g/ml CsCl solutions in VB) and centrifuged at 81,000 × g (average) in a TLS-55 rotor (Beckman) for 2 h at 4°C. The fractions were collected from the top with wide-mouth pipette tips, and equal volumes of each fraction were analyzed by SDS-PAGE and Western blotting with a combination of T1L-virion-specific serum (55) (1:2,000 dilution) and T1L-σ1-specific serum (10) (1:2,000 dilution), followed by secondary antibody and chemiluminescence detection as described above.
CD.
Empty phosphatidylcholine liposomes were made as described above, except that lipids were resuspended in phosphate buffer (PB) (10 mM phosphate [pH 7.2]), a 0.1-μm filter was used for extrusion (to minimize scattering effects), and there was no gel filtration step. Samples (3 ml) containing 30 μl liposomes, 30 μg/ml synthetic peptide, and 5% DMSO in PB were incubated at 37°C for 10 min and then chilled on ice. DMSO was removed by extensive dialysis against PB in the cold. Spectra were recorded with a 62DS circular dichroism (CD) spectrometer (Aviv), in a 1-cm path-length stirred quartz cuvette at 4°C, with a constant 1.5-nm bandwidth and 10-s averaging time.
RESULTS
Membrane disruption accompanies rearrangement of particle-free μ1. (i) Hemolytic activity of μ1*.
WT and mutant μ13σ33 heterohexamers were expressed in insect cells and purified as previously described (58). Hemolysis reactions were performed with bovine RBCs, and the extent of hemolysis was measured by the release of hemoglobin (Fig. 2A). As expected, untreated heterohexamers did not mediate hemolysis. To prime μ1 for conformational change, σ3 was removed from purified heterohexamers by limited digestion with chymotrypsin in the presence of RBCs, followed by addition of PMSF to stop digestion (58). The μ1 trimer, kept at 4°C after σ3 removal, was also essentially inactive. When the μ1→μ1* transition was induced by incubation at 37°C in the presence of Cs+ (10, 58), there was extensive hemolysis, with the release of ∼80% of the total hemoglobin. Incubation of the μ1 trimer in buffer containing Na+ (instead of Cs+) also led to hemoglobin release, but only about half as much as in buffer containing Cs+.
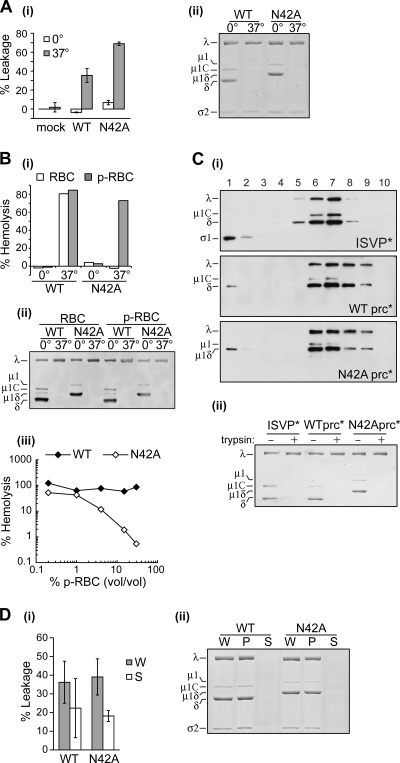
FIG. 2.
Membrane disruption activity of WT and mutant μ1*. (A) Purified μ13σ33 heterohexamers were mixed with bovine RBCs, and σ3 was removed by chymotrypsin digestion. Reactions were incubated for 30 min at the indicated temperatures in buffer containing Cs+ or Na+ ions, and hemolysis was measured by the A415 of the supernatant after pelleting unlysed cells. Control experiments without μ13σ33 but with protease and protease inhibitor (mock) or omitting protease (μ1σ3) are shown in columns 1 and 2. Data are normalized with respect to samples containing only buffer and RBCs (0%) and containing RBCs with detergent to mediate complete lysis (100%). The mean ± standard deviation of at least three determinations is shown. (B) In the presence of CF-loaded liposomes, purified μ13σ33 heterohexamers were digested with chymotrypsin to remove σ3, and conformational change was induced by addition of Cs+. CF leakage was measured by fluorescence dequenching at 1-min intervals. The additions of μ13σ33, chymotrypsin, PMSF, Cs+, and Triton X-100 detergent are indicated by arrows. Note that the time points of addition of detergent are slightly different for each experiment, but the complete dequenching of the fluorescent signal was always immediate. Data are normalized to the level of complete dequenching upon addition of detergent (100%).
Autocleavage of μ1, generating the N-terminal myristoylated fragment μ1N, is required both for hemolytic activity and for infectivity (42). The N42A mutation, which prevents μ1 autocleavage both on virus particles and in particle-free μ1 trimers, without blocking conformational rearrangement (42, 58), eliminated hemolytic activity as expected (Fig. 2A).
(ii) Liposome disruption by μ1*.
To study the membrane-perforating activity of μ1 in a simpler model system, suitable for future structural studies, we prepared phosphatidylcholine liposomes containing CF at a self-quenching concentration. Membrane disruption and consequent dye release were measured by fluorescence dequenching (Fig. 2B). Addition of WT μ13σ33 to liposomes at 37°C led to a moderate increase in fluorescence, probably indicating some leakage due to nonspecific interactions between the proteins and the membrane. Addition of chymotrypsin to remove σ3 induced a further moderate increase in some reactions with both WT and mutant proteins. Digestion of σ3 was confirmed by SDS-PAGE (data not shown). When PMSF and Cs+ ions were introduced, to halt chymotrypsin digestion and to promote μ1 structural rearrangement, respectively, there was first a sudden drop in fluorescence, followed by a rapid increase and leveling off to a plateau. The initial drop in fluorescence is due to quenching of leaked CF by Cs+, as confirmed by control experiments (data not shown). The rise is then a measure of μ1*-induced liposome disruption. Similar results were achieved using liposomes made from a lipid mixture (see Materials and Methods), suggesting that lipid composition does not strongly influence the membrane interaction (data not shown).
