Abstract
“Dehalococcoides” sp. strain CBDB1 in pure culture dechlorinates a wide range of PCB congeners with three to eight chlorine substituents. Congener-specific high-resolution gas chromatography revealed that CBDB1 extensively dechlorinated both Aroclor 1248 and Aroclor 1260 after four months of incubation. For example, 16 congeners comprising 67.3% of the total PCBs in Aroclor 1260 were decreased by 64%. We confirmed the dechlorination of 43 different PCB congeners. The most prominent dechlorination products were 2,3′,5-chlorinated biphenyl (25-3-CB) and 24-3-CB from Aroclor 1248 and 235-25-CB, 25-25-CB, 24-25-CB, and 235-236-CB from Aroclor 1260. Strain CBDB1 removed flanked para chlorines from 3,4-, 2,4,5-, and 3,4,5-chlorophenyl rings, primarily para chlorines from 2,3,4,5-chlorophenyl rings, primarily meta chlorines from 2,3,4- and 2,3,4,6-chlorophenyl rings, and either meta or para chlorines from 2,3,4,5,6-chlorophenyl rings. The site of attack on the 2,3,4-chorophenyl ring was heavily influenced by the chlorine configuration on the opposite ring. This dechlorination pattern matches PCB Process H dechlorination, which was previously observed in situ both in the Acushnet Estuary (New Bedford, MA) and in parts of the Hudson River (New York). Accordingly, we propose that Dehalococcoides bacteria similar to CBDB1 are potential agents of Process H PCB dechlorination in the environment. This is the first time that a complex naturally occurring PCB dechlorination pattern has been reproduced in the laboratory using a single bacterial strain.
Polychlorinated biphenyls (PCBs) are widespread priority pollutants that pose a threat to the health of humans and ecosystems (5, 6, 33). The PCBs most commonly used and spilled, for example, Aroclors, were commercial PCB mixtures composed of 60 to 100 different congeners (17, 21). Microbially mediated reductive dechlorination under anaerobic conditions is the only known process that has a significant impact on complex PCB mixtures in the environment (8, 12, 35). Details of many such anaerobic dechlorination reactions catalyzed by microbial communities in aquatic sediments have been described (reviewed in references 12 and 35). However, little is known about the bacteria involved in these processes.
The first pure bacterial strain shown to reductively dechlorinate PCBs was “Dehalococcoides ethenogenes” strain 195, which dechlorinated two PCB congeners chlorinated on a single ring (19). A second pure strain, DF-1, can grow by respiratory dechlorination of several PCB congeners chlorinated on a single ring (28). Strain DF-1 is a member of the dechlorinating Chloroflexi, as are “Dehalococcoides” species, and it resembles Dehalococcoides species in many morphological and physiological aspects (28). The type strain of Desulfitobacteria dehalogenans, JW/IU-DC, is able to dechlorinate hydroxylated PCBs (36), but the hydroxyl group is essential for dechlorination, and it cannot dechlorinate PCBs (J. Wiegel, unpublished data). To date, the dechlorinating Chloroflexi, including Dehalococcoides spp., are the only bacteria that have been reported to reductively dechlorinate PCBs (7).
In contrast to the PCB dechlorination specificity thus far observed with strains 195 and DF-1, in which exclusively doubly flanked chlorine substituents are removed, dechlorination of complex commercial PCB mixtures in the environment and in sediment microcosms matches one or more of seven multifaceted microbial PCB dechlorination processes. Full descriptions of these processes have been published stating which congeners are substrates, which are products, which chlorophenyl rings are attacked, and which chlorine substituents are removed, as well as listing suites of dechlorination pathways (11, 12, 14, 31). Many of the PCB dechlorination processes observed in the environment have been replicated and studied in detail in sediment microcosms, but the bacterial agents responsible for the dechlorination have not been identified. Several studies have reported the presence of Dehalococcoides populations in PCB-dechlorinating mixed cultures and sediment microcosms (reviewed in reference 23). Two recent studies have linked Dehalococcoides bacteria to Process N dechlorination of Aroclor 1260 and demonstrated that they metabolically dechlorinate Aroclor 1260 (13, 18). However, until now no PCB dechlorination process has been reproduced by a single organism.
Dehalococcoides sp. strain CBDB1 was isolated for its ability to grow using chlorinated benzenes as respiratory electron acceptors (4). This strain is particularly noted for its ability to dehalogenate chlorinated aromatics, and it dehalogenates all highly chlorinated benzenes (4, 24), all highly chlorinated phenols (1), and several chlorinated dibenzodioxins (15). Strain CBDB1 has been studied biochemically and the complete genome has been sequenced and annotated (26). Recently, a chlorobenzene reductive dehalogenase gene, cbrA (2), and a tetrachloroethene reductive dehalogenase gene, pceA, that appears to be also responsible for chlorophenol dehalogenation, were identified in CBDB1 (1, 22, 29). However, 30 additional reductive dehalogenase homologous genes (rdh genes) in the CBDB1 genome remain uncharacterized.
The objectives of the present study were to test the ability of strain CBDB1 to dechlorinate commercial PCB mixtures, to characterize the dechlorination pattern, and to compare it with known microbial PCB dechlorination processes. We report conclusive proof that Dehalococcoides sp. strain CBDB1 in pure culture extensively dechlorinates a broad spectrum of PCBs in Aroclor 1260 according to microbial Process H dechlorination.
MATERIALS AND METHODS
PCB nomenclature.
PCB congeners are named here by listing the substituted positions on each ring separated by a hyphen and followed by CB (chlorobiphenyl). Thus, 2345-245-CB is the congener chlorinated at positions 2, 2′, 3, 4, 4′, 5, and 5′ in the IUPAC nomenclature.
Cultivation and experimental setup.
Strain CBDB1 was cultivated as previously described (3, 4) in purely synthetic, carbonate buffered mineral medium containing Ti(III) citrate as a reducing agent, vitamins, 5 mM acetate as the carbon source, and hydrogen (7.5 mM nominal concentration) as the electron donor. Trichlorobenzenes (TCBz) (4) or PCBs were used as electron acceptors. When TCBz were used, an equimolar mixture of 1,2,3- and 1,2,4-TCBz was added by injection with a Hamilton microliter syringe to a final concentration of 15 μM each. Cell titers were measured by direct cell counting on agarose-coated slides as previously described (1). Cultures grown on TCBz typically reach a cell density of about 5 × 106 cells per ml. All PCBs were purchased from AccuStandard (New Haven, CT). Individual congeners were ≥99% purity. Commercial PCB mixtures including Aroclors were manufactured by catalytically chlorinating biphenyl to a specified weight percent chlorine; hence, different batches (lots) vary in composition (21). The Aroclor 1248 and Aroclor 1260 used in the present study were lots 106-248 and 021-020, respectively, from AccuStandard.
