Abstract
Nested chromosomal deletions are powerful genetic tools. They are particularly suited for identifying essential genes in development either directly or by screening induced mutations against a deletion. To apply this approach to the functional analysis of mouse chromosome 2, a strategy for the rapid generation of nested deletions with Cre recombinase was developed and tested. A loxP site was targeted to the Notch1 gene on chromosome 2. A targeted line was cotransfected with a second loxP site and a plasmid for transient expression of Cre. Independent random integrations of the second loxP site onto the targeted chromosome in direct repeat orientation created multiple nested deletions. By virtue of targeting in an F1 hybrid embryonic stem cell line, F1(129S1×Cast/Ei), the deletions could be verified and rapidly mapped. Ten deletions fell into seven size classes, with the largest extending six or seven centiMorgans. The cytology of the deletion chromosomes were determined by fluorescent in situ hybridization. Eight deletions were cytologically normal, but the two largest deletions had additional rearrangements. Three deletions, including the largest unrearranged deletion, have been transmitted through the germ line. Several endpoints also have been cloned by plasmid rescue. These experiments illustrate the means to rapidly create and map deletions anywhere in the mouse genome. They also demonstrate an improved method for generating nested deletions in embryonic stem cells.
Nested chromosomal deletions have a number of uses. They can identify essential genes by the phenotype of deletion homozygotes. They can be used in mutagenesis screens where induced mutations are screened against a deletion to reveal a phenotype in two generations rather than three. They can be used to localize mutations and markers (1). They can be used in genetic tests to distinguish null alleles from hypomorphic alleles. Deletion endpoints can provide a starting point for the positional cloning of genes (2). All of these uses have been beautifully exemplified by a set of nested deletions over the albino locus on mouse chromosome 7, which has been used to define essential genes, conduct mutagenesis, and map and clone genes (2–4). Complementation analysis was used to first assign functions to regions of the chromosome covered by the deletions (5–7). N-ethyl-N-nitrosourea-induced mutations were recovered with a large 11-centiMorgan (cM) deletion and assigned into complementation groups (8). The deletions were used to map the induced mutations (8). Deletion endpoints were cloned and then used for positional cloning of genes (3). One of the genes cloned was eed, the mouse homologue of Drosophila extra sex-combs, an important regulator of axial patterning (9).
Making nested deletions directly in mice, however, generally has been limited to regions containing visible markers (2, 10). In addition, large numbers of mice need to be screened to recover multiple deletions at the same locus (11). The ability to induce deletions in embryonic stem (ES) cells and to select specific cell lines for germ-line transmission has provided an attractive alternative. As a result, a number of laboratories have created large deletions in mice (12–21).
Currently there are a number of methods available for making deletions in ES cells. A review of them reveals several shortcomings. One type of strategy using Cre recombinase depends on targeting Cre recognition sequences (loxP) to both ends of a deletion (12, 13). Considerable effort is required for each deletion: both endpoints have to be cloned in advance, up to four targeting vectors have to be built, and three transfection steps are required. Also germ-line transmission was tested for all intermediates. The efficiency of this approach has been improved by the development of libraries of premade insertion targeting vectors and the repair of a point mutation in one of the selection cassettes (22, 23). It is also possible to cotransfect the second targeting vector and a Cre expression plasmid, eliminating the third transfection step (13). But that still only generates one deletion with endpoints that have been previously cloned.
Because of these limitations more efficient techniques to generate multiple nested deletions have been developed. One set of methods uses radiation to induce deletions in an ES cell line targeted with a negative selectable marker (14, 16, 24). These are efficient at generating nested deletions. However, the endpoints of deletions made by radiation are harder to clone and cannot be tagged with visible markers (22). There are also data suggesting a 40% frequency of secondary rearrangements with radiation (24).
Recently, a Cre recombinase-based strategy was extended with the use of retroviral insertions to induce multiple deletions with DNA-tagged endpoints (19). However, that requires three transfection steps and the insertion lines are passaged in pools. As a result, the clonality of any given deletion cell line is unknown and needs to be determined (19).
With the goal of improving available techniques, an efficient method for inducing nested deletions with DNA-tagged endpoints was developed. This method was applied to the proximal region of mouse chromosome 2, starting at the Notch1 gene. The chief advantage of the method described here is that it is rapid. Two transfection steps are required. The deletions can be mapped directly in ES cells through the use of an F1 hybrid cell line (24). A DNA tag permits cloning of the deletion endpoints. The germ-line deletions described in this report will provide tools for the functional analysis of this region of mouse chromosome 2, and by extension the syntenic region of the human genome, 9q34.
Materials and Methods
DNA Constructs.