Structural rearrangement of μ1 on ISVPs is required for hemolysis (10), as well as for the infection of cells (1, 11). We examined the effect of two stabilizing mutations in μ1, K459E and A319E, on liposome disruption by particle-free μ1 trimers (Fig. 2B). These mutations were initially selected as heat- or ethanol-resistant mutants (36, 56), and both inhibit the conformational change in ISVP-associated μ1 (11, 36). In particle-free μ1 trimers, both mutations also eliminated liposome disruption. These results with particle-free μ1 parallel those reported for ISVPs, providing further evidence that the μ1→μ1* transition is functionally similar to the ISVP→ISVP* transition in how it releases the μ1 membrane-disruption machinery.
Cleavage of μ1N is not required for membrane disruption. (i) Particle-free μ1(N42A) mediates liposome disruption.
In the liposome disruption assay with particle-free μ1, the N42A mutant induces liposome disruption to an extent similar to that of the WT protein (Fig. 2B). The N42A mutation eliminates hemolysis activity, however, both of free μ1 trimers (Fig. 2A) and recoated cores (42). We suggest that the structural rearrangement of μ1(N42A) leads to exposure of the μ1N region and that the surface of naked liposome, unlike the protein- and glycan-rich surface of an RBC, can interact with exposed μ1N even when tethered to the body of μ1.
(ii) N42A-recoated cores mediate liposome disruption.
Because particle-free μ1(N42A) can disrupt liposomes (Fig. 2B), we investigated the disruption activity of cores recoated with μ1(N42A) (12, 13). Previous work has shown that recoating with μ1(N42A) yields correctly assembled virion-like particles, which undergo normal disassembly steps, including the structural rearrangement of μ1 to a protease-sensitive form (42). Recoated cores were made with WT μ13σ33 or μ1(N42A)3σ33, treated with chymotrypsin to yield ISVP-like particles (WT-pRCs and N42A-pRCs, respectively), and incubated with CF-loaded liposomes. As shown in Fig. 3A, both WT μ1 and μ1(N42A) on pRCs became protease sensitive when warmed to 37°C in the presence of Cs+, indicating ISVP* conversion, and both mediated substantial liposome disruption.
FIG. 3.
Membrane disruption activity of N42A-pRCs. (A) Cores recoated with WT μ1 or μ1(N42A), or buffer alone (mock), were incubated with chymotrypsin to yield WT-pRCs, N42A-pRCs, or cores, respectively, and then mixed with CF-loaded liposomes in buffer containing Cs+. After incubation on ice or at 37°C for 20 min, the fluorescence of each reaction was measured (i), and ISVP* conversion was assayed by trypsin treatment, followed by SDS-PAGE and Coomassie blue staining (ii). Means ± standard deviations of five replicates using two different recoated core preparations are shown, normalized with respect to the 0°C mock sample (0%) and to a sample containing detergent to induce complete dequenching (100%). (B) WT- or N42A-pRCs were mixed with untreated RBCs or RBCs pretreated with trypsin and proteinase K (p-RBCs) in buffer containing Cs+. (i) After incubation for 30 min at 37°C, hemoglobin release was measured by the A405 of the supernatant after pelleting unlysed cells. (ii) An aliquot of supernatant was subjected to trypsin treatment and Western blotting with serum raised against virions to assay ISVP* conversion. (iii) The extent of hemolysis after incubation was determined with various concentrations of pRBCs. Data are normalized as in Fig. 2A. (C) ISVPs and WT- or N42A-pRCs were preconverted in buffer containing Cs+ and banded on CsCl density gradients. (i) Fractions (1, top; 10, bottom) were subjected to Western blotting with serum raised against virions and serum raised against σ1. (ii) A Coomassie blue-stained gel of trypsin-treated samples, confirming ISVP* conversion prior to loading on the gradient, is shown. (D) WT- or N42A-pRCs were preconverted at 37°C in buffer containing Cs+. Parallel samples were either centrifuged at 4°C to pellet particles or kept on ice. CF-loaded liposomes were mixed with the whole, unpelleted reaction (W) or with the supernatant from the pelleted sample (S). Reactions were incubated for 5 min at 37°C, and the fluorescence measured. (i) Means ± standard deviations of the results from three or four experiments are shown. (ii) Equivalent amounts of pellet (P), supernatant (S), and whole unpelleted sample (W) were subjected to SDS-PAGE and Coomassie blue staining, confirming the efficient pelleting of particles.
(iii) N42A-recoated cores mediate hemolysis of treated RBCs.
To test the hypothesis that RBC surface proteins and glycans prevent hemolysis by inhibiting access to the membrane by μ1(N42A) following its conformational rearrangement, RBCs were treated with a combination of trypsin and proteinase K and used for hemolysis assays with WT- and N42A-pRCs (Fig. 3Bi). Protease treatment caused the RBCs to become sensitive to lysis by N42A-pRCs when incubated at 37°C. WT-pRCs mediated efficient lysis of both mock- and protease-treated RBCs. Trypsin treatment of an aliquot of each sample confirmed ISVP* conversion in all cases (Fig. 3Bii). These hemolysis experiments were performed at a final RBC concentration of ∼1% (vol/vol). To test the efficiency of lysis by N42A, we carried out hemolysis reactions with various concentrations of protease-treated RBCs (Fig. 3Biii). At the particle concentration used, WT-pRCs mediated nearly complete lysis over the full range of RBC concentrations tested, indicating that μ1N remained in excess in these reactions. The activity of N42A-pRCs, on the other hand, dropped rapidly as the membrane concentration increased. These results suggest either that there are substantially fewer copies of the membrane-active component in N42A-pRC reactions or that fewer of them are able to access the membrane, despite protease treatment of the RBCs.
A released component of N42A-pRCs mediates liposome disruption.
Following the ISVP* conversion of native, WT ISVPs, δ remains stably associated with particles (3, 10, 28, 29). If the same is true of uncleaved μ1δ, does the membrane-disrupting activity of N42A-pRCs imply that tethering of μ1N to the virus particle is compatible with the μ1N oligomerization necessary to form a membrane pore? One alternative explanation of our observations is that uncleaved μ1δ is less stably associated with particles following ISVP* conversion than δ and that a released μ1δ fraction mediates membrane disruption.
