Abstract
Apart from its regulatory role in protease (PR) activation, little is known about the function of the human immunodeficiency virus type 1 transframe protein p6* in the virus life cycle. p6* is located between the nucleocapsid and PR domains in the Gag-Pol polyprotein precursor and is cleaved by PR during viral maturation. We have recently reported that the central region of p6* can be extensively mutated without abolishing viral infectivity and replication in vitro. However, mutagenesis of the entire p6*-coding sequence in the proviral context is not feasible without affecting the superimposed frameshift signal or the overlapping p1-p6gag sequences. To overcome these limitations, we created a novel NL4-3-derived provirus by displacing the original frameshift signal to the 3′ end of the gag gene, thereby uncoupling the p6* gene sequence from the p1-p6gag reading frame. The resulting virus (AL) proved to be replication competent in different cell cultures and thus represents an elegant tool for detailed analysis of p6* function. Hence, extensive deletions or substitutions were introduced into the p6* gene sequence of the AL provirus, and effects on particle release, protein processing, and viral infectivity were evaluated. Interestingly, neither the deletion of 63% of all p6* residues nor the partial substitution by a heterologous sequence affected virus growth and infectivity, suggesting that p6* is widely dispensable for viral in vitro replication. However, the insertion of a larger reporter sequence interfered with virus production and maturation, implying that the length or conformation of this spacer region might be critical for p6* function.
The transframe protein p6*, also referred to as TFP-p6pol, is one of the least-characterized gene products in human immunodeficiency virus type 1 (HIV-1), consistently raising controversial discussions on its main function in the viral life cycle. p6* is located at the amino terminus of the Pol moiety within the Gag-Pol precursor, which is synthesized following a programmed ribosomal −1 frameshift during translation (Fig. 1). In contrast, the Gag precursor is generated by conventional translation at a 20-fold-higher rate (summarized in reference 13). The characterization of recombinant p6* protein by nuclear magnetic resonance analysis revealed a highly flexible structure (3), suggesting that p6* might serve as an adjustable hinge within the large Gag-Pol precursor to facilitate folding of the bulky protein domains during viral assembly. Proper folding of the embedded homodimeric protease (PR) is an essential prerequisite for full activity following sequential autocleavage from the full-length Gag-Pol polyprotein (summarized in reference 13). Indeed, the p6* residues have been demonstrated to stabilize the PR dimer and dictate the folding propensities of PR precursors, thereby influencing the rate of PR maturation (6, 10, 21). Interestingly, the autoprocessing of a truncated Gag-PR precursor was clearly accelerated when the p6* sequence was deleted, leading to the proposal that the active site of the PR might be less accessible in the presence of p6* (26). Further observations that p6* itself is sequentially cleaved by the PR (1, 7, 22, 23, 24, 30, 31, 32, 41) strengthened the hypothesis that the transframe protein contributes to spatiotemporal activation of the PR. Indeed, we and others have previously shown that the PR needs to be released from the flanking p6* residues to be capable of completing all virus-associated processing steps (24, 27, 36). The potential of p6* to regulate PR activity has been further corroborated by our finding that recombinant p6* protein comprising a free accessible carboxyl terminus is a potent inhibitor of the mature PR in vitro (28). Apart from the carboxyl-terminal tetrapeptide mainly contributing to the observed inhibition, the amino-terminal octapeptide of p6* (TFP) has been reported to inhibit PR activity in vitro (21).
FIG. 1.
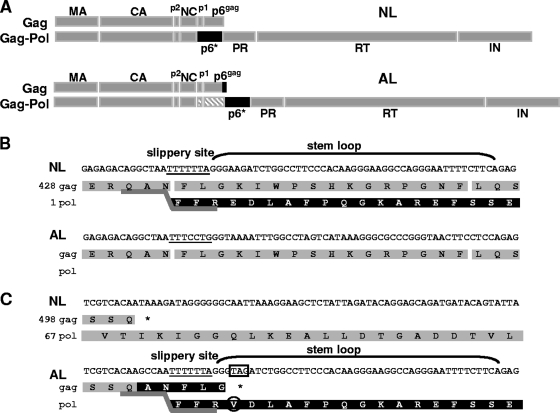
Gag and Gag-Pol polyproteins encoded by NL4-3 (NL) and AL proviruses. (A) Schematic representation of the Gag and Gag-Pol polyprotein precursors. The p6* domain within Gag-Pol is highlighted in black, and the p1-p6gag region inserted in the Gag-Pol precursor of AL is shaded gray; the five residues appended to p6gag in Gag are indicated by a black vertical line. MA, matrix protein; NC, nucleocapsid protein; IN, integrase. (B) The Gag (gray) and Pol (black) products translated in the presence of the original frameshift site in NL are indicated on top, and the corresponding passage synthesized in AL after destruction of the slippery site is shown below. The active (NL) and inactive (AL) slippery sites are underlined, and the 3′ nucleotides contributing to the stem-loop structure are in parentheses. (C) An artificial frameshift site was introduced at the 3′ end of the gag gene in AL, resulting in the synthesis of a Gag-Pol polyprotein with p6* directly fused to p6gag. The gag reading frame in AL is terminated by an artificial stop codon (boxed) and is extended by the residues ANFLG. The E4→V4 substitution in p6* is marked by a circle, and numbers at the left indicate amino acid positions within Gag and Gag-Pol with respect to the Gag start codon.
The fact that p6* has a substantial size of 55 to 72 amino acids, depending on the isolate, raises the question of whether the transframe protein exerts other functions apart from PR regulation. In this regard, p6* has also been proposed to be a possible binding partner of the viral Nef protein, the major pathogenicity factor during HIV infection (9). However, the target site within p6* for Nef interaction has not been mapped so far.
Although it is completely overlapped by the p1-p6gag domains, the p6* sequence offers much room for natural polymorphisms (5). Furthermore, amino acid insertions or duplications, as well as deletions of up to 13 residues, in p6* sequences of infectious isolates have been reported, some of which are associated with viral drug resistance (4, 29, 35, 38). We have recently shown that nonconservative substitutions of up to 70% of the p6* residues in a provirus clone did not abolish viral growth or infectivity in various cell lines, suggesting that the central p6* region is widely dispensable for viral in vitro replication (19, 27).
Unlike the variable center, the p6* amino terminus is highly conserved owing to the overlapping p1gag sequence (14) and the RNA frameshift signal composed of a heptanucleotide slippery site (UUUUUUA) and a 3′ pseudoknot structure (11). This complex sequence context imposes major restrictions on mutational analysis of the p6* amino terminus in the viral background (24). To allow for a more in-depth functional analysis of the conserved p6* residues in the viral context, we have established a novel NL4-3-based provirus with p1-p6gag and p6* reading frames uncoupled via a dislocated frameshift site. Thereby, we could demonstrate that neither the deletion of the bulk of p6* nor the insertion of a short unrelated sequence significantly affected replication and infectivity of the virus in different cell cultures. However, the insertion of a five times larger green fluorescent protein (GFP)-encoding sequence into the p6* reading frame was associated with a clear loss of particle production and infectivity. Therefore, we conclude that p6* is a spacer protein of limited length which is widely dispensable for viral replication in vitro.
MATERIALS AND METHODS
Generation of provirus AL and AL-derived p6* mutants.