Except where specified otherwise, PCBs were added from PCB/silica stock solutions which permitted a fed-batch approach with multiple additions of PCBs. PCB/silica stock solutions were prepared by using a modification of a previously described method (9). Floated silica powder (about 240 mesh; Fisher Scientific) was weighed out, sealed in closed crimp vials equipped with Teflon-lined butyl rubber septa, purged with anoxic N2 gas, and then autoclaved. Then, small volumes of highly concentrated sterile-filtered acetone stock solutions of PCBs were injected with a sterile syringe and mixed with the silica. Subsequently, the bottles were rotated on a vortex mixer to distribute the PCB/silica on the walls of the bottles, and a gentle stream of sterile nitrogen gas was used to evaporate the acetone. Finally, sterile reduced anaerobic mineral medium lacking only vitamins and acetate was injected, and the bottles were vigorously vortex mixed until all silica was in suspension. With this procedure, PCBs were homogeneously distributed on the silica powder and within the suspension and could be used for reproducible replenishment of cultures by injection of small volumes. The PCB/silica stock solutions contained 10 mg of silica per ml and final nominal concentrations of 200 μM of individual PCB congeners or 20 μg of Aroclor per ml. Note that PCBs are extremely insoluble in water and that all of the PCB concentrations given here are expressed in μmol/liter of culture or μg/ml of culture and not as aqueous concentrations.
Aroclor-dechlorinating cultures of strain CBDB1 were set up from cultures initially grown on a mixture of 15 μM 1,2,3-TCBz and 15 μM 1,2,4-TCBz to a cell density of about 5 × 106 cells per ml. Then Aroclor 1260 or Aroclor 1248 was gradually added in exponentially increasing amounts every 2 to 3 days to match anticipated growth. The initial concentration of Aroclors in the cultures was 1.2 ng/ml, while the final concentrations were ∼2.3 μg/ml for the initial experiments and 3.7 μg/ml for the second quantitative experiment. Parallel incubations without inoculum served as abiotic controls.
Additional experiments were carried out with individual PCB congeners that contained three to five chlorine substituents on one ring but none on the other ring at initial nominal concentrations of 0.5 μM and final concentrations of ∼5 μM. For these experiments the PCBs were added in acetone to fresh cultures inoculated to a cell density of about 8.5 × 104 cells per ml with CBDB1 grown on the TCBz mixture.
Three hexachlorinated PCB congeners that contained chlorine substituents on both rings were also studied to determine their dechlorination pathways and to determine whether the chlorine substituents on one ring influence the dechlorination of the opposite ring. For these experiments 245-245-CB, 234-245-CB, or 234-234-CB was added as a PCB/silica solution to a concentration of 10 μM to cultures inoculated to a cell density of about 1.7 × 105 cells per ml with CBDB1 grown on the TCBz mixture. Three parallel cultures were set up for each congener. Again, parallel incubations without inoculum served as abiotic controls.
PCB analysis.
Aliquots (0.5 to 2 ml) from the cultures were collected with syringes and transferred to 8-ml glass vials fitted with Teflon-lined screw caps. PCBs were extracted by shaking on a platform shaker for 2 h with ultra resi-analyzed diethyl ether (J. T. Baker) (1 to 2 volumes for Aroclor analyses and 4 volumes for single congener analyses).
Congener-specific splitless PCB analysis for the initial experiments with Aroclors 1248 and 1260, and with 234-CB or 245-CB, was carried out on an Agilent 6890 gas chromatograph equipped with a Ni63 microelectron capture detector and autosampler. The injection temperature was 250°C, and the detector temperature was 340°C. Samples were chromatographed by using electronic pressure control at a constant pressure of 0.993 bar. Initial column flow was 1.4 ml of nitrogen gas per min. We used a two-stage temperature program: the initial temperature was 50°C; the temperature was then raised at 30°C per min to 160°C and then raised at 2.5°C/min to 300°C. The data were collected by using Chemstation version A.09.03 software (Agilent Technologies). We used a DB-XLB column (30 m, 0.25-μm phase thickness, 0.25-mm inner diameter; J&W Scientific; Agilent Technologies) which gives excellent resolution of PCBs with two to four chlorine substituents. For example, it allowed us to distinguish 24-CB from 25-CB, whereas these congeners coelute on many other columns. However, it was not optimal for analysis of PCBs with five or more chlorine substituents because many of the most important components were not resolved but coeluted with other congeners. Therefore, for quantitative experiments with Aroclor 1260, we used a DB-1 column which gives far better resolution of highly chlorinated PCBs.
Congener-specific splitless PCB analysis for all other experiments was carried out on a Hewlett-Packard 5890 series II gas chromatograph equipped with a Ni63 electron capture detector, an autosampler, and a DB-1 column (30 m, 0.25-μm phase thickness, 0.25-mm inner diameter; J&W Scientific; Agilent Technologies). The injection temperature was 270°C, and the detector temperature was 300°C. Samples were chromatographed by using electronic pressure control. The initial pressure was 0.67 bar; this was raised at 6.83 bar/min to 3.76 bar, held for 5.5 min, and then decreased to 1.6 bar and held. The initial column flow was 0.873 ml of helium gas per min. The makeup gas was nitrogen. We used a multistage temperature program. The initial temperature was 31°C; the temperature was then raised at 20°C/min to 160°C, held for 3 min, raised at 2°C/min to 200°C, and finally raised at 8°C/min to 260°C and held for 5 min. The data were collected by using a Perkin-Elmer Network Chromatography Interface (model 901) and TotalChrom version 6.3.1 software (Perkin-Elmer).
The elution position of all 209 PCB congeners was determined by analyzing dilutions of PCB congener mixes 1 through 9 prepared by AccuStandard (New Haven, CT) for that purpose. The relative elution positions of the PCBs in these mixtures have been published for both DB-1 and DB-XLB columns (20). PCBs were quantified by using a customized calibration standard prepared from Aroclor 1260 plus 43 additional congeners that are known to be frequent dechlorination products which were added to the standard in known amounts (32). The concentrations of the PCB congeners in the Aroclor 1260 in this standard were calculated from the weight percent distributions for the G5 Aroclor 1260 (21), which is believed to be the same lot of Aroclor used in our customized standard. Additional congeners were quantified from standards prepared from the AccuStandard PCB congener mixes. In all cases we used a seven-point second-order calibration curve.
Data were exported to an Excel spreadsheet, and a macro was designed and used to calculate the mole percent distribution of all congeners and of the PCB homologs, as well as the number of ortho, meta, and para chlorine substituents per biphenyl.
RESULTS
Initial PCB dechlorination experiments with Dehalococcoides sp. strain CBDB1.