To assay for the release of μ1 species during ISVP* conversion, native ISVPs and WT- and N42A-pRCs were converted by incubation at 37°C in the presence of Cs+ ions, and released components were separated from particles by CsCl density gradient centrifugation (Fig. 3Ci). Trypsin treatment of an aliquot prior to loading on the gradient confirmed ISVP* conversion (Fig. 3Cii). This control also confirmed that μ1N remains uncleaved from δ after the ISVP* conversion of N42A-pRCs. Conversion of native ISVPs to ISVP*s resulted in complete elution of the attachment protein σ1, which remained at the top of the gradient as expected (10). All large μ1 species comigrated with particles in the gradient (Fig. 3Ci), and no release was detected. In contrast, both WT- and N42A-pRCs released a small fraction of their μ1 proteins. As expected, the recoated cores did not contain σ1. The μ1N peptide, which is released from WT particles, and the C-terminal φ fragment, which is released from both N42A and WT particles (29), are too small to be resolved on this gel. Nearly identical results were obtained with two different preparations of recoated cores.
Previously, components released from N42A-pRCs during ISVP* conversion were found to have no membrane-disrupting activity (29), but since these studies were done with RBC membranes, they would not have revealed the activity of μ1δ. To determine whether components released by N42A-pRCs are responsible for the disruption activity, particles were preconverted at 37°C and then pelleted by centrifugation, and the supernatant was incubated with CF-loaded liposomes (Fig. 3Di). A parallel sample omitting the spin and transfer, and instead kept on ice for the duration of these steps, was included as a measure of the total activity that remains after the delay required for pelleting particles following preconversion. The supernatants from WT-pRCs mediated efficient liposome disruption, as expected (29). The large error bars in these results are consistent with the previous observation that the efficiency of transfer of μ1N after preconversion is highly sensitive to timing (2, 29). The supernatants from N42A-pRCs also mediated efficient liposome disruption: the activity in the supernatant was ∼50% of that observed without pelleting particles. Gels of each sample confirmed efficient pelleting of both the WT- and N42A-pRCs and release of a small fraction of δ and μ1δ, respectively, into the supernatant (Fig. 3Dii). Since the majority of μ1δ is cleared by pelleting, yet a significant fraction of the activity is present in the supernatant, we conclude that particle-associated μ1δ is likely not responsible for liposome disruption mediated by N42A-pRCs. The data are consistent with membrane activity of the small fraction of μ1δ that is released, although they do not rule out the possibility that φ, or some region of μ1 other than the μ1N segment, participates.
Synthetic myr-μ1N peptide forms size-selective pores in liposome membranes.
Ivanovic et al. (29) have shown recently that μ1N is sufficient for pore formation in RBC membranes. To investigate the activity of μ1N in liposomes, we synthesized a myristoylated form of μ1N peptide as well as a modified form of this myr-μ1N, with the polar sequence RGKGR added to the C terminus to enhance solubility. As the noncleaving N42A mutant retained full liposome-disruption activity (Fig. 2B), the C-terminal tag was not expected to impair the function of the peptide.
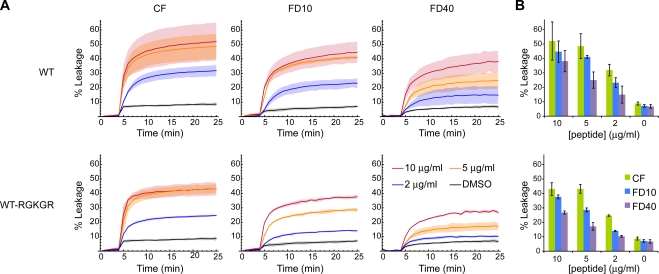
To demonstrate the formation of size-selective pores by myr-μ1N in liposome membranes, we made phosphatidylcholine liposomes that contained self-quenching concentrations of CF, FD10, or FD40 (Fig. 4). The myr-μ1N or myr-μ1N-RGKGR peptide was dissolved in DMSO and added to liposomes in buffer at 37°C to final peptide concentrations of 2, 5, or 10 μg/ml and 5% DMSO. Leakage of encapsulated molecules was measured by fluorescence dequenching (Fig. 5). The myr-μ1N and myr-μ1N-RGKGR peptides yielded similar results. More CF leakage was induced by 5 μg/ml than 2 μg/ml of peptide, but concentrations of 10 μg/ml or greater were not more effective than 5 μg/ml, probably because of solubility limitations. At each peptide concentration, the amount of content leakage negatively correlated with the size of the encapsulated molecule, suggesting the presence of size-selective pores comparable to those observed in previous studies (3, 29).
FIG. 5.
(A) Liposome disruption time courses with WT myr-μ1N (top) and myr-μ1N-RGKGR (bottom). Liposome disruption was measured at 1-min intervals by fluorescence dequenching of encapsulated CF, FD10, or FD40. Between the 4- and 5-min time points, peptide dissolved in DMSO was added to a final concentration of 10, 5, or 2 μg/ml and 5% DMSO. Alternatively, DMSO alone was added as a vehicle control. Between the 25- and 26-min time points, addition of detergent induced complete dequenching. Data are shown normalized with respect to the background fluorescence before addition of peptide (0%) and the total signal after complete dequenching (100%). Curves are the means of the results from three experiments performed with at least two different liposome preparations, and each shaded region represents the mean ± one standard deviation. (B) Endpoints (at 25 min) of the results shown in panel A. DMSO controls are duplicated in the top and bottom panels for clarity.
Synthetic myr-μ1N peptide mediates L-cell membrane disruption.
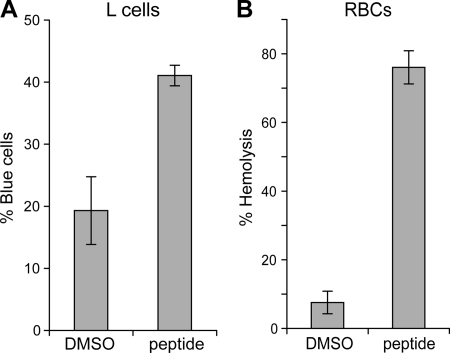
To test the membrane-disruption activity of μ1N in L-cell plasma membranes, cells were incubated with 10 μg/ml myr-μ1N-RGKGR peptide, and membrane disruption and subsequent cell death were assayed by trypan blue uptake (Fig. 6A). Although DMSO alone led to some cell death, the fraction of dead cells was substantially increased in the presence of peptide. These results indicate that μ1N can mediate disruption of L-cell plasma membranes. As a control, parallel experiments were conducted with chicken RBCs (Fig. 6B). The extent of hemolysis is comparable to previously reported experiments with myr-μ1N peptide and bovine RBCs (29).