Proviruses AL and AL Nef− were generated from a recombinant NL4-3-derived virus clone (34) and a derivative thereof containing a deficient nef reading frame by replacing the ApaI-BclI fragments (nucleotide positions 1217 to 1645 with respect to Gag ATG, comprising the complete p1-p6gag gene region of the gag reading frame and the p6* gene plus two-thirds of the PR-encoding sequence of the superimposed pol reading frame) with a synthetically produced fragment, thereby destroying the original slippery site UUUUUUA at position 1296 and the 3′ stem-loop structure by point mutations to abrogate frameshifting. In lieu thereof, the complete functional frameshift signal, including the slippery site and stem-loop, was introduced at position 1501 5′ to the Gag stop codon. In the resulting provirus AL, the gag and pol genes were extended by 15 and 210 nucleotides, respectively, compared to those of the parent NL4-3 provirus clone. AL derivatives containing amino-terminal p6* substitutions (ALn), central deletions (ALΔ), or insertions (ALΔ-myc) in the truncated central p6* sequence were generated by replacing the ApaI-BclI fragment of AL with corresponding synthetic gene fragments. All synthetic genes were purchased from Geneart AG (Regensburg, Germany), and the entire ApaI-BclI region of the resulting provirus clones was verified by sequencing with specific primers. For generation of ALΔ-GFP, the GFP-encoding sequence from the vector pc-huGFP (12) (GenBank accession number AX511259) was PCR amplified and inserted into the ALΔ-myc construct via single RsrII and SnaBI restriction sites, thereby replacing the 30-bp myc sequence.
Generation of frameshift reporter constructs.
The firefly luciferase gene of pGL2-Control vector (Promega) was extended 5′ by the corresponding frameshift regions of the various provirus constructs, resulting in luciferase reporter genes fs-NL (original NL4-3-derived frameshift site at position 1296), fs-inact (inactivated frameshift site of AL at position 1296), fs-AL (artificial frameshift site of AL at position 1501), and fs-ALn and fs-ALΔ (comprising substitutions in the stem-loop region) allowing for frameshift-dependent expression of luciferase from the −1 frame. All frameshift regions were generated by Geneart AG and were inserted into pGL2-Control via HindIII and XbaI restriction sites. An additional reporter construct, fs-luc, with a destroyed slippery site driving constitutive luciferase expression from the −1 frame was generated accordingly.
Cell culture.
Adherent 293T human kidney epithelial and TZM-bl HeLa indicator cells (NIH AIDS Research & Reference Reagent Program) were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 100 U/ml penicillin, and 100 μg/ml streptomycin. Peripheral blood mononuclear cells (PBMCs) were isolated from healthy human donors by Pancoll (Pan Biotech GmbH, Aidenbach, Germany) density gradient centrifugation of whole blood (950 × g, 30 min) and were stimulated with phytohemagglutinin (0.5 μl/ml) for 2 days. PBMCs and the permissive human T-cell lines CEM and MT-4 (18) were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum, 100 U/ml penicillin, and 100 μg/ml streptomycin.
Virus production and CA quantification.
For analysis of virus production and infection experiments, 5 × 106 293T cells were transiently transfected with 20 μg of the various provirus plasmids by using polyethylenimine (PolySciences, Inc.) according to the manufacturer's instructions. To inhibit the viral PR, Saquinavir (Roche Diagnostics) was added to the medium at 4 h posttransfection at a final concentration of 10 μM. Virus-containing supernatants were collected after 48 or 72 h, filtered through a 0.45-μm-pore-size filter, and quantified for total amounts of particle-associated capsid (CA) protein by enzyme-linked immunosorbent assay (ELISA) by using CA-specific antibodies from Polymun (Vienna, Austria) as recently described in detail (24).
To determine the amount of precursor-associated CA proteins, pelleted particles from culture supernatants of transfected 293T cells were resuspended in phosphate-buffered saline and CA protein amounts were quantified by ELISA. Particles were then diluted 1:50 in PR assay buffer (100 mM Na acetate [pH 5.0], 0.5 M NaCl, 4 mM EDTA, 5 mM dithiothreitol, 0.1% Triton X-100 [wt/vol]), solubilized by vortexing, and subsequently incubated with 250 nM of recombinant HIV-1 PR dimer (Bachem) for 2 h at 37°C. CA amounts in PR-treated samples were quantified by ELISA and compared to the values obtained prior to PR treatment.
Analysis of virus proteins by Western blotting.
Transfected 293T cells and extracellular particles in culture supernatants were harvested at 24, 48, or 72 h posttransfection as specified in Results and were prepared as described previously (27). The total amount of protein within cell lysates and particle preparations was determined using the Bio-Rad protein assay (Munich, Germany) according to the manufacturer's protocol, and equal amounts of total protein were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose. Viral proteins were visualized using the CA-specific monoclonal antibody 13/5 (39), the PR-specific sheep antiserum ARP 413 (D. Bailey and M. Page, NIH AIDS Research & Reference Reagent Program), the reverse transcriptase (RT)-specific mouse ascites 4F8 (P. Chandra, Gustav-Embden-Center of Biological Chemistry, University of Frankfurt, Germany), the c-myc-specific monoclonal antibody 9E10 (Roche Diagnostics), and the Nef-specific sheep antiserum ARP 444 (MRC AIDS Reagent Project) together with horseradish peroxidase-conjugated antibodies, followed by enhanced chemiluminescence detection (SuperSignal; Pierce Chemical, Rockford, IL).
Infectivity assay.
The adherent TZM-bl cell line expresses the β-galactosidase and firefly luciferase reporter genes under the control of the HIV-1 long terminal repeat promoter. To determine virus infectivity on this cell line, supernatants of transfected 293T cells were filtered through a 0.45-μm-pore-size filter and analyzed for total amounts of particle-associated CA antigen by ELISA. A total of 1.3 × 104 TZM-bl cells were seeded in 96-well plates and infected 24 h later with different dilutions of the virus-containing supernatants. At 48 hours after infection, cells were fixed and stained with the β-galactosidase substrate X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) for 4 h, and blue cell nuclei were counted. Alternatively, infected cells were lysed in luciferase lysis buffer (luciferase assay; Promega), and luciferase activity was quantified in a Lumat 9501 luminometer (Berthold, Bad Wildbach, Germany) following addition of luciferin according to the manufacturer's instructions.
Analysis of virus replication.
Virus-containing supernatants from transfected 293T cells were normalized for total CA protein content by ELISA, and particle amounts corresponding to 250 ng CA protein were used to infect 1 million logarithmically growing CEM or MT-4 cells in 1 ml medium at 37°C. After 6 h, infected cells were washed to remove residual virus, resuspended in 4 ml culture medium, and then further incubated at 37°C. Likewise, PBMCs were infected with 50 ng CA protein equivalents and the medium was supplemented with 20 U/ml interleukin-2. Cultures were diluted with 1 volume of fresh medium every 48 h, and samples were collected to monitor virus growth via quantification of CA amounts in the culture supernatants over a period of 2 weeks.
Frameshift luciferase reporter assay.
A total of 5 × 106 293T cells were seeded and 16 h later transiently transfected with 20 μg of the luciferase reporter constructs by using polyethylenimine according to the manufacturer's protocol. At 48 hours posttransfection, cells were harvested and analyzed for luciferase activity with the luciferase assay from Promega as described previously (27) after adequate dilution of the samples.
RESULTS
Design of an NL4-3-derived HIV-1 provirus with a displaced ribosomal frameshift site.
