Abstract
We investigated whether a 28-day course of potent antiretroviral therapy, initiated at a time point (48 h postinoculation) following simian immunodeficiency virus (SIV) inoculation when the acquisition of a viral infection was virtually assured, would sufficiently sensitize the immune system and result in controlled virus replication when treatment was stopped. The administration of tenofovir 48 h after SIV inoculation to six Mamu-A*01-negative rhesus macaques did, in fact, potently suppress virus replication in all of the treated rhesus macaques, but plasma viral RNA rapidly became detectable in all six animals following its cessation. Unexpectedly, the viral set points in the treated monkeys became established at two distinct levels. Three controller macaques had chronic phase virus loads in the range of 1 × 103 RNA copies/ml, whereas three noncontroller animals had set points of 2 × 105 to 8 × 105 RNA copies/ml. All of the noncontroller monkeys died with symptoms of immunodeficiency by week 60 postinfection, whereas two of the three controller animals were alive at week 80. Interestingly, the three controller macaques each carried major histocompatibility complex class I alleles that previously were reported to confer protection against SIV, and two of these animals generated cytotoxic T-lymphocyte escape viral variants during the course of their infections.
Acute human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV) infections are characterized by the extensive and rapid killing of memory CD4+ T lymphocytes at multiple effector sites, including the gastrointestinal tract (1, 5, 10, 17, 18, 31). Previous studies have utilized transient early postinoculation antiretroviral therapy (ART) to blunt the acute infection and to preserve the integrity of the immune system. Initiating a 28-day course of the reverse transcriptase (RT) inhibitor tenofovir (PMPA) at 24 h post inoculation (p.i.) of rhesus macaques with either SIVmac239 or SIVsmE660 potently suppressed virus replication during and following drug administration (11, 12). In contrast, extending the time of treatment start from 24 to 72 h after SIVsmE660 infection resulted in two of four animals becoming viremic during the 28-day course of therapy and generating measurable levels of plasma viral RNA in all four monkeys following the cessation of ART (12).
Based on these reports, which suggested that ART started at 24 h potently aborted the SIV infection, we elected to delay the treatment after virus inoculation until 48 h p.i. to permit a controlled establishment of the viral infection and possibly enable a sensitized immune system to control virus replication when PMPA treatment was discontinued. Accordingly, we treated six Mamu-A*01-negative Indian-origin rhesus macaques with the RT inhibitor beginning at 48 h p.i. Although virus replication (monitored by RT-PCR and cell-associated DNA PCR) was effectively suppressed and memory CD4+ T cells in the blood and at an effector site (lung) were preserved, plasma viremia became detectable in all six of the treated animals immediately following the cessation of ART. Interestingly, by 15 weeks p.i., the plasma viral RNA set points of the treated monkeys had become established at two distinct levels. Three animals (termed noncontrollers) had plasma viral RNA loads in the range of 105 to 106 viral RNA copies/ml (similar to untreated SIV-inoculated macaques), and three monkeys (termed controllers) had viral loads in the range of 103 viral RNA copies/ml. Analyses of the major histocompatibility complex (MHC) class I alleles in the tenofovir-treated cohort revealed that the three controllers carried genes (Mamu-B*08, Mamu-B*29, and Mamu-A*02) previously associated with the suppression of virus replication in SIV-inoculated rhesus macaques (15, 20, 25). Nevertheless, two of the controller macaques began to experience the loss of memory CD4+ T cells at months 9 and 11 postchallenge, respectively. This was accompanied by marked increases in the levels of their set point viremia, from approximately 103 RNA copies/ml to 105 to 106 RNA copies/ml. Changes in immunodominant restricting CD8+ T-cell epitopes were observed in two of the controller macaques. At week 80 p.i., only two tenofovir-treated animals, both bearing protective MHC alleles, remained alive.
MATERIALS AND METHODS
Virus and animal experiments.
The origin and preparation of the tissue culture-derived SIVmac239 stocks from molecular clones have been described previously (7, 21). Adult rhesus macaques (Macaca mulatta), weighing between 4.7 and 6.2 kg, were maintained in accordance with the guidelines of the Committee on Care and Use of Laboratory Animals (3) and were housed in a biosafety level 2 facility; biosafety level 3 practices were followed. All rhesus macaques were Mamu-A*01 negative.
Tenofovir treatment of SIVmac239-infected rhesus macaques.
Tenofovir {9-[2-(R)-(phosphonometoxy)propyl]adenine} was generously provided by Norbert Bischofberger, Gilead Science, Inc. (Foster City, CA). An aqueous solution (200 mg/ml) of PMPA was prepared after adjusting the pH to 7.0 with NaOH and filtration through a membrane filter (pore size, 0.22 μm) as previously described (30). Tenofovir was intramuscularly administered (30 mg/kg of body weight) every 24 h for 4 weeks, starting at 48 h p.i. with 100 50% tissue culture infectious doses (TCID50) of SIVmac239.
Quantitation of proviral DNA and plasma viral RNA levels.
The number of viral DNA copies in peripheral blood mononuclear cells was measured by quantitative DNA PCR (29). Viral RNA levels in plasma were determined by real-time reverse transcription-PCR (ABI Prism 7700 sequence detection system; Applied Biosystems, Foster City, CA) as previously reported, using reverse-transcribed viral RNA in plasma samples from SIVmac239-inoculated rhesus macaques (4).
Amplification of viral RNA to identify SIV escape variants.
To analyze SIV variants bearing mutations affecting epitopes restricted by MHC class I alleles, plasma viral RNA was extracted using a QIAamp viral RNA mini-kit (Qiagen) and then reverse transcribed using SuperScript III First-Strand Synthesis SuperMix (Invitrogen). PCR amplifications were performed using primer sets SIVmac239(1407-1436)-F (TGTAGTATGGGCAGCAAATGAATTAGATAG) and SIVmac239(2701-2681)-R (GGTCCTCTGGGGGAGCAGTTG) for the GY9 Mamu-A*02 epitope, SIVmac239(5093-5116)-F (ATTGGCAGGCAGATGGCCTATTAC) and SIVmac239(6454-6431)-R (CCATGGTTCCCTTTGTGGTCCTTC) for the RL8 and RL9 Mamu-B*08 epitopes, SIVmac239(7005-7027)-F (GGGATACTTGGGGAACAACTCAG) and SIVmac239(9380-9358)-R (CAGATCTCCAGACGGCCTGGACC) for the RY8 Mamu-A*02 epitope, SIVmac239(9381-9405)-F (CGACAGAGACTCTTGCGGGCGCGTG) and SIVmac239(10129-10104)-R (CTGTTTCAGCGAGTTTCCTTCTTGTC) for YY9 of the Mamu-A*02 epitope, and the RL9 and RL10 Mamu-B*08 epitopes. The PCRs were performed using 10 pmol of each forward and reverse primer, Platinum PCR SuperMix High Fidelity (Invitrogen), and 1 μl of viral cDNA in a final volume of 50 μl. The reaction mixtures were heated to 94°C for 2 min, followed by 32 cycles of denaturing at 94°C for 20 s, annealing at 58°C for 30 s, and extension at 68°C for 2 min. A final extension was conducted at 68°C for 7 min. PCR products were gel extracted using a Qiaquick gel extraction kit (Qiagen), cloned into pCR2.1 TOPO cloning vector (Invitrogen), and then sequenced.
Lymphocyte immunophenotyping in monkey BAL fluid and peripheral blood.