FIG. 6.
L-cell membrane disruption (A) and hemolytic activity (B) of the myr-μ1N-RGKGR peptide were assayed in parallel experiments. L cells and chicken RBCs were incubated for 15 min with 10 μg/ml peptide. L-cell membrane disruption was measured by counting the percentage of blue cells after incubation with trypan blue, and hemolysis was determined as described in the legend to Fig. 2. Data represent the means ± standard deviations of the results from three experiments.
Substitutions V13N and L36D both impair μ1N liposome-disruption activity.
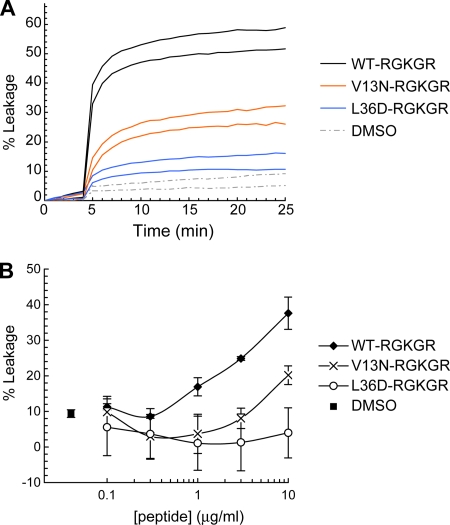
If μ1N alone forms pores, it probably does so by insertion into the lipid bilayer. Two regions of alternating hydrophobic residues separated by a polar region, similar to β-barrel toxins, suggest that μ1N may insert as a β-hairpin (see Discussion and Fig. 8B). We synthesized two myr-μ1N peptides, each modified in one of the putative transmembrane regions by an amino acid substitution: V13N or L36D. Since results with the WT myr-μ1N-RGKGR peptide were more reproducible than those with the WT myr-μ1N peptide (Fig. 5), the variants were made with the C-terminal RGKGR sequence. The level of liposome disruption with the V13N substitution was about half that with the WT peptide; the L36D substitution reduced the disruption more substantially (Fig. 7). At concentrations of up to 10 μg/ml, the L36D peptide mediated little or no liposome disruption. These data suggest that Leu36, and perhaps Val13, may be in regions that interact with the hydrophobic interior of the bilayer. The mutations might prevent membrane insertion altogether, or they might simply alter the capacity of μ1N to form a pore-like structure.
FIG. 8.
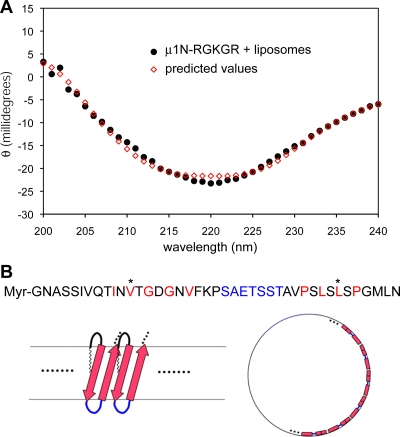
(A) CD spectrum of liposome disruption reactions. Empty liposomes were incubated with 30 μg/ml myr-μ1N-RGKGR peptide and 5% DMSO for 10 min at 37°C, then chilled on ice, and dialyzed extensively against cold PB. The spectrum of a parallel sample, containing 5% DMSO in PB without peptide, was essentially flat, and the difference spectrum of the peptide sample after subtracting the DMSO control is shown. A predicted spectrum after curve fitting using the multilinear regression algorithm (25) is superimposed. (B) Model of pore formation by μ1N. Top, amino acid sequence of μ1N, showing segments of alternating hydrophobic (red) and polar residues connected by a polar sequence (blue). The three prototype reovirus strains (T1L, type 2 Jones, and type 3 Dearing) have identical μ1N amino acid sequences. Bottom left, proposed model for μ1N insertion as a β-hairpin, with a number of such hairpins forming a β-barrel. The strands of the hairpin are shown as arrows, and the polar connector is shown as a blue loop; the limits of the hydrophobic parts of the membrane bilayer are represented by gray lines. The N-terminal myristoyl group is assumed to insert into the membrane. Bottom right, schematic en face view to illustrate the formation of a pore ∼5 nm in diameter by 14 to 16 such hairpins.
FIG. 7.
(A) Liposome disruption time courses with CF-loaded liposomes and 5 μg/ml myr-μ1N-RGKGR, myr-μ1N(V13N)-RGKGR, or myr-μ1N(L36D)-RGKGR. Data are normalized as described in the legend to Fig. 5. (B) Liposome disruption after 20-min incubations with various concentrations of peptide. Data are normalized with respect to a buffer control (0%) and a sample containing detergent (100%). Means ± standard deviations of the results from three experiments are shown.
Liposome-associated μ1N adopts a conformation with substantial β-strand content.
In the μ1 trimer prior to conformational change, μ1N is in a relatively extended conformation, with little well-defined secondary structure (31) (Fig. 1). Virtually nothing is known about the structure of μ1N following μ1 conformational change or upon insertion into the membrane. The CD spectrum of reactions containing empty liposomes, 30 μg/ml myr-μ1N-RGKGR peptide, and 5% DMSO, incubated at 37°C followed by extensive dialysis in the cold to remove DMSO, is shown in Fig. 8A. We were unable to determine an accurate protein concentration following liposome disruption and dialysis, preventing accurate deconvolution into secondary-structure elements by most algorithms. An analysis with a nonconstrained multilinear regression (25), which does not require correction for the concentration, using a basis set derived from 32 proteins of known structure plus poly-l- glutamate (53), led to estimates of ∼30% β-strand and ∼10% α-helix content (Fig. 8A). Similar results were obtained with the CONTIN program (43), assuming 30 μg/ml protein in the sample (data not shown). The consistency of the constrained and unconstrained fits (25) supports the qualitative conclusion that μ1N in liposomes has substantial β-strand content but little or no α-helix.