Mutational analysis of p6* in the viral context is restricted by (i) the RNA frameshift signal at the 5′ end of the pol gene inducing Gag-Pol synthesis and (ii) the overlapping p1 and p6gag reading frame. To allow for independent and specific mutagenesis of the p6* domain, we constructed a derivative of the NL4-3 provirus (NL) with uncoupled gag and pol reading frames. For this purpose, the original slippery site located at the respective 5′ ends of the p1 and p6* genes and the adjacent stem-loop structure were inactivated via point mutations without altering the p1 amino acid sequence, thereby abolishing ribosomal frameshifting from the gag to the pol reading frame (Fig. 1A). Instead, a novel frameshift signal composed of the heptanucleotide sequence (TTTTTTA) and the entire nucleotide stretch building the contiguous RNA pseudoknot structure was introduced at the 3′ end of the p6gag gene. Because translation of the Pol domains was now expected to start from the 3′ end of the p6gag gene, the resulting Gag-Pol polyprotein would be extended by the 7.8-kDa p1-p6gag domains directly fused to the downstream p6* protein, which is normally attached to the NC domain in the wild-type precursor (Fig. 1A). Furthermore, five additional amino acids (Ala-Asn-Phe-Leu-Gly) are added to the carboxyl terminus of the p6gag protein in the Gag precursor encoded by the inserted frameshift region. In addition, the artificial frameshift signal brings along a stop codon to terminate p6gag translation, which causes a Glu4→Val4 substitution in p6* (Fig. 1C). To allow for correct processing of the modified Gag-Pol precursor by the viral PR, the original NC-p6* PR cleavage site (QAN/FFR) was reestablished at the novel p6gag-p6* junction in Gag-Pol. A schematic representation of the gag and pol sequences modified to obtain the altered NL provirus (AL) is given in Fig. 1B and C.
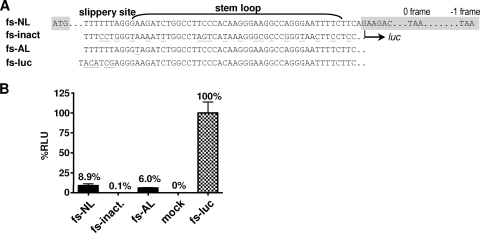
To prove whether (i) the original frameshift site was inactivated by modifications in AL and (ii) the novel displaced frameshift signal was functional, we performed a frameshift reporter assay (24, 27). Therefore, the original NL-derived frameshift signal comprising the slippery sequence and the 3′ stem-loop region was cloned 5′ of a firefly luciferase gene (fs-NL) allowing for synthesis of luciferase exclusively from the −1 frame, whereas translation is preterminated at a stop codon in the 0 frame (Fig. 2A). For comparison, the luciferase gene was fused to the corresponding AL-derived sequences representing either the inactivated frameshift site (fs-inact) or the dislocated active signal (fs-AL), the latter comprising a single nucleotide substitution in the lower stem. As a positive control, we used a reporter harboring a modified slippery site to trigger constitutive expression of the luciferase from the −1 frame (fs-luc). The various reporter constructs were then used to transfect 293T cells, and frameshift efficiencies of the respective reporter genes were evaluated via quantification of cell-associated luciferase activity (Fig. 2B). As expected, luciferase activity was at the background level (mock) in cells transfected with fs-inact, confirming that the original frameshift signal had been successfully destroyed. In contrast, functional luciferase was produced from the fs-AL construct, corresponding to a frameshift efficiency of 6%, which was, however, slightly lower than the frameshift rate of 8.9% determined for the wild-type fs-NL sequence.
FIG. 2.
Functional analysis of the modified frameshift sites in a luciferase reporter assay. (A) The nucleotide sequence representing the original NL4-3-derived frameshift site (fs-NL), the inactivated frameshift site (fs-inact), and the artificial frameshift site (fs-AL) in provirus AL were cloned 5′ of a firefly luciferase reporter gene to allow for frameshift-dependent expression of luciferase from the −1 frame. The luciferase reading frame is depicted in gray, and nucleotide positions altered with respect to the original frameshift site are underlined. An additional construct with a modified slippery sequence driving frameshift-independent reporter synthesis from the −1 frame was used as positive control (fs-luc). (B) 293T cells were transiently transfected with the indicated reporter constructs or an empty vector (mock), and cell-associated luciferase activity was quantified after 48 h. Luciferase activity (relative light units [RLU]) determined for fs-luc was set to 100%, and all other values were related accordingly. Results from one representative blot comprising mean values and standard deviations from three independent transfection experiments are shown.
AL-derived virions harbor extended Gag-Pol precursors and are capable of productive replication in various cell lines.
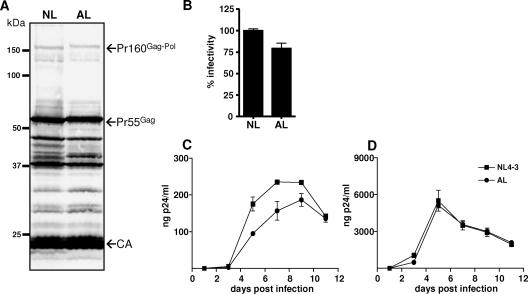
Because the original provirus sequence was extensively modified in order to separate the gag and pol reading frames, we first asked to what extent viral protein synthesis and particle production might be affected by the corresponding mutations. For this purpose, 293T cells were transiently transfected with NL and AL provirus constructs, and released virus particles were harvested from supernatants at 72 h posttransfection and subjected to Western blot analysis with a CA-directed antibody. As illustrated in Fig. 3A, comparable amounts of Gag-specific products were found in both particle preparations, with the mature CA protein representing the dominant species. Furthermore, specific signals corresponding to molecular masses of approximately 160 kDa and 55 kDa were detectable in both virus samples, indicating full-length production and packaging of viral Gag-Pol and Gag polyprotein precursors. In accordance with our expectations, the band likely representing the Gag-Pol precursor was slightly shifted in AL particles owing to translation of the artificial 7.8-kDa p1-p6gag region. Likewise, the extension of the p6gag carboxyl terminus by five additional amino acids (0.5 kDa) was nicely reflected by a slight shift of the AL-specific Pr55Gag precursor. The modifications in AL also seemed to affect the processing order of Gag or Gag-Pol-related intermediates, as suggested by the deviating signals in the range of 37 to 50 kDa. Nevertheless, these observations strongly argue for the functionality of the displaced frameshift signal in the provirus AL, resulting in viral Gag-Pol precursors with rearranged protein domains. Together, these findings suggest that virus production and packaging and maturation of the precursor proteins were equally efficient in NL and AL species.
FIG. 3.
Influence of frameshift displacement on virus maturation, infectivity, and replication. (A) 293T cells were transiently transfected with the indicated viral plasmids. For analysis of virus composition, particles released into cell supernatants were harvested at 72 h posttransfection and sedimented through a 20% sucrose cushion. Viral proteins were separated by SDS-PAGE and analyzed by Western blotting using a monoclonal CA-specific antibody. Positions of molecular mass markers are shown at the left, and arrows at the right indicate Gag-specific protein bands. (B) To evaluate viral infectivity, virus-containing supernatants from 293T cells were harvested at 72 h posttransfection, normalized for CA content by ELISA, and used to infect CD4-positive TZM-bl indicator cells. At 48 hours postinfection, blue cells were counted and the values obtained were related to NL infections (100%). Error bars indicate the standard deviations from triplicate infections. (C and D) To monitor viral replication kinetics, PBMCs (C) and MT4 cells (D) were infected in duplicate with the virus-containing supernatants, and spreading infections were monitored by quantification of CA amounts in culture supernatants by ELISA. Results from one representative experiment are shown, with error bars indicating the standard deviations for duplicate infections.