Initial experiments with Aroclor 1248 or Aroclor 1260 at a nominal final concentration of 2.7 μg/ml resulted in dechlorination of many different congeners in both Aroclor 1248 and Aroclor 1260. In Aroclor 1248, congeners containing 234-, 245-, and 34-chlorophenyl rings were preferentially dechlorinated. Specifically, large decreases were observed for 234-23-CB, 234-24-CB, 234-25-CB, 234-34-CB, 245-34-CB, 23-34-CB, 24-34-CB, 25-34-CB, 234-4-CB, 245-2-CB, and 245-4-CB. The most prominent dechlorination products of Aroclor 1248 were 25-3-CB and 24-3-CB. The 25-3-CB was likely formed by para dechlorination of 245-34-CB and 25-34-CB. The 24-3-CB was likely formed by dechlorination of 234-34-CB and 24-34-CB. Hence, the 34-chlorophenyl rings were dechlorinated from the para position, but the 234-chlorophenyl ring was apparently dechlorinated at the doubly flanked meta position. 25-4-CB was also a dechlorination product in Aroclor 1248, most likely from dechlorination of 245-4-CB. The most prominent dechlorination products of Aroclor 1260 were 25-25-CB, 24-25-CB, and 235-25-CB, which were accompanied by corresponding decreases in 245-245-CB, 234-245-CB, and 2345-245-CB, respectively.
Dechlorination of Aroclor 1260.
In order to reproduce these results and to obtain a more detailed and quantitative analysis of PCB dechlorination by strain CBDB1, we optimized our PCB analysis for the observed dechlorination pattern and focused additional experiments on the more highly chlorinated mixture Aroclor 1260. After 118 days of incubation with Aroclor 1260, seven of nine parallel cultures showed dechlorination products. The medium in the remaining two had turned pink, indicating the presence of O2, which kills strain CBDB1, and the PCBs in those cultures showed no evidence of dechlorination. Abiotic controls which were not inoculated also showed no dechlorination. Additional samples were taken from the cultures at later times, but when all samples were ultimately quantified it became apparent that the tri- and tetrachlorobiphenyl products were increasingly being lost into the headspace and septa (because the Teflon coating had been injured by piercing during sampling) and hence were not being measured. Therefore, we used data only from the first samples collected.
Table 1 shows the PCB homolog distribution in uninoculated controls of Aroclor 1260 and in CBDB1 cultures incubated with Aroclor 1260. The heptachlorobiphenyls decreased by nearly one-half, and the hexa- and octachlorobiphenyls decreased by one-third. The products were predominantly tetra- and pentachlorobiphenyls, although some tri- and hexachlorinated biphenyls were also products. These data provide conclusive evidence of extensive dechlorination of the most highly chlorinated PCBs in Aroclor 1260.
TABLE 1.
Effect of CBDB1 on PCB homolog distribution in Aroclor 1260
| PCB homolog | mol% of total PCBs
|
% Decrease | ||
|---|---|---|---|---|
| Aroclor 1260a | Dechlor. Aroclor 1260b | SD | ||
| Tri-CB | 0.16 | 0.88 | 0.31 | |
| Tetra-CB | 0.33 | 17.68 | 2.30 | |
| Penta-CB | 10.32 | 25.95 | 2.19 | |
| Hexa-CB | 50.31 | 33.92 | 1.61 | 32.6 |
| Hepta-CB | 33.91 | 18.05 | 2.31 | 46.7 |
| Octa-CB | 4.77 | 3.25 | 0.57 | 31.9 |
| Nona-CB | 0.19 | 0.24 | 0.02 | |
Data are the means for two uninoculated controls.
Dechlor., dechlorinated. Data are the means for the three CBDB1 cultures that showed the most extensive dechlorination of Aroclor 1260 after 118 days of incubation (Fig. 1).
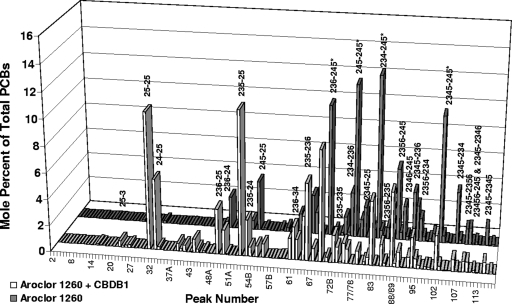
The complete PCB congener distribution of Aroclor 1260 incubated in uninoculated controls and in CBDB1 cultures is shown in Fig. 1. The controls showed no difference from Aroclor 1260. In contrast, nearly all hexa- through octachlorinated congener peaks were significantly decreased in cultures incubated with CBDB1. The most prominent dechlorination products were 235-25-CB, 25-25-CB, 24-25-CB, and 235-236-CB.
FIG. 1.
PCB congener distribution of Aroclor 1260 (3.7 μg/ml, equivalent to 10 μM) in uninoculated controls and in cultures inoculated with CBDB1. The data shown for CBDB1 are the means for the three cultures that showed the greatest dechlorination after 118 days of incubation. Uninoculated controls showed no changes from Aroclor 1260. The predominant congeners in major peaks are indicated. Congeners marked with asterisks are the four congeners that accumulate to the highest levels in humans (37).
Table 2 lists all major constituents of Aroclor 1260, i.e., all congeners that are present at 1 mol% or higher, in decreasing order of their prominence in Aroclor 1260. These 23 congeners constitute ∼86% of the total PCBs in Aroclor 1260. Sixteen of them were substantially dechlorinated by strain CBDB1, four showed little or no change, and three, which were apparently intermediates or final products of other congeners, significantly increased. The 16 congeners that were dechlorinated decreased from 67.3 mol% of the total PCBs to 24.2 mol%, a 64% decrease. Furthermore, three of the four most abundant congeners were decreased by 73 to 90% (Table 2).
TABLE 2.