DISCUSSION
Pore formation by μ1N.
The pores formed by synthetic myr-μ1N peptide in phosphatidylcholine liposomes appear to be size selective, allowing some degree of discrimination between the three fluorescent labels CF, FD10, and FD40. Decreasing the myr-μ1N concentration from 10 to 2 μg/ml decreased the amount of leakage of each label. One explanation is that the pore size changes with the peptide concentration, consistent with results from hemolysis reactions in which pore size varies with ISVP concentration (3). The fluorescence dequenching assay does not, however, distinguish between that explanation and the alternative that at lower peptide concentrations, a smaller fraction of the liposomes contain pores.
The size selectivity detected here agrees with the pore diameter of ∼5 nm estimated in assays using RBCs (3, 29). To make a transmembrane pore of this size, μ1N must oligomerize, and the best precedents for such a structure are the homo-oligomeric β-barrels, for example, the toxins that insert one or more transmembrane β-hairpins per subunit. The cholesterol-dependent toxins, such as perfringolysin O and listeriolysin O, form pores with a very large diameter, in which each subunit contributes a pair of β-hairpins (47, 48); anthrax toxin and its relatives form heptameric pores, to which each subunit contributes a single hairpin (30, 50). The hairpin segments form quite unrelated structures in the preinsertion form of these proteins, such as short α-helices in the cholesterol-dependent toxins. The sequences of these β-hairpins have two regions with nonpolar residues at alternating positions, each about 10 residues long, and separated by a polar region. Reovirus μ1N also has two such alternating segments with a polar sequence between them (Fig. 8B). If membrane-inserted μ1N were to make a β-barrel structure, a pore with a 5-nm diameter would require roughly 15 copies. The myristoyl group would presumably help to anchor the N terminus of the peptide in the outer leaflet of the bilayer. Because neither terminus would need to traverse the membrane, additional C-terminal residues would not be expected to prevent hairpin formation, consistent with the observed liposome-disrupting activity of both uncleaved μ1* and the myr-μ1N-RGKGR peptide. The detrimental effect of substitutions V13N and L36D in myr-μ1N-RGKGR, as well as the significant β-strand content of myr-μ1N-RGKGR in the presence of liposomes, is consistent with this model.
Because myr-μ1N is not soluble in aqueous buffer, it is unlikely that a prepore complex forms in solution prior to membrane insertion. Rather, we propose that the peptide associates directly with the membrane, perhaps initially through insertion of the myristoyl group, and that lateral diffusion allows oligomerization and pore formation. The observation that an excess of RBCs can inhibit hemolysis (data not shown) supports this suggestion. As insertion of a single β-hairpin peptide monomer is not expected to be favorable, the peptide may associate initially with the membrane in a different conformation than the one it ultimately adopts when in a pore.
Deployment of μ1N by the μ1 trimer.
Prior to conformational change, μ1N is largely buried near the base of the μ1 trimer (Fig. 1) (31). The μ1 conformational change associated with ISVP→ISVP* conversion allows release of the μ1N peptide (3, 29, 42). The experiments in our previous report (58) show that mild heating in the presence of K+ or Cs+ ions will induce free μ1 trimer, prepared in vitro by the treatment of purified recombinant μ13σ33 with chymotrypsin, to undergo a conformational rearrangement similar to the one that particle-associated μ1 undergoes during ISVP→ISVP* conversion. We have extended those observations here, demonstrating that the transition of particle-free μ1 to μ1* can induce lysis of RBCs or leakage of a small amount of dye from liposomes (Fig. 2).
Cleavage of μ1N is not required for liposome disruption. Both free μ1(N42A) and recoated cores containing this mutant protein, while defective at hemolysis as reported previously (42), can mediate dye release from liposomes (Fig. 2 and 3). Treatment of RBCs with proteases allows (somewhat inefficient) hemolysis by uncleaved μ1, consistent with our suggestion that the liposome-disrupting activity reflects the unhindered accessibility of the liposome surface to μ1N as it projects from conformationally rearranged μ1*, while the barrier presented by the glycoprotein-covered surface of RBCs allows only the free μ1N peptide to access the underlying bilayer. Pore formation by N42A-pRCs and by the μ1(N42A) protein probably proceeds by similar mechanisms, as our data show that a released component, rather than particle-associated μ1δ, is responsible. This result is consistent with our model for an oligomeric pore; that is, the small fraction of μ1δ that is released from N42A-pRCs following ISVP→ISVP* conversion, like the μ1N released from WT-pRCs, can presumably form pores by the same sequence of insertion, diffusion, and oligomerization of the μ1N region. The apparently disproportionate efficiency of N42A-pRCs may be due to the chaperoning effect of the δ part of μ1, which can effectively keep μ1N available in solution. Aggregation or adherence to the tube will ordinarily sequester a large fraction of μ1N (29). It is possible that membrane-disrupting properties of another region of μ1 could contribute to the activity of μ1(N42A) with naked liposomes. One candidate is the C-terminal φ fragment, which is recruited to RBC membranes in the presence of μ1N (29) and which localizes to internal cellular membranes and lipid droplets in transfected cells (16). A double mutant containing both N42A and L36D may resolve this issue.
Cell entry by reovirus and related nonenveloped viruses.
A pore with an ∼5-nm diameter is, of course, too small to allow passage of the reovirus ISVP* or core. In principle, such a pore might make the endosome leaky enough to allow free passage of nucleotides and nascent RNA transcripts between the virus particle and the cytosol, but fluorescence imaging of entering virus suggests that cores do not remain associated with endo- or lysosomal markers (11). Formation of a small pore could thus be merely a first step in endosomal disruption, after which additional cellular factors might participate (28). Alternatively, the very high concentration of μ1N in the enclosed space of an endosome or related structure could lead to more efficient insertion and formation of much larger pores or a large number of small pores. The release of all 600 copies of μ1N from a virus particle within an endosome 250 nm in diameter would lead to a peptide concentration of about 0.1 mM, 20 to 50 times the concentrations used in this work. A β-barrel pore 70 nm in diameter, large enough to admit a reovirus particle, would require about 250 μ1N hairpins.