The neomycin resistance (neo)-loxP-thymidine kinase (tk) cassette was created by cloning the neo and tk genes from pPNT (25) into Bluescript (Stratagene). The Notch1 targeting vector consisted of 2.1 kb of 5′ homology, neo-loxP-tk, 6.3 kb of 3′ homology (26), and a β-actin diphtheria toxin cassette (27). tk-loxP-puromycin resistance (puro) was made by cloning tk from pPNT (25) and a puro gene (28) into Bluescript. To protect tk from exonucleolytic attack, a 1.5-kb fragment of nonessential bacterial DNA was inserted 5′ of tk, and the vector was linearized 5′ of this insert. The Cre expression plasmid pOG231 was a gift from Stephen O'Gorman, Salk Institute, San Diego, CA.
Cell Culture.
CAST no. 1 ES cells are an F1 hybrid between the 129/Sv+Tyr+p (abbreviated 129S1; refs. 29 and 30) and CAST/Ei subspecies of mice (24) and were grown on gelatinized plastic in media supplemented with leukemia inhibitory factor. Electroporations followed standard protocols (31), unless noted. Cell lines with a targeted insertion of neo-loxP-tk at Notch1 were screened by Southern blot with external probes both 5′ and 3′ of the targeted event. Targeted lines also were screened with internal probes against secondary insertions, and the chromosome number was determined. Three targeted lines were tested for their ability to contribute to chimeric animals. One cell line, NALT #3, was selected for further derivation. Targeted and deletion cell lines were maintained in 100 μg/ml G418 (GIBCO) throughout all experiments. To generate deletions, 5.6 × 106 neo-loxP-tk-targeted ES cells were electroporated with 25 μg of supercoiled pOG231 and 50 μg linearized tk-loxP-puro per cuvette. Each cuvette was plated onto two 100-mm dishes. Selection with 0.75 μg/ml puromycin (Sigma) was begun 24 h after electroporation and replenished daily for 3 days. Selection with 0.2 μM FIAU (1–2′-deoxy-2′-fluoro-β-d-arabinofuranosyl-5-iodouracil) was begun 5 days after electroporation and replenished daily for 3 days. At this point colonies were picked into 24-well dishes in media supplemented with 100 μg/ml G418 and grown up for both freezing and confirmation by Southern blot. After deletion characterization (see below), deletion lines without additional rearrangements were selected for transmission through the germ line, and transmission was confirmed by Southern blot.
Deletion Characterization.
Simple sequence length polymorphisms and single-strand conformation polymorphisms were detected by standard methods (D.M.C., unpublished work; refs. 32 and 33). Fluorescent in situ hybridization (FISH) was performed with 22 different yeast artificial chromosomes (YACs) mapping to chromosome 2. Of these, seven (188E3, 144F11, 376A1, 304E7, 456A8, 158D10, and 350B3) were obtained by screening the Whitehead Large Insert Library (Research Genetics, Huntsville, AL) with primers to genes in the vicinity of Notch1 (D.M.C., unpublished work). The remaining 15 YACs were chosen to span the length of chromosome 2 and were identified from the Massachusetts Institute of Technology/Whitehead Genome Center on-line database (www.genome.wi.mit.edu) and obtained from Research Genetics. The approximate chromosomal locations (in cM) associated with each of the 22 YACs are provided in Table 1. FISH was performed on unstained slides, using standard methodology (34), with minor modifications. In each of the hybridization experiments, at least two YACs were used, one in the vicinity of Notch1 (test probe) and one from distal chromosome 2 (control probe). For each cell line, a minimum of seven hybridization experiments were conducted, one for each of seven test probes. In each instance, at least 10–15 metaphases were analyzed. Chromosome counts were made to ensure that the cell lines had 40 chromosomes, and the cells were scanned to determine the presence or absence of the test and control probes on each of the two chromosomes 2. Digital images were captured with a Zeiss epifluorescence microscope equipped with a Photometrics Sensys camera, using Vysis quipsmfish software.
Table 1.
Summary of FISH mapping
| Line | Chromosome no. | Number of chromosome 2 FISH signals with each YAC (cM)
|
|||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 384A4, (2) | 443A7, (8) | 188E3, (9) | 144F11, (9) | 376A1, (12) | 304E7, (13) | 456A8, (13) | 158D10, (15) | 350B3, (20) | 303C2, (32) | 406F11, (38) | 110E6, (45) | 88C11, (48) | 215F12, (54) | 387G3, (60) | 328B10, (66) | 426C1, (68) | 345E6, (73) | 329A12, (85) | 150F4, (85) | 279C5, (87) | 188A3, (98) | ||
| 7 | 40 | 1* | 2 | 1 | 1? | 2 | 2 | 2 | 2 | ||||||||||||||
| 10 | 40 | 1* | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 3 | 3 | 3 | 3 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | ||
| 9 | 40 | 2 | 2 | 1 | 1 | 1 | 2 | 2 | 2 | ||||||||||||||
| 4 | 40 | 2 | 2 | 1 | 1 | 1 | 2 | 2 | 2 | ||||||||||||||
| 3 | 40 | 2 | 2 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | |||||||||||||
| 5 | 40 | 2 | 2 | 2 | 1 | 1 | 2 | 2 | 2 | ||||||||||||||
| 2 | 40 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 2 | ||||||||||||||
| 6 | 40 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | ||||||||||||||
| 1 | 40 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | ||||||||||||||
| 8 | 40 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | ||||||||||||||
| Targeted | 40 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | ||||||||||||||
? indicates ambiguous
An additional signal on another chromosome consistent with a translocation was observed.