Bronchoalveolar lavage (BAL) lymphocytes were prepared from uninfected donor animals and SIV-inoculated animals by using a pediatric bronchoscope (BF3C40; Olympus America, Inc., Melville, NY), as previously described (7). The BAL fluid was filtered through a 70-μm-pore-size cell strainer (BD Falcon, Bedford, MA) and centrifuged, and the cell pellet was washed three times with 1% bovine serum albumin (Sigma-Aldrich, St. Louis, MO)-phosphate buffered saline (1% BSA-PBS). The cells were resuspended in mouse immunoglobulin (Ig) (1 mg/ml)-1% BSA-PBS. EDTA-treated blood samples and cells from BAL specimens were stained for flow-cytometric analysis as described previously (22, 23) using combinations of the following fluorochrome-conjugated monoclonal antibodies (MAbs): CD3 (phycoerythrin [PE]), CD4 (peridinin chlorophyll protein-Cy5.5 [PerCP-Cy5.5]), CD8 (PerCP or allophycocyanin [APC]), CD28 (fluorescein isothiocyanate [FITC] or PE), or CD95 (APC). All antibodies were obtained from BD Biosciences (San Diego, CA), samples were analyzed by four-color flow cytometry (FACSCalibur; BD Biosciences Immunocytometry Systems), and data analysis was performed using CellQuest Pro (BD Biosciences, San Diego, CA). In this study, naïve CD4+ T cells were identified by their CD95low CD28high phenotype, whereas memory CD4+ T cells were CD95high CD28high or CD95high CD28low in the CD4+ small lymphocyte gate (27).
Intracellular cytokine assays.
Stimulation was performed on frozen lymphocytes as described previously (28). Freshly thawed lymphocytes were resuspended (106/ml) in RPMI medium supplemented with antibiotics and glutamine. Anti-CD28 conjugated to Alexa 594-PE was used for costimulation. Staphylococcus enterotoxin B (1 μg/ml; Sigma-Aldrich, St. Louis, MO) was used to stimulate T cells mitogenically through the T-cell receptor as a positive control. A negative control (cells treated only with costimulatory anti-CD28) was included in every experiment. Peptides used to stimulate SIV-specific T cells were 15 amino acids in length, overlapping by 11 amino acids, and encompassed SIVmac239 Gag (New England Peptide, Gardner, MA). The concentration of each peptide was 2 μg/ml for stimulations, which were performed in the presence of brefeldin-A (BFA; 1 μg/ml; Sigma-Aldrich, St. Louis, MO) for 16 h at 37°C. All cells were surface stained with the dead cell exclusion dye Aqua Blue (Invitrogen Corporation, Carlsbad, CA), followed by staining with anti-CD3 Alexa700 (BD), anti-CD4 Cy5.5-PE (eBioscience Inc., San Diego, CA), anti-CD8 Pacific Blue (BD), and anti-CD95 Cy5-PE (BD). Cells then were fixed, permeabilized, stained with anti-gamma interferon (IFN-γ) Cy7-PE (BD), anti-interleukin-2 (IL-2) APC (BD), tumor necrosis factor (TNF) FITC (BD), and Mip1-β PE (BD) and analyzed by flow cytometry (FACSAria; BD Biosciences Immunocytometry Systems). SIV-specific CD8 T-cell responses are reported as the frequency of memory CD8 T cells gated by characteristic light scatter properties, and then as Aqua blue-negative, CD3+, CD8+, CD4−, CD95+, and by the production of either TNF or Mip-1β. All data are reported after background subtraction.
Cells also were purified from freshly collected BAL specimens as described above and resuspended in RPMI 1640 medium (Cambrex Bio Science, Walkersville, MD) supplemented with 10% fetal bovine serum (HyClone, Loagan, UT), 2 mM l-glutamine, 1 mM sodium pyruvate, and 100 U/ml penicillin-0.1 mg/ml streptomycin (Sigma-Aldrich, St. Louis, MO). The cells then were stimulated with the SIVmac239 Gag peptide pool at a concentration of 10 μM. After 2 h of incubation, BFA (Sigma) was added to block protein transport, and following four additional hours of incubation, the cells were stained for flow cytometry using combinations of the following fluorochrome-conjugated MAbs: CD3 (FITC) and CD8 (PerCP). For IFN-γ staining, cells were treated with fluorescence-activated cell sorting permeabilization buffer 2 (Becton Dickinson) and stained with CD69 (PE) and IFN-γ (APC) MAbs. All antibodies were obtained from BD Biosciences (San Diego, CA). IFN-γ production in CD8+ T cells was analyzed with a FACSCalibur. The percentage of IFN-γ-producing memory CD8+ T cells in BAL samples was calculated after subtracting values obtained with contemporaneously analyzed mock-infected cells.
MHC class I cDNA cloning and sequencing.
The cloning of Mamu-A and Mamu-B cDNA from rhesus macaques was performed by RT-PCR amplification as described previously (9). Briefly, total cellular RNA was extracted from activated rhesus peripheral blood mononuclear cells using TRI reagent (Molecular Research Center, Cincinnati, OH). Complete Mamu-A and Mamu-B cDNAs were generated using the 3′ rapid amplification of cDNA ends (RACE) adapter from the First Choice RLM RACE kit (Ambion, Austin, TX). PCR amplifications were performed using sense primer Mane5UA (GATTCTCCGCAGACGCCCA), Mane5UA20 (GATTCTCCGCAGACGCCAA), Mane5UB2 (AAAGTCTCCTCAGACGCCGA), or Mane5UB3 (AGAGTCTCCTCAGACCCCAA), oligonucleotides annealing in the 5′-untranslated region of Mamu-A or -B cDNA, and the 3′ RACE outer reverse primer. The PCR mixtures contained 50 mM potassium acetate, 1.5 mM MgSO4, 10 mM Tris-HCl, pH 9.0, 0.2 mM each dGTP, dCTP, dATP, and dTTP, 20 pmol of each sense and reverse primers, and 5 U of Super Taq Plus DNA polymerase (Ambion, Austin, TX). Each reaction mixture contained 2 μl of cDNA in a final volume of 50 μl. The reaction mixtures were heated at 95°C for 3 min, and then amplification was conducted for 30 cycles as follows: denatured for 30 s at 95°C, annealed for 30 s at 59°C, and extended for 90 s at 72°C. A final extension was conducted for 7 min at 72°C. PCR products were gel purified using a Qiaquick gel extraction kit (Qiagen, Valencia, CA) and then cloned into pCR2.1 TOPO cloning vector (Invitrogen, Carlsbad, CA). Insert-containing clones were identified after restriction analysis using EcoRI and then were sequenced using an Applied Biosystems 3130XL genetic analyzer.
Sequence analysis and allele identification.
Sequences were aligned using the Clustal W program of MacVector 10.0.2 software (MacVector Inc., Cary, NC) with a panel of 56 Mamu-A and 157 Mamu-B published alleles. Phylogenetic trees were constructed based on the alignment using the neighbor-joining method of the software. Genetic distances were estimated using Kimura's two-parameter method. Alleles were identified based on the close clustering of cloned sequences with known alleles.
Western blot analysis.
For immunoblotting, SIVmac239 particles were concentrated by ultracentrifugation (100,000 × g for 75 min by using a Type 55 rotor in an Optima XL-100K Ultracentrifuge; Beckman Coulter, Fullerton, CA). The virus pellet was resuspended in PBS and subjected to ultracentrifugation through 20% sucrose in PBS (200,000 × g for 75 min in an SW55 rotor using an Optima XL-100K Ultracentrifuge; Beckman Coulter). After incubation with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, viral proteins were resolved by SDS-PAGE and transferred to a polyvinylidene difluoride membrane (Invitrolon PVDF; Invitrogen, Carlsbad, CA). The membranes were incubated with serially collected plasma samples (diluted 1:500), which had been preadsorbed with sonicated lysate of Escherichia coli and MT-4 cells, and then with goat anti-monkey IgG conjugated with horseradish peroxidase (Santa Cruz Biotechnology, Santa Cruz, CA). Protein bands were visualized on X-ray film (Kodak Biomax MR) after incubation with chemiluminescent substrate (Western Blotting Luminol Reagent; Santa Cruz Biotechnology, Santa Cruz, CA).