To evaluate whether the novel virus was capable of infecting cultured cells, NL and AL particles released from transfected 293T cells were normalized by CA-specific ELISA and were used to infect TZM-bl indicator cells containing β-galactosidase and luciferase reporter genes under the control of a long terminal repeat promoter. As summarized in Fig. 3B, the infectivity of AL particles, reflected by the number of β-galactosidase-positive cells, was slightly decreased to approximately 80% of NL wild-type infectivity. Read-out based on luciferase activity yielded comparable results (data not shown), indicating that the reduced infectivity of AL might be due to compromised frameshifting or to the rearranged protein domains.
To further characterize the AL phenotype in spreading infections, we compared the growth rates of AL and NL viruses in permissive cells. Thus, PBMCs and MT4 lymphocytes were infected with CA-normalized amounts of virus particles, and viral replication was monitored by quantification of CA amounts in the culture supernatants over a period of 2 weeks. Whereas AL-specific replication curves were slightly delayed compared to NL growth and reached lower maximum titers in PBMCs (Fig. 3C), both viruses replicated equally efficiently in MT4 cells (Fig. 3D), suggesting that the impact of the AL-specific sequence modifications on viral fitness depended on the cell culture system employed. In sum, these data provide evidence that the NL derivative AL is an infectious and replication-competent virus mutant representing a suitable tool for further mutagenesis approaches.
Construction of AL-based mutants with extended modifications in the p6* reading frame.
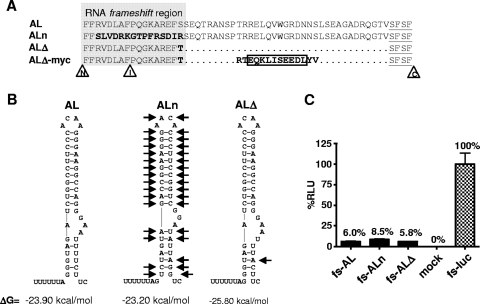
On the basis of the uncoupled gag and pol reading frames, the AL provirus offered new possibilities to specifically modify the p6* sequence at so-far inaccessible sites. Hence, a first mutant was constructed to alter the amino-terminal region of p6* (Phe1-Ser17) (Fig. 4A), which is no longer overlapped by the p1-p6gag-encoding domains. However, to preserve the functionality of the still-underlying frameshift signal, residues Phe1-Phe2 were maintained and the nucleotides building the RNA pseudoknot were mirrored, thereby creating a novel stem-loop with a predicted thermodynamic stability (ΔG value) similar to that of the original structure (Fig. 4B). As a consequence of this nucleotide inversion, residues Arg3-Ser17 were completely replaced in p6*, yielding mutant ALn (Fig. 4A).
FIG. 4.
Influence of p6*-specific mutations in AL on frameshift efficiency. (A) On the basis of provirus AL, further modifications were introduced into the p6* sequence, resulting in virus mutants ALn (substitution of the amino-terminal residues in boldface), ALΔ (deletion of the central p6* sequence indicated by dots), and ALΔ-myc (carrying a 10-amino-acid myc tag instead of the central p6* region [indicated by a box] and flanking substitutions in p6* [boldface]). Those amino acids overlapping the RNA region involved in frameshifting are highlighted by a gray box, and p6* residues contributing to carboxyl-terminal p6*-PR cleavage are underlined. Amino-terminal (N), internal (I), and carboxyl-terminal (C) cleavage sites in p6* are indicated by arrowheads. (B) Modifications in the stem-loop regions of ALn and ALΔ compared to the AL-specific structure are indicated by black arrows. Stem-loop structures were predicted as described by Zuker and coworkers (37, 40), with the calculated free energy values (ΔG) shown below. (C) Functional analysis of frameshift efficiency in a luciferase reporter assay. The frameshift regions of the indicated AL derivatives were cloned 5′of a firefly luciferase reporter gene to allow for frameshift-dependent expression of luciferase from the −1 frame as illustrated in Fig. 2A. The corresponding reporter constructs fs-AL, fs-ALn, and fs-ALΔ; the control vector fs-luc; or an empty vector (mock) was used to transfect 293T cells, and cell-associated luciferase activity was quantified after 48 h. Luciferase activity (relative light units [RLU]) determined for fs-luc was set to 100%, and all other values were related accordingly. Results for one representative blot comprising mean values and standard deviations from three independent transfection experiments are shown.
With a second modification, we aimed to investigate whether a severe truncation of the p6* region affects viral precursor processing and replication competence. For this purpose, a provirus construct lacking the majority of the p6* residues (Ser18→Val52) was designed. The resulting provirus ALΔ was expected to express only the amino-terminal 17 residues (with an additional Ser17→Thr17 mutation) directly fused to the carboxyl-terminal tetrapeptide, thus encoding only 21 of the total 56 amino acids of p6*.
Finally, a third mutant was generated to analyze the influence of short unrelated spacer sequences within the truncated p6* region. To allow at the same time specific detection of the inserted sequence, a 10-amino-acid encoding a c-myc tag (myc) was inserted to replace part of the deleted p6* sequence, resulting in construct ALΔ-myc (Fig. 4A) In addition, two p6* residues flanking the myc sequence at either site (RT-myc-YV) were modified to introduce single restriction sites. In order to guarantee efficient cleavage of the viral PR from the modified p6* proteins, the carboxyl-terminal tetrapeptide SFSF remained unaltered in all mutants.
To provide evidence that the inverted stem-loop in mutant ALn (Fig. 4B) supported ribosomal pausing, we generated luciferase reporter constructs containing the frameshift regions of mutants ALn (fs-ALn) and ALΔ (fs-ALΔ) and performed the luciferase reporter assay as described above (Fig. 2). As depicted in Fig. 4C, the single point mutation in fs-ALΔ did not significantly alter frameshift efficiency compared to the parent fs-AL construct, although it did rearrange base pairing in the lower stem. Interestingly, the inversion of the stem-loop structure even enhanced the frameshift rate to 8.5%, which corresponds to the value obtained for the wild-type NL-derived frameshift site (Fig. 2B). Taken together, these results suggest that the modified frameshift signals are capable of driving efficient Gag-Pol polyprotein synthesis.
The p6* sequence is not essential for in vitro viral replication and infectivity.
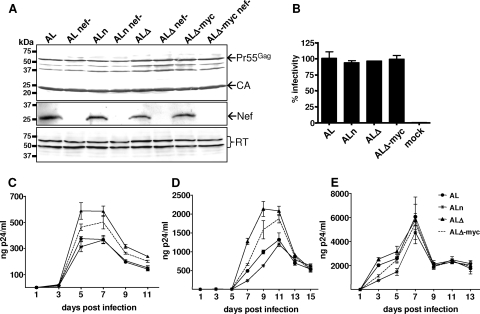
To assess the impact of the extensive p6* mutations on virus production and processing of viral precursor proteins, 293T cells were transiently transfected with the various provirus mutants. In addition, derivatives of the AL provirus species containing a nef-deficient reading frame (Nef−) were tested to detect possible effects of the p6* mutations on the proposed interaction of Nef and p6* (9). Virus particles released from transfected cells were harvested, and the protein content was analyzed with CA-, RT-, and Nef-specific antibodies. Western blot analysis (Fig. 5A) revealed comparable compositions of all virus preparations with readily processed CA and RT products, indicating that neither virus production nor PR-mediated maturation of the precursor proteins was affected by the modifications in p6*. A clear signal corresponding to a molecular size of approximately 27 kDa detected with a Nef-specific antibody in all nef-expressing virus mutants further indicates that packaging of the viral Nef protein was not affected in either mutant.
