Abstract
A Lactobacillus sakei strain named FLEC01 was isolated from human feces and characterized genotypically. Comparison of the genetic features of this strain with those of both the meat-borne L. sakei strain 23K and another human isolate, LTH5590, showed that they belong to different but closely related clusters. The three L. sakei strains did not persist and only transited through the gastrointestinal tracts (GITs) of conventional C3H/HeN mice. In contrast, they all colonized the GITs of axenic mice and rapidly reached a population of 109 CFU/g of feces, which remained stable until day 51. Five days after mice were fed, a first subpopulation, characterized by small colonies, appeared and reached 50% of the total L. sakei population in mice. Fifteen to 21 days after feeding, a second subpopulation, characterized by rough colonies, appeared. It coexisted with the two other populations until day 51, and its cell shapes were also affected, suggesting a dysfunction of the cell division or cell wall. No clear difference between the behaviors of the meat-borne strain and the two human isolates in both conventional and axenic mice was observed, suggesting that L. sakei is a food-borne bacterium rather than a commensal one and that its presence in human feces originates from diet. Previous observations of Escherichia coli strains suggest that the mouse GIT environment could induce mutations to increase their survival and colonization capacities. Here, we observed similar mutations concerning a food-grade gram-positive bacterium for the first time.
Although initially characterized from rice wine (28), the lactic acid bacterium species Lactobacillus sakei is considered the main representative flora of meat products, representing the major population of many fermented meat products and of raw meat stored under vacuum-packaged conditions (10, 12, 13). L. sakei is naturally present in many fish and meat products that are traditionally processed without the use of starter cultures (33). In addition, when small-scale facilities producing traditional dry fermented sausage were searched, L. sakei was detected only in the meat matrix, suggesting that meat is contaminated by this species mainly during the early processing steps (certainly by hide or feces of the animals) and not later on or by contact with the environment or materials within the facilities (2).
L. sakei shows high degrees of phenotypic and genomic diversity (11-13) that may explain the difficult detection and misidentification of it in the past. For instance, although the human gut microbiota has been intensively investigated by different microbial and molecular methods for many years, the presence of L. sakei in the feces of healthy humans was reported only recently (16, 17, 26, 39). The presence of the meat-borne species L. sakei in human feces, similar to that of several other lactobacilli, could be correlated to human diet, including raw and fermented meat (or fish), for millennia (37). Considering its relatively high concentration in human feces (106 per g) that was previously reported (16), L. sakei was considered as one of the predominant food-associated Lactobacillus species present in human feces. Its natural reservoir and its origin prior to meat contamination are still not known. One can hypothesize that it belongs to the intestinal microbiota of animals used for meat production, although its presence has not yet been reported in mammals and has been reported only recently in the intestines of salmonids (5).
Most of the available literature on L. sakei deals with its physiology in relation to preservation, fermentation, or spoilage of meat products (see references 10 and 13 and the references therein). Since its use as an ingredient or additive bioprotective culture, to ensure microbial safety of nonfermented meat products, has been proposed (8, 10), information on its behavior in the gastrointestinal tract (GIT) after ingestion of foodstuffs is required. The purpose of this study was thus to evaluate the ability of L. sakei to survive and transit in the GIT. Therefore, we compared two independent L. sakei strains isolated from human feces to the meat-associated L. sakei 23K model strain and analyzed their behaviors in the GITs of both conventional and axenic mice.
MATERIALS AND METHODS
Media, growth conditions, and isolation from feces.
Bacteria were routinely grown at 30°C in MRS medium (18) or in the chemically defined medium MCD (29) for physiological studies. When required, plates were incubated under anaerobiosis for 2 days, either in anaerobic jars (Gaspak; BBL) with anaerobic bags (Oxoid) or in incubators with a modified atmosphere (90% N2, 5% CO2, and 5% H2).
For mutant or transformant propagation, MRS medium was supplemented with the following concentrations of antibiotics: 5 mg liter−1 erythromycin, 10 mg liter−1 chloramphenicol, or 100 mg liter−1 rifampin (rifampicin). In order to isolate L. sakei from human feces, we used a modified MRS medium derived from those in the studies of Dal Bello et al. in 2006 (17) and Najjari et al. in 2008 (33), containing 10 g liter−1 polypeptone (Difco), 8.0 g liter−1 beef extract (Difco), 5 g liter−1 yeast extract (Difco), 2 g liter−1 K2HPO4, 2 g liter−1 diammonium citrate, 0.1 g liter−1 MnSO4, 0.05 g liter−1 MgSO4, 0.1% (vol/vol) Tween 80 (Merck), 12% (wt/vol) agar (Gibco), 20 g liter−1 glucose, and 0.025 g liter−1 bromocresol green, pH 6.2. The same medium, with the addition of rifampin (100 mg liter−1) and nalidixic acid (40 mg liter−1), was used to select and determine the L. sakei population present in the feces of conventional mice. Spores of Bacillus subtilis were counted by plating serial dilutions of fecal samples on brain heart infusion medium and incubating at 55°C for 48 h.
Fecal samples from 13 different healthy subjects living in France, males and females between 25 and 60 years of age, were collected and refrigerated at 4°C. Ten grams of feces was diluted in 90 ml of phosphate buffer (0.1 M K2HPO4, pH 6.8) and homogenized in a stomacher for 2 min. Serial dilutions in tryptone salt buffer were plated in duplicate on the modified MRS medium described above and incubated anaerobically for 48 h at 30°C.
A Bioscreen C system (Labsystems) was used to study the growth behavior of various clones when cultivated in liquid MRS medium. Fifteen microliters of bacterial precultures in early stationary phase was inoculated in 275 μl of fresh medium. The temperature was 30°C, and the cell density was measured every 20 min for 44 h.
Bacterial strains and plasmids.