Anti-SIV Env ELISA.
Anti-SIV gp130 Env-specific IgGs in monkey plasma were detected and quantified by end point dilution enzyme-linked immunosorbent assay (ELISA). Flat-bottomed 96-well plates (Corning, Lowell, MA) were coated with purified recombinant SIVmac239 gp130 (6) that had been obtained from the NIH AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, NIH. The coated plates first were blocked with 4% nonfat milk in PBS and then incubated with serially diluted (in 4% nonfat milk-PBS) samples of macaque plasma (with a starting dilution of 1:100) for 1 h at 37°C, followed by an overnight incubation at 4°C. Horseradish peroxidase-conjugated polyclonal goat anti-monkey IgG (Nordic Immunological Laboratories, The Netherlands) (1:2,000 dilution in 4% nonfat milk-PBS) was used as the secondary antibody. Incubated plates were developed with 2,2′-azino-bis liquid substrate (Sigma-Aldrich, St. Louis, MO), and the endpoint titers, defined as the reciprocal of the highest dilution giving an absorbance reading of more than three times the mean of the preimmune serum at the same dilution, were determined (8).
Virus neutralization assays.
Plasma samples (1:20 dilution) from PMPA-treated and untreated SIV-infected macaques were incubated with SIVmac239 in quadruplicate in a total volume of 50 μl for 1 h at 37°C in 96-well flat-bottomed culture plates. Samples from uninfected animals served as controls. Freshly trypsinized TZM-bl cells (33) (1 × 104 in 150 μl Dulbecco's modified essential medium containing 20 μg/ml DEAE dextran) were added to each well, and the cultures were maintained in a 37°C incubator for 28 h. The amount of virus-induced luciferase activity present in cell lysates was determined using a commercially available luciferase assay kit (Invitrogen Corporation, Carlsbad, CA), and the average neutralization activity for each plasma sample was determined. Any sample resulting in a 50% reduction of luciferase activity compared to that obtained with the uninfected control sample was considered positive for neutralizing antibodies (NAbs).
RESULTS
The aim of this study was to administer a proven ART regimen very early during acute SIV infection that would effectively suppress virus replication, and to ascertain whether immune responses would be sufficiently activated during drug administration to control virus replication following treatment cessation. As indicated in Table 1, six Mamu-A*01-negative Indian-origin rhesus macaques were inoculated intravenously with 100 TCID50 of SIVmac239; treatment with PMPA commenced at 48 h p.i. This time for treatment initiation was selected based on two previous reports: the first showed that tenofovir administration beginning at 24 h p.i. resulted in no measurable plasma viremia during, or 6 weeks following, ART in three of four animals infected with SIVsmE660 (12); and the second showed that treatment initiated 24 h after SIVmac239 inoculation led to low to unmeasurable plasma viral RNA levels during 28 days of PMPA therapy or for an additional 6 weeks following the discontinuation of ART (11). When tenofovir treatment was delayed until 72 h p.i., SIVsmE660 viremia was detectable during the 4-week course of therapy in two of four monkeys and in four of four animals following the cessation of drug administration (12). Thus, the start of ART at 48 h p.i. would ensure SIV acquisition and the possible stimulation of the immune system while limiting injury to the CD4+ T-cell population. Two age-matched untreated but SIV-infected macaques served as controls (Table 1).
TABLE 1.
Experimental protocol
| Animal | SIVmac239 inoculum size (TCID50) | Tenofovir dose (mg/kg/day) | Tenofovir initiation time point (h p.i.) | Tenofovir duration (days) |
|---|---|---|---|---|
| CL8P | 100 | 30 | 48 | 28 |
| DA14 | 100 | 30 | 48 | 28 |
| DA21 | 100 | 30 | 48 | 28 |
| DA63 | 100 | 30 | 48 | 28 |
| DA87 | 100 | 30 | 48 | 28 |
| DB05 | 100 | 30 | 48 | 28 |
| CK2G | 100 | None | ||
| CL33 | 100 | None |
PMPA potently suppresses virus replication during acute SIV infection.
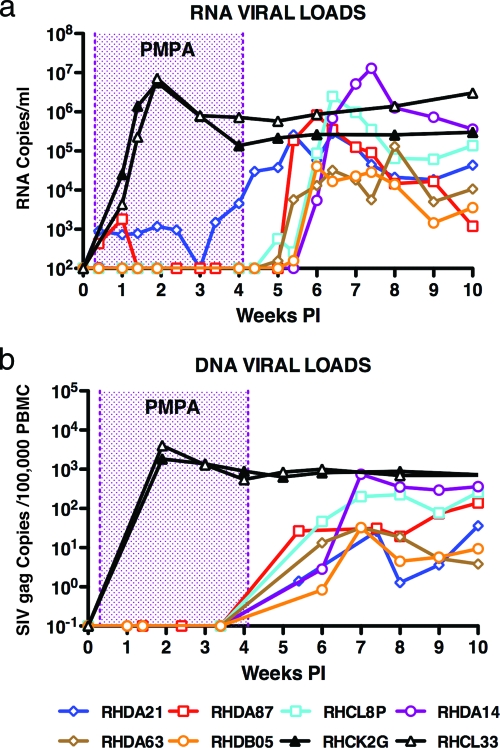
As expected, the two untreated animals inoculated with 100 TCID50 SIVmac239 developed peak levels of plasma viremia (5.6 × 106 and 7.1 × 106 RNA copies/ml, respectively) and high frequencies of cell-associated viral DNA on day 14 p.i. (Fig. 1). In contrast, no plasma viral RNA was measurable in four of the six tenofovir-treated macaques (the limit of detection is 100 viral RNA copies/ml) during the 4-week course of antiretroviral administration. One treated monkey (DA87) had detectable SIV RNA in its plasma on days 3 (4.3 × 102 copies/ml) and 7 (1.8 × 103 copies/ml) p.i. and none thereafter; a second (DA21) had persistent low levels of viremia (730 to 4,600 RNA copies/ml) throughout the period of tenofovir administration. The level of cell-associated viral DNA was below the threshold of detection in all six of the SIV-infected and PMPA-treated monkeys (the limit of detection is 1 infected cell per 106 cells). Taken together, these results indicate that the p.i. ART was highly effective; plasma viremia during the first 4 weeks after SIV inoculation had been reduced at least 3 to 4 logs in the cohort of treated infected macaques.
FIG. 1.
Plasma viral RNA and cell-associated DNA levels during acute SIV infections. Sequential viral RNA (a) and DNA (b) loads are shown. Tenofovir-treated and untreated animals are indicated in Table 1.
Within 7 to 14 days of stopping PMPA therapy, however, SIV replication became detectable in all of the treated monkeys (Fig. 1a). The levels of peak plasma viremia ranged from 3.2 × 104 to 1.3 × 107 RNA copies/ml, with a geometric mean (6.7 × 105) that was approximately 10-fold lower than that observed in the untreated control macaques 4 to 5 weeks earlier. Cell-associated viral DNA, which was not detected during tenofovir administration in any of the treated monkeys, became measurable only after the cessation of ART (Fig. 1b). The two treated animals (CL8P and DA14) with the highest levels of peak plasma viremia also had the highest levels of cell-associated viral DNA.
Viral set points in PMPA-treated monkeys were established at two distinct levels.