Major components of Aroclor 1260 and their dechlorination by CBDB1
| Congenera | DB1 Pk | IUPAC no. | mol% of total PCBs
|
% Decrease | Inferred pathway and ultimate productsd | |||
|---|---|---|---|---|---|---|---|---|
| Aroclor 1260b | Dechlor. Aroclor 1260c | SD | Change | |||||
| 245-245-CB* | 75 | 153 | 11.66 | 1.12 | 0.48 | -10.54 | 90 | 245-25-CB → 25-25-CB |
| 2345-245-CB* | 102 | 180 | 9.71 | 2.61 | 1.07 | -7.10 | 73 | 235-245-CB → 235-25-CB |
| 236-245-CB* | 69 | 149 | 9.47 | 8.41 | 0.48 | -1.06 | 11 | 236-25-CB |
| 234-245-CB* | 82 | 138 | 8.53 | 1.49 | 0.25 | -7.04 | 83 | 234-25-CB → 24-25-CB + 23-25-CB |
| 2356-245-CB | 88 | 187 | 5.50 | 5.61 | 0.29 | 0.11 | ||
| 2345-234-CB | 106 | 170 | 3.97 | 0.88 | 0.40 | -3.09 | 78 | 235-234-CB → 235-24-CB + 235-23-CB |
| 245-25-CB | 53 | 101 | 3.79 | 0.70 | 0.04 | -3.09 | 82 | 25-25-CB |
| 2345-236-CB | 93 | 174 | 3.77 | 0.79 | 0.33 | -2.98 | 79 | 235-236-CB |
| 234-236-CB | 74 | 132 | 3.39 | 1.52 | 0.22 | -1.87 | 55 | 24-236-CB + 23-236-CB |
| 2356-34-CB | 82 | 163 | 3.09 | 3.09 | 0.00 | 0.00 | ||
| 2356-25-CB | 64 | 151 | 3.00 | 3.38 | 0.13 | 0.38 | ||
| 2356-234-CB | 94 | 177 | 2.73 | 2.33 | 0.15 | -0.40 | 15 | 2356-24-CB |
| 2346-245-CB | 90 | 183 | 2.59 | 1.15 | 0.25 | -1.44 | 56 | 246-245-CB → 246-25-CB |
| 236-25-CB | 48B | 95 | 2.44 | 3.44 | 0.13 | 1.00 | ||
| 236-34-CB | 61 | 110 | 1.69 | 2.50 | 0.17 | 0.81 | ||
| 235-236-CB | 65 | 135 | 1.65 | 5.87 | 0.37 | 2.53 | ||
| 236-236-CB | 60 | 136 | 1.47 | 1.62 | 0.03 | 0.15 | ||
| 235-245-CB | 73 | 146 | 1.47 | 1.08 | 0.35 | -0.39 | 27 | 235-25-CB |
| 2345-2345-CB | 115 | 194 | 1.37 | 0.83 | 0.19 | -0.54 | 39 | 235-2345-CB → 235-235-CB |
| 2345-25-CB | 77 | 141 | 1.36 | 0.00 | 0.00 | -1.36 | 100 | 235-25-CB |
| 2345-2356-CB | 109 | 199 | 1.21 | 0.81 | 0.16 | -0.40 | 33 | 235-2356-CB |
| 2346-234-CB | 95 | 171 | 1.19 | 0.50 | 0.11 | -0.69 | 58 | 246-234-CB→ 246-24-CB + 246-23-CB |
| 2346-25-CB | 66 | 144 | 1.10 | 0.00 | 0.00 | -1.10 | 100 | 246-25-CB |
The congeners marked with asterisks are the four congeners that accumulate to the highest levels in humans (37).
Data are the means for two uninoculated controls.
Dechlor., dechlorinated. Data are the means for the three CBDB1 cultures that showed the most extensive dechlorination.
When there is more than one pathway, only the dominant pathway is shown because of space limitations.
The eight most abundant congeners in Aroclor 1260 all have 245- or 2345-chlorophenyl rings, and it is these that account for the appearance of para dechlorination products with 25- or 235-chlorophenyl rings. The congeners that were not dechlorinated were composed mainly of 25-, 235-, 236-, and 2356-chlorophenyl rings which later experiments confirmed were not substrates.
All dechlorination products that increased by 0.45 mol% or more are listed in Table S1 in the supplemental material (this table lists the 19 most abundant dechlorination products produced from Aroclor 1260 by strain CBDB1). The three most abundant dechlorination products, the tetra- and pentachlorobiphenyls 25-25-CB, 235-25-CB, and 24-25-CB, together constitute 26% of the total PCBs after dechlorination. Other prominent products include 235-236-CB, 235-24-CB, 236-24-CB, 235-235-CB and 245-246-CB which together account for 11% of the total PCBs after dechlorination. The products were for the most part composed of 25-, 235-, 236-, and 24-chlorophenyl rings and, to a lesser extent, 246- and 35-chlorophenyl rings. para dechlorination of 245- and 2345-chlorophenyl rings accounted for most of the observed dechlorination. However, the dechlorination also targeted doubly flanked meta chlorines on 234- and 2346-chlorophenyl rings to generate 24- and 246-chlorophenyl rings, respectively, and doubly flanked meta or para dechlorination of 23456-chlorophenyl rings to generate 2346- and 2356-chlorophenyl rings. The 236-chlorophenyl ring is abundant in Aroclor 1260, comprising 14.43 mol% of all chlorophenyl rings (21), and was likely not formed by dechlorination. In contrast, the 23- and 35-chlorophenyl rings are extremely rare in Aroclor 1260, 0.51 and 0.00 mol%, respectively (21), and were likely formed by para dechlorination of 234- and 345-chlorophenyl rings, respectively. The 25-3-CB was likely formed by para dechlorination of 245-34-CB and 25-34-CB, as seen in our preliminary experiments with Aroclor 1248.
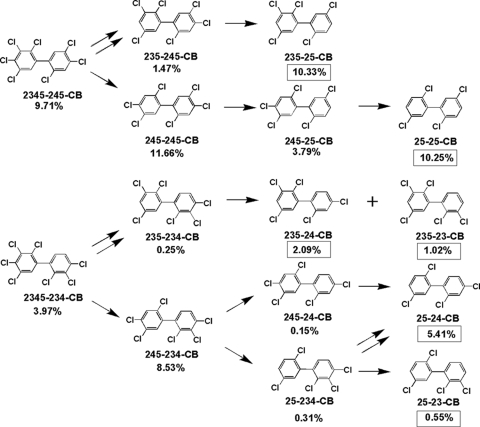
Taking into account congener disappearance and appearance over time, and the considerations just mentioned, dechlorination pathways and products were inferred for the most abundant congeners in Aroclor 1260 (Table 2). Figure 2 shows an example of how the dechlorination pathways were deduced for major components in Aroclor 1260 and compares the initial mole percent of the individual PCB substrates with the mole percent increases of the individual products derived from them by dechlorination. It is clear that 2345-245-CB was primarily dechlorinated via 235-245-CB to 235-25-CB. However, 2345-234-CB was dechlorinated via 235-234-CB to both 235-23-CB and 235-24-CB, i.e., both meta and para dechlorination of the 234-chlorophenyl group occurred. Likewise the 234-chlorophenyl ring of 245-234-CB was dechlorinated from both positions to yield the ultimate products 24-25-CB and 23-25-CB, albeit in a 10:1 ratio. In contrast, 245-245-CB was exclusively dechlorinated via 245-25-CB to 25-25-CB.
FIG. 2.
Inferred dechlorination pathways for several major components of Aroclor 1260 dechlorinated by strain CBDB1. Where there is more than one route of dechlorination, double arrows indicate the major pathway. The mole percent of the total PCBs in Aroclor 1260 for each substrate before dechlorination is given. The mole percent values of total PCBs for dechlorination products formed by CBDB1 are indicated in boxes.