FIG. 5.
Characterization of particle composition, infectivity, and replication of the AL-derived p6* mutants. (A) 293T cells were transiently transfected with the indicated proviral plasmids, and particles released into cell supernatants were harvested at 72 h posttransfection and sedimented through a 20% sucrose cushion. Particle-associated proteins were separated by SDS-PAGE and analyzed by Western blotting using CA-, Nef-, and RT-specific antisera. Positions of molecular mass markers are shown at the left, and arrows at the right indicate specific protein bands. (B) To evaluate viral infectivity, virus-containing supernatants from 293T cells were harvested at 72 h posttransfection, normalized for CA content by ELISA, and used to infect CD4-positive TZM-bl indicator cells. At 48 hours postinfection, blue cells were counted and the values obtained were related to AL infections (100%). Error bars indicate the standard deviations from triplicate infections. To analyze viral replication kinetics, PBMCs (C), CEM cells (D), and MT-4 cells (E) were infected in duplicate with the virus-containing supernatants, and spreading infections were monitored by quantification of CA amounts in culture supernatants by ELISA. Results from one representative experiment are shown, with error bars indicating the standard deviations from duplicate infections.
Next, the capacities of the different AL virus species to infect cultured cells were compared in a TZM-bl reporter assay as described above. The results indicate that the mutant ALn containing the amino-terminal substitutions as well as the p6*-truncated species ALΔ and ALΔ-myc were as efficient as the parent AL virus in infecting TZM-bl indicator cells (Fig. 5B).
Finally, the panel of virus mutants was tested for replication competence in different permissive cell systems. To monitor the individual growth curves, PBMCs, CEM cells, and MT4 cells were infected with normalized amounts of the various mutants, and the CA content in the culture supernatants was quantified over a period of 2 weeks. It is noteworthy that all mutants performed similarly to AL viruses in MT-4 cells (Fig. 5E), whereas clear differences in virus production were observed in PBMC (Fig. 5C) and CEM (Fig. 5D) cultures. In the latter systems, those variants carrying the truncated p6* sequence (ALΔ) reached approximately 60% higher maximum titers than the parent AL virus, whereas AL-myc titers where still increased by 20 to 40% in both cell cultures compared to AL and ALn viruses, both of which showed similar replication profiles. The observation that particle production was most successful in the presence of the shortest p6* sequence in two out of three tested cell systems led us to speculate that p6* is widely dispensable for viral replication. These findings further imply that the length and context of the native p6* sequence are mainly determined by the overlapping Gag domains and the frameshift signal. This assumption was further substantiated by the finding that no significant effects on viral replication associated with ALn-specific substitutions were observed in either cell line, suggesting that the amino-terminal region in p6* exerts no specific function in the HIV life cycle.
Heterologous reporter sequences expressed from the p6* reading frame are incorporated into virus particles.
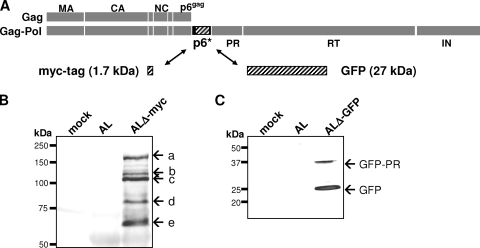
The experiments described above have shown that AL might serve as a platform for heterologous protein expression within the p6* reading frame, as the ALΔ-myc mutant is capable of producing infectious viruses which replicate to high titers in various cell lines. As we found it intriguing to develop a replication-competent reporter virus by replacing p6* with a reporter sequence, we further asked what maximum sequence length would be tolerated within the p6* reading frame. To address this issue, a widely established reporter gene encoding the easily detectable GFP, which is approximately five times larger than the full-length p6* sequence (789 versus 168 bp), was inserted into the central p6* region, thereby adding another 27 kDa to the resulting Gag-Pol polyprotein (Fig. 6A). To test whether the myc and GFP reporter sequences were correctly expressed from the p6* reading frame and subsequently packaged into budding particles, 293T cells were transfected with AL, ALΔ-myc, or ALΔ-GFP constructs and released virions were analyzed by Western blotting with myc- and GFP-specific antibodies. Correct processing of the ALΔ-myc Gag-Pol precursor proteins at the internal (I) and carboxyl-terminal (C) cleavage sites of p6* was expected to result in a 3.2-kDa myc product flanked by p6* residues (Fig. 4A). Since we had difficulties in detecting this small peptide (PQGKAREFTRT-myc-YVSFSF) in virus particles, transfections were performed in the presence of the PR inhibitor Saquinavir to reveal precursor-associated myc. In fact, we found several myc-specific signals in ALΔ-myc particles with molecular masses ranging between 60 and 170 kDa (Fig. 6B, bands a to e), indicating that the myc insert was part of the sequentially cleaved, virus-associated Gag-Pol precursors (Fig. 6B) (20, 31, 32).
FIG. 6.
Packaging of heterologous sequences expressed from the p6* reading frame. (A) A schematic representation of the AL Gag and Gag-Pol precursors is shown, with the p6* central region highlighted by a shaded box. A myc tag and a GFP reporter gene were inserted into the p6* reading frame, yielding ALΔ-myc and ALΔ-GFP provirus constructs. (B) 293T cells were transfected with AL-, ALΔ-myc, or an empty vector (mock) in the presence of the PR inhibitor Saquinavir, and myc-specific products (a to d) were detected in particles released into culture supernatants at 48 h posttransfection by Western blotting. (C) 293T cells were transfected the indicated constructs, and particles were analyzed with a GFP-specific antibody. GFP-specific products are indicated by arrows at the right.
Likewise, a specific signal corresponding to a molecular mass of approximately 27 kDa was detected in ALΔ-GFP particles (Fig. 6C), suggesting that GFP was also properly expressed from the p6* reading frame and incorporated into released virions. However, the presence of a larger GFP-specific product likely representing a GFP-PR intermediate indicated that GFP was not quantitatively released from the Gag-Pol precursor. Nevertheless, the above data suggest that AL might serve as a new tool to channel heterologous reporter proteins into virus particles.
Large p6* insertions interfere with the production of infectious virus.
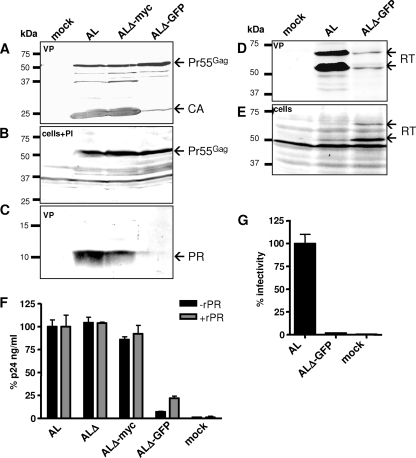
Since GFP was expressed from the p6* reading frame and successfully packaged into budding particles, we next aimed to clarify whether this large insertion affected the maturation of infectious viruses. Whereas comparable Gag processing patterns were observed in AL and ALΔ-myc particles (Fig. 7A and 5A), ALΔ-GFP virions contained significantly diminished amounts of mature CA protein (Fig. 7A), pointing to a severe processing defect or inefficient virus release. To exclude possible effects of the GFP insertion on cell-associated protein expression, we analyzed transfected cells in the presence of Saquinavir, thereby providing evidence that equal amounts of Pr55Gag precursor proteins were produced from all proviruses (Fig. 7B). However, only small amounts of mature PR (Fig. 7C) and RT (Fig. 7D) proteins could be detected in ALΔ-GFP virions, suggesting inefficient packaging of the Pol proteins in the presence of the 27-kDa GFP spacer sequence. This assumption is supported by the substantial amounts of RT proteins detected in ALΔ-GFP-transfected cells after the bulk of particles had been released (Fig. 7E).