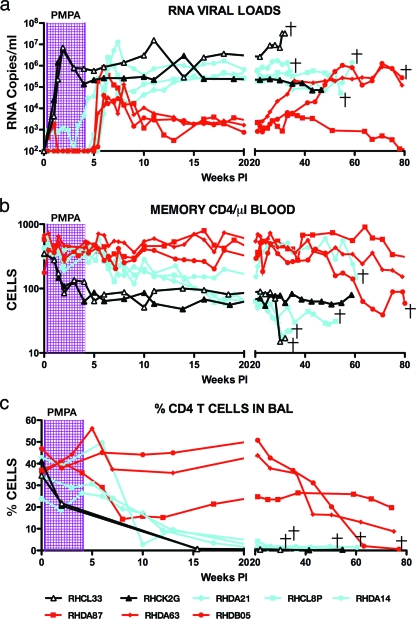
The six recipients of tenofovir resolved their acute SIV infections by week 15 p.i. (Fig. 2a). Interestingly, three of these monkeys (DA87, DA63, and DB05) controlled their plasma viral loads to relatively low levels, whereas the other three drug-treated macaques (DA21, CL8P, and DA14) had viral set points that were several hundredfold higher. At week 22 p.i., the geometric mean plasma virus load for the three controller animals was 1.33 × 103 RNA copies/ml. In contrast, the comparable set point value for the three noncontroller monkeys was 3.6 × 105 RNA copies/ml. The latter level was in the same general range as that observed for the two untreated SIV-infected control animals.
FIG. 2.
SIV controller and noncontroller animals establish two distinct levels of set point viremia. Levels of plasma viral RNA (a) and circulating memory CD4+ T cells (b) and the percentage of CD4+ T cells in BAL specimens (c) during the 80 weeks of SIV infection are shown. Controller (red), noncontroller (blue), and untreated (black) monkeys are indicated.
Not unexpectedly, the untreated macaques experienced depletions of memory CD4+ T cells both in the blood and at an effector site (lung) during acute SIV infection (Fig. 2b and c). In contrast, this CD4+ T-cell subset was preserved in all six of the infected animals during PMPA administration. However, two patterns of memory CD4+ T-cell dynamics were observed in the drug-treated cohort after the discontinuation of ART that correlated with the two distinct levels of set point viremia. This is best seen with the CD4+ T lymphocytes recovered by BAL (Fig. 2c). The three noncontroller monkeys experienced a nearly complete loss of CD4+ T cells in BAL specimens by week 22, which was delayed relative to that observed in the untreated macaques. Two of the noncontrollers also sustained depletions of this T-lymphocyte subset in the blood (Fig. 2b). In contrast, two of the controller animals (DA63 and DB05) experienced no depletion of CD4+ T cells in BAL samples during the first 6 months of their SIV infections, while the third controller (DA87) sustained a moderate loss of this lymphocyte subset. The loss of BAL fluid CD4+ T cells in macaque DA87 may reflect its 10- to 20-fold-higher levels of peak viremia following the cessation of tenofovir therapy compared to that of the two other controller animals (DA63 and DB05) (Fig. 1a), which experienced no decline of this T-cell subset.
Virus-specific immune responses in tenofovir-treated animals.
A major premise underlying the administration of PMPA at 48 h p.i. was that by potently suppressing SIV replication and preventing severe injury to the memory CD4+ T-lymphocyte population during acute SIV infection, the immune system would be stimulated by low residual levels of virus production and generate a protective antiviral response when ART was stopped. We therefore initially examined whether T-cell-mediated immunity was induced in the treated macaques during the period in which they received ART. SIV Gag-specific CD8+ T-cell responses were measured by intracellular staining for the production of either TNF or Mip-1β after stimulation with a peptide pool containing 125 15mer peptides (overlapping by 11 amino acids) spanning SIVmac239 Gag. As shown in Table 2, the frequency of virus-specific CD8+ T cells remained at background levels during the period of tenofovir therapy, except for macaque DA14. As noted earlier, DA14 generated the highest levels of peak plasma viremia (1.3 × 107 RNA copies/ml) following drug treatment, possibly accounting for the elevated frequency of SIV-specific CD8+ T cells measured in this animal at week 4.1. All five of the PMPA recipients, tested at week 16 p.i., generated increased frequencies of virus-specific CD8+ T lymphocytes, presumably reflecting several months of antigen stimulation. In four of these animals, the responses at late times exceeded those measured for the untreated macaques.
TABLE 2.
Virus-specific immune responses measured in circulating CD8+ T cells
| Monkey | Status | Wk p.i. | SIV Gag-specific responsea |
|---|---|---|---|
| DB05 | Controller | 2.0 | 0.18 |
| DB05 | Controller | 6.0 | 0.50 |
| DB05 | Controller | 16.0 | 0.82 |
| DA87 | Controller | 3.4 | 0.18 |
| DA87 | Controller | 5.4 | 0.21 |
| DA87 | Controller | 16.0 | 0.92 |
| DA63 | Controller | 2.0 | 0.16 |
| DA63 | Controller | 6.0 | 0.20 |
| DA63 | Controller | 16.0 | 0.32 |
| DA14 | Noncontroller | 4.1 | 1.07 |
| DA14 | Noncontroller | 8.0 | 1.20 |
| DA14 | Noncontroller | 16.0 | 2.70 |
| DA21 | Noncontroller | 3.4 | 0.37 |
| DA21 | Noncontroller | 6.4 | 0.11 |
| DA21 | Noncontroller | 16.0 | 1.20 |
| CL8P | Noncontroller | 2.0 | 0.00 |
| CL8P | Noncontroller | 6.0 | 0.04 |
| CL33 | Untreated | 3.0 | 0.97 |
| CL33 | Untreated | 5.0 | 0.10 |
| CL33 | Untreated | 14.0 | 0.27 |
| CL2G | Untreated | 2.0 | 0.16 |
| CL2G | Untreated | 6.0 | 0.20 |
| CL2G | Untreated | 16.0 | 0.32 |
The frequency of CD8+ CD4− memory T cells that produce TNF and/or Mip-1β after subtracting the frequency of memory CD8+ T cells that spontaneously produce TNF and/or Mip-1β.
SIV Gag-specific CD8+ T-cell responses also were assessed in specimens recovered by BAL of four tenofovir-treated macaques by intracellular staining for IFN-γ following stimulation with the same SIVmac239 Gag peptide pool (Table 3). Increased frequencies of virus-specific CD8+ T lymphocytes were detected both during and following the cessation of PMPA therapy. In general, the SIV Gag-specific responses were higher in the two noncontroller macaques (DA14 and CL8P), particularly during and immediately following drug administration.
TABLE 3.
Virus-specific immune responses measured in BAL CD8+ T cells
| Monkey | Status | Wk p.i. | SIV Gag-specific responsea |
|---|---|---|---|
| DA14 | Noncontroller | 2.0 | 1.90 |
| DA14 | Noncontroller | 6.0 | 3.69 |
| DA14 | Noncontroller | 10.0 | 1.18 |
| CL8P | Noncontroller | 3.0 | 3.32 |
| CL8P | Noncontroller | 5.0 | 0.36 |
| CL8P | Noncontroller | 7.0 | 0.71 |
| CL8P | Noncontroller | 13.0 | 0.38 |
| DB05 | Controller | 2.0 | 0.84 |
| DB05 | Controller | 6.0 | 0.77 |
| DB05 | Controller | 10.0 | 1.56 |
| DA63 | Controller | 3.0 | 1.31 |
| DA63 | Controller | 5.0 | 1.18 |
| DA63 | Controller | 7.0 | 1.64 |
| DA63 | Controller | 13.0 | 1.36 |
The frequency of CD8+ T cells in BAL fluid that produce IFN-γ and CD69 after subtracting the frequency of CD8+ T cells that spontaneously produce IFN-γ and CD69.