Plasmid Rescue.
DNA flanking deletion endpoints was cloned by plasmid rescue (35) from XbaI- or HindIII-digested cell line DNA. Confirmation that a cloned insert flanked its respective endpoint was made by Southern blot as follows. Each deletion creates a unique puro-Bluescript-tagged junction on Southern blots with XbaI or HindIII. Coincidence with this puro-Bluescript-tagged junction was demonstrated when these blots were reprobed with rescued plasmid inserts from deletion lines 1, 5, 6, 8, and 9, indicating that these cloned inserts are their respective deletion endpoints. One or both ends of the rescue plasmids from deletion lines 1, 5, 6, and 9 were sequenced.
Results
Outline of the Deletion Strategy.
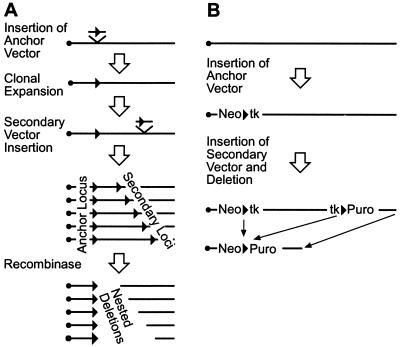
To catalyze deletions, Cre recombinase requires a pair of its recognition sequences, called loxP sites, on the same chromosome in the same orientation. Colocalization of two directly repeated loxP sites was achieved by the sequential insertion of two loxP vectors (Fig. 1A). The first vector, called the anchor, was inserted by gene targeting in ES cells, creating an anchor cell line. This line was expanded and a second loxP vector was inserted at random, creating a number of secondary insertion cell lines. Among the secondary vector insertions were some on the anchor chromosome that were both distal to and in the same orientation as the anchor loxP. These then were deleted by Cre recombinase generating multiple nested deletions from a fixed starting point (Fig. 1A). The two vectors contain a loxP site flanked by selectable markers (Fig. 1B). Positive selectable markers for neo resistance and puro resistance were used to select for the insertion events. When a second loxP vector insertion occurs on the targeted chromosome both distal to and in the same orientation as the anchor loxP, then two copies of a negative selectable marker, tk, are contained within the DNA that is to be deleted. Negative selection against the two tk genes therefore was used to select against cell lines that had not deleted (Fig. 1B). Because of the order of the selectable markers, only deletions distal to the anchor loxP were expected to survive selection. Inversion or translocation events would not delete the tk genes and therefore were not expected to survive selection. The deletion chromosomes retain plasmid sequences, permitting the cloning of deletion endpoints. These manipulations were performed in an F1 hybrid ES cell line, which made possible the rapid verification and mapping of deletions. In this ES cell line (CAST no. 1), the homologous chromosomes are highly polymorphic because the cell line is an F1 hybrid of two subspecies of mice, 129S1 and CAST/Ei (24). The 129S1 chromosome was targeted with an isogenic targeting construct. Deletions therefore were mapped by assaying for loss of the 129S1 markers along a deletion.
Figure 1.
Strategy for generating deletions. (A) To catalyze a deletion Cre recombinase requires a pair of loxP sites (arrows) on the same chromosome in the same orientation. One loxP vector, called the anchor, is inserted by targeting. The other loxP vector, called the secondary vector, is inserted at random. Cre recombinase (shown separately here but in practice introduced into cells at the same time as the secondary insertions) catalyzes the deletions. (B) The targeted anchor vector contains a neo-loxP-tk cassette. The secondary insertion vector, containing a tk-loxP-puro cassette, then is inserted at random. In the configuration illustrated, the two tk genes are internal to the deleted segment and removed: deletions therefore survive negative selection against the two tk genes. The neo and puro genes are included to select for the targeted and the random insertions, respectively.
Generation of Nested Deletions in ES Cells on Mouse Chromosome 2.