L. sakei 23K is a plasmid-free strain obtained from a natural isolate originating from fermented sausage (7). L. sakei LTH5588, LTH5589, and LTH5590 (16), as well as L. sakei FLEC01 (isolated in this study), were isolated from feces of healthy humans. Spontaneous mutants resistant to rifampin (FLEC01rifR, LTH5590rifR, and 23KrifR) were obtained from corresponding L. sakei strains by plating 0.5 and 1 ml of an overnight culture onto MRS plates containing 100 mg liter−1 rifampin. Resistant clones appearing after 4 to 5 days of incubation were subcultured and then stored. Plasmids pRV566, carrying an erythromycin resistance gene (1), and pRV620, derived from pRV566 and carrying a chloramphenicol resistance cassette (15), were introduced by electroporation as previously described (7) into LTH5590 and 23K strains, respectively.
Clones presenting different morphotypes after passage through the GIT were collected and named following a nomenclature indicating their parent strain, the time after transit, the colony morphotype, and the isolation incubation conditions. For instance, L. sakei 23K J10SN2 (day 10, small [S] phenotype, isolated under anaerobic conditions) and L. sakei 23K J30RO2 (day 30, rough [R] phenotype, isolated under aerobic conditions) were issued from strain 23K. All strains were stored frozen at −80°C in MRS medium in the presence of 25% glycerol.
Experiments with conventional mice.
Twelve male adult C3H/HeN conventional mice, 6 to 8 weeks of age, born and bred in the facilities of Unité Expérimentale Animalerie Rongeurs at the INRA center of Jouy en Josas, France, were used following established methods (14). Mice were fed with powdered ground rodent chow (food no. R03-40; SAFE, Augy, France) in powdered food hoppers for mice (UAR, Epinay/Orge, France). The animals were permanently maintained in Trexler-type isolators (La Calhène, Vélizy, France). Autoclaved tap water and pelleted standard chow sterilized by gamma irradiation at 45 kGy were given ad libitum. The room housing the isolators was maintained at a constant temperature (21°C ± 1°C) and constant humidity (50% ± 5%) with a cycle consisting of 12 h of light and 12 h of darkness. Four mice were housed in each cage. L. sakei LTH5590rifR, RV2002rifR, and FLEC01rifR strains were cultivated in MRS liquid medium containing 100 mg liter−1 of rifampin. Fifty milliliters of 24-h bacterial cultures was washed with the same volume of peptone water, resuspended in 0.5 ml of phosphate-buffered saline (PBS) buffer, and mixed with 600 μl of a solution of B. subtilis spores. Two hundred microliters of this bacterial suspension (corresponding to an L. sakei population of approximately 109 CFU) was administered to mice by a unique intragastric gavage at time zero. Fresh fecal samples (between 0.1 g and 0.3 g) were collected from each mouse every 12 h until day 5, 10-fold diluted in LCY medium (2 g liter−1 N-Z Amine A [Sigma-Aldrich], 5 g liter−1 NaCl, 2 g liter−1 yeast extract, 1 g liter−1 K2HPO4), mixed, and homogenized with a sterile pipette. Serial dilutions were prepared in LCY medium, and 100-μl volumes of 10−3, 10−4, 10−5, and 10−6 dilutions were plated on MRS medium containing rifampin and nalidixic acid and on brain heart infusion medium (Difco).
Experiments in germfree mice.
Twenty-four male adult C3H/HeN mice, 6 to 8 weeks of age, born and bred in germfree conditions in the facilities of Unité Ecologie et Physiologie du Système Digestif, were used and treated as described above. In order to prevent contaminations, the animals were permanently maintained in sterilized Trexler-type isolators (La Calhène, Vélizy, France). Mice were acclimatized for 1 week (including PBS pretreatment during the last 4 days). Bacteria from 50 ml of 24-h MRS cultures were washed in peptone water and resuspended in 1 ml of PBS buffer. Two hundred microliters of the bacterial suspension (corresponding to a population of approximately 109 CFU) was administered to mice via a unique intragastric gavage at time zero. Fecal samples were collected and plated on MRS medium as described above every 12 h for 7 days and then on days 10, 15, 21, 30, and 40. After 51 days, mice were killed by cervical dislocation, and the GIT was sectioned in five parts (duodenum, jejunum, ileum, cecum, and colon). The luminal content of each section was weighed, diluted, and plated on MRS medium as described for feces.
All procedures were carried out in accordance with the institutional guidelines for the care and use of laboratory animals.
Scanning electron microscopy sample preparation.
Scanning electron microscopy was performed in the Laboratoire Bioadhésion et Hygiène des Matériaux, UMR/INRA-AgroParisTech, Massy, France. Samples for scanning electron microscopy were treated as described in reference 25. Briefly, samples were immersed in a fixative solution containing 2% glyceraldehyde for 48 h and put onto sterile tinfoil discs. Samples were rinsed with cacodylate buffer (0.2 M, pH 7.4) and then dehydrated in a graded ethanol series, air dried, placed on aluminum stubs with graphite paint, and coated with gold-palladium sputtering coater (Polaron SC7640; Elexience, Verrières-le-Buisson, France) for 140 s at 10 mA and 80 mtorr. Samples were visualized by field emission gun scanning electron microscope, and the resulting secondary electron images were analyzed (8 kV) by use of Hitachi S4500 equipment (Elexience).
DNA manipulations.
Purification of genomic DNA to be used in PCR experiments was performed as described in reference 4. DNA of small or medium-sized plasmids (<6 kb) was isolated from L. sakei by using the GenElute plasmid miniprep kit (Sigma-Aldrich) according to the recommendations of the manufacturer and following the modifications suggested by Hüfner et al. (27). Standard methods were used for electrophoresis and PCR DNA fragment purification (36). The presence of specific PCR products and of plasmids was monitored by electrophoresis on gels containing 1.5% and 0.8% agarose, respectively.