Despite their liposome disruption activity in vitro, cores recoated with μ1(N42A) have a severe infectivity defect (42). Possible explanations are that intact μ1* or μ1δ* makes pores of insufficient size or number to allow endosome escape; that pore docking of the virus particle, as recently described (29), is aberrant and does not permit subsequent entry steps; or that the endosomal membrane, like the RBC membrane, is sufficiently crowded to inhibit pore formation. It is not clear which of the membranes used in this study—liposome, RBC, proteolysed RBC, or L-cell surface—is the best model for the endosomal lumen. Similar results with the peptide in liposomes, L-cells, and RBCs suggest, however, that the pore-forming activity of μ1N is context independent.
Our results show that myristoylated μ1N will form pores in pure lipid bilayers when added at concentrations well below those likely to be achieved during cell entry. While we cannot rule out the participation of other parts of μ1 at some stage in membrane penetration, it is clear that μ1N is the principal agent of bilayer perforation. Intact μ1 is thus a reservoir that delivers μ1N when appropriately triggered, as well as a structural component of the virion outer capsid.
The structural homolog of μ1 in rotaviruses is VP6. Both are trimeric proteins with similarly folded subunits—a radially oriented, β-jelly-roll domain flanked in the polypeptide chain by N- and C-terminal extensions, which form a largely α-helical “pedestal” (35). VP6 is not a penetration effector, however, and it probably has a purely structural role as a connecting element between the actual penetration effectors of the outer layer (VP4 and VP7) and the RNA-enclosing VP2 shell (32, 57, 59). The pedestal of μ1 is substantially larger than that of VP6: the additional structure is primarily the part that sequesters μ1N and that unfolds in order to release it (31). In other words, a key difference between VP6 and μ1 is that the latter has acquired additional structural elements to retain and release the myristoylated penetration peptide μ1N.
Picornaviruses such as poliovirus also release a myristoylated peptide, VP4, during cell entry, and there is some evidence that VP4, like μ1N, is the critical agent of membrane perforation (54). The structural parallels are also suggestive. VP4 is the N-terminal segment of the picornavirus polyprotein, and it separates by autolytic cleavage from the precursor (26). In the virion, it winds into a heterotrimeric assembly of VP1, VP2, and VP3, each of which is based on a central β-jelly-roll domain with N- and C-terminal extensions; the release of VP4 requires loosening of the interfaces among VP1 to -3, just as the release of μ1N requires dissociation of the μ1 trimer. The molecular mechanisms of penetration by reoviruses and picornaviruses may therefore be quite similar, perhaps even more closely related to each other than those of reoviruses and rotaviruses.
Acknowledgments
We thank Marina Babyonyshev and Elaine Freimont for technical support; Marcelo Berardi, Jawdat Al-Bassam, Kelly Arnett, Michael Eck, Daniel Floyd, Stephen Blackwell, and Thomas Walz for assistance and for use of equipment; and Irene Kim for helpful comments on the manuscript.
L.Z. acknowledges postdoctoral fellowship from the Helen Hay Whitney Foundation, and M.A.A. acknowledges National Institutes of Health graduate training fellowship AI064142. The research was also supported by National Institutes of Health grants AI046440 (to M.L.N.) and CA013202 (to S.C.H.). S.C.H. is an investigator at the Howard Hughes Medical Institute.
Footnotes
Published ahead of print on 13 May 2009.
REFERENCES
- 1.Agosto, M. A., J. K. Middleton, E. C. Freimont, J. Yin, and M. L. Nibert. 2007. Thermolabilizing pseudoreversions in reovirus outer-capsid protein μ1 rescue the entry defect conferred by a thermostabilizing mutation. J. Virol. 817400-7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agosto, M. A., K. S. Myers, T. Ivanovic, and M. L. Nibert. 2008. A positive-feedback mechanism promotes reovirus particle conversion to the intermediate associated with membrane penetration. Proc. Natl. Acad. Sci. USA 10510571-10576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agosto, M. A., T. Ivanovic, and M. L. Nibert. 2006. Mammalian reovirus, a nonfusogenic, nonenveloped virus, forms size-selective pores in a model membrane. Proc. Natl. Acad. Sci. USA 10316496-16501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baer, G. S., and T. S. Dermody. 1997. Mutations in reovirus outer-capsid protein σ3 selected during persistent infections of L cells confer resistance to protease inhibitor E64. J. Virol. 714921-4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bodkin, D., M. L. Nibert, and B. N. Fields. 1989. Proteolytic digestion of reovirus in the intestinal lumens of neonatal mice. J. Virol. 634676-4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borsa, J., D. G. Long, M. D. Sargent, T. P. Copps, and J. D. Chapman. 1974. Reovirus transcriptase activation in vitro: involvement of an endogenous uncoating activity in the second stage of the process. Intervirology 4171-188. [DOI] [PubMed] [Google Scholar]
- 7.Borsa, J., D. G. Long, T. P. Copps, M. D. Sargent, and J. D. Chapman. 1974. Reovirus transcriptase activation in vitro: further studies on the facilitation phenomenon. Intervirology 315-35. [DOI] [PubMed] [Google Scholar]
- 8.Borsa, J., M. D. Sargent, D. G. Long, and J. D. Chapman. 1973. Extraordinary effects of specific monovalent cations on activation of reovirus transcriptase by chymotrypsin in vitro. J. Virol. 11207-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandran, K., and M. L. Nibert. 1998. Protease cleavage of reovirus capsid protein μ1/μ1C is blocked by alkyl sulfate detergents, yielding a new type of infectious subvirion particle. J. Virol. 72467-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandran, K., D. L. Farsetta, and M. L. Nibert. 2002. Strategy for nonenveloped virus entry: a hydrophobic conformer of the reovirus membrane penetration protein μ1 mediates membrane disruption. J. Virol. 769920-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chandran, K., J. S. L. Parker, M. Ehrlich, T. Kirchhausen, and M. L. Nibert. 2003. The δ region of outer-capsid protein μ1 undergoes conformational change and release from reovirus particles during cell entry. J. Virol. 7713361-13375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chandran, K., S. B. Walker, Y. Chen, C. M. Contreras, L. A. Schiff, T. S. Baker, and M. L. Nibert. 1999. In vitro recoating of reovirus cores with baculovirus-expressed outer-capsid proteins μ1 and σ3. J. Virol. 733941-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chandran, K., X. Zhang, N. H. Olson, S. B. Walker, J. D. Chappell, T. S. Dermody, T. S. Baker, and M. L. Nibert. 2001. Complete in vitro assembly of the reovirus outer capsid produces highly infectious particles suitable for genetic studies of the receptor-binding protein. J. Virol. 755335-5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang, C. T., and H. J. Zweerink. 1971. Fate of parental reovirus in infected cell. Virology 46544-555. [DOI] [PubMed] [Google Scholar]