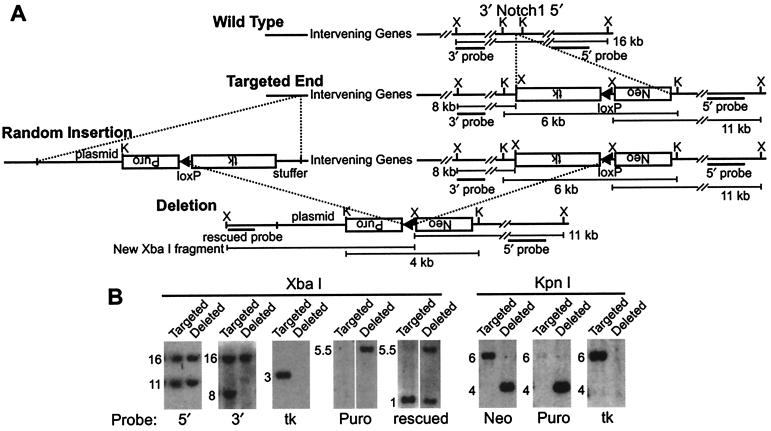
To test the strategy a cassette consisting of neo, a loxP site, and tk (neo-loxP-tk) was targeted into the Notch1 gene on mouse chromosome 2 (Fig. 2A). The targeting was confirmed on both sides by Southern blot with external probes, and cell lines with multiple insertions were eliminated by screening with internal probes (data not shown). Three targeted lines were tested for their ability to make strong chimeras (data not shown). The targeted cell lines were not expected to transmit through the germ line because of germ-line toxicity of the tk gene (36), which proved to be the case. Because tk is removed by deletions, this does not create problems in establishing deletion mice (see below). A targeted cell line was expanded and coelectroporated with two vectors: (i) a random insertion vector consisting of tk, loxP, puro, and a plasmid backbone (tk-loxP-puro), and (ii) a plasmid for transient expression of Cre recombinase (Fig. 2A). A small fraction of the tk-loxP-puro random insertions will be downstream of the targeted neo-loxP-tk, creating a pair of directly repeated loxP sites (Fig. 2A). Cre recombinase can catalyze a deletion between the loxP sites. In this configuration, both tk genes will be deleted along with the intervening DNA, providing strong selection for cell lines that have undergone the Cre-catalyzed deletion (Fig. 2A). Cell lines triply resistant to G418, puromycin, and FIAU (1–2′-deoxy-2′-fluoro-β-d-arabinofuranosyl-5-iodouracil) were recovered and tested for the expected deletion and loss of the tk genes by Southern blot (Fig. 2B). Correct deletion also generates a neo-loxP-puro recombination junction. This junction was verified by determining that neo and puro probes hybridized to the same bands on Southern blots from each of the 10 deletions (Fig. 2B). Of the 10 deletion lines recovered, one of them, line 7, had an additional insertion of the puro gene (data not shown). Based on the average of five independent experiments, one bona fide Cre-catalyzed deletion was recovered in 20 cell lines that survived selection. These 20 cell lines were generated from 2 × 107 coelectroporated cells. Some of the nondeleted cell lines may be targeted insertions of tk-loxP-puro into the tk of neo-loxP-tk, subsequently undergoing Cre-catalyzed deletion, resulting in inactivation of both tks (data not shown).
Figure 2.
Strategy for generation of nested deletions on mouse chromosome 2 by a combined targeted/random approach. (A) The Notch1 gene was targeted to insert a cassette consisting of neo, a loxP site, and tk. A targeted line was selected, and a linearized vector consisting of a stuffer fragment of bacterial DNA, tk, a loxP site, puro, and Bluescript plasmid (tk-loxP-puro) was introduced by electroporation along with a plasmid to transiently express Cre (the Cre plasmid is not shown). In a small fraction of random integrations, tk-loxP-puro is expected to insert in the orientation shown, generating a chromosome that could delete the genes between the two loxP sites. Both tk genes are lost with the deletion, generating cells that are resistant to selection for the neo (G418R) and puro (puromycinR) genes and against the tk genes (FIAUR). Cells that haven't integrated the tk-loxP-puro vector, or cells that have integrated the second vector but have not deleted the two tk genes, will not survive selection with all three drugs. Probes and predicted fragments for confirmation of targeting, deletion and plasmid rescue of the deletion junction are shown. (B) Verification of targeting, deletion, and junction fragment rescue by Southern blot. Analysis of a targeted line and a deleted line with a probe from the 5′ end of Notch1 shows that the 5′ end of the wild-type (16 kb) and targeted (11 kb) Notch1 alleles are present in both the targeted and deleted lines. However, the 3′ end of the targeted Notch1 allele is missing in the deleted line as shown by the absence of an 8-kb XbaI fragment. The tk gene is present in the targeted line (3-kb XbaI fragment), but is absent from the deleted line, as expected. A probe to puro detects a 5.5-kb fragment in the deleted line, and an XbaI fragment of the same size is detected with a probe cloned by plasmid rescue. The rescued fragment probe from deletion 5 detects a 1-kb XbaI fragment in the targeted line, and fragments of 1 kb and 5.5 kb of half intensity in the deleted line, consistent with the proposed deletion event. The neo and puro genes are present on the same 4-kb KpnI fragment in the deletion line and tk is absent in the deletion line, consistent with the predicted Cre-catalyzed recombination between the loxP sites of the two vectors. (K: KpnI; X: XbaI)
Deletion Characterization.
