Abstract
Constructed subsurface flow (SSF) and free-surface flow (FSF) wetlands are being increasingly implemented worldwide into wastewater treatments in response to the growing need for microbiologically safe reclaimed waters, which is driven by an exponential increase in the human population and limited water resources. Wastewater samples from four SSF and FSF wetlands in northwestern Ireland were tested qualitatively and quantitatively for Cryptosporidium spp., Giardia duodenalis, and human-pathogenic microsporidia, with assessment of their viability. Overall, seven species of human enteropathogens were detected in wetland influents, vegetated areas, and effluents: Cryptosporidium parvum, C. hominis, C. meleagridis, C. muris, G. duodenalis, Encephalitozoon hellem, and Enterocytozoon bieneusi. SSF wetland had the highest pathogen removal rate (i.e., Cryptosporidium, 97.4%; G. duodenalis, 95.4%); however, most of these values for FSF were in the negative area (mean, −84.0%), meaning that more pathogens were discharged by FSF wetlands than were delivered to wetlands with incoming wastewater. We demonstrate here that (i) the composition of human enteropathogens in wastewater entering and leaving SSF and FSF wetlands is highly complex and dynamic, (ii) the removal and inactivation of human-pathogenic microorganisms were significantly higher at the SSF wetland, (iii) FSF wetlands may not always provide sufficient remediation for human enteropathogens, (iv) wildlife can contribute a substantial load of human zoonotic pathogens to wetlands, (v) most of the pathogens discharged by wetlands were viable, (vi) large volumes of wetland effluents can contribute to contamination of surface waters used for recreation and drinking water abstraction and therefore represent a serious public health threat, and (vii) even with the best pathogen removal rates achieved by SSF wetland, the reduction of pathogens was not enough for a safety reuse of the reclaimed water. To our knowledge, this is the first report of C. meleagridis from Ireland.
Demand for high-quality drinking and recreational waters rises exponentially due to global demographic growth in the human population, reinforcing an urgent need for microbiologically safe reclaimed waters (12). Wastewater discharges are worldwide risk factors for the introduction of human pathogens into surface waters used as drinking and recreational resources. Cryptosporidium parvum, C. hominis, Giardia duodenalis, and human-virulent microsporidia (i.e., Encephalitozoon intestinalis, E. hellem, E. cuniculi, and Enterocytozoon bieneusi) are waterborne enteropathogens inflicting considerable morbidity in healthy people and mortality (e.g., Cryptosporidium and microspora) in immunodeficient individuals (34, 44). Their transmissive stages, i.e., oocysts, cysts, and spores, are resistant to environmental stressors and are therefore long-lasting and relatively ubiquitous in the environment (13, 27, 45). These pathogens are category B biodefense agents on the U.S. National Institutes of Health list, and microsporidian spores are on the Contaminant Candidate List of the U.S. Environmental Protection Agency (29) because spore identification, removal, and inactivation in drinking water are technologically challenging. Surface water is not routinely monitored for these pathogens, despite evidence demonstrating environmental contamination derived from wastewater discharges (12). Environmentally, all aforementioned pathogens (except C. hominis) have a broad zoonotic reservoir (13, 27, 34).
Constructed wetlands of either vertical or horizontal flow are increasingly used worldwide for secondary or tertiary treatment of municipal wastewater due to minimum electric requirements and low maintenance costs (6, 32). The wetland concept has become an attractive wastewater treatment alternative to conventional tertiary treatment processes for (i) municipal wastewater, (ii) on-site domestic wastewater treatment, and (iii) concentrated animal feeding operations (24). In wetlands, human-pathogenic microorganisms are physically removed and biodegraded by sedimentation (2, 23), filtration and evapotranspiration-driven attachment to plant roots (10, 43), natural die-off (28), UV radiation, straining and sorption by biofilm (31), and protozoan predation (37). It is thought that the performance of subsurface flow (SSF) wetlands in removing human pathogens is superior to that of secondary wastewater treatment, i.e., conventional sewage sludge activation (40). Horizontal wetlands usually discharge to surface waters that are frequently used for recreation or drinking water production (6). It is commonly assumed that human pathogens identified in wetland effluents originate from the wastewater (39). However, this was never proven because studies of human pathogens in wetlands (10, 23, 28, 31, 32, 39, 40) did not utilize molecular epidemiology techniques to differentiate pathogen species or assess their viability.
In general, wastewater can be injected under the wetland surface for plug flow hydraulics, i.e., SSF (43), or be delivered to the wetland surface for free-surface flow (FSF). Because the wastewater resides in wetlands, these areas can act as endemic sites supporting both propagation and transmission of human zoonotic pathogens (15). Sizing reed-bed systems for a residence time of 5 days has become a standard practice (6, 31, 39), leaving plenty of time for the propagation and spreading of wastewater-derived pathogens in wetland habitats via a wide variety of wildlife (12, 15). Any temporal or permanent malfunctioning caused by clogged inlet pipes can cause (i) hydraulic short circuits that bypass part of the filtration area in FSF wetlands or (ii) chance SSF wetland filtration dynamics to FSF dynamics (31, 40). This can additionally increase wastewater retention time in wetlands.
The purposes of the present study were to (i) determine species of human protozoan and fungal enteropathogens entering, residing, and leaving constructed horizontal wetlands used for tertiary treatment of municipal wastewater; (ii) determine the efficacy for removal of Cryptosporidium oocysts, G. duodenalis cysts, and human-virulent microsporidian spore species by wetlands from wastewater subjected to secondary treatment; and (iii) compare pathogen removal efficacies between SSF and FSF wetlands. We used a multiplexed fluorescence in situ hybridization (FISH) assay with immunofluorescence antibody (IFA) to identify C. parvum and C. hominis oocysts and microsporidian spores and to assess their viability in a quantitative manner. Since multiplexed FISH specifically identifies C. parvum and C. hominis oocysts but does not differentiate between these species (36), we used PCR-restriction fragment length polymorphism (RFLP) to identify other potential oocyst species. In addition, we used PCR to confirm species of microsporidian spores identified by FISH.