The analysis of large plasmids (>6 kb) was performed by pulsed-field gel electrophoresis (PFGE) after digestion with S1 nuclease as described in reference 6. Bacteria were collected from 1-ml exponential MRS cultures by centrifugation at room temperature, suspended in 0.5 ml TEE buffer (10 mM Tris-HCl, pH 9.0, 100 mM EDTA, 10 mM EGTA), and embedded in an equal volume of 2% low-melting-point agarose (Sigma). Plugs were then incubated 1 h at 37°C with 1 U S1 nuclease from Aspergillus oryzae (Sigma), and migration was carried out at 14°C for 15 h at 200 V with an angle of 120°. The switch time was 50 s to 90 s. After this, a second migration was performed for 4 h with the same parameters, except there was a switch time of 1 s to 12 s.
The size and structure of a chromosome was determined by PFGE after digestion with I-CeuI (New England Biolab), an enzyme that cuts a DNA sequence specific for rrn genes. Lysis and digestion were performed as described previously (20). Restriction digestion was performed using 2 U I-CeuI per plug. After PFGE migration, gels were stained with Vistra-Green (Amersham) according to the recommendations of the manufacturer and analyzed on a FluorImager (Molecular Dynamics), with a filter at 488 nm. Three independent gel electrophoreses were performed.
PCR conditions and DNA sequencing.
The 16S rrn genes were amplified by PCR as 1.5-kb DNA fragments with universal primers pA and pH* (9). PCR amplification was performed with a PCT-200 thermocycler (MJ Research). The 50-μl PCR mixture contained PCR buffer, 1.5 nmol liter−1 MgCl2, 0.2 nmol liter−1 of each deoxynucleoside triphosphate, 0.5 μmol liter−1 of each primer, 1 μg ml−1 of chromosomal DNA, and 1 U of Taq polymerase (Fermentas). The program used was as follows: 95°C for 4 min, 35 cycles of 94°C for 45 s, 55°C for 45 s, and 72°C for 1 min, and a final elongation step at 72°C for 5 min.
To detect the presence of the L. sakei katA gene, primers 702F and 310R (3) were used to amplify a 410-bp DNA fragment. Ten microliters of an overnight culture of L. sakei in MRS broth were deposed on FTA membranes (Whatman). Small discs (2-mm diameter) were cut and washed twice for 3 min with Tris-EDTA buffer, dried for 1 h and used as chromosomal DNA templates in a 50-μl PCR mixture containing PCR buffer, 1.5 nmol liter−1 MgCl2, 0.2 nmol liter−1 of each deoxynucleoside triphosphate, 0.8 μmol liter−1 of each primer, and 2 U of Taq polymerase. An initial denaturation step was performed at 94°C for 5 min, followed by 30 cycles of 94°C for 1 min, 56°C for 1 min, and 72°C for 1 min, and a final extension step at 72°C for 5 min.
For 16S rRNA gene product sequencing, the PCR fragment was treated with 0.1 U shrimp alkaline phosphatase (USB) and 1 U exonuclease I (Biolabs) in 20 mM Tris-HCl (pH 8.0)-10 mM MgCl2 buffer for 1 h at 37°C and then sequenced in both directions. Genotyping of strain FLEC01 and creation of a clustering dendrogram were carried out as previously described (11).
Nucleotide sequence accession number.
The sequence of the FLEC01 16S rRNA gene has been deposited in GenBank under accession number EU867793.
RESULTS
Isolation of L. sakei FLEC01 from human feces.
In order to compare human and meat-borne L. sakei isolates, we used strains L. sakei 23K (7), issued from a sausage isolate, and L. sakei LTH5588, LTH5589, and LTH5590, isolated from human feces (16). In addition, we isolated a new L. sakei human strain. For that purpose, the feces of 13 healthy humans were collected and lactobacilli were isolated by using a method derived from previously described protocols (17, 33). Clones showing a putative L. sakei morphology were purified, and the presence of the specific L. sakei katA gene was determined by PCR amplification. Only one positive clone was found. Its 16S rRNA gene was then amplified and sequenced. The 1,438-nucleotide sequence showed 99% identity to the 16S rRNA gene of L. sakei and this clone, hereafter called FLEC01, was thus considered to belong to this species. From the feces dilutions used to isolate L. sakei FLEC01, its presence was estimated as approximately 106 per g of feces.
Comparison of genetic features of human and meat-borne L. sakei strains.
We first determined the plasmid contents of the two human isolates: LTH5590 and FLEC01 strains carry two (66- and 14-kb) and three (29-, 14-, and 11-kb) plasmids, respectively. We then compared their genome sizes and organizations. PFGE analysis showed that L. sakei FLEC01 harbored seven rrn operons distributed on the chromosome, as previously observed for L. sakei (11, 20, 32). The genome size of L. sakei FLEC01 was evaluated to be 1,966 ± 5 kb, thus, comparable to those of L. sakei 23K (1,885 kb) (20) and LTH5590 (1,915 ± 22 kb) (11).
Currently, the natural L. sakei population can be divided into 10 genotypic clusters (A to J), and new L. sakei isolates can be rapidly genotyped by using PCR-based detection of a few genetic markers (11). This method was used to examine the genotypic clustering of L. sakei FLEC01 regarding the two other strains. We observed that FLEC01, like LTH5588, another GIT isolate, and two well-characterized industrial starters for sausage (strains L110 and 205) belonged to cluster B. In contrast, L. sakei 23K belongs to cluster A, and LTH5590 and LTH5589 belong to cluster C (see Fig. S1 in the supplemental material). The three strains belong, thus, to three different but closely related clusters.