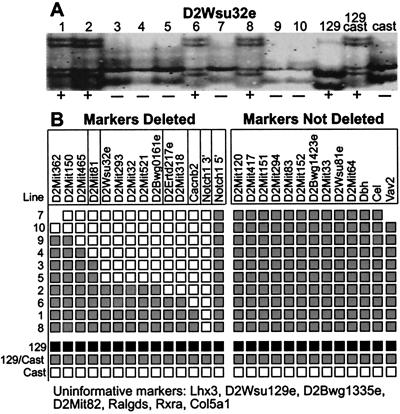
Because the parent ES cell line is an F1 hybrid of 129S1 and CAST/Ei, deletions can be mapped by assaying for loss of 129S1 alleles (24). The 129S1 alleles were lost in all cases, consistent with targeting of the Notch1 locus on the 129S1 chromosome, and with subsequent deletion of alleles on that same chromosome (Fig. 3A). A total of 26 polymorphic markers in the region of Notch1 were analyzed (Fig. 3 and see Fig. 5). Deletions were used to order the markers, assuming simple deletions with one fixed endpoint (Fig. 3B). This analysis distinguished the 10 deletions into seven different size classes. Comparison of marker order on the Whitehead radiation hybrid map shows exact correspondence to the ordering of markers by the deletions, although two markers distinguished by radiation hybrid mapping were not ordered by the deletions (37). Deletion number 9 is the largest deletion that does not have additional rearrangements (see below). The distance between the closest nondeleted markers for this line, D2Mit150 and D2Mit417, is 7.7 cM on the Whitehead genetic map (Fig. 3B). The same calculation based on the Whitehead radiation hybrid map yields an estimate of 6.2 cM for deletion line 9, which corresponds to approximately 10.4 Mb (33, 37, 38).
Figure 3.
Ten chromosome 2 deletions of seven different size classes were recovered. Verification and mapping of the deletions was performed by analysis of marker loss in cell line DNA. Because the deletions were made in an ES cell line derived from an F1 hybrid embryo (129S1 × Cast/Ei), deletions could be mapped with polymorphic markers. (A) The 129S1 allele of D2Wsu32e was lost (−) in six of the 10 lines, indicating deletions on the 129S1 chromosome 2 of these cell lines. (B) The results for markers in the vicinity of Notch1 in the 10 cell lines are summarized and the deletions are arranged from largest to smallest. Note that only 129S1 alleles were lost. The vertical lines in B separate bins of markers. Markers within bins are not ordered relative to each other within the bins.
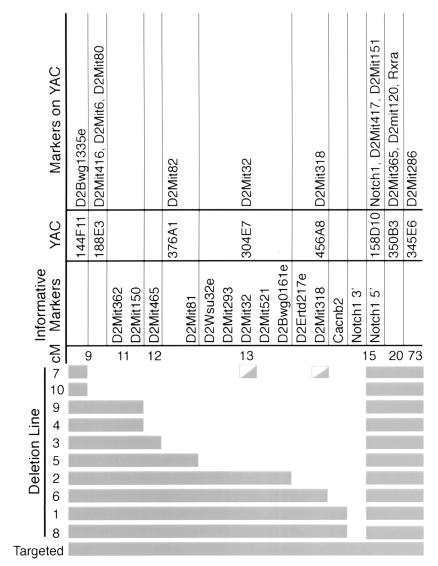
Figure 5.
Summary of chromosome 2 deletions. The deletions are arrayed from largest to smallest, and the markers are shown at the top. Line 7 had the only discrepancies between the results obtained with polymorphic markers and the results obtained by FISH. Line 7 was deleted for marker D2Mit318 but retained the YAC that is associated with it, 456A8. Line 7 also was deleted for D2Mit32 and could not be unambiguously scored for its YAC, 304E7. All lines have 40 chromosomes.
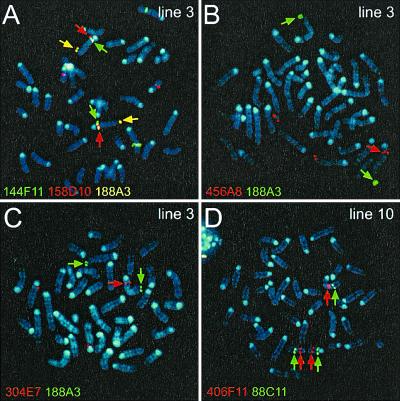
The deletions were confirmed and additional rearrangements were screened for by FISH (Fig. 4 and Table 1). Pairs of YACs were hybridized so that chromosome 2 could be unambiguously identified with a nonchimeric YAC (usually 345E6 at 73 cM). Seven of the 10 cell lines showed patterns of FISH signals consistent with the expected deletions (Figs. 4 and 5; Table 1). For the three cell lines with the smallest predicted deletions, probes mapping to the deleted portion were not available and thus these deletions were not confirmed by FISH, but the order of markers along these chromosomes was otherwise normal (Table 1). In the two cell lines with the largest predicted deletions, there were rearrangements in addition to deletions. In lines 7 and 10 proximal chromosome 2 was translocated to another unidentified chromosome (Table 1). Line 10 also has an inverted duplication involving the part of the chromosome from 32 to 48 cM (Fig. 4D and Table 1). These results indicate that the majority of deletions occurred without other rearrangements.