MATERIALS AND METHODS
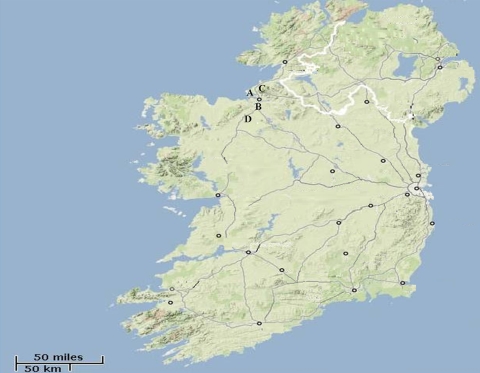
Samples originated from four constructed horizontal wetlands, i.e., wetlands A (53°40′41"N, 08°34′24"W), B (53°03′12"N, 08°08′57"W), C (54°04′07"N, 08°12′12"W), and D (53°41′11"N, 08°45′17"W), in northwestern Ireland (Fig. 1). All wetlands received unchlorinated municipal wastewater subjected to secondary treatment after sewage sludge activation and secondary sedimentation. Wetland A had two components; the first FSF component discharged to SSF component with a final effluent released to groundwater (Table 1). Both components represented monoculture systems with emergent vegetation, i.e., the common reed, Phragmites australis. All remaining wetlands were small-scale FSF wetlands discharging to surface waters. Wetlands B, C, and D were multispecies systems with both emergent and submerged plants overwhelmed by P. australis, which was the dominant vegetation type at wetland B. The influent rates were similar at all four wetlands and varied from ca. 46 to 56 liters min−1. The inflow, outflow, and vegetation densities were similar at all four wetlands, and the influent and effluent flow rates were relatively constant. Two grab samples (2 liters) of both wetland influents and effluents were collected in addition to three to four samples from the wetland longitudinal transect in regular intervals (Table 1). As seasonal differences in removal of Cryptosporidum oocysts and Giardia cysts by constructed wetland were not statistically significant (32), although they can vary from year to year, the samples for the present study were collected in a spatial manner within 1 week during the spring. Each constructed wetland was sampled in full on a different day during that week period, and the pathogen load data are presented in Table 1. Fecal coliform results related to wetland influent and effluent were provided by wastewater treatment plants. Samples were transported to the laboratory in a cooler and processed by gravity sedimentation (15). Briefly, samples were vortex mixed, transferred to 1-liter Imhoff settlement cones, and left overnight at 4°C. Portions (50 ml) of the top sediment layer were transferred to plastic 50-ml tubes and centrifuged (3,000 × g, 10 min). The supernatant was discarded, and the pellet was transferred to a 1.5-ml tube and preserved with 75% ethanol. The recovery efficacy of human waterborne pathogens from wastewater matrices was determined previously to be ca. 77% (15).
FIG. 1.
Location of four wastewater treatment plants (i.e., A, B, C, and D) in northwestern Ireland. The effluent from plant A was discharged to Sligo Bay. The effluents from plants B, C, and D were released to local rivers.
TABLE 1.
Average concentrations of C. parvum and C. hominis oocysts, G. duodenalis cysts, and microsporidian sporesa
| Wetland | No. of samples | Avg concn ([oo]cysts/liter ± SD)
|
|||
|---|---|---|---|---|---|
| C. parvum and C. hominis | G. duodenalis | E. hellem | E. bieneusi | ||
| A (SSF and FSF)b | |||||
| Influent | 2 | 118 ± 4.0 | 241 ± 4.0 | 0 | 0 |
| Wetland transect | 4 | 101 ± 77.4 | 28 ± 11.5 | 4 ± 3.2 | 3 ± 3.2 |
| First effluent | 2 | 4 ± 1.0 | 121 ± 113.5 | 119 ± 103 | 0 |
| Final effluent | 3 ± 1.5 | 11 ± 1.0 | 68 ± 51.0 | 0 | |
| B (FSF) | |||||
| Influent | 2 | 10 ± 2.5 | 8 ± 6.5 | 0 | 0 |
| Wetland transect | 3 | 50 ± 7.6 | 78 ± 22.0 | 10 ± 3.2 | 3 ± 3.0 |
| Effluent | 2 | 63 ± 26.5 | 140 ± 27.5 | 33 ± 3.5 | 12 ± 2.0 |
| C (FSF) | |||||
| Influent | 2 | 32 ± 6.5 | 17 ± 7.0 | 0 | 0 |
| Wetland transect | 4 | 48 ± 17.0 | 6 ± 2.1 | 8 ± 1.8 | 0 |
| Effluent | 2 | 22 ± 5.0 | 111 ± 57.0 | 9 ± 4.0 | 6 ± 1.0 |
| D (FSF) | |||||
| Influent | 2 | 17 ± 6.5 | 9 ± 3.5 | 0 | 0 |
| Wetland transect | 4 | 44 ± 13.7 | 92 ± 15.9 | 13 ± 6.5 | 8 ± 4.7 |
| Effluent | 2 | 78 ± 11.0 | 90 ± 55.5 | 9 ± 1.5 | 2 ± 2.0 |
That is, Encephalitoozon hellem and Enterocytozoon bieneusi, in constructed SSF and FSF horizontal wetlands.
Wetland A had two components: the first FSF component discharged to the SSF component (first effluent), which discharged to groundwater (final effluent).