FIG. 7.
Influence of p6* insertions on viral maturation and infectivity. 293T cells were transfected with the indicated provirus constructs or an empty vector (mock), and virus-containing supernatants (VP) (A, C, and D) or cells (B and E) were harvested at 48 h posttransfection and analyzed by Western blotting using PR-, CA-, or RT-specific antisera. (B) Cells were transfected in the presence of the PR inhibitor Saquinavir (PI) and were harvested at 24 h posttransfection. Positions of molecular mass markers are shown at the left, and arrows at the right indicate specific protein bands. (F) Virus particles released into cell supernatants were harvested at 72 h posttransfection and sedimented through a 20% sucrose cushion. To determine precursor-associated CA amounts, virus preparations were quantified for CA content by ELISA prior to (black bars) or after (gray bars) processing with an excess of HIV-1 rPR. (G) To measure viral infectivity, virus-containing supernatants from 293T cells were harvested at 72 h posttransfection, normalized for CA content by ELISA, and used to infect CD4-positive TZM-bl indicator cells. At 48 hours postinfection, blue cells were counted, and the values obtained were related to AL infections (100%). Error bars indicate the standard deviations from triplicate infections.
To further assess specific effects of the various p6* insertions on virus production, particles released into supernatants of transfected cells were quantified by CA ELISA. Whereas those viruses harboring a truncated p6* region contained AL-like CA amounts, the ALΔ-GFP-specific CA content was significantly reduced to approximately 7% of AL-specific particle preparations (Fig. 7F). Because the ELISA detects primarily the processed CA species, virion preparations were incubated with an excess of recombinant HIV-1 PR (rPR) as recently described (19) to cleave all residual Gag precursor proteins, thereby determining the degree of CA maturation. Following processing in trans, CA amounts were quantified and compared to the values obtained prior to rPR treatment. Whereas the CA contents of AL, ALΔ, and ALΔ-myc samples was not further elevated in the presence of rPR, indicating a high degree of particle maturation, rPR treatment of ALΔ-GFP particles clearly increased total CA amounts by threefold, which were, however, still fourfold lower than all other values. These results demonstrate that the insertion of the large GFP reporter sequence interfered with (i) viral particle production and (ii) quantitative maturation of the Gag proteins. As expected, the reduced production of processing-deficient ALΔ-GFP viruses resulted in a complete loss of infectivity on TZM-bl reporter cells (Fig. 7G). Together, these findings imply that large insertions into the p6* reading frame might interfere with efficient production and maturation of infectious viruses.
DISCUSSION
The p6* transframe domain may be the most variable region in the HIV-1 Gag-Pol precursor, displaying a remarkable degree of naturally occurring and partly drug-associated polymorphisms, including insertions and deletions (2, 4, 15, 16, 35, 38). The so-far-largest natural deletion of 13 amino acids was found in the p6*/p6gag region of the Chinese isolate HIV-1 CRF07_BC and was reported to have no significant effect on viral fitness (35). Likewise, artificial deletions of 20 amino acids in the NL4-3-derived p6*/p6gag region encompassing 36% of the p6* sequence (R28-D47) did not have an impact on autoprocessing of the PR or viral infectivity (4). However, this deletion mutant showed a compromised replication profile on PBMCs which could not be clearly attributed to modifications in p6* or p6gag due to the overlapping reading frames.
Owing to its intricate position, p6* has mainly been studied in vitro by using recombinant proteins or truncated Gag-Pol precursors for mutational analysis (21, 22, 23, 26, 41, 42). However, these studies are often difficult to interpret in terms of structural and kinetic conditions directing precursor maturation, which might differ significantly in a live virus. To allow for a more independent mutagenesis of p6* without leaving the viral background, we developed a novel virus platform by displacing the original frameshift site from the 5′ end of the p6* gene to the 3′ end of the p6gag gene, thereby uncoupling the gag and pol reading frames. Based on this artificial AL virus, which proved to be infectious and replication competent on different cell cultures, we deleted 35 of the 56 p6* residues, which corresponds to 63% of the entire p6* sequence and is the largest p6* deletion in a viral context reported to date. However, on the basis of our latest investigations on the importance of the p6* cleavage sites for PR activation (24), we retained the carboxyl-terminal tetrapeptide of p6* to allow for efficient autorelease of the PR. The resulting virus, ALΔ, showed no deficits with regard to particle maturation, replication, or infectivity. Interestingly, the truncated virus showed an improved replication in some cell cultures, which might either be due to the smaller size of the corresponding provirus or, more likely, be indicative for a faster autoactivation of the PR in the absence of the p6* spacer, as suggested previously (26). In sum, this study together with our former observations (19, 27) contribute to the assumption that the major central part of p6*, which is overlapped and thus widely determined by the p6gag domain, is dispensable for the viral life cycle.
Recently, Chiu and colleagues sought to shed light on p6* function via coexpression of Gag and p6*-deleted Gag-Pol precursors, deleting 60 of the total 66 p6* residues (8). Whereas the large p6* deletion did not affect Gag-Pol incorporation, the resulting virus particles displayed reduced processing of the Gag polyproteins, leading the authors to speculate that the presence of p6* is essential for PR-mediated Gag cleavage. However, it has not been considered whether the leftover p6* residues allowed for autorelease of the PR, as p6* deletion was associated with a destruction of the original PR cleavage sites.
As p6* deletion in the ALΔ mutant was restricted for the sake of a functional frameshift site, we had to adopt a new strategy to analyze the conserved amino terminus of p6*. Hence, mutagenesis of p6* residues R3-S17 was achieved by a complete reversion of the stem-loop structure. Although the novel stem-loop triggered wild-type-like frameshift rates, it was surprising that these severe modifications had no significant effects on the maturation, in vitro replication, and infectivity of the resulting ALn virus. Therefore, we conclude that the amino-terminal p6* residues lack a specific function in the virus life cycle and are determined solely by the overlapping p1gag domain and the underlying frameshift signal.
Although it was not the focus of this study, we analyzed the influence of the p6* truncations or substitutions on Nef incorporation to address the recently proposed interaction of Nef and p6* (9). In fact, none of the modifications in p6* did affect Nef incorporation, nor did they result in a compromised replication, which would have been expected when almost all possible sites in p6* relevant for Nef interaction were destroyed. A possible interaction of Nef with the carboxyl terminus of p6* is also unlikely, since we could not observe any impact on Nef packaging following substitution of the carboxyl-terminal tetrapeptide of p6* (19).
The truncation of a large part of p6* provided room for inserting unrelated sequences, which appeared to be interesting in light of developing a replication-competent reporter virus. Indeed, the insertion of a 10-amino-acid myc tag into the deleted p6* domain allowed for detection of the Gag-Pol-associated myc tag in released virus particles. Notably, the replication efficiency of the myc-tagged virus containing a p6* region of 35 amino acids ranged between the AL and ALΔ growth efficiencies, which points to a direct relation of p6* size and viral fitness.