L. sakei only transits and does not persist in gastrointestinal tract of conventional mice.
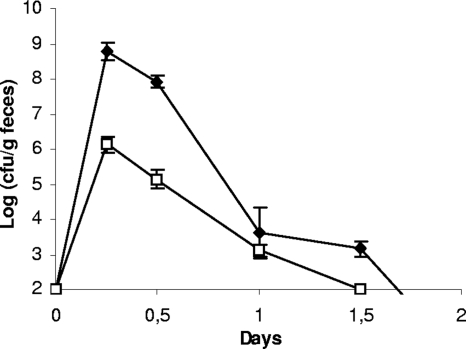
Spontaneous rifampin-resistant mutants, derived from the meat-borne strain L. sakei 23K and from the two human isolates, L. sakei LTH5590 and FLEC01, were inoculated independently in conventional mice in order to determine their ability to survive in the GIT by monitoring their presence in feces. The bacterial population of fecal samples before the mice were fed with L. sakei was determined on MRS selective medium containing rifampin and nalidixic acid. After 48 h of incubation at 30°C, no colonies were detected. Six hours after gavage, the L. sakei population reached ∼108 CFU per g of feces, depending on the strain, and then rapidly decreased (104 CFU per g of feces after 24 h) and became undetectable after 2 days (Fig. 1). The three strains showed similar behaviors: all survived in the GIT, but none was able to durably persist and colonize it. Consequently, their survival in axenic mice was therefore investigated.
FIG. 1.
Transit of L. sakei in the GITs of conventional C3H/HeN mice. Counts of L. sakei strain FLEC01 (black symbols) at different time points are shown. B. subtilis spores (white symbols) were used as a marker. Similar results were obtained with L. sakei 23K and LTH5590 (not shown). Data are the means of the cell counts obtained by plating feces excreted by four mice. Error bars indicate standard deviations.
Administration of L. sakei strains in axenic mice led to the appearance of subpopulations.
As our aim was to compare human and meat-borne strains, with the idea of measuring their competitiveness when mixed together, we used various antibiotic-resistant derivative strains. Several attempts to transform LTH5588, LTH5589, and FLEC01 strains with different plasmids were unsuccessful; however, LTH5590 could be transformed with pRV566, leading to erythromycin resistance. Therefore, L. sakei 23K carrying pRV620 (Cmr), LTH5590 carrying pRV566 (Emr), and a FLEC01rifR spontaneous mutant were first administered independently in axenic mice to compare their behaviors. Fecal samples were plated either on MRS medium or on MRS medium containing antibiotics. Colonies of L. sakei 23K and LTH5590 strains were not detected on MRS medium with antibiotics at 2 weeks after the mice were fed, indicating that the two strains had grown but lost the plasmid in the absence of antibiotic-selective pressure. In contrast, counts of L. sakei FLEC01rifR on MRS medium and MRS medium with rifampin remained identical until the end of the experiment (data not shown). Therefore, we considered only the CFU obtained on MRS plates without antibiotic, and when repeated, experiments were performed by using MRS medium without antibiotics.
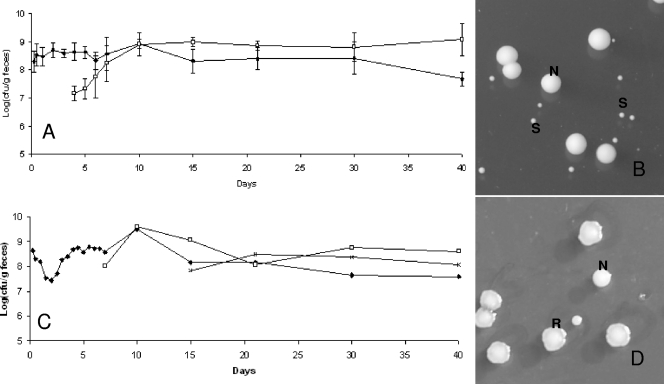
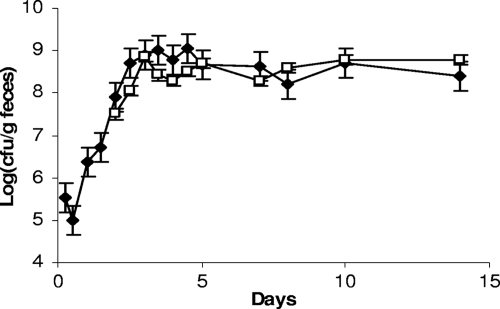
We observed that the three strains were rapidly detected in feces of germfree mice and that the populations remained stable over 51 days after ingestion (Fig. 2A). The population rapidly reached ∼109 per g for each strain. However, we observed that the colony morphology observed on MRS plates became heterogeneous between days 4 and 5: a subpopulation appeared and reached approximately half of the total population, and it persisted until day 40 (Fig. 2A). The colonies of this subpopulation are smaller than the average colony size for L. sakei (Fig. 2B). Similar results were observed for the three strains in all treated mice. At days 15 to 21, a second population appeared, characterized by R colonies on MRS medium (Fig. 2C and D). The emergence of this subpopulation occurred for all three strains (L. sakei 23K, LTH5590, and FLEC01rifR) but not in all treated mice, in contrast to the subpopulation harboring an S colony phenotype. One hundred twelve clones of different morphologies (normal [N], S, and R) were collected at various times and stored for further studies. The presence of the specific L. sakei katA gene was checked by PCR amplification on those clones. All were positive, attesting that they were not contaminants but indeed derived from the initial L. sakei strains that were fed to the mice. In addition, PFGE analysis, performed on one S clone and one R clone issued from mice fed with L. sakei 23K, LTH5590, and FLEC01 showed that the patterns obtained for S and R colonies were identical to that of the original strains (data not shown). The R colony morphotype was stable after several plating steps. However, for some S clones, we could observe that plating on MRS medium led to the appearance of colonies with an N phenotype at a low frequency (∼10−6), suggesting a reversion to the normal size and, thus, that these morphological changes might result from mutations.