- 15.Chow, M., J. F. Newman, D. Filman, J. M. Hogle, D. J. Rowlands, and F. Brown. 1987. Myristylation of picornavirus capsid protein VP4 and its structural significance. Nature 327482-486. [DOI] [PubMed] [Google Scholar]
- 16.Coffey, C. M., A. Sheh, I. S. Kim, K. Chandran, M. L. Nibert, and J. S. Parker. 2006. Reovirus outer capsid protein μ1 induces apoptosis and associates with lipid droplets, endoplasmic reticulum, and mitochondria. J. Virol. 808422-8438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dermody, T. S., M. L. Nibert, J. D. Wetzel, X. Tong, and B. N. Fields. 1993. Cells and viruses with mutations affecting viral entry are selected during persistent infections of L cells with mammalian reoviruses. J. Virol. 672055-2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drayna, D., and B. N. Fields. 1982. Activation and characterization of the reovirus transcriptase: genetic analysis. J. Virol. 41110-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drayna, D., and B. N. Fields. 1982. Genetic studies on the mechanism of chemical and physical inactivation of reovirus. J. Gen. Virol. 63149-159. [DOI] [PubMed] [Google Scholar]
- 20.Dryden, K. A., G. Wang, M. Yeager, M. L. Nibert, K. M. Coombs, D. B. Furlong, B. N. Fields, and T. S. Baker. 1993. Early steps in reovirus infection are associated with dramatic changes in supramolecular structure and protein conformation: analysis of virions and subviral particles by cryoelectron microscopy and image reconstruction. J. Cell Biol. 1221023-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ebert, D. H., J. Deussing, C. Peters, and T. S. Dermody. 2002. Cathepsin L and cathepsin B mediate reovirus disassembly in murine fibroblast cells. J. Biol. Chem. 27724609-24617. [DOI] [PubMed] [Google Scholar]
- 22.Furlong, D. B., M. L. Nibert, and B. N. Fields. 1988. Sigma 1 protein of mammalian reoviruses extends from the surfaces of viral particles. J. Virol. 62246-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Golden, J. W., J. A. Bahe, W. T. Lucas, M. L. Nibert, and L. A. Schiff. 2004. Cathepsin S supports acid-independent infection by some reoviruses. J. Biol. Chem. 2798547-8557. [DOI] [PubMed] [Google Scholar]
- 24.Golden, J. W., and L. A. Schiff. 2005. Neutrophil elastase, an acid-independent serine protease, facilitates reovirus uncoating and infection in U937 promonocyte cells. Virol. J. 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greenfield, N. J. 25 January 2007. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 12876-2890. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hindiyeh, M., Q. H. Li, R. Basavappa, J. M. Hogle, and M. Chow. 1999. Poliovirus mutants at histidine 195 of VP2 do not cleave VP0 into VP2 and VP4. J. Virol. 739072-9079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hooper, J. W., and B. N. Fields. 1996. Role of the μ1 protein in reovirus stability and capacity to cause chromium release from host cells. J. Virol. 70459-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ivanovic, T., M. A. Agosto, K. Chandran, and M. L. Nibert. 2007. A role for molecular chaperone Hsc70 in reovirus outer-capsid disassembly. J. Biol. Chem. 28212210-12219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ivanovic, T., M. A. Agosto, L. Zhang, K. Chandran, S. C. Harrison, and M. L. Nibert. 2008. Peptides released from reovirus outer capsid form membrane pores. EMBO J. 271289-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lacy, D. B., and R. J. Collier. 2002. Structure and function of anthrax toxin. Curr. Top. Microbiol. Immunol. 27161-85. [DOI] [PubMed] [Google Scholar]
- 31.Liemann, S., K. Chandran, M. L. Nibert, and S. C. Harrison. 2002. Structure of the reovirus membrane-penetration protein, μ1, in a complex with its protector protein, σ3. Cell 108283-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liprandi, F., Z. Moros, M. Gerder, J. E. Ludert, F. H. Pujol, M. C. Ruiz, F. Michelangeli, A. Charpilienne, and J. Cohen. 1997. Productive penetration of rotavirus in cultured cells induces coentry of the translation inhibitor α-sarcin. Virology 237430-438. [DOI] [PubMed] [Google Scholar]
- 33.Lucia-Jandris, P., J. W. Hooper, and B. N. Fields. 1993. Reovirus M2 gene is associated with chromium release from mouse L cells. J. Virol. 675339-5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mao, Z. X., and W. K. Joklik. 1991. Isolation and enzymatic characterization of protein λ2, the reovirus guanylyltransferase. Virology 185377-386. [DOI] [PubMed] [Google Scholar]
- 35.Mathieu, M., I. Petitpas, J. Navaza, J. Lepault, E. Kohli, P. Pothier, B. V. Prasad, J. Cohen, and F. A. Rey. 2001. Atomic structure of the major capsid protein of rotavirus: implications for the architecture of the virion. EMBO J. 201485-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Middleton, J. K., M. A. Agosto, T. F. Severson, J. Yin, and M. L. Nibert. 2007. Heat-resistance mutations in outer-capsid protein μ1 selected by heat inactivation of infectious subvirion particles of mammalian reovirus. Virology 361412-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Middleton, J. K., T. F. Severson, K. Chandran, A. L. Gillian, J. Yin, and M. L. Nibert. 2002. Thermostability of reovirus disassembly intermediates (ISVPs) correlates with genetic, biochemical, and thermodynamic properties of major surface protein μ1. J. Virol. 761051-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nibert, M. L. 1993. Structure and function of reovirus outer capsid proteins as they relate to early steps in infection. Ph.D. thesis, Harvard University, Cambridge, MA.