Figure 4.
Most deletion chromosomes do not have secondary rearrangements. Verification of cytology and mapping of deletions was performed by FISH with YAC probes to chromosome 2. Hybridization signals on chromosome 2 are indicated with arrows; signals without arrows are on chromosomes other than 2 and are caused by YAC chimerism. (A) Deletion line 3 has two chromosomes 2, both with signals for each of YACs 144F11 (green, 9 cM), 158D10 (red, 15 cM), and 188A3 (yellow, 98 cM). (B) Deletion line 3 has signal for YAC 188A3 (green, 98 cM) on both chromosomes 2, but is missing signal for 456A8 (red, 14 cM) on one. (C) Deletion line 3 again has two signals for 188A3 (green, 98 cM), but only one chromosome 2 signal for YAC 304E7 (red, 13 cM). (D) In deletion line 10, one chromosome 2 has an inverted duplication containing 406F11 (red, 38 cM) and 88C11 (green, 48 cM).
Plasmid Rescue and Germ-Line Transmission.
Because the random insertion vector included plasmid sequences, plasmid rescue was used to clone deletion endpoints. Putative junction fragments were recovered for seven of the 10 deletion cell lines (data not shown). The junction clones recovered for deletion lines 1, 5, 6, 8, and 9 hybridize back to the appropriate junction fragments in Southern analyses of deletion cell lines (Fig. 2B and data not shown). The remaining two junction clones are either too small or largely repetitive and thus are not effective probes for Southern analysis. Sequence homology searches with blast 2.0 have revealed no matches (39).
Three deletions (lines 3, 8, and 9), including the largest cytologically normal deletion, line 9 (Table 1), have been transmitted through the germ line (data not shown). Heterozygous mice carrying deletions 3, 8, and 9 are viable, fertile, and display no obvious phenotypes. Because deletions generally define null alleles, this indicates that no haploinsufficient loci with obvious phenotypes have been deleted in these mice.
Discussion
This deletion strategy has a number of advantages over other methods. First, deletion generation is rapid. Only two transfection steps are required. Few cell lines survive selection and require screening. On average, one deletion per 20 colonies screened by Southern blot was observed. A number of other methods exist for making nested deletions in ES cells. All of these methods use a selection scheme to enrich for deletion events, and then screen through the surviving cell lines to identify independent deletion lines. Because picking and screening colonies is the most labor-intensive aspect of making deletions, comparison is made to other methods by calculating the frequency of independent deletions by the number of colonies screened. This indicates the effort required with each technique.
Nested deletions can be generated with retroviral vectors (19). First, a loxP site was targeted. The targeted line was expanded and infected with a loxP retrovirus. The secondary insertions then were passaged and transfected with Cre, followed by positive selection. Hundreds of lines survived selection and these were first screened by sib selection to identify deletion events. Because passaging amplifies each retroviral insertion many of the deletion lines will have the exact same endpoint. Because it would be wasteful to put the same deletion through the germ-line multiple times, independent deletions were identified by Southern blot. In total, 49 independent deletions were identified from 1,395 surviving cell lines: an efficiency of one deletion per 28 colonies screened (19).
Multiple deletions also can be created by X or γ radiation in a two-step procedure: one for targeting a negative selectable marker and the second for irradiation and negative selection (1, 14, 16, 24). Many cell lines survived selection, and these were screened by sib selection followed by Southern blotting or PCR assay. Using γ radiation, 23 independent deletions were recovered from 142 surviving cell lines: one deletion per six colonies screened (14). Among deletions induced with X radiation, 26 independent deletions were identified from 59 surviving cell lines: one deletion per two colonies screened (24). This higher efficiency with radiation, however, is offset by lack of a DNA tag and an increased likelihood of additional rearrangements (see below).
A second advantage of the method described here is that deletions can be rapidly verified and mapped through the use of an F1 hybrid ES cell line, CAST no. 1 (24). The deletions made with the retroviral technique were first put through the germ line. They then were bred into the CAST/Ei background to fully map the extents of deletion (19). Breeding to CAST/Ei also was used to map a set of deletions made with radiation (14). It should be noted that phenotypic differences caused by the mixed genetic background of mice derived from hybrid ES cells can occur (14). However, this drawback is outweighed by the ease of mapping deletions, and once in the germ line a deletion can be bred into a desired genetic background.
A third advantage of the deletion strategy is that deletion endpoints can be rapidly cloned by virtue of a DNA tag. This is also an advantage of the retroviral technique (19). The endpoints of deletions generated with radiation require positional cloning of the breakpoint junction to characterize them. In addition, deletions created with radiation cannot be tagged with visible markers (22).