FIG. 2.
Implantation of L. sakei in the GITs of axenic mice and appearance of a subpopulation. (A) Counts of L. sakei 23K (black symbols) at different time points are shown. White symbols represent a subpopulation showing an S colony morphology. Data are the means of the cell counts obtained by plating feces excreted by four mice. Error bars indicate standard deviations. (B) L. sakei S and N colony morphotypes on MRS plates. (C) Counts of the original L. sakei LTH5590 strain (black symbols) and the appearance of two subpopulations characterized by S (white symbols) and R (multiplication signs) colony morphologies. The R population appeared in only some mice. (D) An example of the R population in one mouse is shown. The image shows an L. sakei LTH5590 R colony morphotype on MRS medium.
Distribution of L. sakei in the lumen of the GITs of axenic mice.
We investigated the segmental distribution of strains 23K, FLEC01, and LTH5590 in the murine GIT, in order to identify the putative preferred colonization sites for L. sakei. Mice were killed 51 days after being fed, and the luminal contents of the small intestine (divided into duodenum, jejunum, and ileum), cecum, and colon were spread on MRS medium. The bacterial CFU obtained for each segment ranged from 104 to 109 CFU/g, and the highest cell counts were found in the cecum and colon (Fig. 3), indicating that these are the preferred colonization sites for L. sakei. Amounts of bacterial CFU in the small intestine (especially the duodenum and jejunum) differed significantly between the different mice. This suggests that the presence of L. sakei in these segments is transitory and probably due to the ingestion of feces and recontamination of the mouse. In addition, we noticed that S and R clones were recovered from all segments.
FIG. 3.
Distribution of L. sakei FLEC01 (lanes A), 23K (lanes B), and LTH5590 (lanes C) strains along the GITs of axenic mice. Cell counts obtained by plating the intestinal content of duodenum, jejunum, ileum, colon, and cecum of four mice (⋄, mouse 1; ▪, mouse 2; ▵, mouse 3; ×, mouse 4) are shown for the three strains. In the duodenum, jejunum, and ileum, the presence of L. sakei strains was sometimes detected in only two or three mice.
Characterization of S and R L. sakei morphotypes.
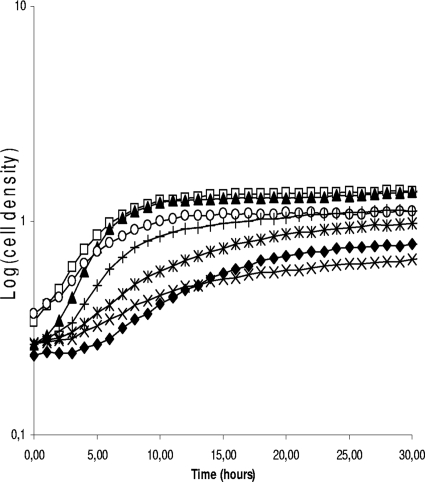
We further analyzed phenotypic traits of S and R clones. The growth of both S and R colonies in MRS medium was affected compared to that of the original strains (Fig. 4). Even after 44 h of incubation, S clones could not reach the cell density of parent strains, whereas R clones could. All the 112 clones were screened and, although the growth was always slower in S and R clones, a wide variability of curves could be noticed. This suggests that these clones were affected in different ways.
FIG. 4.
Growth curves of L. sakei 23K and of various S and R clones in MRS medium. Wild type (□) and L. sakei 23K N clone J7NN2 (▴), S clone J7SO2 (⧫), S clone J40SO2 (×), S clone J10SN2 (*), R clone J40RN2 (○), and R clone J30RO2 (+).
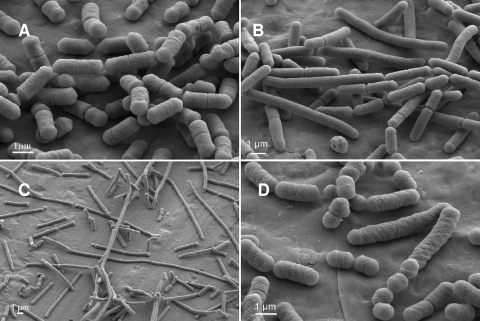
Images obtained by scanning electron microscopy of the cultures showed that for the three strains, cells issued from S clones were longer than wild-type cells. Cells issued from R clones of L. sakei strains FLEC01 and LTH5590 showed a filament structure, whereas cells issued from 23K R clones showed a mix of short and long cells (Fig. 5).
FIG. 5.
Morphologies of cells issued from S and R clones of L. sakei LTH5590 or 23K observed by scanning electron microscopy. L. sakei LTH5590 wild type (A) and S (B) or R (C) clones and the L. sakei 23K rough clone (D) grown in MRS liquid medium.
According to the growth curves and the stability of the colony morphotype, one S clone (23K J10SN2) and one R clone (23K J30RO2) showing a stable phenotype were chosen for an investigation of their behavior in mice. Both strains colonize the GITs of axenic mice, and both reached 109 CFU/g at 2 to 3 days after the feeding, as shown Fig. 6 for clone 23K J10SN2. None of the S and R clones returned to N colonies after GIT passage. Interestingly, in mice fed with the S strain 23K J10SN2, R colonies appeared after only 2 days and persisted until the end of the experiment (Fig. 6). These results indicate that R colonies are probably issued from S colonies.
FIG. 6.
Implantation of small clones in the GITs of germfree mice. Counts of L. sakei 23K J10SN2 at different time points (S colony, black symbols) are shown. White symbols represent R colony morphotypes. Data are the means of the cell counts obtained after plating feces excreted by four mice. Error bars indicate standard deviations.