The ethanol was washed with phosphate-buffered saline (pH 7.4) and centrifuged (5,000 × g, 10 min), and the pellet was purified by sugar flotation; a 2.5 M sucrose solution with a specific gravity of 1.34 was used (22). The resulting pellet was divided evenly into three aliquots. The first was processed for C. parvum, C. hominis, and G. duodenalis by multiplexed FISH in combination with an immunofluorescence antibody assay and processed a second time for human-virulent microsporidia (i.e., E. intestinalis, E. hellem, E. cuniculi, and E. bieneusi) by multiplexed FISH (15). FISH-reactive pathogen cells were enumerated (15), and their numbers were adjusted for the separation into aliquots. The third aliquot was assayed by PCR using primers based on the 18S rRNA gene for the detection of E. intestinalis, E. hellem, E. cuniculi, and E. bieneusi (3-5, 7, 42). Briefly, 0.3 μM concentrations of each primer and AmpliTaq Gold PCR Master Mix (Applied Biosystems, Foster City, CA) were mixed in a 50-μl final volume. The cycling parameters were 95°C for 5 min, followed by 40 cycles of 95°C for 15 s, 60°C for 15 s, and 72°C for 1 min, with a final extension of 72°C for 7 min. Negative controls and the spores of other microsporidian species were included in the PCR testing. All PCR products were analyzed on a 2% agarose gel (agarose GTG/LE; American Bioanalytical, Natick, MA) and stained with ethidium bromide for visualization. For identification of Cryptosporidium spp., samples were analyzed by utilizing a small-subunit rRNA-based nested PCR-RFLP with restriction enzymes SspI and VspI as described previously (20, 46). Briefly, each sample was assayed using 2 μl of the DNA template per PCR mixture. Nonacetylated bovine serum albumin (400 ng/μl; Sigma-Aldrich, St. Louis, MO) was used in all primary PCRs to neutralize residual PCR inhibitors in the extracted DNA. Portions (10 μl) of the secondary PCR products were digested at 37°C overnight in a 40-μl reaction mixture. The digested products were visualized by 2% agarose gel electrophoresis.
Precipitation and air temperature data for the month of sample collection and two preceding months were obtained electronically from the local weather station.
Statistical analysis was carried out with Statistix 9.0 (Analytical Software, St. Paul, MN). Variables were tested by using Wilk-Shapiro ranking plots to determine whether their distribution conformed to a normal distribution and, if not, nonparametric tests were used. Differences in pathogen concentrations were assessed by the Wilcoxon signed-rank test, and differences in pathogen fractions were assessed by the chi-square test (χ2). Mean values were associated with the standard deviation and were considered statistically significant at a P value of <0.05, and all P values for the Wilcoxon signed-rank test were two-tailed. Pathogen removal efficiency was calculated as the percentage of pathogens remaining in the samples after the treatment (Fig. 1).
RESULTS
Overall, seven species of human protozoan and fungal enteropathogens were detected: four species in wetland influents (C. parvum, C. hominis, C. meleagridis, and G. duodenalis), seven in wetlands (C. parvum, C. hominis, C. muris, C. meleagridis, G. duodenalis, E. hellem, and E. bieneusi), and five in wetland outfalls (i.e., C. parvum, C muris, G. duodenalis, E. hellem, and E. bieneusi) (Tables 1 and 2). Only C. parvum and G. duodenalis were detected in both wetland influents and effluents (Tables 1 and 2). C. hominis oocysts were identified in all wetland influents and in only one of four wetlands, i.e., wetland A (Table 2 and Fig. 2). Similarly, C. meleagridis was common in wetland influents but absent in the wetland effluents (except wetland D), indicating that C. hominis and C. meleagridis oocysts were removed by wetland-associated mechanisms (Table 2). In contrast, C. muris oocysts and E. hellem and E. bieneusi spores were not detected in any of the wetland influents, but these species were common in the wetland outfalls (Tables 1 and 2). This may indicate that C. muris oocysts and microsporidian spores were not delivered by the wetland-incoming wastewater but originated from other sources. The concentration of human enteropathogens did not vary along the transect.
TABLE 2.
Cryptosporidium species identified in constructed SSF and FSF horizontal wetlands used for polishing municipal wastewater subjected to secondary treatment
| Wetland | Cryptosporidium species |
|---|---|
| A (FSF and SSF)a | |
| Influent | C. hominis, C. meleagridis |
| Wetland area | C. parvum, C. hominis, C. muris |
| First effluent | C. parvum, C. muris |
| Final effluent | Negative |
| B (FSF) | |
| Influent | C. hominis, C meleagridis |
| Wetland area | C. parvum, C. meleagridis |
| Effluent | C. parvum, C. muris |
| C (FSF) | |
| Influent | C. parvum, C. hominis, C. meleagridis |
| Wetland area | C. parvum |
| Effluent | C. parvum, C. muris |
| D (FSF) | |
| Influent | C. hominis, C. parvum |
| Wetland area | C. muris |
| Effluent | C. parvum, C. muris, C. meleagridis |
Wetland A had two components: the first FSF component discharged to the SSF component (first effluent), which discharged to groundwater (final effluent).
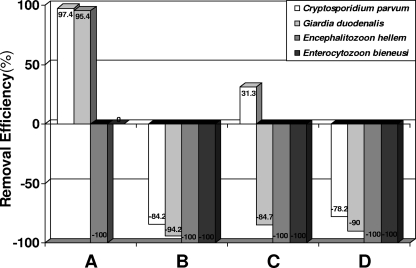
FIG. 2.
Removal efficacy (%) of human enteropathogens from municipal wastewater subjected to secondary treatment by constructed SSF (A) and FSF (B, C, and D) wetlands.
Most pathogens detected by the FISH assays were potentially viable; a fraction of nonviable cells represented <2%. Potentially viable G. duodenalis cysts versus nonviable and non-G. duodenalis cysts were clearly differentiated by color as a result of FISH and monoclonal antibody (MAb) labeling. Nonviable cysts were represented by (i) shells with apparently structurally damaged walls and (ii) intact cells with a very small amount of internal structures with diffused appearance. In comparison, potentially viable intact cysts were filled out completely with cytoplasm without the gap between the internal structures and the wall. Oocysts labeled by FISH and MAb were predominantly intact and revealed a small gap between the oocyst wall and internal structures, and in most of them, the sporozoites were visible. In comparison, dead oocysts, i.e., oocyst shells, frequently had discernible damage to their walls. Rarely, FISH-positive potentially viable oocysts had noticeable ruptures in their walls which was clearly revealed by MAb staining.