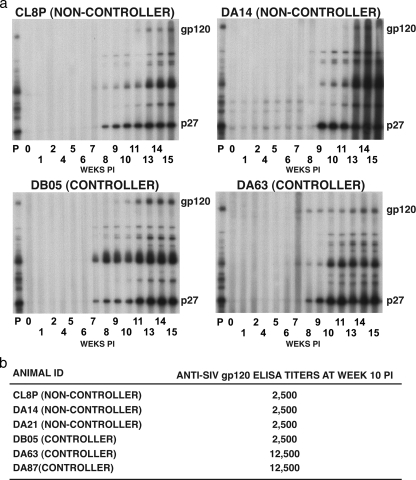
SIV-specific humoral responses initially were assessed by Western blot analyses using plasma samples from two controller and two noncontroller animals. As shown in Fig. 3a, antibody reacting with gp130 was delayed (weeks 11 to 13) in noncontrollers CL8P and DA14 compared to its earlier appearance (weeks 7 to 9) in controllers DA63 and DB05. Antiviral binding antibody, as measured by ELISA in all six treated macaques at week 10 p.i., revealed that two of the controller monkeys (DB05 and DB87) had generated the highest (1:12,500) anti-gp130 titers (Fig. 3b).
FIG. 3.
Profiles of anti-SIV antibody responses of noncontroller and controller monkeys. (a) SIV proteins, prepared from detergent-treated pelleted particles, were transferred to PVDF membranes following PAGE and incubated with a 1:500 dilution of serially collected plasma samples from two noncontroller monkeys (upper) and two controller monkeys (bottom). The positions of SIV gp120 envelope glycoproteins and p27 CA are indicated. A plasma sample (diluted 1:500) collected from a chronically SIVmac239-infected monkey was used as a positive control in lane P. (b) Anti-SIV gp120 ELISA titers.
Virus neutralization assays were performed using TZM-bl cells and intact SIVmac239 virions in combination with plasma specimens recovered at weeks 10 and 22. No anti-SIVmac239 NAb activity was detected in any of the treated or untreated macaques at either time point with samples diluted 1:20 (data not shown).
The three controller monkeys express protective MHC class I alleles.
The establishment of two distinct levels of viral set points in the six Mamu-A*01-negative, SIV-infected, tenofovir-treated recipients raised the possibility that this outcome might be genetically determined. It is now recognized that the expression of certain MHC class I alleles is strongly associated with reduced plasma viremia and slower disease progression in HIV- and SIV-infected humans and macaques, respectively (2, 15, 19, 20, 25, 34). In the rhesus macaque, MHC class I alleles associated with the control of SIV replication include Mamu-A*01, Mamu-A*02, Mamu-B*08, Mamu-B*17, and Mamu-B*29 (13, 15, 20, 25, 34). The MHC class I alleles expressed by the controller and noncontroller animals were determined following the RT-PCR amplification, cloning, and sequencing of full-length cDNAs for Mamu-A and Mamu-B alleles. Individual alleles were identified by comparing each cloned sequence to a database of published rhesus MHC class I sequences representing 56 Mamu-A and 157 Mamu-B alleles. This exhaustive approach was used to obtain a broad view of the MHC class I allele repertoire expressed in each macaque and to avoid limiting the analysis to only a few previously identified MHC alleles.
The repertoire of Mamu-A and Mamu-B alleles expressed by the six tenofovir-treated monkeys was very heterogeneous and included 10 Mamu-A and 17 Mamu-B alleles that had been identified previously (Table 4). No single MHC class I allele determined the segregation of our cohort into two distinct groups. Some MHC class I alleles (Mamu-A*0602 and Mamu-A*1403) that were present in both controller and noncontroller monkeys clearly were not involved in suppressing SIV replication in the former group. Two noncontroller macaques had a combination of Mamu-B alleles known to be present on the same haplotype (DA21, Mamu-B*12, Mamu-B*38, and Mamu-B*30; CL8P, Mamu-B*19 and Mamu-B*24) (26). Interestingly, two of the three controllers (DA87 and DA63) carried MHC alleles (Mamu-B*08 and Mamu-B*290101) previously reported to suppress SIV replication (15, 34). The third controller (DB05) expressed Mamu-A*02, an allele found at increased frequency within an elite controller cohort and associated with a partial reduction of plasma viral load in both the acute and the chronic phases of SIV infection (15). In contrast, none of the MHC class I alleles associated with reduced plasma viremia and slower disease progression were found in the three noncontroller monkeys.
TABLE 4.
MHC class I alleles expressed by each SIV-infected tenofovir-treated rhesus macaquea
| Animal | Mamu-A allele(s) | Mamu-B allele(s) | |||||
|---|---|---|---|---|---|---|---|
| Noncontroller | |||||||
| RhDA21 | A*08 | A*1302 | A*1303 | B*12 | B*38 | B*71 | B*30-like |
| B*46-like | B*74-like | B*09-like | |||||
| RhDA14 | A*1403 | B*12 | B*74-like | new B | new B | ||
| RhCL8P | A*0602 | B*19 | B*24 | B*52 | B*55-like | ||
| Controller | |||||||
| RhDA87 | A*06 | A*07-like | B*01 | B*08 | new B | ||
| RhDB05 | A*02 | A*04 | A*1403 | B*7301 | B*31-like | B*74-like | |
| RhDA63 | A*0602 | A*28-like | B*290101 | B*61 | B*55-like | B*09-like | |
Long-term effects of PMPA therapy.
After week 20, the three noncontroller macaques continued to experience high set point plasma viremia levels (105 to 106 RNA copies/ml) and barely detectable CD4+ T cells in BAL specimens (Fig. 2). These animals subsequently became symptomatic and were euthanized at weeks 34, 54, and 60, respectively, because of intractable diarrhea, anorexia, and marked weight loss. The control of plasma viremia varied greatly among the three controller monkeys. As shown in Fig. 2a, the plasma viral load in animal DA63 began to increase after week 27 p.i., reaching a set point of 2 × 105 RNA copies/ml by week 36 p.i. This increase in plasma viremia was accompanied by modest declines of memory CD4+ T lymphocytes in blood and BAL samples in DA63 (Fig. 2b and c). Similarly, animal DB05 began to lose control of SIV replication after week 35 and experienced severe losses of memory CD4+ T cells from both the blood and BAL specimens. Macaque DB05 had to be euthanized at week 80 p.i. because of its deteriorating clinical status. The third SIV controller (DA87) has maintained remarkably low levels of plasma viremia. Its plasma virus load at week 78 p.i. was 127 RNA copies/ml, and levels of memory CD4+ T lymphocytes, in both the blood and BAL fluid, have been durably maintained.
Antiviral cytotoxic T-lymphocyte (CTL) responses in SIV-infected macaques are known to select for viral escape variants bearing mutations in, or close to, epitopes restricted by the selecting MHC class I alleles. We investigated if efficient immune responses restricted by protective MHC alleles led to viral evolution in controller macaques. The analysis was limited to virus present in RhDB05 and RhDA87, animals carrying Mamu-A*02 and Mamu-B*08, respectively, since several CTL epitopes have been characterized for these two MHC alleles but not for Mamu-B*290101 (14, 16, 32). Viral sequences encoding Gag, Env, Vif, or Nef were amplified from plasma samples. In viral sequences obtained from the Mamu-A*02 controller macaque RhDB05 at week 80 p.i., amino acid substitutions were identified in three Mamu-A*02-restricted epitopes (Gag GY9, Env RY8, and Nef YY9) (Fig. 4). Similarly, viral escape mutants in three Mamu-B*08-restricted epitopes (Vif RL9, Vif RL8, and Nef RL9) were detected in samples from the Mamu-B*08-positive macaque RhDA87 at week 78 p.i. Additionally, an alanine-to-proline substitution also was found one amino acid upstream from the Mamu-B*08 restricted epitope Nef RL10. This mutation, observed previously in SIV-infected Mamu-B*08-positive macaques, likely affects the proper generation of Mamu-B*08 binding peptide by the proteasome (14). Therefore, the presence of protective MHC alleles in two controller macaques was associated with the selection of CTL escape variants, suggesting that CTL activity restricted by protective MHC alleles played a role in controlling plasma viremia.