DISCUSSION
The presence of several food-associated lactic acid bacteria (Lactobacillus curvatus, Leuconostoc mesenteroides, Leuconostoc argentinum, Pediococcus pentosaceus, and Pediococcus acidilactici), including L. sakei, in human feces has been previously demonstrated (26, 39) but not further investigated. Although L. sakei could be detected in human fecal samples by using PCR-specific primers for lactobacilli coupled with denaturing gradient gel electrophoresis analysis, these human L. sakei strains were not characterized. In 2003, Dal Bello et al. (16) isolated L. sakei strains from human feces (at ∼106 CFU per g of feces) and showed that this species was actually the dominant lactobacillus in some subjects when alternative incubation conditions were applied to isolate strains. In this study, we tried to better understand the behavior of L. sakei in the GIT.
We first isolated L. sakei strain FLEC01 from human feces by using incubation conditions similar to those previously described (16) and adapted from a phenotypical screening method (33). L. sakei FLEC01 was found to be present at ∼106 CFU per g of feces in one subject. The presence of this species in only some subjects is not unexpected, since differences between subjects were previously reported (26, 39). However, its detection in only one subject out of 13 might be due to the different incubation conditions used in our protocol (90% N2, 5% CO2, and 5% H2), suggesting that a low percentage of oxygen (2%) could be important for the isolation of L. sakei.
The plasmid contents of the two human L. sakei strains are different, and they also differ from that of L. sakei 23, the parent strain of 23K, isolated from sausage and harboring a unique large (>42-kb) plasmid (7). In this study, the genomic analysis and the phylogenomic classification of three L. sakei strains revealed that although the L. sakei 23K, LTH5590, and FLEC01 strains were isolated from different environments, they possess chromosomes of similar sizes and belong to closely related genotypic clusters. The three strains have relatively small genomes indeed, considering that genome sizes in the species L. sakei can range from 1,814 ± 12 kb to 2,308 ± 79 kb (11).
Interestingly, we noticed that strains L. sakei L110 and 205, used as starters for the production of French dry sausages and their genotypic clusters previously determined (11), belong to the same group as FLEC01 and show highly similar genetic fingerprints. In addition, LTH5588, a human isolate, also clustered in the same group, whereas another human isolates LTH5589 and LTH5590 cluster tag either in a different group. The observation that human isolates and commercial meat starter strains belong to closely related clusters suggests that the presence of L. sakei in human feces may result from ingestion of dry sausages.
So far, little is known about the behavior of L. sakei in the GIT. It was recently shown that L. sakei could persist in the intestines of brown trout (5). In the present study, we evaluated whether the fitness in the mouse GIT of strains isolated from human feces, thus, from the GIT, was higher than that of a strain isolated from food. The three strains showed the same behavior: they all transited in the GITs of conventional CH3/HeN mice (as fast as the spores of B. subtilis, used as a transit control), but none was able to persist in the presence of a preexisting microbiota. It is thus probable that L. sakei belongs to the transient microbiota of humans rather than to the resident one.
In contrast, the three strains of L. sakei were able to colonize the GITs of axenic mice, and as already shown for other lactic acid bacteria such as Lactobacillus acidophilus (34), Lactobacillus johnsonii (19), Lactobacillus plantarum (31), and Lactococcus lactis (35), the favorite sites of implantation are generally the cecum (intestine) and the proximal colon.
Mutants of the gram-negative bacterium Escherichia coli better adapted to the GIT environment were previously isolated after transit in conventional mice (30), and it was reported that E. coli mutates during the transit through the GITs of streptomycin-treated mice, giving rise to a different colony morphology (23, 24). Similarly, in axenic mice, we observed the appearance of subpopulations characterized by a different colony morphotype. Until now, this phenomenon has never been described for gram-positive bacteria. The occurrence of this phenomenon was observed with the three L. sakei strains, indicating that it can be considered a feature of the species. The subpopulations characterized by abnormal morphotypes had a selective advantage in the mouse GIT environment, since they could rapidly reach an equilibrium with the preexisting L. sakei population. However, they could not completely displace it. In a study of adaptive evolution of bacteria in the mouse gut, and based on previous results (21, 38), it was proposed that in addition to accumulating rare adaptive mutations, hypermutator bacteria rapidly accumulate numerous detrimental mutations (22). Many of the accumulated mutations may not affect growth in the GIT but may reduce bacterial fitness in a secondary environment. In our case, although well adapted to the GIT environment, S and R colonies grew poorly in MRS medium, supporting what was described for E. coli.
The S morphotype was conserved for subcultures of most but not all S clones in MRS medium. In contrast, the R colony phenotype was stable for all R clones tested, and R clones appeared rapidly after axenic mice were fed with S clones. The reversion of some of the S clones could indicate that the S phenotype is due to a single mutation or at least requires only a single suppressor mutation, whereas others are the result of a number of sequential mutational events providing a selective advantage in this environment.
The necessity for an accumulation of mutations to adapt to the GIT conditions provides one more indication that this environment is not the preferred niche for L. sakei.
Finally, scanning electron microscopy observations showed that bacterial cells issued from S and R clones had a strongly modified cell shape. We can thus hypothesize that either cell wall composition or the cell division system is affected. Experiments to deeply investigate S and R clone physiology are in progress in order to understand the reasons for the modified phenotype and to identify eventual adaptive mutations that occurred in L. sakei strains during transit through the GITs of axenic mice.
In conclusion, our results established a basis to better understand how the representative species of bacteria in meat, L. sakei, behaves during its transit to the GIT. We confirm previous results showing that L. sakei is indeed present in the human GIT (23), and we isolated a new strain from human feces. Human- and food-originating strains which are very close genotypically did not display relevant differences in either the survival or the colonization of the GITs of conventional and axenic mice. In addition, our human strain, similar to some other human isolates, belongs to genomic strain clusters that also encompass industrial starters used for the fermentation of dry sausages. Finally, the observation of phenotypic changes in the colony morphology and in the cell shape of L. sakei during GIT colonization, leading to a selective advantage in this environment, suggests that the GIT is not its preferred niche and the presence of this species in the human GIT is probably diet associated. The genetic and proteomic characterization of S and R clones will give us useful information to understand how L. sakei adapts to the GIT environment and to identify some of the functions implicated in its survival.