Most of the Aroclor 1260 dechlorination catalyzed by strain CBDB1 was from the para position of PCBs and is reflected in a 47% decrease in the number of para chlorines per biphenyl from 1.32 to 0.71. In contrast, the number of meta chlorines per biphenyl decreased by only 5%. No ortho dechlorination was observed.
Dechlorination of PCB congeners chlorinated on a single ring.
CBDB1 cultures were also incubated with single PCB congeners as sole electron acceptors to confirm and further clarify the dechlorination patterns described above. The first series of congeners tested included all congeners with three to five chlorine substituents on a single ring. Of these, 235-CB, 236-CB, 246-CB, and 2356-CB were not dechlorinated after 97 days of incubation, and no products were seen. In contrast, all congeners with three or more adjacent chlorines were dechlorinated, but exclusively by loss of a doubly flanked chlorine substituent. Hence, 234-CB was dechlorinated to 24-CB, 345-CB was dechlorinated to 35-CB, and 2346-CB was dechlorinated to 246-CB. All of the doubly flanked meta and para chlorine substituents on 2345-CB and 23456-CB were amenable to dechlorination. 23456-CB was dechlorinated to 2356-CB and to 2346-CB, which was in turn dechlorinated to 246-CB. In the case of 2345-CB, 86.3 mol% was dechlorinated to 235-CB and 13.7 mol% was converted via 245-CB to 25-CB; hence, for the 2345-chlorophenyl ring, doubly flanked para dechlorination was favored over doubly flanked meta dechlorination in a ratio of 7:1. Only one of the tested PCB congeners lacking three adjacent chlorine substituents, 245-CB, was dechlorinated by strain CBDB1. This congener was dechlorinated in the singly flanked para position to yield 25-CB. The products 2356-CB, 246-CB, 235-CB and 24-CB, 25-CB, and 35-CB were not transformed, even after prolonged incubations of more than 3 months. No dechlorination of ortho substituents was ever observed.
These results confirmed our deductions for the dechlorination pathways in Aroclor 1260. The single exception was the observation that 234-CB was exclusively dechlorinated to 24-CB from the doubly flanked meta position, whereas the 23-chlorophenyl groups observed on several dechlorination products of Aroclor 1260 (see Table S1 in the supplemental material) most likely came from para dechlorination of 234-chlorophenyl groups.
Dechlorination of individual hexachlorinated biphenyls.
To further clarify the dechlorination pathways of congeners that were dechlorinated in Aroclor 1260, we studied the dechlorination of several hexachlorobiphenyls that are significant constituents of Aroclor 1260.
(i) Dechlorination of 245-245-CB.
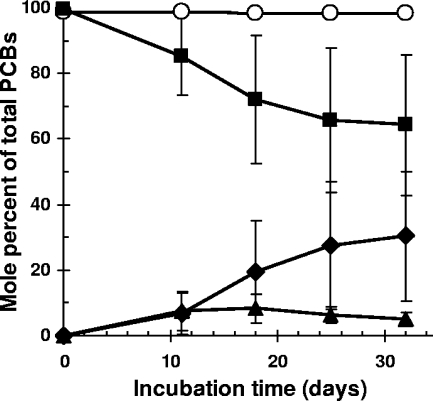
The 245-245-CB congener is one of the most abundant congeners in Aroclor 1260, comprising 11.7 mol% of the total PCBs. CBDB1 dechlorinated 90% of this congener in Aroclor 1260 (Table 2). When 245-245-CB was incubated separately with CBDB1, both 245-25-CB and 25-25-CB were detected at the first sampling time at 11 days (Fig. 3). Two of three parallel cultures dechlorinated ∼65 mol% of 245-245-CB in 66 days; the third dechlorinated 21 mol% in the same period.
FIG. 3.
Dechlorination of the hexachlorinated biphenyl congeners 234-234-CB or 245-245-CB by strain CBDB1. Parallel cultures were inoculated with about 1.65 × 105 cells from three parallel cultures grown with the TCBz mixture. The inocula for both sets of cultures were the same. Shown are results of three parallel cultures ± the standard deviation. While less than 1% of 234-234-CB (○) was dechlorinated, 245-245-CB (▪) was extensively dechlorinated via 245-25-CB (▴) to 25-25-CB (⧫).
(ii) Dechlorination of 234-245-CB.
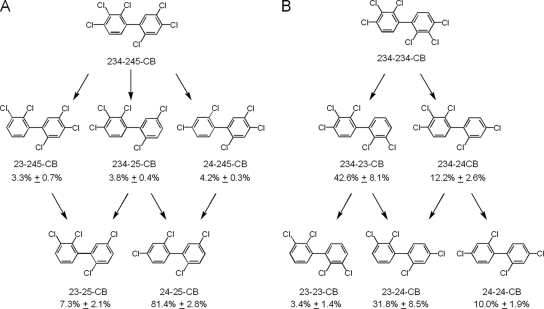
The congener 234-245-CB is the fourth most abundant congener in Aroclor 1260, comprising 8.5 mol% of the total PCBs. When 234-245-CB was incubated separately with CBDB1, dechlorination products were again observed at the first sampling time, in this case after 8 (data not shown). Several different intermediates were detected, demonstrating that the first attack could occur on either ring (Fig. 4A). The 245-chlorophenyl ring was again exclusively dechlorinated from the para position. In contrast, dechlorination of the 234-chlorophenyl ring occurred at either the meta or the para position. Ultimately, 24-25-CB and 23-25-CB were formed in a 10:1 ratio (Fig. 4A), the same ratio observed for dechlorinated Aroclor 1260 (Fig. 2). These data confirm that the preferred dechlorination of the 234-chlorophenyl ring for this congener was at the doubly flanked meta position, as seen with 234-CB, and confirm that the 23-25-CB observed as dechlorination product in Aroclor 1260 (see Table S1 in the supplemental material) came from para dechlorination of both rings of 234-245-CB.
FIG. 4.
Quantitative analysis of product formation of two hexachlorinated biphenyls incubated separately with CBDB1 for 32 days. In each case, the PCB concentration was 10 μM. (A) 234-245-CB; (B) 234-234-CB. The data show the mole percent distributions of the total products ± the standard deviations.
(iii) Dechlorination of 234-234-CB.
Cultures of strain CBDB1 also dechlorinated 234-234-CB to multiple products (Fig. 4B). However, unlike the other two hexachlorobiphenyls, which were significantly dechlorinated within 1 month and almost completely transformed to tetrachlorobiphenyls within 3 months, only 1% of the 234-234-CB was dechlorinated after 32 days (Fig. 3), and no further dechlorination was seen even after 145 days of incubation.
(iv) Influence of the chlorine configuration of the opposite ring on dechlorination specificity.