C. parvum and C. hominis oocysts and G. duodenalis cysts were detected by FISH in 32 of 33 samples (prevalence of 97%) (Table 1). The concentrations of C. parvum and C. hominis oocysts in wetland influents varied from 10 to 118 oocysts/liter (mean, 44 ± 16.5) and from 3 to 78 oocysts/liter (mean, 41 ± 12.7) in effluents (Table 1). Concentrations of the oocysts in wetland areas varied from 44 to 101 oocysts/liter (mean, 61 ± 20.3) and were higher than in the influents (Table 1). However, because the FISH reaction was not specific for C. meleagridis and C. muris oocysts (as identified by PCR-RFLP) (Table 2), it is possible that the concentration levels presented in Table 1 were underestimated. The most common Cryptosporidium species in wetland influents was C. hominis (44.4%), C. parvum was most common in the wetland area (60.6%), and C. muris was a predominant species in wetland effluents (50.0%) (Table 2). Overall, C. parvum was the most common species identified (i.e., 36.4%), followed by C. hominis and C. meleagridis (22.7% each) and C. muris (18.2%).
Concentrations of G. duodenalis cysts in wetland influents ranged from 8 to 241 cysts/liter (mean, 69 ± 37.7), from 6 to 92 cysts/liter in wetland areas (mean, 49 ± 11.2), and from 11 to 140 cysts/liter (mean, 88 ± 24.0) in wetland effluents (Table 1).
Multiplex FISH identified E. hellem and E. bieneusi spores, and of all the microsporidian species tested, only E. hellem and E. bieneusi DNA was amplified with the 18S rRNA PCR. The overall prevalence of E. hellem-positive samples (75.8%) was significantly higher (chi-square test; χ2 = 9.21, P < 0.01) than that of E. bieneusi-positive samples (51.5%). Also, the average concentration of E. hellem spores in wetland effluents of 29 ± 13.3 spores/liter was significantly higher than that of E. bieneusi (7 ± 1.9 spores/liter) (Wilcoxon signed-rank test; t = 2.09, P < 0.03) (Table 1) in wetland influents. Encephalizotoon hellem was the predominant microsporidian species (59.5%), with a lower percentage of E. bieneusi (40.4%).
On average, 56 pathogen transmissive stages per liter were delivered to the wetlands, 38 pathogens/liter were found in the wetland areas, and 50 pathogens/liter were discharged to receiving waters. The concentrations of C. parvum and C. hominis oocysts discharged to surface water by wetlands B, C, and D (mean, 55 ± 12.9) were significantly higher (Wilcoxon rank-sum test; t = 1.7, P < 0.05) than the concentrations of oocysts coming to these wetlands with wastewater (mean, 19 ± 4.8) (Table 1). Similarly, wetlands B, C, and D discharged significantly (Wilcoxon rank-sum test; t = 2.45, P < 0.01) more G. duodenalis cysts (mean, 78 ± 18.4) than the wetland incoming wastewater (mean, 11 ± 3.2).
Most of the values of pathogen removal rates by constructed wetlands were in the negative area (Fig. 1). The average pathogen removal rates were 30.9%, −96.6%, −63.4%, and −92.1% for wetlands A, B, C, and D, respectively (Fig. 1). SSF wetland A had significantly higher pathogen removal rates (30.9%) than the remaining three FSF wetlands (mean, −84.0%) (chi-square test; χ2 = 4.28, P < 0.03). The pathogen removal efficacy reached positive values for Cryptosporidium oocysts at SSF wetland A and at wetland C, i.e., 97.4 and 31.3%, respectively, and for G. duodenalis cysts at SSF wetland A, i.e., 95.4% (Fig. 1). Interestingly, although SSF wetland A had the highest concentration of pathogens in incoming wastewater, the lowest pathogen level occurred in its effluents (Table 1).
The precipitation level during the week of sample collection was 56 mm, and the precipitation levels were 54 and 92 mm for the two preceding months, respectively. The average daily temperature for the month of collection was 6.9°C but was 6.3 and 6.8°C for the two preceding months, respectively.
The levels of fecal indicators in influent and effluent did not vary significantly between influent and effluent samples for all four wetlands.
DISCUSSION
We demonstrated here that (i) the composition of human pathogen species in wastewater subjected to secondary treatment entering constructed wetlands and in wastewater subjected to tertiary treatment is highly complex and quite dynamic; (ii) removal and inactivation of human-pathogenic microorganisms were influenced by wetland type, i.e., SSF or FSF, reaching significantly higher levels at a SSF wetland; (iii) small-scale FSF wetlands may not provide sufficient remediation for human zoonotic protozoa and fungi; (iv) most of the pathogens discharged by wetlands were viable and thus capable of causing human infections; (v) large volumes of wetland effluents can substantially contribute to contamination of surface waters used for recreation and drinking water abstraction and therefore represent a serious public health threat; and (vi) even with the best pathogen removal rates at the SSF wetland, the reduction of pathogens was not enough for a safety reuse of the reclaimed water; this confirms previous reports (32).