Supplementary Material
Acknowledgments
Fabrizio Chiaramonte is the recipient of a fellowship in the framework of the EC EST project “LABHEALTH,” Marie Curie contract MEST-2-CT-2004-514428.
We gratefully thank Christian Hertel for providing the L. sakei strains LTH5588, LTH5589, and LTH5590 and Thierry Meylheuc from the MIMA2 platform (Microscopie et Imagerie des Micro-organismes, Animaux et Aliments), INRA, Massy, France, for scanning electron microscopy analysis. We thank the germfree rodent facilities of UR910, Ecologie et Physiologie du Système Digestif, for providing germfree animals and for assistance with experiments.
Footnotes
Published ahead of print on 15 May 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alpert, C. A., A. M. Crutz-Le Coq, C. Malleret, and M. Zagorec. 2003. Characterization of a theta-type plasmid from Lactobacillus sakei: a potential basis for low-copy-number vectors in lactobacilli. Appl. Environ. Microbiol. 69:5574-5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ammor, S., C. Rachman, S. Chaillou, H. Prévost, X. Dousset, M. Zagorec, E. Dufour, and I. Chevallier. 2005. Phenotypic and genotypic identification of lactic acid bacteria isolated from a small-scale facility producing traditional dry sausage. Food Microbiol. 22:373-382. [Google Scholar]
- 3.Ammor, S., E. Dufour, M. Zagorec, S. Chaillou, and I. Chevallier. 2005. Characterization and selection of Lactobacillus sakei strains isolated from traditional dry-sausage. Food Microbiol. 22:529-538. [Google Scholar]
- 4.Anderson, D. G., and L. L. McKay. 1983. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl. Environ. Microbiol. 46:549-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balcázar, J. L., I. de Blas, I. Ruiz-Zarzuela, D. Vendrell, A. C. Calvo, I. Marquez, O. Girones, and J. L. Muzquiz. 2007. Changes in intestinal microbiota and humoral immune response following probiotic administration in brown trout (Salmo trutta). Br. J. Nutr. 97:522-527. [DOI] [PubMed] [Google Scholar]
- 6.Barton, B. M., G. P. Harding, and A. J. Zuccarelli. 1995. A general method for detecting and sizing large plasmids. Anal. Biochem. 226:235-240. [DOI] [PubMed] [Google Scholar]
- 7.Berthier, F., M. Zagorec, M. Champomier-Vergès, S. D. Ehrlich, and F. Morel-Deville. 1996. High-frequency transformation of Lactobacillus sake by electroporation. Microbiology 142:1273-1279. [DOI] [PubMed] [Google Scholar]
- 8.Bredholt, S., T. Nesbakken, and A. Holck. 2001. Industrial application of an antilisterial strain of L. sakei as a protective culture and its effect on the sensory acceptability of cooked, sliced, vacuum-packaged meat products. Int. J. Food Microbiol. 66:191-196. [DOI] [PubMed] [Google Scholar]
- 9.Broda, D. M., P. A. Lawson, R. G. Bell, and D. R. Musgrave. 1999. Clostridium frigidicans sp. nov., a psychrotolerant bacterium associated with ‘blown pack’ spoilage of vacuum-packed meats. Int. J. Syst. Bacteriol. 49:1539-1550. [DOI] [PubMed] [Google Scholar]
- 10.Chaillou, S., M. C. Champomier-Vergès, M. Cornet, A. M. Crutz-Le Coq, A. M. Dudez, V. Martin, S. Beaufils, R. Bossy, E. Darbon-Rongère, V. Loux, and M. Zagorec. 2005. Complete genome sequence of the meat-borne lactic acid bacterium Lactobacillus sakei 23K. Nat. Biotechnol. 23:1527-1533. [DOI] [PubMed] [Google Scholar]
- 11.Chaillou, S., M. Daty, F. Baraige, A.-M. Dudez, P. Anglade, R. Jones, C. A. Alpert, M. C. Champomier-Vergès, and M. Zagorec. 2009. Intraspecies genomic diversity and natural population structure of the meat-borne lactic acid bacterium Lactobacillus sakei. Appl. Environ. Microbiol. 75:970-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Champomier-Vergès, M. C., M. C. Montel, F. Grimont, and P. A. D. Grimont. 1987. Genomic identification of meat lactobacilli of meat as Lactobacillus sake. Ann. Inst. Pasteur Microbiol. 138:751-758. [DOI] [PubMed] [Google Scholar]
- 13.Champomier-Vergès, M. C., S. Chaillou, M. Cornet, and M. Zagorec. 2002. Lactobacillus sakei: recent development and future prospects. Res. Microbiol. 153:115-123. [DOI] [PubMed] [Google Scholar]
- 14.Coates, M. E. 1968. The germ-free animal in research. Academic Press, London, United Kingdom.