Whereas the truncation of the natural p6* sequence had a rather positive effect on in vitro viral replication, the insertion of a 240-amino-acid GFP reporter sequence into the deleted p6* reading frame significantly interfered with virus production and protein maturation, although GFP was packaged into released virions and was partly released from the Gag-Pol precursor. Nonetheless, we observed a dramatic reduction in virus-associated Pol proteins in the presence of GFP, which might be due to the bulky conformation of the extended Gag-Pol polyprotein affecting the process of assembly of viral precursor proteins. Since the p6* GFP fusion was five times larger than full-length p6* alone, we inserted another reporter sequence into the truncated p6* reading frame, encoding a protein which extends full-length p6* by only 12 amino acids (data not shown). Surprisingly, even this small elongation decreased viral infectivity by 50%. However, we cannot exclude that these effects were due to the conformation or nature rather than the length of the heterologous insertions.
Interestingly, Müller and colleagues have previously demonstrated that the insertion of a GFP gene between the MA and CA domains of a viral Gag precursor was well tolerated, exhibiting wild-type-like particle release and a modest effect on viral infectivity (25), suggesting that the Gag precursor was the better choice for artificial extensions. At this point it should be mentioned that the strained AL Gag-Pol precursor already carried additional p1-p6gag sequences, which might limit the capacity to accept further insertions.
In this regard, sequences adjacent to the PR region have been demonstrated to influence the dimerization efficiency of the Gag-Pol precursors and proper folding of the embedded PR domains, thereby triggering PR autoprocessing (6, 10, 21, 22, 23, 30, 33, 42). Accordingly, dimerization defects of PR mutants carrying an inactivated dimer interface were successfully compensated for when the PR was expressed as part of the Gag-Pol precursor (30). Whereas adjacent p6* residues have been reported to facilitate the dimerization of a drug-resistant and proteolytically inactive PR variant, p6* appears to have rather little influence on dimer formation of wild-type PR (10, 42). While p6* seems to play a supportive or at least a neutral role in precursor dimerization, the amino-terminal extension of the PR by p6* residues has been demonstrated to affect the folding preferences within the PR domain, thereby destabilizing the folded PR structure, which results in a retarded PR maturation (6, 17, 23). This might explain the observation that Gag-PR precursors lacking the p6* domain are characterized by a faster autoactivation of the PR (26), whereas elongated or bulky p6*-GFP fusions might in return even prevent the formation of functional Pol dimers.
In sum, this study has provided evidence that 50 of the total of 56 residues in p6* could be specifically mutated in the virus context by using of novel virus platform comprising uncoupled gag and pol reading frames. It is remarkable, though, that an extensive reorganization of Gag and Pol domains did not disturb vital functions of the virus. In this regard the novel provirus might represent a valuable tool to examine other HIV protein domains which would be difficult to access in a natural virus context.
Acknowledgments
The provirus clone NL4-3R71 was kindly provided by K. Saksela (Tampere, Finland), and permissive CEM cells were obtained from J. C. Guatelli (San Diego, CA). We thank the contributors to the NIH AIDS Research and Reference Reagent Program for antisera and cell lines.
Part of this work was supported by the Bill & Melinda Gates Foundation (grant 38637 to R.W.) and the National Institutes of Health (grant 5P01AI066287-02 to R.W.).
Footnotes
Published ahead of print on 29 April 2009.
REFERENCES
- 1.Almog, N., R. Roller, G. Arad, L. Passi-Even, M. A. Wainberg, and M. Kotler. 1996. A p6Pol-protease fusion protein is present in mature particles of human immunodeficiency virus type 1. J. Virol. 707228-7232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrie, K. A., E. E. Perez, S. L. Lamers, W. G. Farmerie, B. M. Dunn, J. W. Sleasman, and M. M. Goodenow. 1996. Natural variation in HIV-1 protease, Gag p7 and p6, and protease cleavage sites within gag/pol polyproteins: amino acid substitutions in the absence of protease inhibitors in mothers and children infected by human immunodeficiency virus type 1. Virology 219407-416. [DOI] [PubMed] [Google Scholar]
- 3.Beissinger, M., C. Paulus, P. Bayer, H. Wolf, P. Rosch, and R. Wagner. 1996. Sequence-specific resonance assignments of the 1H-NMR spectra and structural characterization in solution of the HIV-1 transframe protein p6. Eur. J. Biochem. 237383-392. [DOI] [PubMed] [Google Scholar]
- 4.Bleiber, G., S. Peters, R. Martinez, D. Cmarko, P. Meylan, and A. Telenti. 2004. The central region of human immunodeficiency virus type 1 p6 protein (Gag residues S14-I31) is dispensable for the virus in vitro. J. Gen. Virol. 85921-927. [DOI] [PubMed] [Google Scholar]
- 5.Candotti, D., C. Chappey, M. Rosenheim, P. M'Pele, J. M. Huraux, and H. Agut. 1994. High variability of the gag/pol transframe region among HIV-1 isolates. C. R. Acad. Sci. III 317183-189. [PubMed] [Google Scholar]
- 6.Chatterjee, A., P. Mridula, R. K. Mishra, R. Mittal, and R. V. Hosur. 2005. Folding regulates autoprocessing of HIV-1 protease precursor. J. Biol. Chem. 28011369-11378. [DOI] [PubMed] [Google Scholar]
- 7.Chen, N., A. Morag, N. Almog, I. Blumenzweig, O. Dreazin, and M. Kotler. 2001. Extended nucleocapsid protein is cleaved from the Gag-Pol precursor of human immunodeficiency virus type 1. J. Gen.Virol. 82581-590. [DOI] [PubMed] [Google Scholar]
- 8.Chiu, H. C., F. D. Wang, Y. M. Chen, and C. T. Wang. 2006. Effects of human immunodeficiency virus type 1 transframe protein p6* mutations on viral protease-mediated Gag processing. J. Gen. Virol. 872041-2046. [DOI] [PubMed] [Google Scholar]
- 9.Costa, L. J., Y. H. Zheng, J. Sabotic, J. Mak, O. T. Fackler, and B. M. Peterlin. 2004. Nef binds p6* in GagPol during replication of human immunodeficiency virus type 1. J. Virol. 785311-5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dautin, N., G. Karimova, and D. Ladant. 2003. Human immunodeficiency virus (HIV) type 1 transframe protein can restore activity to a dimerization-deficient HIV protease variant. J. Virol. 778216-8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dulude, D., M. Baril, and L. Brakier-Gingras. 2002. Characterization of the frameshift stimulatory signal controlling a programmed −1 ribosomal frameshift in the human immunodeficiency virus type 1. Nucleic Acids Res. 305094-5102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graf, M., C. Ludwig, S. Kehlenbeck, K. Jungert, and R. Wagner. 2006. A quasi-lentiviral green fluorescent protein reporter exhibits nuclear export features of late human immunodeficiency virus type 1 transcripts. Virology 352295-305. [DOI] [PubMed] [Google Scholar]
- 13.Hill, M., G. Tachedjian, and J. Mak. 2005. The packaging and maturation of the HIV-1 Pol proteins. Curr. HIV Res. 373-85. [DOI] [PubMed] [Google Scholar]