The results pertinent to three FSF wetlands investigated in the present study showed that the concentrations of all seven pathogen species were higher in wetland outfalls than in the influents (Table 1). Even the SSF wetland (i.e., wetland A), with the highest removal rates for Cryptosporidium oocysts and Giardia cysts, discharged E. hellem spores despite the fact that they were absent in the wastewater entering this wetland (Table 1). The presence of pathogens at higher concentrations in wetland-polished wastewater, as well as the different pathogen species composition in influents, vegetated areas, and outfalls (Tables 1 and 2), may be explained by the facts that some of these pathogens, i.e., C. parvum, C. muris, C. meleagridis, E. hellem, and E. bieneusi, (i) were propagated in the wetlands by residing wildlife; (ii) contributed to the wetland water by visiting wildlife; or (iii) originated from other sources, e.g., surface runoff from wetland banks utilized by rodents as habitats or visiting areas. Aquatic birds and mammalian wildlife that frequently inhabit wetlands can disseminate human-virulent species of Cryptosporidium, Giardia, and microsporidia, i.e., E. hellem and E. bieneusi (14, 17, 35, 38). It has been estimated that a single visitation of an average-size waterfowl flock can introduce into the water approximately 9.3 × 106 C. parvum oocysts, 1.1 × 107 G. duodenalis cysts, and 9.1 × 108 E. hellem spores (17). Interestingly, in the present study all of the human pathogen species known to be disseminated by birds (17) were detected in wetland effluents. Fecal samples collected from local wildlife will be screened for human pathogen species detected in the wetland samples for further study.
Wildlife that inhabit or visit constructed wetlands have previously been demonstrated to significantly contribute fecal coliforms (e.g., Escherichia coli and Klebsiella pneumonia) to wetlands (39). It has been suggested that wildlife play an important role in the elevation of total and fecal coliform levels in wetland effluents due to their fecal deposition (39) and the spontaneous multiplication of wildlife-derived coliforms in wetlands during summer months (9). The pathogens identified in the present study cannot multiply in the environment without their hosts. We conclude that, in addition to fecal coliform bacteria, wildlife can also substantially contribute human-pathogenic protozoan and fungal microorganisms to wetland systems.
The present study indicates a high complexity of human pathogens in wetland habitats used for tertiary wastewater treatment. Because of the host adaptation nature within the Cryptosporidium genus, results of the present study demonstrate that humans (C. hominis, C. parvum, and C meleagridis) and wildlife such as rodents and birds (C. parvum, C. meleagridis, and C. muris) contributed to the contamination of wastewater. The predominance of C. hominis in wastewater charging the wetlands indicates the importance of anthroponotic transmission in northwestern Ireland, which may play a predominant role in the epidemiology of cryptosporidiosis in Ireland; these findings are consistent with the reports originating from this country (25, 48). In Ireland, C. hominis is constantly present in the human population at low prevalence levels throughout the year (48) and has been reported in higher proportion in older age group people (48); related outbreaks occur throughout the year, whereas C. parvum outbreaks coincide with calving or lambing periods (48). In Ireland, C. hominis is most prevalent in outbreak situations (48), being reported in many epidemics (1, 30), including at least five epidemics of waterborne etiology (11). C. parvum was also identified in waterborne outbreaks in Ireland; however, the number of people affected by C. hominis was always almost four times higher (48). The overall prevalence of 22.7% of C. meleagridis in the present study is of great interest. Although common in the United Kingdom (25, 48), this species has not been reported from Ireland. Thus, the present study is the first report of C. meleagridis from Ireland. Regarding the microsporidian spore species, both E. hellem and E. bieneusi have been reported from Ireland as contaminating coastal water receiving untreated sewage (26).
The observed wastewater concentrations of Cryptosporidium oocysts and Giardia cysts (Table 1) confirm previous reports (23, 28, 31, 32, 40). However, the results of the present study are in some contrast to existing data on pathogen removal efficiencies (10, 23, 24, 28, 31, 32, 39, 40). Horizontal SSF engineered wetlands have been shown to remove Cryptosporidium oocysts and Giardia cysts from wastewater with efficiencies of 64.2 and 87.8% (39); 100 and 100% (31), 99.9% (24), 44.3 and 98.6% (28), 1.5- to 2.5-log and 1.5- to 2.5-log removal (40), and 98.9 and 97.6% (32), respectively. However, the reported removal efficiency of Cryptosporidium oocysts and Giardia cysts was lower for an FSF wetland (i.e., 47.8%) than for an SSF wetland (i.e., 63.1%) (32), and this was confirmed by the present study (Fig. 1). Our study did not confirm better removal rates for Giardia cysts than for Cryptosporidium oocysts observed previously in constructed wetlands (39). None of the aforementioned studies reported negative values of pathogen recovery rates (Fig. 2), assessed the viability of pathogens in wetland effluents to indicate their epidemiological importance, or considered Cryptosporidium and Giardia as zoonotic pathogens that can be propagated within reed-bed areas.
There are several possibilities for why the levels of Cryptosporidium oocysts, Giardia cysts, and microsporidian spores in the FSF wetland outfalls were considerably higher than in the influents. All wetlands operated without implemented means to prevent animal access. The vegetation density in constructed wetlands has been shown not to influence the removal rates of Cryptosporidium oocysts and Giardia cysts (28). However, in FSF wetland B, robust vegetation (i.e., P. australis) and tall trees around the wetland reduced exposure to sunlight and prevented heating and full exposure to UV light. In all wetlands, precipitation potentially caused (i) inflow of runoff water to the wetland from wetland banks inhabited by rodents, which may explain the presence of C. muris, and (ii) surface runoff from other sources. Potential malfunctioning caused by a clogged inlet pipe(s) could cause temporal hydraulic short circuits that bypass part of the wetland filtration area, consequently resulting in the reduction or collapse of removal performances (Fig. 2) (31, 40). The concentration of human pathogens in wetland samples may also show diurnal fluctuation. Irrespective of the causative mechanism, we conclude that FSF small-scale constructed wetlands may not provide sufficient remediation for human enteropathogens present in wastewater subjected to secondary or tertiary treatment, although such systems are excellent in the absorption, removal, and storage of nitrogen and phosphorus from the wastewater (21, 47). We further conclude that the large volume of effluents discharged by FSF wetlands can contribute to contamination of surface waters used for recreation and drinking water abstraction and thereby represent a serious public health threat. Since the recovery efficacy of human waterborne pathogens from wastewater matrices determined previously was ca. 77% (15), the values of pathogen concentrations reported in the present study are most likely underestimated.