- 15.Crutz-Le Coq, A. M., and M. Zagorec. 2008. Vectors for lactobacilli and other Gram-positive bacteria based on the minimal replicon of pRV500 from Lactobacillus sakei. Plasmid 60:212-230. [DOI] [PubMed] [Google Scholar]
- 16.Dal Bello, F., J. Walter, W. P. Hammes, and C. Hertel. 2003. Increased complexity of the species composition of lactic acid bacteria in human feces revealed by alternative incubation condition. Microb. Ecol. 45:455-463. [DOI] [PubMed] [Google Scholar]
- 17.Dal Bello, F., and C. Hertel. 2006. Oral cavity as natural reservoir for intestinal lactobacilli. Syst. Appl. Microbiol. 29:69-76. [DOI] [PubMed] [Google Scholar]
- 18.De Man, J. C., M. Rogosa, and M. E. Sharpe. 1960. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 23:130-135. [Google Scholar]
- 19.Denou, E., B. Berger, C. Barretto, J. M. Panoff, F. Arrigoni, and H. Brussof. 2007. Gene expression of commensal Lactobacillus johnsonii strain NCC533 during in vitro growth and in the murine gut. J. Bacteriol. 189:8109-8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dudez, A. M., S. Chaillou, L. Hissler, R. Stentz, M. Champomier-Vergès, C. A. Alpert, and M. Zagorec. 2002. Physical and genetic map of the Lactobacillus sakei 23K chromosome. Microbiology 148:421-431. [DOI] [PubMed] [Google Scholar]
- 21.Funchain, P., A. Yeung, J. L. Stewart, R. Lin, M. M. Slupska, and J. H. Miller. 2000. The consequences of growth of a mutator strain of Escherichia coli as measured by loss of function among multiple gene targets and loss of fitness. Genetics 154:959-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giraud, A., M. Radman, I. Matic, and F. Taddei. 2001. The rise and fall of mutator bacteria. Curr. Opin. Microbiol. 4:582-585. [DOI] [PubMed] [Google Scholar]
- 23.Giraud, A., I. Matic, O. Tenaillon, A. Clara, M. Radman, M. Fons, and F. Taddei. 2001. Costs and benefits of high mutation rates: adaptive evolution of bacteria in the mouse gut. Science 291:2606-2608. [DOI] [PubMed] [Google Scholar]
- 24.Giraud, A., S. Arous, V. Gaboriau-Routhiau, M. De Paepe, J. C. Bambou, S. Rakotobe, A. B. Lindner, F. Taddei, and N. Cerf-Bensussan. 2008. Dissecting the genetic components of adaptation of Escherichia coli to the mouse gut. PLoS Genet. 4:e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guyard-Nicodème, M., A. Bazire, G. Hémery, T. Meylheuc, D. Mollé, N. Orange, L. Fito-Boncompte, M. Feuilloley, D. Haras, A. Dufour, and S. Chevalier. 2008. Outer membrane modifications of Pseudomonas fluorescens MF37 in response to hyperosmolarity. J. Proteome Res. 7:1218-1225. [DOI] [PubMed] [Google Scholar]
- 26.Heilig, H. G., E. G. Zoetendal, E. E. Vaughan, P. Marteau, A. D. Akkermans, and W. M. de Vos. 2002. Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA. Appl. Environ. Microbiol. 68:114-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hüfner, E., T. Markieton, S. Chaillou, A. M. Crutz-Le Coq, M. Zagorec, and C. Hertel. 2007. Identification of Lactobacillus sakei genes induced in meat fermentation and their role in survival and growth. Appl. Environ. Microbiol. 73:2522-2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katagiri, H., K. Kitahara, and K. Fukami. 1934. The characteristics of the lactic acid bacteria isolated from moto, yeast mashes for sake manufacture. IV. Classification of the lactic acid bacteria. Bull. Agr. Chem. Soc. Jpn. 10:156-157. [Google Scholar]
- 29.Lauret, R., F. Morel-Deville, F. Berthier, M. C. Champomier-Vergès, P. Postma, S. D. Ehrlich, and M. Zagorec. 1996. Carbohydrate utilization in Lactobacillus sake. Appl. Environ. Microbiol. 62:1922-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leatham, M. P., S. J. Stevenson, E. J. Gauger, K. A. Krogfelt, J. J. Lins, T. L. Haddock, S. M. Autieri, T. Conway, and P. S. Cohen. 2005. Mouse intestine selects nonmotile flhDC mutants of Escherichia coli MG1655 with increased colonizing ability and better utilization of carbon sources. Infect. Immun. 73:8039-8049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marco, M. L., R. S. Bongers, W. M. de Vos, and M. Kleerebezem. 2007. Spatial and temporal expression of Lactobacillus plantarum genes in the gastrointestinal tracts of mice. Appl. Environ. Microbiol. 73:124-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McLeod, A., O. L. Nyquist, L. Snipen, K. Naterstad, and L. Axelsson. 2008. Diversity of Lactobacillus sakei strains investigated by phenotypic and genotypic methods. Syst. Appl. Microbiol. 31:393-403. [DOI] [PubMed] [Google Scholar]
- 33.Najjari, A., I. Ouzari, A. Boudabous, and M. Zagorec. 2008. Method for reliable isolation of a collection of Lactobacillus sakei strains originating from Tunisian seafood and meat products. Int. J. Food Microbiol. 121:342-351. [DOI] [PubMed] [Google Scholar]
- 34.Norin, K. E., A. K. Persson, H. Saxerholt, and T. Midtvedt. 1991. Establishment of Lactobacillus and Bifidobacterium species in germfree mice and their influence on some microflora-associated characteristics. Appl. Environ. Microbiol. 57:1850-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roy, K., M. Meyrand, G. Corthier, V. Monnet, and M. Y. Mistou. 2008. Proteomic investigation of the adaptation of Lactococcus lactis to the mouse digestive tract. Proteomics 8:1661-1676. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Tannock, G. W. 2004. A special fondness for lactobacilli. Appl. Environ. Microbiol. 70:3189-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tenaillon, O., B. Toupance, H. Le Nagard, F. Taddei, and B. Godelle. 1999. Mutators, population size, adaptive landscape and the adaptation of asexual populations of bacteria. Genetics 152:485-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walter, J., C. Hertel, G. W. Tannock, C. M. Lis, K. Munro, and W. P. Hammes. 2001. Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group specific PCR primers and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67:2578-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.