- 14.Hill, M. K., M. Shehu-Xhilaga, S. M. Crowe, and J. Mak. 2002. Proline residues within spacer peptide p1 are important for human immunodeficiency virus type 1 infectivity, protein processing, and genomic RNA dimer stability. J. Virol. 7611245-11253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ho, S. K., R. M. Coman, J. C. Bunger, S. L. Rose, P. O'Brien, I. Munoz, B. M. Dunn, J. W. Sleasman, and M. M. Goodenow. 2008. Drug-associated changes in amino acid residues in Gag p2, p7(NC), and p6(Gag)/p6(Pol) in human immunodeficiency virus type 1 (HIV-1) display a dominant effect on replicative fitness and drug response. Virology 378272-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ibe, S., N. Shibata, M. Utsumi, and T. Kaneda. 2003. Selection of human immunodeficiency virus type 1 variants with an insertion mutation in the p6(gag) and p6(pol) genes under highly active antiretroviral therapy. Microbiol. Immunol. 4771-79. [DOI] [PubMed] [Google Scholar]
- 17.Ishima, R., D. A. Torchia, and J. M. Louis. 2007. Mutational and structural studies aimed at characterizing the monomer of HIV-1 protease and its precursor. J. Biol. Chem. 28217190-17199. [DOI] [PubMed] [Google Scholar]
- 18.Koyanagi, Y., Y. Hinuma, J. Schneider, T. Chosa, G. Hunsmann, N. Kobayashi, M. Hatanaka, and N. Yamamoto. 1984. Expression of HTLV-specific polypeptides in various human T-cell lines. Med. Microbiol. Immunol. 173127-140. [DOI] [PubMed] [Google Scholar]
- 19.Leiherer, A., C. Ludwig, and R. Wagner. 2009. Influence of extended mutations of the HIV-1 transframe protein p6 on Nef-dependent viral replication and infectivity in vitro. Virology 387200-210. [DOI] [PubMed] [Google Scholar]
- 20.Lindhofer, H., K. von der Helm, and H. Nitschko. 1995. In vivo processing of Pr160gag-pol from human immunodeficiency virus type 1 (HIV) in acutely infected, cultured human T-lymphocytes. Virology 214624-627. [DOI] [PubMed] [Google Scholar]
- 21.Louis, J. M., F. Dyda, N. T. Nashed, A. R. Kimmel, and D. R. Davies. 1998. Hydrophilic peptides derived from the transframe region of Gag-Pol inhibit the HIV-1 protease. Biochemistry 372105-2110. [DOI] [PubMed] [Google Scholar]
- 22.Louis, J. M., N. T. Nashed, K. D. Parris, A. R. Kimmel, and D. M. Jerina. 1994. Kinetics and mechanism of autoprocessing of human immunodeficiency virus type 1 protease from an analog of the Gag-Pol polyprotein. Proc. Natl. Acad. Sci. USA 917970-7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Louis, J. M., G. M. Clore, and A. M. Gronenborn. 1999. Autoprocessing of HIV-1 protease is tightly coupled to protein folding. Nat. Struct. Biol. 6868-875. [DOI] [PubMed] [Google Scholar]
- 24.Ludwig, C., A. Leiherer, and R. Wagner. 2008. Importance of protease cleavage sites within and flanking human immunodeficiency virus type 1 transframe protein p6* for spatiotemporal regulation of protease activation. J. Virol. 824573-4584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Müller, B., J. Daecke, O. T. Fackler, M. T. Dittmar, H. Zentgraf, and H. G. Kräusslich. 2004. Construction and characterization of a fluorescently labeled infectious human immunodeficiency virus type 1 derivative. J. Virol. 7810803-10813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Partin, K., G. Zybarth, L. Ehrlich, M. DeCrombrugghe, E. Wimmer, and C. Carter. 1991. Deletion of sequences upstream of the proteinase improves the proteolytic processing of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 884776-4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paulus, C., C. Ludwig, and R. Wagner. 2004. Contribution of the Gag-Pol transframe domain p6* and its coding sequence to morphogenesis and replication of human immunodeficiency virus type 1. Virology 330271-283. [DOI] [PubMed] [Google Scholar]
- 28.Paulus, C., S. Hellebrand, U. Tessmer, H. Wolf, H. G. Kräusslich, and R. Wagner. 1999. Competitive inhibition of human immunodeficiency virus type-1 protease by the Gag-Pol transframe protein. J. Biol. Chem. 27421539-21543. [DOI] [PubMed] [Google Scholar]
- 29.Peters, S., M. Munoz, S. Yerly, V. Sanchez-Merino, C. Lopez-Galindez, L. Perrin, B. Larder, D. Cmarko, S. Fakan, P. Meylan, et al. 2001. Resistance to nucleoside analog reverse transcriptase inhibitors mediated by human immunodeficiency virus type 1 p6 protein. J. Virol. 759644-9653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pettit, S. C., S. Gulnik, L. Everitt, and A. H. Kaplan. 2003. The dimer interfaces of protease and extraprotease domains influence the activation of protease and the specificity of GagPol cleavage. J. Virol. 77366-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pettit, S. C., L. E. Everitt, S. Choudhury, B. M. Dunn, and A. H. Kaplan. 2004. Initial cleavage of the human immunodeficiency virus type 1 GagPol precursor by its activated protease occurs by an intramolecular mechanism. J. Virol. 788477-8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pettit, S. C., J. C. Clemente, J. A. Jeung, B. M. Dunn, and A. H. Kaplan. 2005. Ordered processing of the human immunodeficiency virus type 1 GagPol precursor is influenced by the context of the embedded viral protease. J. Virol. 7910601-10607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quillent, C., A. M. Borman, S. Paulous, C. Dauguet, and F. Clavel. 1996. Extensive regions of pol are required for efficient human immunodeficiency virus polyprotein processing and particle maturation. Virology 21929-36. [DOI] [PubMed] [Google Scholar]
- 34.Saksela, K., G. Cheng, and D. Baltimore. 1995. Proline-rich (PxxP) motifs in HIV-1 Nef bind to SH3 domains of a subset of Src kinases and are required for the enhanced growth of Nef+ viruses but not for down-regulation of CD4. EMBO J. 14484-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song, Y. H., Z. F. Meng, H. Xing, Y. H. Ruan, X. P. Li, R. L. Xin, P. F. Ma, H. Peng, and Y. Shao. 2007. Analysis of HIV-1 CRF07_BC gag p6 sequences indicating novel deletions in the central region of p6. Arch. Virol. 1521553-1558. [DOI] [PubMed] [Google Scholar]
- 36.Tessmer, U., and H. G. Krausslich. 1998. Cleavage of human immunodeficiency virus type 1 proteinase from the N-terminally adjacent p6* protein is essential for efficient Gag polyprotein processing and viral infectivity. J. Virol. 723459-3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walter, A. E., D. H. Turner, J. Kim, M. H. Lyttle, P. Muller, D. H. Mathews, and M. Zuker. 1994. Coaxial stacking of helixes enhances binding of oligoribonucleotides and improves predictions of RNA folding. Proc. Natl. Acad. Sci. USA 919218-9222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whitehurst, N., C. Chappey, C. Petropoulos, N. Parkin, and A. Gamarnik. 2003. Polymorphisms in p1-p6/p6* of HIV type 1 can delay protease autoprocessing and increase drug susceptibility. AIDS Res. Hum. Retroviruses 19779-784. [DOI] [PubMed] [Google Scholar]
- 39.Wolf, H., S. Modrow, M. Soutschek, M. Motz, R. Grunow, H. Döbl, and R. von Baehr. 1990. Production, mapping and biological characterization of monoclonal antibodies against core protein (p24) of the human immunodeficiency virus. AIDS-Forsch. 116-18. [Google Scholar]
- 40.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 313406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zybarth, G., H. G. Kräusslich, K. Partin, and C. Carter. 1994. Proteolytic activity of novel human immunodeficiency virus type 1 proteinase proteins from a precursor with a blocking mutation at the N terminus of the PR domain. J. Virol. 68240-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zybarth, G., and C. Carter. 1995. Domains upstream of the protease (PR) in human immunodeficiency virus type 1 Gag-Pol influence PR autoprocessing. J. Virol. 693878-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]