The minimal levels of nonviable pathogens in the present study indicate that (i) some pathogens represented species nonreactive with FISH or (ii) the pathogen walls become permeable to compounds and microorganisms present in large quantities in wastewater, and they undergo fast biodegradation. Such a phenomenon was observed previously for human-pathogenic microorganisms in wastewater matrices (16, 18). Loss of pathogen viability in constructed wetlands was attributed to the lytic action of bacteria and bacteriophages, oxidation reactions, adsorption, and exposure to plant and microbial toxins (39).
The approach taken by previous studies (10, 23, 24, 31, 32, 39) to assess removal rates of human protozoan pathogens by engineered wetland was strictly based on comparison of pathogen concentration levels in influent samples to the concentration levels in the effluent. This approach was not taking into account the contribution of protozoan pathogens to the wetland by the residing wildlife. The results of the present study indicate that the final outcome of wetland performance is more complex and represents an outcome of (i) pathogen decay in the wetland and (ii) the contribution of pathogens to the wetland by local wildlife. The genetic results of the present study (Table 2) indicate that the residing wildlife shed the species and genotypes of pathogens that were originally present in the incoming wastewater and also novel human-virulent pathogen genotypes indigenously carried out by mammals and birds inhabiting the wetlands. Since pathogens coming into the wetland with treated wastewater can be differentiated only by genetic analysis of pathogens originating from the wildlife, we conclude that the identification of pathogen genotypes and assemblages must be applied in an analysis of the performance of the wetland used in tertiary wastewater treatment.
Because Cryptosporidium, Giardia, and microsporidia can infect a variety of nonhuman hosts, identification of human-pathogenic species represents a challenge. Another challenge is the determination of viability of these pathogens since they may be nonviable and thus not of epidemiological importance. Both challenges are addressed by the FISH technique used here. FISH uses fluorescently labeled oligonucleotide probes targeted to species-specific sequences of 18S rRNA, and therefore the identification of pathogens is species specific (15). Since rRNA has a short half-life and is present only in numerous copies in viable organisms, FISH allows for differentiation between potentially viable and nonviable pathogens (8, 19, 41).
Improving reclaimed water quality by lowering fecal coliforms is not a sound solution for human protozoan and fungal parasites (12), since multiple studies have shown the inadequacy of standard fecal coliforms (i.e., E. coli, enterococci, and fecal and total coliforms) as indicators for the contamination of wastewater with pathogenic protozoa (33). This reinforces a need for better water quality indicators or, alternatively, for testing of reclaimed water for Cryptosporidium, Giardia, and human-virulent microsporidia. Source-tracking of fecal coliform indicators does not offer a satisfactory solution to the safety of reclaimed waters (12); however, Clostridium perfringens spores offer some hope as an alternative indicator for quality of reclaimed water (24, 40). The provision of safe and high microbiological quality reclaimed waters through constructed wetland systems should be an outcome of a partnership between wastewater engineering, environmental health, and epidemiological sciences and research-sponsoring institutions and be reinforced by the relevant regulatory agencies.
Acknowledgments
The study was supported by the Fulbright Senior Specialist Fellowship (grant 2225 [T.K.G.]); the Johns Hopkins Center in Urban Environmental Health (grant no. P30 ES03819); the School of Science, Institute of Technology, Sligo, Ireland; and the U.S. Environmental Protection Agency Science to Achieve Results (STAR) program (grant RD83300201).
The views expressed herein have not been subjected to U.S. EPA review and therefore do not necessarily reflect the views of the agency, and no official endorsement should be inferred.
We acknowledge Irish local authorities for providing access to wastewater treatment plants.
Footnotes
Published ahead of print on 1 May 2009.
REFERENCES
- 1.Carlow County Council. 2008. Report on cryptosporidiosis outbreak in Carlow town and environs, October 2005. http://www.carlow.ie/PublicNotices/Pages/ReportsandPublications.
- 2.Dai, X., and J. Boll. 2006. Settling velocity of Cryptosporidium parvum and Giardia lamblia. Water Res. 40:1321-1325. [DOI] [PubMed] [Google Scholar]
- 3.da Silva, A. J., D. A. Schwartz, G. S. Visvesvara, H. de Moura, S. B. Slemenda, and N. J. Pieniazek. 1996. Sensitive PCR diagnosis of infections by Enterocytozoon bieneusi (microsporidia) using primers based on the region coding for small-subunit rRNA. J. Clin. Microbiol. 34:986-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.da Silva, A. J., S. B. Slemenda, G. S. Visvesvara, D. A. Schwartz, C. M. Wilcox, S. Wallace, and N. J. Pieniazek. 1997. Detection of Septata intestinalis (microsporidia) Cali et al. 1993 using polymerase chain reaction primers targeting the small subunit rRNA coding region. Mol. Diagn. 2:47-52. [DOI] [PubMed] [Google Scholar]
- 5.da Silva, A. J., F. J. Bornay-Llinares, I. N. S. Moura, S. B. Slemenda, T. L. Tuttlem, and N. J. Pieniazek. 1999. Fast and reliable extraction of protozoan parasite DNA from fecal specimens. Mol. Diagn. 4:57-63. [DOI] [PubMed] [Google Scholar]
- 6.Davidson, L., T. Headley, and K. Pratt. 2005. Secondary treatment by reed-bed: eight year experience in northeastern New South Wales. Water Sci. Technol. 51:129-138. [PubMed] [Google Scholar]
- 7.De Groote, M. A., G. S. Visvesvara, M. L. Wilson, N. J. Pieniazek, S. B. Slemenda, A. J. daSilva, G. J. Leitch, R. T. Bryan, and R. Reeves. 1995. Polymerase chain reaction and culture confirmation of disseminated Encephalitozoon cuniculi in patient with AIDS: successful therapy with albendazole. J. Infect. Dis. 171:1375-1378. [DOI] [PubMed] [Google Scholar]
- 8.Dorsch, M. R., and D. A. Veal. 2001. Oligonucleotide probes for specific detection of Giardia lamblia cysts by fluorescent in situ hybridization. J. Appl. Microbiol. 90:836-842. [DOI] [PubMed] [Google Scholar]
- 9.Geldreich, E. E. 1996. Microbial quality of water supply in distribution systems. Lewis Publishers, New York, NY.