FIG. 4.
Amino acid changes associated with SIV escape variants in Mamu-B*-08- and Mamu-A*02-bearing rhesus macaques. The number of independent clones sequenced is indicated in parentheses. The Nef RL10 sequence contains an amino acid residue immediately N-terminal to the epitope, alanine (A), which has undergone mutation.
Thus, of the original six tenofovir-treated monkeys, only two remained alive at week 80 p.i. Both carried protective MHC class I alleles. One (DA63), however, had high levels of plasma viremia and declining levels of CD4+ T cells and was destined to develop immunodeficiency.
DISCUSSION
In these experiments, we have investigated whether a 28-day course of potent ART, initiated at a time (48 h p.i.) following SIV inoculation when the acquisition of a viral infection was virtually assured, would sufficiently sensitize the immune system to result in controlled virus replication when treatment was stopped. The administration of tenofovir 48 h after SIV inoculation did, in fact, delay virus replication in all of the treated rhesus macaques. However, the three animals deriving the greatest benefit (lowering the viral set point by nearly 3 logs for 6 months) were genetically privileged; they carried MHC class I alleles associated with the suppression of virus replication. The three PMPA-treated animals bearing nonprotective MHC alleles were not as fortunate; they generated levels of set point viremia comparable to that observed in control animals, and all three have died with symptoms of immunodeficiency.
This study is an extension of two previous reports describing ART initiated very early during acute SIV infection. When Lifson et al. initiated tenofovir treatment at 24 h p.i., three of four SIVsmE660-infected macaques experienced sterilizing protection both during and following drug administration (12). If PMPA was started at 72 h p.i., two of four animals became viremic during the 28-day course of therapy, and virus replication was measurable in all four monkeys following the cessation of ART. In a separate study using SIVmac239-inoculated animals also treated with PMPA beginning at 24 h p.i., the levels of viremia were low to undetectable during or following ART (11). However, in contrast to the SIVsmE660 macaque recipients of tenofovir, which generated SIV-specific proliferative responses in the absence of detectable virus replication during ART administration and resisted rechallenge, the treated SIVmac239 monkeys failed to produce measurable immune responses and readily became infected following rechallenge with homologous virus.
Therefore, 48 h after SIVmac239 infection seemed to be the appropriate time to begin tenofovir therapy. The establishment of an SIV infection would be ensured, and the possible development of effective immune responses might occur at a time when virus-induced damage to the immune system had been minimized. Our results show that PMPA was effective in suppressing SIV replication during the 4 weeks of ART. Cell-associated viral DNA was undetectable in all six treated monkeys, and plasma viremia was below detection limits in four of the six macaques. In addition, memory CD4+ T cells in the blood and at an effector site (lungs) were maintained at preinfection levels in all six animals during drug administration. Thus, the experimental regimen used had permitted infection acquisition, suppressed virus replication, and prevented injury to the immune system.
Unfortunately, however, all six of the PMPA-treated monkeys produced modest to high levels of plasma viremia within 1 to 2 weeks of ART cessation, suggesting that the potent suppression of virus replication mediated by 4 weeks of tenofovir treatment resulted in insufficient immune stimulation despite the preservation of the CD4+ T-lymphocyte population. As had been reported previously by Lifson et al. for PMPA therapy initiated at 24 h p.i. in SIVmac239-inoculated monkeys (11), we also observed low to background levels of anti-SIVmac239 immune responses in circulating CD8+ T cells during the 28-day course of treatment that was started at 48 h p.i. (Table 2). However, SIV Gag-specific responses were measurable in CD8+ T cells present in BAL specimens obtained from most of the macaques during the period of PMPA therapy (Table 3), but this immune sensitization appeared to have minimal long-term effects.
Given that Mamu-A*01-negative and otherwise randomly selected macaques were used in this study, initially we were intrigued by the resolution of viral set points at two discrete levels, differing by 300-fold, in controller and noncontroller animals at week 15 p.i. In the three controller animals, set points were maintained at a range of 1 × 103 RNA copies/ml plasma for the initial 6 months of their viral infections. This degree of control rarely is observed in SIVmac239-infected Indian-origin rhesus macaques. To determine whether any virologic or immunologic parameter could predict controller or noncontroller status, data collected during and immediately following tenofovir treatment was evaluated retrospectively. Clearly, peak viremia following ART was not a predictor of the subsequent clinical course. For example, and not unexpectedly, the mean level of peak plasma viremia immediately following the cessation of PMPA treatment was 20-fold lower in controller than in noncontroller animals (1 × 105 and 2.1 × 106 RNA copies/ml, respectively) (Fig. 1b). Nevertheless, one controller macaque (DB05), with a relatively low peak virus load (4.0 × 104 RNA copies/ml), eventually died from immunodeficiency at week 80 p.i., whereas another controller monkey (DA87), with the highest peak viremia (8.3 × 105 RNA copies/ml at week 6 p.i.), was asymptomatic, with a plasma load of only 127 RNA copies/ml at week 78 p.i.
Similarly, the frequencies of circulating virus-specific CD8+ T lymphocytes present during or immediately after PMPA treatment did not predict which animals were destined to become controllers or noncontrollers. The levels of these SIV-specific CD8+ T cells were low to undetectable during the 4-week course of tenofovir therapy and increased in both groups following the cessation of ART. As noted earlier, SIV Gag-specific CD8+ T-cell responses in BAL samples were higher in two noncontroller animals during and immediately following drug administration, most likely reflecting somewhat higher levels of antigen production in these macaques rather than foretelling clinical outcomes.
ELISA and immunoblotting experiments both revealed that the controller macaques generated earlier and higher-titer anti-SIV gp130 antibody responses compared to those of the noncontrollers. However, because virus-specific NAbs were undetectable at weeks 10 and 22 in both controller and noncontroller macaques, the significance of the binding antibody results presently is unclear.
As noted earlier, the establishment of viral set points at either high or low levels in the treated monkeys correlated with the presence of MHC class I alleles that are reported to control SIV replication. Only controller animals carried protective MHC alleles, and every one suppressed its set point viremia at or below the level of 103 viral RNA copies/ml for at least 6 months. The emergence of CTL escape variants in two of the controller macaques suggested that CTL activity restricted by protective MHC alleles played a role in controlling plasma viremia when ART was discontinued. One could ask whether the suppression of virus replication observed in the controller group of treated animals simply reflected the presence of protective MHC alleles rather than a consequence of early postinoculation PMPA therapy. At present, we have no direct experimental data supporting the latter suggestion, although some previously published reports pertaining to SIV elite controllers are relevant to this question. The Mamu-B*08 allele previously has been reported to be overrepresented in a significant fraction (38%) of SIV elite controllers (15). Although SIV-infected Mamu-B*08+ rhesus macaques do not experience significant reductions in peak viremia during acute infections, they are able to reduce set point virus loads by 7.3-fold compared to the loads of other elite controllers. In this regard, the suppression of plasma viremia to 127 RNA copies/ml at week 78 p.i. in monkey DA87 (carrying the Mamu-B*08 allele) would appear to be quite unusual. In addition, although Mamu-A*02+ and Mamu-B*29+ macaques also are overrepresented in elite controller cohorts, their capacity to suppress set point viremia is quite limited compared to that of the more potent control reported for Mamu-B*08 and Mamu-B*17 (15). It was surprising, therefore, that controller macaques DB05 and DA63, carrying the Mamu-A*02 and Mamu-B*29 alleles, respectively, were able to effectively control plasma viremia to the range of 103 viral RNA copies/ml for 6 to 9 months. In the context of previously published studies, the potent suppression of virus replication in these two animals may not be due solely to the modest protective effects of the Mamu-A*02 and Mamu-B*29 alleles.