The deletions were screened against additional rearrangements by FISH. The incidence observed was low: two of 10. One of the two lines, line 7, had an additional insertion of the puro gene on Southern blots, but the rearrangements in line 10 were detected only by FISH. This suggests that screening for rearrangements is advisable when manipulating chromosomes in this way. The incidence of aberrations with deletions made with radiation appears to be higher, two of five, although more radiation-induced deletions need to be analyzed to determine whether this is a general problem (24). No FISH analysis was reported for the retroviral technique (19).
In summary, the strategy described in this report has the advantages of rapid creation of multiple deletions, rapid verification and mapping, clonable endpoints, and the means to include genes for visible traits for following the deletions in mice. Vectors that incorporate coat color markers or fluorescent proteins and random integration of the first vector are among the improvements that can be made to make the approach still more powerful. The method of making deletions is efficient enough that it could easily be used to generate deletions covering a chromosome or to map critical regions for microdeletion syndromes.
Acknowledgments
We thank J. Gasparini and S. Kendall for vector construction, B. Johnson for support, and C. LaMantia and T. Magnuson for ES cell lines and advice. D.F.L. was supported by a National Institutes of Health training grant in developmental biology. This work was supported by a grant from the Cuyahoga County Unit of the American Cancer Society to R.A.C.
Abbreviations
- cM
centiMorgan
- ES
embryonic stem
- FISH
fluorescent in situ hybridization
- YAC
yeast artificial chromosome
- tk
thymidine kinase gene
- neo
neomycin resistance gene
- puro
puromycin resistance gene
Footnotes
References
- 1.You Y, Browning V L, Schimenti J C. Methods. 1997;13:409–421. doi: 10.1006/meth.1997.0547. [DOI] [PubMed] [Google Scholar]
- 2.Rinchik E, Russell L B. In: Genome Analysis. Davies K, Tilghman S, editors. Vol. 1. Plainview, NY: Cold Spring Harbor Lab. Press; 1990. pp. 121–158. [Google Scholar]
- 3.Holdener-Kenny B, Sharan S, Magnuson T. BioEssays. 1992;14:831–838. doi: 10.1002/bies.950141208. [DOI] [PubMed] [Google Scholar]
- 4.Justice M J, Zheng B, Woychik R P, Bradley A. Methods. 1997;13:423–436. doi: 10.1006/meth.1997.0548. [DOI] [PubMed] [Google Scholar]
- 5.Russell L B. Mutat Res. 1971;11:107–123. doi: 10.1016/0027-5107(71)90036-4. [DOI] [PubMed] [Google Scholar]
- 6.Niswander L, Yee D, Rinchik E M, Russell L B, Magnuson T. Development (Cambridge, UK) 1988;102:45–53. doi: 10.1242/dev.102.1.45. [DOI] [PubMed] [Google Scholar]
- 7.Niswander L, Yee D, Rinchik E M, Russell L B, Magnuson T. Development (Cambridge, UK) 1989;105:175–182. doi: 10.1242/dev.105.1.175. [DOI] [PubMed] [Google Scholar]
- 8.Rinchik E M, Carpenter D A. Genetics. 1999;152:373–383. doi: 10.1093/genetics/152.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schumacher A, Faust C, Magnuson T. Nature (London) 1996;383:250–253. doi: 10.1038/383250a0. [DOI] [PubMed] [Google Scholar]
- 10.Cattanach B M, Burtenshaw M D, Rasberry C, Evans E P. Nat Genet. 1993;3:56–61. doi: 10.1038/ng0193-56. [DOI] [PubMed] [Google Scholar]
- 11.Rinchik E. Trends Genet. 1991;7:15–21. doi: 10.1016/0168-9525(91)90016-j. [DOI] [PubMed] [Google Scholar]
- 12.Ramirez S R, Liu P, Bradley A. Nature (London) 1995;378:720–724. doi: 10.1038/378720a0. [DOI] [PubMed] [Google Scholar]
- 13.Li Z W, Stark G, Gotz J, Rulicke T, Muller U, Weissmann C. Proc Natl Acad Sci USA. 1996;93:6158–6162. doi: 10.1073/pnas.93.12.6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.You Y, Bergstrom R, Klemm M, Lederman B, Nelson H, Ticknor C, Jaenisch R, Schimenti J. Nat Genet. 1997;15:285–288. doi: 10.1038/ng0397-285. [DOI] [PubMed] [Google Scholar]