Collectively, these data indicate that the dechlorination of a 245-chlorophenyl ring was the same regardless of whether the second ring was a 23-, 24-, 25-, 234-, or 245-chlorophenyl ring. In sharp contrast, dechlorination of the 234-chlorophenyl ring was strongly influenced by the chlorine configuration on the opposite ring. The presence of a 245- or 25-chlorophenyl ring favored meta dechlorination of the 234-chlorophenyl ring by a ratio of 10:1 (Fig. 4A). The situation was quite different in the case of 234-234-CB. Analysis of the distribution of the intermediates and products of this congener (Fig. 4B) shows that 234-23-CB comprised 42.6 mol% of the total products. This means that at least 42.6% of the dechlorination of the first 234-chlorophenyl ring was from the singly flanked para chlorine. In addition, the distribution of the tetrachlorobiphenyl products reveals that 23-24-CB was heavily favored over both 24-24-CB and 23-23-CB. This indicates that the dechlorination of the second 234-chlorophenyl ring was strongly influenced by the 23- or 24- chlorophenyl ring of the intermediate such that the 23-chlorophenyl ring favored meta dechlorination of 234-23-CB to 23-24-CB and the 24-chlorophenyl ring favored para dechlorination of 234-24-CB to 23-24-CB. These findings are in contrast to the results with 234-CB, where exclusively meta dechlorination occurred, and demonstrate that the dechlorination of the 234-chlorophenyl ring was strongly influenced by the chlorine substitution pattern or lack thereof on the opposite ring.
DISCUSSION
Dechlorination of commercial PCB mixtures.
This study is the first documentation that a single organism in pure culture can reductively dechlorinate complex commercial PCB mixtures. We have shown that strain CBDB1 dechlorinated 16 of the 23 most abundant PCBs with five or more chlorines in Aroclor 1260. Half of these were decreased by 73% or more. Corresponding increases were observed in less-chlorinated PCBs, and we were able to link substrates to products and to infer dechlorination pathways. The appearance of dechlorination products and intermediates from congeners incubated separately in additional experiments confirmed and further elucidated dechlorination pathways.
Including the single congeners tested in the present study and intermediates observed, we have confirmed unequivocal dechlorination of 43 PCB congeners. This is undoubtedly an underestimate of the range of PCB substrates for CBDB1 because many potential PCB substrates are not present in Aroclor 1260 and were not tested here. In addition, many of the dechlorination intermediates were likely not detected in Aroclor 1260 because they were further dechlorinated without accumulating. As an extrapolation of the present study we hypothesize that strain CBDB1 is able to dechlorinate additional congeners containing a 23456-, 2345-, 2346-, 234-, 245-, 345-, or 34-chlorophenyl ring that were not tested here. We also conclusively identified 39 dechlorination products and 10 dechlorination intermediates from Aroclor 1260 and the single congeners studied.
Dechlorination specificity.
Experiments with congeners chlorinated on a single ring demonstrated that all doubly flanked meta and para chlorines on 234-, 345-, 2345-, 2346-, and 23456-chlorophenyl rings are amenable to dechlorination. However, for Aroclor 1260, most dechlorination resulted from the para dechlorination of 2345- and 245-chlorophenyl rings. These are the two most common chlorophenyl ring substitutions for Aroclor 1260, comprising 15.04 and 30.26 mol%, respectively, of the total chlorophenyl ring substitutions (21). Nevertheless, doubly flanked meta dechlorination of some congeners in Aroclor 1260 also occurred and accounted for ∼8% of the total chlorine removal. In Aroclor 1248, para dechlorination of 34-chlorophenyl rings led to the most prominent dechlorination products. Isolated chlorine substituents were never removed nor were ortho chlorine substituents.
The chlorine substitution pattern on the second ring clearly affected the route of dechlorination of 234-chlorophenyl rings but not that of 245-chlorophenyl rings. Previous studies have shown that the chlorine configuration on the unattacked ring can affect the extent of oxidation (10) or dechlorination (11) of PCBs, but to our knowledge, this is the first demonstration that the distribution of products is affected by the unattacked ring.
Identification of the PCB dechlorination process.
Based on the chlorophenyl rings attacked, the position of chlorines removed on those rings, the chlorophenyl rings produced, and the congeners produced by dechlorination, we have identified the dechlorination exhibited by CBDB1 as Process H. This PCB dechlorination process was first described by Brown and Wagner on the basis of congener-specific PCB analyses of in situ dechlorination of Aroclor 1254 in sediments of the Acushnet Estuary (New Bedford, MA) (14). They further reported that the same dechlorination process was occurring in situ in some areas of the Hudson River (NY) (14). In their description of Process H, Brown and Wagner stated that the only chlorophenyl rings dechlorinated were those chlorinated at positions 23456-, 2345-, 2346-, 234-, 245-, and 34-, exactly the same chlorophenyl rings that were dechlorinated by CBDB1 in our experiments. The most prominent dechlorination products of the Aroclor 1254 in the Acushnet Estuary were 25-3-CB and 24-3-CB, the same products that we saw in our incubations of Aroclor 1248 with CBDB1. Furthermore, the dechlorination pathways for individual congeners that Brown and Wagner proposed showed the same dominant chlorine removal pattern that we observed: removal of para chlorines from 2345-, 245-, and 34-chlorophenyl rings and removal of doubly flanked meta chlorines from 234- and 2346-chlorophenyl rings. Quensen et al. subsequently observed Process H dechlorination in sediment microcosms spiked with Aroclor 1260 and inoculated with microorganisms from Hudson River sediment (31). These researchers found the same sets of congeners decreasing and increasing that we observed with CBDB1, as well as the same three primary products that we observed: 235-25-CB, 25-25-CB, and 24-25-CB (31).
Based on microcosm studies, it has been proposed that complex commercial PCB mixtures could only be dechlorinated by the combined action of at least several different dechlorinating organisms. That is clearly not the case. Our data conclusively prove that a single pure strain of Dehalococcoides can transform Aroclor 1260 by the same complex pattern of dechlorination, Process H, observed in situ at the two sites with the highest levels of PCB contamination in the United States: the Acushnet Estuary and the Hudson River (14, 17).
The PCB dechlorination specificity exhibited by CBDB1 is completely different from that exhibited by the Dehalococcoides bacteria in the sediment-free mixed JN cultures (13). The JN cultures, which were derived from the Housatonic River (Lenox, MA), manifest Process N dechlorination, which is characterized by predominantly flanked meta dechlorination of 23456-, 2345-, 2346, 2356-, 234-, 235-, 236-, and 245-chlorophenyl rings (12). Some dechlorination of doubly flanked para chlorines of 23456- and 2345-chlorophenyl rings also occurs (9, 13). Process N is the dominant pattern of PCB dechlorination observed in the Housatonic River (7). Thus, two major PCB dechlorination processes observed in situ at PCB-contaminated sites can be linked to the action of Dehalococcoides bacteria.