Clearly, the prevention of immediate virus-induced damage to the immune system mediated by p.i. ART by itself did not lead to augmented immunologic responses capable of controlling SIV in monkeys not bearing protective MHC alleles. These results emphasize the incredibly narrow window of opportunity for controlling de novo SIV and, by extension, HIV infections in individuals not bearing protective MHC alleles. Starting PMPA treatment at 24 h did confer the durable control of SIV replication in most, but not all, inoculated animals. ART commencing at 48 h p.i. clearly was too late; the suppression of virus replication at 48 h p.i. did not sensitize the immune system for subsequent control SIV replication but merely delayed the emergence of detectable virus. In this context, we have reported previously that the transfer of high titers of antiviral NAbs at 6 h p.i., but not at 24 h p.i., potently controlled virus replication in the macaque model (24), compressing even further the time interval between lentivirus exposure and successful antiviral intervention.
Acknowledgments
We are indebted to Boris Skopets and Rahel Petros for their diligence and assistance in the maintenance of our animals; Ronald Willey for performing virus neutralization assays; Ranjini Iyengar and Robin Kruthers for determining plasma viral RNA levels; Martha Vazquez and Norbert Bischofberger, Gilead Science, Inc., for providing tenofovir; and John Loffredo for providing information about protective MHC class I alleles.
This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
Published ahead of print on 6 May 2009.
REFERENCES
- 1.Brenchley, J. M., T. W. Schacker, L. E. Ruff, D. A. Price, J. H. Taylor, G. J. Beilman, P. L. Nguyen, A. Khoruts, M. Larson, A. T. Haase, and D. C. Douek. 2004. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J. Exp. Med. 200749-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carrington, M., and S. J. O'Brien. 2003. The influence of HLA genotype on AIDS. Annu. Rev. Med. 54535-551. [DOI] [PubMed] [Google Scholar]
- 3.Committee on the Care and Use of Laboratory Animals. 1985. Guide for the care and use of laboratory animals. Department of Health and Human Services publication no. NIH 85-23. National Institutes of Health, Bethesda, MD.
- 4.Endo, Y., T. Igarashi, Y. Nishimura, C. Buckler, A. Buckler-White, R. Plishka, D. S. Dimitrov, and M. A. Martin. 2000. Short- and long-term clinical outcomes in rhesus monkeys inoculated with a highly pathogenic chimeric simian/human immunodeficiency virus. J. Virol. 746935-6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guadalupe, M., E. Reay, S. Sankaran, T. Prindiville, J. Flamm, A. McNeil, and S. Dandekar. 2003. Severe CD4+ T-cell depletion in gut lymphoid tissue during primary human immunodeficiency virus type 1 infection and substantial delay in restoration following highly active antiretroviral therapy. J. Virol. 7711708-11717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hill, C. M., H. Deng, D. Unutmaz, V. N. Kewalramani, L. Bastiani, M. K. Gorny, S. Zolla-Pazner, and D. R. Littman. 1997. Envelope glycoproteins from human immunodeficiency virus types 1 and 2 and simian immunodeficiency virus can use human CCR5 as a coreceptor for viral entry and make direct CD4-dependent interactions with this chemokine receptor. J. Virol. 716296-6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Igarashi, T., O. K. Donau, H. Imamichi, M. J. Dumaurier, R. Sadjadpour, R. J. Plishka, A. Buckler-White, C. Buckler, A. F. Suffredini, H. C. Lane, J. P. Moore, and M. A. Martin. 2003. Macrophage-tropic simian/human immunodeficiency virus chimeras use CXCR4, not CCR5, for infections of rhesus macaque peripheral blood mononuclear cells and alveolar macrophages. J. Virol. 7713042-13052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jayaraman, P., T. Zhu, L. Misher, D. Mohan, L. Kuller, P. Polacino, B. A. Richardson, H. Bielefeldt-Ohmann, D. Anderson, S. L. Hu, and N. L. Haigwood. 2007. Evidence for persistent, occult infection in neonatal macaques following perinatal transmission of simian-human immunodeficiency virus SF162P3. J. Virol. 81822-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lafont, B. A., C. M. McGraw, S. A. Stukes, A. Buckler-White, R. J. Plishka, R. A. Byrum, V. M. Hirsch, and M. A. Martin. 2007. The locus encoding an oligomorphic family of MHC-A alleles (Mane-A*06/Mamu-A*05) is present at high frequency in several macaque species. Immunogenetics 59211-223. [DOI] [PubMed] [Google Scholar]
- 10.Li, Q., L. Duan, J. D. Estes, Z. M. Ma, T. Rourke, Y. Wang, C. Reilly, J. Carlis, C. J. Miller, and A. T. Haase. 2005. Peak SIV replication in resting memory CD4+ T cells depletes gut lamina propria CD4+ T cells. Nature 4341148-1152. [DOI] [PubMed] [Google Scholar]
- 11.Lifson, J. D., M. Piatak, Jr., A. N. Cline, J. L. Rossio, J. Purcell, I. Pandrea, N. Bischofberger, J. Blanchard, and R. S. Veazey. 2003. Transient early postinoculation anti-retroviral treatment facilitates controlled infection with sparing of CD4+ T cells in gut-associated lymphoid tissues in SIVmac239-infected rhesus macaques, but not resistance to rechallenge. J. Med. Primatol. 32201-210. [DOI] [PubMed] [Google Scholar]
- 12.Lifson, J. D., J. L. Rossio, R. Arnaout, L. Li, T. L. Parks, D. K. Schneider, R. F. Kiser, V. J. Coalter, G. Walsh, R. J. Imming, B. Fisher, B. M. Flynn, N. Bischofberger, M. Piatak, Jr., V. M. Hirsch, M. A. Nowak, and D. Wodarz. 2000. Containment of simian immunodeficiency virus infection: cellular immune responses and protection from rechallenge following transient postinoculation antiretroviral treatment. J. Virol. 742584-2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loffredo, J. T., A. T. Bean, D. R. Beal, E. J. Leon, G. E. May, S. M. Piaskowski, J. R. Furlott, J. Reed, S. K. Musani, E. G. Rakasz, T. C. Friedrich, N. A. Wilson, D. B. Allison, and D. I. Watkins. 2008. Patterns of CD8+ immunodominance may influence the ability of Mamu-B*08-positive macaques to naturally control simian immunodeficiency virus SIVmac239 replication. J. Virol. 821723-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loffredo, J. T., T. C. Friedrich, E. J. Leon, J. J. Stephany, D. S. Rodrigues, S. P. Spencer, A. T. Bean, D. R. Beal, B. J. Burwitz, R. A. Rudersdorf, L. T. Wallace, S. M. Piaskowski, G. E. May, J. Sidney, E. Gostick, N. A. Wilson, D. A. Price, E. G. Kallas, H. Piontkivska, A. L. Hughes, A. Sette, and D. I. Watkins. 2007. CD8+ T cells from SIV elite controller macaques recognize Mamu-B*08-bound epitopes and select for widespread viral variation. PLoS ONE 2e1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loffredo, J. T., J. Maxwell, Y. Qi, C. E. Glidden, G. J. Borchardt, T. Soma, A. T. Bean, D. R. Beal, N. A. Wilson, W. M. Rehrauer, J. D. Lifson, M. Carrington, and D. I. Watkins. 2007. Mamu-B*08-positive macaques control simian immunodeficiency virus replication. J. Virol. 818827-8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loffredo, J. T., J. Sidney, C. Wojewoda, E. Dodds, M. R. Reynolds, G. Napoe, B. R. Mothe, D. H. O'Connor, N. A. Wilson, D. I. Watkins, and A. Sette. 2004. Identification of seventeen new simian immunodeficiency virus-derived CD8+ T cell epitopes restricted by the high frequency molecule, Mamu-A*02, and potential escape from CTL recognition. J. Immunol. 1735064-5076. [DOI] [PubMed] [Google Scholar]