- 39.Nibert, M. L., and B. N. Fields. 1992. A carboxy-terminal fragment of protein μ1/μ1C is present in infectious subvirion particles of mammalian reoviruses and is proposed to have a role in penetration. J. Virol. 666408-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nibert, M. L., A. L. Odegard, M. A. Agosto, K. Chandran, and L. A. Schiff. 2005. Putative autocleavage of reovirus μ1 protein in concert with outer-capsid disassembly and activation for membrane permeabilization. J. Mol. Biol. 345461-474. [DOI] [PubMed] [Google Scholar]
- 41.Nibert, M. L., L. A. Schiff, and B. N. Fields. 1991. Mammalian reoviruses contain a myristoylated structural protein. J. Virol. 651960-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Odegard, A. L., K. Chandran, X. Zhang, J. S. L. Parker, T. S. Baker, and M. L. Nibert. 2004. Putative autocleavage of outer capsid protein μ1, allowing release of myristoylated peptide μ1N during particle uncoating, is critical for cell entry by reovirus. J. Virol. 788732-8745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Provencher, S. W., and J. Glöckner. 1981. Estimation of protein secondary structure from circular dichroism. Biochemistry 2033-37. [DOI] [PubMed] [Google Scholar]
- 44.Reinisch, K. M., M. L. Nibert, and S. C. Harrison. 2000. Structure of the reovirus core at 3.6 Å resolution. Nature 404960-967. [DOI] [PubMed] [Google Scholar]
- 45.Sahli, R., R. Freund, T. Dubensky, R. Garcea, R. Bronson, and T. Benjamin. 1993. Defect in entry and altered pathogenicity of a polyoma virus mutant blocked in VP2 myristylation. Virology 192142-153. [DOI] [PubMed] [Google Scholar]
- 46.Shatkin, A. J., and J. D. Sipe. 1968. RNA polymerase activity in purified reoviruses. Proc. Natl. Acad. Sci. USA 611462-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shatursky, O., A. P. Heuck, L. A. Shepard, J. Rossjohn, M. W. Parker, A. E. Johnson, and R. K. Tweten. 1999. The mechanism of membrane insertion for a cholesterol-dependent cytolysin: a novel paradigm for pore-forming toxins. Cell 99293-299. [DOI] [PubMed] [Google Scholar]
- 48.Shepard, L. A., A. P. Heuck, B. D. Hamman, J. Rossjohn, M. W. Parker, K. R. Ryan, A. E. Johnson, and R. K. Tweten. 1998. Identification of a membrane-spanning domain of the thiol-activated pore-forming toxin Clostridium perfringens perfringolysin O: an α-helical to β-sheet transition identified by fluorescence spectroscopy. Biochemistry 3714563-14574. [DOI] [PubMed] [Google Scholar]
- 49.Simon, L. D., and T. F. Anderson. 1967. The infection of Escherichia coli by T2 and T4 bacteriophages as seen in the electron microscope. I. Attachment and penetration. Virology 32279-297. [DOI] [PubMed] [Google Scholar]
- 50.Song, L., M. R. Hobaugh, C. Shustak, S. Cheley, H. Bayley, and J. E. Gouaux. 1996. Structure of staphylococcal α-hemolysin, a heptameric transmembrane pore. Science 2741859-1866. [DOI] [PubMed] [Google Scholar]
- 51.Sturzenbecker, L. J., M. L. Nibert, D. B. Furlong, and B. N. Fields. 1987. Intracellular digestion of reovirus particles requires a low pH and is an essential step in the viral infectious cycle. J. Virol. 612351-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tao, Y., D. L. Farsetta, M. L. Nibert, and S. C. Harrison. 2002. RNA synthesis in a cage—structural studies of reovirus polymerase λ3. Cell 111733-745. [DOI] [PubMed] [Google Scholar]
- 53.Toumadje, A., S. W. Alcorn, and W. C. Johnson, Jr. 1992. Extending CD spectra of proteins to 168 nm improves the analysis of secondary structures. Anal. Biochem. 200321-331. [DOI] [PubMed] [Google Scholar]
- 54.Tuthill, T. J., D. Bubeck, D. J. Rowlands, and J. M. Hogle. 2006. Characterization of early steps in the poliovirus infection process: receptor-decorated liposomes induce conversion of the virus to membrane-anchored entry-intermediate particles. J. Virol. 80172-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Virgin, H. W., IV, R. Bassel-Duby, and K. L. Tyler. 1988. Antibody protects against lethal infection with the neurally spreading reovirus type 3 (Dearing). J. Virol. 624594-4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wessner, D. R., and B. N. Fields. 1993. Isolation and genetic characterization of ethanol-resistant reovirus mutants. J. Virol. 672442-2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yeager, M., J. A. Berriman, T. S. Baker, and A. R. Bellamy. 1994. Three-dimensional structure of the rotavirus haemagglutinin VP4 by cryo-electron microscopy and difference map analysis. EMBO J. 131011-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang, L., K. Chandran, M. L. Nibert, and S. C. Harrison. 2006. The reovirus μ1 structural rearrangement that mediates membrane penetration. J. Virol. 8012367-12376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang, X., E. Settembre, C. Xu, P. R. Dormitzer, R. Bellamy, S. C. Harrison, and N. Grigorieff. 2008. Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proc. Natl. Acad. Sci. USA 1051867-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang, X., Y. Ji, L. Zhang, S. C. Harrison, D. C. Marinescu, M. L. Nibert, and T. S. Baker. 2005. Features of reovirus outer capsid protein μ1 revealed by electron cryomicroscopy and image reconstruction of the virion at 7.0 Å resolution. Structure 131545-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]