- 15.Liu P, Zhang H, McLellan A, Vogel H, Bradley A. Genetics. 1998;150:1155–1168. doi: 10.1093/genetics/150.3.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kushi A, Edamura K, Noguchi M, Akiyama K, Nishi Y, Sasai H. Mamm Genome. 1998;9:269–273. doi: 10.1007/s003359900747. [DOI] [PubMed] [Google Scholar]
- 17.Lindsay E A, Botta A, Jurecic V, Carattini-Rivera S, Cheah Y C, Rosenblatt H M, Bradley A, Baldini A. Nature (London) 1999;401:379–383. doi: 10.1038/43900. [DOI] [PubMed] [Google Scholar]
- 18.Kimber W L, Hsieh P, Hirotsune S, Yuva-Paylor L, Sutherland H F, Chen A, Ruiz-Lozano P, Hoogstraten-Miller S L, Chien K R, Paylor R, et al. Hum Mol Genet. 1999;8:2229–2237. doi: 10.1093/hmg/8.12.2229. [DOI] [PubMed] [Google Scholar]
- 19.Su H, Wang X, Bradley A. Nat Genet. 2000;24:92–95. doi: 10.1038/71756. [DOI] [PubMed] [Google Scholar]
- 20.Schlake T, Schupp I, Kutsche K, Mincheva A, Lichter P, Boehm T. Oncogene. 1999;18:6078–6082. doi: 10.1038/sj.onc.1203021. [DOI] [PubMed] [Google Scholar]
- 21.Zhu Y, Jong M C, Frazer K A, Gong E, Krauss R M, Cheng J F, Boffelli D, Rubin E M. Proc Natl Acad Sci USA. 2000;97:1137–1142. doi: 10.1073/pnas.97.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng B, Mills A A, Bradley A. Nucleic Acids Res. 1999;27:2354–2360. doi: 10.1093/nar/27.11.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng B, Sage M, Sheppeard E A, Jurecic V, Bradley A. Mol Cell Biol. 2000;20:648–655. doi: 10.1128/mcb.20.2.648-655.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas J W, LaMantia C, Magnuson T. Proc Natl Acad Sci USA. 1998;95:1114–1119. doi: 10.1073/pnas.95.3.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tybulewicz V L, Crawford C E, Jackson P K, Bronson R T, Mulligan R C. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- 26.Conlon R, Reaume A G, Rossant J. Development (Cambridge, UK) 1995;121:1533–1545. doi: 10.1242/dev.121.5.1533. [DOI] [PubMed] [Google Scholar]
- 27.Yagi T, Ikawa Y, Yoshida K, Shigetani Y, Takeda N, Mabuchi I, Yamamoto T, Aizawa S. Proc Natl Acad Sci USA. 1990;87:9918–9922. doi: 10.1073/pnas.87.24.9918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tucker K L, Beard C, Dausmann J, Jackson-Grusby L, Laird P W, Lei H, Li E, Jaenisch R. Genes Dev. 1996;10:1008–1020. doi: 10.1101/gad.10.8.1008. [DOI] [PubMed] [Google Scholar]
- 29.Simpson E M, Linder C C, Sargent E E, Davisson M T, Mobraaten L E, Sharp J J. Nat Genet. 1997;16:19–27. doi: 10.1038/ng0597-19. [DOI] [PubMed] [Google Scholar]
- 30.Threadgill D W, Yee D, Matin A, Nadeau J H, Magnuson T. Mamm Genome. 1997;8:390–393. doi: 10.1007/s003359900453. [DOI] [PubMed] [Google Scholar]
- 31.Wurst W, Joyner A L. In: Gene Targeting. Joyner A L, editor. Vol. 126. New York: Oxford Univ. Press; 1993. pp. 33–61. [Google Scholar]
- 32.Orita M, Iwahana H, Kanazawa H, Hayashi K, Sekiya T. Proc Natl Acad Sci USA. 1989;86:2766–2770. doi: 10.1073/pnas.86.8.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dietrich W F, Miller J, Steen R, Merchant M A, Damron-Boles D, Husain Z, Dredge R, Daly M J, Ingalls K A, O'Connor T J, et al. Nature (London) 1996;380:149–152. doi: 10.1038/380149a0. [DOI] [PubMed] [Google Scholar]
- 34.Pinkel D, Landegent J, Collins C, Fuscoe J, Segraves R, Lucas J, Gray J. Proc Natl Acad Sci USA. 1988;85:9138–9142. doi: 10.1073/pnas.85.23.9138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hicks G G, Shi E G, Li X M, Li C H, Pawlak M, Ruley H E. Nat Genet. 1997;16:338–344. doi: 10.1038/ng0897-338. [DOI] [PubMed] [Google Scholar]
- 36.Braun R, Lo D, Pinkert C, Widera G, Flavell R, Palmiter R, Brinster R. Biol Reprod. 1990;43:684–693. doi: 10.1095/biolreprod43.4.684. [DOI] [PubMed] [Google Scholar]
- 37.Van Etten W J, Steen R G, Nguyen H, Castle A B, Slonim D K, Ge B, Nusbaum C, Schuler G D, Lander E S, Hudson T J. Nat Genet. 1999;22:384–387. doi: 10.1038/11962. [DOI] [PubMed] [Google Scholar]
- 38.Nusbaum C, Slonim D K, Harris K L, Birren B W, Steen R G, Stein L D, Miller J, Dietrich W F, Nahf R, Wang V, et al. Nat Genet. 1999;22:388–393. doi: 10.1038/11967. [DOI] [PubMed] [Google Scholar]
- 39.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]