- 17.Mattapallil, J. J., D. C. Douek, B. Hill, Y. Nishimura, M. Martin, and M. Roederer. 2005. Massive infection and loss of memory CD4+ T cells in multiple tissues during acute SIV infection. Nature 4341093-1097. [DOI] [PubMed] [Google Scholar]
- 18.Mehandru, S., M. A. Poles, K. Tenner-Racz, A. Horowitz, A. Hurley, C. Hogan, D. Boden, P. Racz, and M. Markowitz. 2004. Primary HIV-1 infection is associated with preferential depletion of CD4+ T lymphocytes from effector sites in the gastrointestinal tract. J. Exp. Med. 200761-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Migueles, S. A., M. S. Sabbaghian, W. L. Shupert, M. P. Bettinotti, F. M. Marincola, L. Martino, C. W. Hallahan, S. M. Selig, D. Schwartz, J. Sullivan, and M. Connors. 2000. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc. Natl. Acad. Sci. USA 972709-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muhl, T., M. Krawczak, P. Ten Haaft, G. Hunsmann, and U. Sauermann. 2002. MHC class I alleles influence set point viral load and survival time in simian immunodeficiency virus-infected rhesus monkeys. J. Immunol. 1693438-3446. [DOI] [PubMed] [Google Scholar]
- 21.Naidu, Y. M., H. W. Kestler III, Y. Li, C. V. Butler, D. P. Silva, D. K. Schmidt, C. D. Troup, P. K. Sehgal, P. Sonigo, M. D. Daniel, et al. 1988. Characterization of infectious molecular clones of simian immunodeficiency virus (SIVmac) and human immunodeficiency virus type 2: persistent infection of rhesus monkeys with molecularly cloned SIVmac. J. Virol. 624691-4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishimura, Y., C. R. Brown, J. J. Mattapallil, T. Igarashi, A. Buckler-White, B. A. Lafont, V. M. Hirsch, M. Roederer, and M. A. Martin. 2005. Resting naive CD4+ T cells are massively infected and eliminated by X4-tropic simian-human immunodeficiency viruses in macaques. Proc. Natl. Acad. Sci. USA 1028000-8005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishimura, Y., T. Igarashi, A. Buckler-White, C. Buckler, H. Imamichi, R. M. Goeken, W. R. Lee, B. A. Lafont, R. Byrum, H. C. Lane, V. M. Hirsch, and M. A. Martin. 2007. Loss of naive cells accompanies memory CD4+ T-cell depletion during long-term progression to AIDS in simian immunodeficiency virus-infected macaques. J. Virol. 81893-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishimura, Y., T. Igarashi, N. L. Haigwood, R. Sadjadpour, O. K. Donau, C. Buckler, R. J. Plishka, A. Buckler-White, and M. A. Martin. 2003. Transfer of neutralizing IgG to macaques 6 h but not 24 h after SHIV infection confers sterilizing protection: implications for HIV-1 vaccine development. Proc. Natl. Acad. Sci. USA 10015131-15136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Connor, D. H., B. R. Mothe, J. T. Weinfurter, S. Fuenger, W. M. Rehrauer, P. Jing, R. R. Rudersdorf, M. E. Liebl, K. Krebs, J. Vasquez, E. Dodds, J. Loffredo, S. Martin, A. B. McDermott, T. M. Allen, C. Wang, G. G. Doxiadis, D. C. Montefiori, A. Hughes, D. R. Burton, D. B. Allison, S. M. Wolinsky, R. Bontrop, L. J. Picker, and D. I. Watkins. 2003. Major histocompatibility complex class I alleles associated with slow simian immunodeficiency virus disease progression bind epitopes recognized by dominant acute-phase cytotoxic-T-lymphocyte responses. J. Virol. 779029-9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otting, N., C. M. Heijmans, R. C. Noort, N. G. de Groot, G. G. Doxiadis, J. J. van Rood, D. I. Watkins, and R. E. Bontrop. 2005. Unparalleled complexity of the MHC class I region in rhesus macaques. Proc. Natl. Acad. Sci. USA 1021626-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pitcher, C. J., S. I. Hagen, J. M. Walker, R. Lum, B. L. Mitchell, V. C. Maino, M. K. Axthelm, and L. J. Picker. 2002. Development and homeostasis of T cell memory in rhesus macaque. J. Immunol. 16829-43. [DOI] [PubMed] [Google Scholar]
- 28.Pitcher, C. J., C. Quittner, D. M. Peterson, M. Connors, R. A. Koup, V. C. Maino, and L. J. Picker. 1999. HIV-1-specific CD4+ T cells are detectable in most individuals with active HIV-1 infection, but decline with prolonged viral suppression. Nat. Med. 5518-525. [DOI] [PubMed] [Google Scholar]
- 29.Shibata, R., F. Maldarelli, C. Siemon, T. Matano, M. Parta, G. Miller, T. Fredrickson, and M. A. Martin. 1997. Infection and pathogenicity of chimeric simian-human immunodeficiency viruses in macaques: determinants of high virus loads and CD4 cell killing. J. Infect. Dis. 176362-373. [DOI] [PubMed] [Google Scholar]
- 30.Van Rompay, K. K., J. M. Cherrington, M. L. Marthas, C. J. Berardi, A. S. Mulato, A. Spinner, R. P. Tarara, D. R. Canfield, S. Telm, N. Bischofberger, and N. C. Pedersen. 1996. 9-[2-(Phosphonomethoxy)propyl]adenine therapy of established simian immunodeficiency virus infection in infant rhesus macaques. Antimicrob. Agents Chemother. 402586-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Veazey, R. S., M. DeMaria, L. V. Chalifoux, D. E. Shvetz, D. R. Pauley, H. L. Knight, M. Rosenzweig, R. P. Johnson, R. C. Desrosiers, and A. A. Lackner. 1998. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science 280427-431. [DOI] [PubMed] [Google Scholar]
- 32.Vogel, T. U., T. C. Friedrich, D. H. O'Connor, W. Rehrauer, E. J. Dodds, H. Hickman, W. Hildebrand, J. Sidney, A. Sette, A. Hughes, H. Horton, K. Vielhuber, R. Rudersdorf, I. P. De Souza, M. R. Reynolds, T. M. Allen, N. Wilson, and D. I. Watkins. 2002. Escape in one of two cytotoxic T-lymphocyte epitopes bound by a high-frequency major histocompatibility complex class I molecule, Mamu-A*02: a paradigm for virus evolution and persistence? J. Virol. 7611623-11636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422307-312. [DOI] [PubMed] [Google Scholar]
- 34.Yant, L. J., T. C. Friedrich, R. C. Johnson, G. E. May, N. J. Maness, A. M. Enz, J. D. Lifson, D. H. O'Connor, M. Carrington, and D. I. Watkins. 2006. The high-frequency major histocompatibility complex class I allele Mamu-B*17 is associated with control of simian immunodeficiency virus SIVmac239 replication. J. Virol. 805074-5077. [DOI] [PMC free article] [PubMed] [Google Scholar]