- 10.Gerba, C. P., J. A. Thurston, J. A. Falabi, P. M. Watt, and M. M. Karpiscak. 1999. Optimization of artificial wetland design for the removal of indicator microorganisms and pathogenic protozoa. Water Sci. Technol. 40:363-368. [Google Scholar]
- 11.Glaberman, S., J. E. Moore, C. J. Lowery, R. M. Chalmers, I. Sulaiman, K. Elwin, P. J. Rooney, B. C. Millar, J. S. Dooley, A. A. Lal, and L. Xiao. 2002. Three drinking-water-associated cryptosporidiosis outbreaks, Northern Ireland. Emerg. Infect. Dis. 8:631-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graczyk, T. K., and F. E. Lucy. 2007. Quality of reclaimed waters; a public health need for source-tracking of wastewater-derived protozoan enteropathogens in engineered wetlands. Trans. R. Soc. Trop. Med. Hyg. 101:532-533. [DOI] [PubMed] [Google Scholar]
- 13.Graczyk, T. K., R. Fayer, and M. R. Cranfield. 1997. Zoonotic potential of Cryptosporidium parvum: implications for waterborne cryptosporidiosis. Parasitol. Today 13:348-351. [DOI] [PubMed] [Google Scholar]
- 14.Graczyk, T. K., D. Sunderland, A. M. Rule, A. J. da Silva, I. N. S. Moura, L. Tamang, A. S. Girouard, K. J. Schwab, and P. N. Breysse. 2007. Urban feral pigeons (Columba livia) as a source for air- and waterborne contamination with Enterocytozoon bieneusi spores. Appl. Environ. Microbiol. 73:4357-4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graczyk, T. K., F. E. Lucy, L. Tamang, and A. Miraflor. 2007. Human enteropathogen load in activated sewage sludge and corresponding sewage sludge end products. Appl. Environ. Microbiol. 73:2013-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graczyk, T. K., M. Kacprzak, E. Neczaj, L. Tamang, H. Graczyk, F. E. Lucy, and A. S. Girouard. 2007. Human-virulent microsporidian spores in solid waste landfill leachate and sewage sludge samples and comparative analysis of effects of various sanitization treatments on their inactivation. Parasitol. Res. 101:569-575. [DOI] [PubMed] [Google Scholar]
- 17.Graczyk, T. K., A. C. Majewska, and K. J. Schwab. 2008. The role of aquatic birds in dissemination of human waterborne enteropathogens. Trends Parasitol. 24:55-59. [DOI] [PubMed] [Google Scholar]
- 18.Graczyk, T. K., M. Kacprzak, E. Neczaj, L. Tamang, H. Graczyk, F. E. Lucy, and A. S. Girouard. 2008. Occurrence of Cryptosporidium and Giardia in sewage sludge and solid waste landfill leachate and quantitative comparative analysis of sanitization treatments on pathogen inactivation. Environ. Res. 106:27-33. [DOI] [PubMed] [Google Scholar]
- 19.Hester, J. D., H. D. Linquist, A. M. Bobst, and F. W. Schaefer III. 2000. Fluorescent in situ detection of Encephalitozoon hellem spores with a 6-carboxyfluorescein-labeled ribosomal RNA-targeted oligonucleotide probe. J. Eukaryot. Microbiol. 47:299-308. [DOI] [PubMed] [Google Scholar]
- 20.Jiang, J., K. A. Alderisio, A. Singh, and L. Xiao. 2005. Development of procedures for direct extraction of Cryptosporidium DNA from water concentrates and for relief of PCR inhibitors. Appl. Environ. Microbiol. 71:1135-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kadlec, R. H. 2005. Phosphorus removal in emergent free surface wetlands. J. Environ. Sci. Health A 40:1293-1306. [DOI] [PubMed] [Google Scholar]
- 22.Kahler, A. M., and J. A. Thurston-Enriquez. 2007. Human pathogenic microsporidia detection in agricultural samples: method development and assessment. Parasitol. Res. 100:529-538. [DOI] [PubMed] [Google Scholar]
- 23.Karim, M. R., F. D. Manshadi, M. M. Karpiscak, and C. P. Gerba. 2004. The persistence and removal of enteric pathogens in constructed wetlands. Water Res. 38:1831-1837. [DOI] [PubMed] [Google Scholar]
- 24.Karpiscak, M. M., L. R. Sanchez, R. J. Freitas, and C. P. Gerba. 2001. Removal of bacterial indicators and pathogens from dairy wastewater by a multi-component treatment system. Water Sci. Technol. 44:183-190. [PubMed] [Google Scholar]
- 25.Lowery, C. J., B. C. Millar, J. E. Moore, J. Xu, L. Xiao, P. J. Rooney, L. Crothers, and J. S. Dooley. 2001. Molecular genotyping of human cryptosporidiosis in Northern Ireland: epidemiological aspects and review. Ir. J. Med. Sci. 170:246-250. [DOI] [PubMed] [Google Scholar]
- 26.Lucy, F. E., T. K. Graczyk, L. Tamang, A. Miraflor, and D. Minchin. 2008. Biomonitoring of surface and coastal water for Cryptosporidium, Giardia and human-virulent microsporidia using molluscan shellfish. Parasitol. Res. 103:1369-1375. [DOI] [PubMed] [Google Scholar]
- 27.Matchis, A., R. Weber, and P. Deplazes. 2005. Zoonotic potential of microsporidia. Clin. Microbiol. Rev. 18:423-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nokes, R. L., C. P. Gerba, and M. M. Karpiscak. 2003. Microbial water quality improvement by small scale on-site subsurface wetland treatment. J. Environ. Sci. Health A 38:1849-1855. [DOI] [PubMed] [Google Scholar]