Process H dechlorination is similar to Process P dechlorination, which has also been described in microcosms of sediment from the Housatonic River. Many of the same products are formed, but Process P dechlorination exclusively removes chlorines from the para position (12). Thus, 234-chlorophenyl rings are solely dechlorinated to 23-chlorophenyl rings, and 2346- and 23456-chlorophenyl rings are exclusively dechlorinated to 236- and 2356-chlorophenyl rings, respectively.
Determining the PCB dechlorination potential of other pure Dehalococcoides strains.
Two previous attempts to determine whether CBDB1 could dechlorinate Aroclors failed (L. Adrian, J. Quensen, and F. E. Löffler, unpublished results; L. Adrian and D. L. Bedard, unpublished results), as did attempts to dechlorinate Aroclor 1260 with four other pure strains of Dehalococcoides and a mixed culture containing Dehalococcoides bacteria (D. Bedard, Q. Wu, and F. E. Löffler, unpublished data). All of those experiments used high PCB concentrations, 20 to 500 μg/ml, that were known to be dechlorinated in sediment microcosms or in sediment-free mixed cultures derived from PCB-contaminated sites. PCBs, like other hydrophobic compounds, can adsorb to and partition into cell membranes causing cell stress as manifested in changes in membrane fluidity, decreased cell surface area, and separation of membrane layers (16, 25, 30). At high PCB-to-cell ratios these effects would presumably be maximal and might be toxic. In this successful demonstration of Aroclor dechlorination by a pure strain we used a different strategy: we began with a higher density of cells, about 5 × 106 cells/ml, and slowly added the PCBs beginning with extremely low PCB concentrations; thus, we used a much lower PCB to cell ratio. Using this fed-batch approach, it should be possible to determine whether other pure strains of Dehalococcoides can dechlorinate PCBs and, if so, what their specificity is.
Implications for detoxification and bioremediation.
The PCB dechlorination carried out by CBDB1 substantially reduced the concentrations of the congeners that are most persistent in humans (Table 2) (37) and reduced the concentration of “dioxin-like” PCBs, which by definition must have at least two meta and two para chlorines and no more than one ortho chlorine. Furthermore, the less-chlorinated products are more readily degradable by other organisms, including humans, because the dechlorination provides open sites for attack by mono- or dioxygenases.
Both biostimulation and bioaugmentation approaches using Dehalococcoides bacteria have been successfully implemented at sites contaminated with chlorinated ethenes (27). Thus, the demonstration that Dehalococcoides bacteria are responsible for extensive dechlorination of commercial PCBs by both Process H and Process N indicates that the development of effective in situ bioremediation technologies for PCBs is feasible. Furthermore, such technologies should be applicable to both estuarine and freshwater sites because Aroclor-dechlorinating Dehalococcoides bacteria have been found in both habitats (13, 18) and because Process H and N dechlorinations occur under both conditions (7, 12, 14, 18, 34). It will be important to identify the reductive dehalogenases responsible for these activities in future studies. This would enable the development of tools to screen for the presence of Dehalococcoides bacteria harboring PCB-dechlorinating enzymes at PCB-contaminated sites.
Supplementary Material
Acknowledgments
We thank Bernd Krostitz, Sarah-Jane Weisberg, Ashwana Fricker, and Sarah LaRoe for excellent technical assistance.
This study was funded by NSF grant 0641743 to D.L.B., ERC starting grant Microflex to L.A., and grants NPVII 2B06156 and MSM 6046137305 to K.D.
Footnotes
Published ahead of print on 8 May 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Adrian, L., S. K. Hansen, J. M. Fung, H. Görisch, and S. H. Zinder. 2007. Growth of Dehalococcoides strains with chlorophenols as electron acceptors. Environ. Sci. Technol. 41:2318-2323. [DOI] [PubMed] [Google Scholar]
- 2.Adrian, L., J. Rahnenführer, J. Gobom, and T. Hölscher. 2007. Identification of a chlorobenzene reductive dehalogenase in Dehalococcoides sp. strain CBDB1. Appl. Environ. Microbiol. 73:7717-7724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adrian, L., U. Szewzyk, and H. Görisch. 2000. Bacterial growth based on reductive dechlorination of trichlorobenzenes. Biodegradation 11:73-81. [DOI] [PubMed] [Google Scholar]
- 4.Adrian, L., U. Szewzyk, J. Wecke, and H. Görisch. 2000. Bacterial dehalorespiration with chlorinated benzenes. Nature 408:580-583. [DOI] [PubMed] [Google Scholar]
- 5.Agency for Toxic Substances and Disease Registry. 2008. CERCLA priority list of hazardous compounds. ATSDR, Washington, DC. http://www.atsdr.cdc.gov/cercla/07list.html.
- 6.Agency for Toxic Substances and Disease Registry. 2000. Toxicological profile for polychlorinated biphenyls (update). U.S. Department of Health and Human Services Agency for Toxic Substances and Disease Registry, Atlanta, GA. [PubMed]
- 7.Bedard, D. L. 2008. A case study for microbial degradation: anaerobic bacterial reductive dechlorination of polychlorinated biphenyls—from sediment to defined medium. Annu. Rev. Microbiol. 62:253-270. [DOI] [PubMed] [Google Scholar]
- 8.Bedard, D. L. 2003. Polychlorinated biphenyls in aquatic sediments: environmental fate and outlook for biological treatment, p. 443-465. In M. M. Häggblom and I. Bossert (ed.), Dehalogenation: microbial processes and environmental applications. Kluwer Press, Boston, MA.
- 9.Bedard, D. L., J. J. Bailey, B. L. Reiss, and G. V. S. Jerzak. 2006. Development and characterization of stable sediment-free anaerobic bacterial enrichment cultures that dechlorinate Aroclor 1260. Appl. Environ. Microbiol. 72:2460-2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bedard, D. L., and M. L. Haberl. 1990. Influence of chlorine substitution pattern on the degradation of polychlorinated biphenyls by eight bacterial strains. Microb. Ecol. 20:87-102. [DOI] [PubMed] [Google Scholar]
- 11.Bedard, D. L., E. A. Pohl, J. J. Bailey, and A. Murphy. 2005. Characterization of the PCB substrate range of microbial Dechlorination Process LP. Environ. Sci. Technol. 39:6831-6839. [DOI] [PubMed] [Google Scholar]
- 12.Bedard, D. L., and J. F. Quensen III. 1995. Microbial reductive dechlorination of polychlorinated biphenyls, p. 127-216. In L. Y. Young and C. E. Cerniglia (ed.), Microbial transformation and degradation of toxic organic chemicals. Wiley-Liss, New York, NY.
- 13.Bedard, D. L., K. M. Ritalahti, and F. E. Löffler. 2007. The Dehalococcoides population in sediment-free mixed cultures metabolically dechlorinates the commercial polychlorinated biphenyl mixture Aroclor 1260. Appl. Environ. Microbiol. 73:2513-2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown, J. F., and R. E. Wagner. 1990. PCB movement, dechlorination, and detoxication in the Acushnet Estuary. Environ. Toxicol. Chem. 9:1215-1233. [Google Scholar]
- 15.Bunge, M., L. Adrian, A. Kraus, M. Opel, W. G. Lorenz, J. R. Andreesen, H. Görisch, and U. Lechner. 2003. Reductive dehalogenation of chlorinated dioxins by an anaerobic bacterium. Nature 421:357-360. [DOI] [PubMed] [Google Scholar]
- 16.Denich, T. J., L. A. Beaudette, H. Lee, and J. T. Trevors. 2003. Effect of selected environmental and physico-chemical factors on bacterial cytoplasmic membranes. J. Microbiol. Methods 52:149-182. [DOI] [PubMed] [Google Scholar]
- 17.Erickson, M. D. 1997. Analytical chemistry of PCBs, 2nd ed. Lewis Publishers, New York, NY.
- 18.Fagervold, S. K., H. D. May, and K. R. Sowers. 2007. Microbial reductive dechlorination of Aroclor 1260 in Baltimore Harbor sediment microcosms is catalyzed by three phylotypes within the phylum Chloroflexi. Appl. Environ. Microbiol. 73:3009-3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fennell, D. E., I. Nijenhuis, S. F. Wilson, S. H. Zinder, and M. M. Häggblom. 2004. Dehalococcoides ethenogenes strain 195 reductively dechlorinates diverse chlorinated aromatic pollutants. Environ. Sci. Technol. 38:2075-2081. [DOI] [PubMed] [Google Scholar]
- 20.Frame, G. M. 1997. A collaborative study of 209 PCB congeners and 6 Aroclors on 20 different HRGC columns. 1. Retention and coelution database. Fresenius J. Anal. Chem. 357:701-713. [Google Scholar]
- 21.Frame, G. M., J. W. Cochran, and S. S. Bøwadt. 1996. Complete PCB congener distributions for 17 Aroclor mixtures determined by 3 HRGC systems optimized for comprehensive, quantitative, congener-specific analysis. J. High Resol. Chromatogr. 19:657-668. [Google Scholar]
- 22.Fung, J. M., R. M. Morris, L. Adrian, and S. H. Zinder. 2007. Expression of reductive dehalogenase genes in Dehalococcoides ethenogenes strain 195 growing on tetrachloroethene, trichloroethene, or 2,3-dichlorophenol. Appl. Environ. Microbiol. 73:4439-4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hiraishi, A. 2008. Biodiversity of dehalorespiring bacteria with special emphasis on polychlorinated biphenyl/dioxin dechlorinators. Microbes Environments 23:1-12. [DOI] [PubMed] [Google Scholar]
- 24.Jayachandran, G., H. Görisch, and L. Adrian. 2003. Dehalorespiration with hexachlorobenzene and pentachlorobenzene by Dehalococcoides sp. strain CBDB1. Arch. Microbiol. 180:411-416. [DOI] [PubMed] [Google Scholar]
- 25.Kim, I. S., H. Lee, and J. T. Trevors. 2001. Effects of 2,2′,5,5′-tetrachlorobiphenyl and biphenyl on cell membranes of Ralstonia eutropha H850. FEMS Microbiol. Lett. 200:17-24. [DOI] [PubMed] [Google Scholar]
- 26.Kube, M., A. Beck, S. H. Zinder, H. Kuhl, R. Reinhardt, and L. Adrian. 2005. Genome sequence of the chlorinated compound respiring bacterium Dehalococcoides species strain CBDB1. Nat. Biotechnol. 23:1269-1273. [DOI] [PubMed] [Google Scholar]
- 27.Löffler, F. E., and E. A. Edwards. 2006. Harnessing microbial activities for environmental cleanup. Curr. Opin. Biotechnol. 17:274-284. [DOI] [PubMed] [Google Scholar]
- 28.May, H. D., G. S. Miller, B. V. Kjellerup, and K. R. Sowers. 2008. Dehalorespiration with polychlorinated biphenyls by an anaerobic ultramicrobacterium. Appl. Environ. Microbiol. 74:2089-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morris, R. M., J. M. Fung, B. G. Rahm, S. Zhang, D. L. Freedman, S. H. Zinder, and R. E. Richardson. 2007. Comparative proteomics of Dehalococcoides spp. reveals strain-specific peptides associated with activity. Appl. Environ. Microbiol. 73:320-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parnell, J. J., J. Park, V. Denef, T. Tsoi, S. Hashsham, J. Quensen III, and J. M. Tiedje. 2006. Coping with polychlorinated biphenyl (PCB) toxicity: physiological and genome-wide responses of Burkholderia xenovorans LB400 to PCB-mediated stress. Appl. Environ. Microbiol. 72:6607-6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quensen, J. F., III, S. A. Boyd, and J. M. Tiedje. 1990. Dechlorination of four commercial polychlorinated biphenyl mixtures (Aroclors) by anaerobic microorganisms from sediments. Appl. Environ. Microbiol. 56:2360-2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smullen, L. A., K. A. DeWeerd, D. L. Bedard, W. A. Fessler, J. C. Carnahan, and R. E. Wagner. 1993. Development of a customized congener specific PCB standard for quantification of Woods Pond sediment PCBs, p. 45-59. In Research and Development Program for the Destruction of PCBs: twelfth progress report. General Electric Company, Corporate Research and Development, Schenectady, NY.
- 33.Stockholm Convention on Persistent Organic Pollutants. 1971. The 12 POPs under the Stockholm Convention. Stockholm Convention on Persistent Organic Pollutants, Stockholm, Sweden. http://www.pops.int/documents/pops/default.htm.
- 34.Van Dort, H. M., L. A. Smullen, R. J. May, and D. L. Bedard. 1997. Priming microbial meta-dechlorination of polychlorinated biphenyls that have persisted in Housatonic River sediments for decades. Environ. Sci. Technol. 31:3300-3307. [Google Scholar]
- 35.Wiegel, J., and Q. Wu. 2000. Microbial reductive dehalogenation of polychlorinated biphenyls. FEMS Microbiol. Ecol. 32:1-15. [DOI] [PubMed] [Google Scholar]
- 36.Wiegel, J., X. Zhang, and Q. Wu. 1999. Anaerobic dehalogenation of hydroxylated polychlorinated biphenyls by Desulfitobacterium dehalogenans. Appl. Environ. Microbiol. 65:2217-2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolff, M. S., J. Thornton, A. Fischbein, R. Lilis, and I. J. Selikoff. 1982. Disposition of polychlorinated biphenyl congeners in occupationally exposed persons. Toxicol. Appl. Pharmacol. 62:294-306. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.