- 29.Nwachcuku, N., and C. P. Gerba. 2004. Emerging waterborne pathogens: can we kill them all? Curr. Opin. Biotechnol. 15:175-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pelly, H., M. Cormican D. O'Donovan, R. H. Chalmers, B. Hanahoe, R. Cloughley, P. McKeowan, and G. Corbett-Feeney. 2007. A large outbreak of cryptosporidiosis in western Ireland linked to public water supply: a preliminary report. Eur. Surveill. 12:E070503.3. http://www.eurosurveillance.org/ew/2007/070503.asp#3. [DOI] [PubMed] [Google Scholar]
- 31.Quinonez-Diaz, M. J., M. M. Karpiscak, E. D. Ellman, and C. P. Gerba. 2001. Removal of pathogenic and indicator microorganisms by a constructed wetland receiving untreated domestic wastewater. J. Environ. Sci. Health A 36:1311-1320. [DOI] [PubMed] [Google Scholar]
- 32.Reinoso, R., L. A. Torres, and E. Becares. 2008. Efficiency of natural systems for removal of bacteria and pathogenic parasites from wastewater. Sci. Total Environ. 395:80-86. [DOI] [PubMed] [Google Scholar]
- 33.Rimhanen-Finne, R., A. Vourinen, S. Marmo, S. Malmberg, and M. L. Hanninen. 2004. Comparative analysis of Cryptosporidium and Giardia and indicator bacteria during sewage sludge hygienization in various composting processes. Lett. Appl. Microbiol. 38:301-305. [DOI] [PubMed] [Google Scholar]
- 34.Savioli, L., H. Smith, and A. Thompson. 2006. Giardia and Cryptosporidium join the “Neglected Disease Initiative.” Trends Parasitol. 22:203-208. [DOI] [PubMed] [Google Scholar]
- 35.Slodkowicz-Kowalska, A., T. K. Graczyk, L. Tamang, A. S. Girouard, and A. Majewska. 2007. Asymptomatic Enterocytozoon bieneusi microsporidiosis in captive mammals. Parasitol. Res. 100:505-509. [DOI] [PubMed] [Google Scholar]
- 36.Smith, J. J., T. S. Gunasekera, C. R. M. Barardi, D. Veal, and G. Vesey. 2004. Determination of Cryptosporidium parvum oocyst viability by fluorescence in situ hybridization using a rRNA-directed probe. J. Appl. Microbiol. 96:409-417. [DOI] [PubMed] [Google Scholar]
- 37.Stott, R., E. May, E. Matsushita, and A. Warren. 2001. Protozoan predation as a mechanism for the removal of Cryptosporidium oocysts from wastewaters in constructed wetlands. Water Sci. Technol. 44:191-198. [PubMed] [Google Scholar]
- 38.Sulaiman, I. M., R. Fayer, A. A. Lal, J. M. Trout, F. W. Schaefer III, and L. Xiao. 2003. Molecular characterization of microsporidia indicates that wild mammals harbor host-adapted Enterocytozoon spp. as well as human-pathogenic Enterocytozoon bieneusi. Appl. Environ. Microbiol. 69:4495-4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thurston, J. A., C. P. Gerba, K. E. Foster, and M. M. Karpiscak. 2001. Fate of indicator microorganisms, Giardia and Cryptosporidium in subsurface flow constructed wetlands. Water Res. 35:1547-1551. [DOI] [PubMed] [Google Scholar]
- 40.Ulrich, H., D. Klaus, F. Irmgard, H. Annette, L. P. Juan, and S. Regine. 2005. Microbiological investigations for sanitary assessment of wastewater treated in constructed wetlands. Water Res. 39:4849-4858. [DOI] [PubMed] [Google Scholar]
- 41.Vesey, G., N. Ashbolt, E. J. Fricker, D. Deere, K. L. William, D. A. Veal, and M. Dorsch. 1998. The use of a ribosomal RNA targeted oligonucleotide probe for fluorescent labelling of viable Cryptosporidium parvum oocysts. J. Appl. Microbiol. 85:429-440. [DOI] [PubMed] [Google Scholar]
- 42.Visvesvara, G. S., A. J. da Silva, G. P. Croppo, N. J. Pieniazek, G. J. Leitch, D. Ferguson, H. de Moura, S. Wallace, S. B. Slemenda, and I. Tyrrell. 1995. In vitro culture and serologic and molecular identification of Septata intestinalis isolated from urine of a patient with AIDS. J. Clin. Microbiol. 33:930-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weaver, R. W., M. C. Stecher, and K. J. McInnes. 2003. Water flow patterns in subsurface flow constructed wetlands designed for on-site domestic wastewater treatment. Environ. Technol. 24:77-86. [DOI] [PubMed] [Google Scholar]
- 44.Weber, R., and R. T. Bryan. 1994. Microsporidial infections in immunodeficient and immunocompetent patients. Clin. Infect. Dis. 19:517-521. [DOI] [PubMed] [Google Scholar]
- 45.Wolfe, M. S. 1992. Giardiasis. Clin. Microbiol. Rev. 5:93-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiao, L., K. A. Alderisio, and J. Jiang. 2006. Detection of Cryptosporidium oocysts in water: effect of the number of samples and analytic replicates on test results. Appl. Environ. Microbiol. 72:5942-5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, Z., Z. Rengel, and K. Meney. 2008. Interactive effects of nitrogen and phosphorus loadings on nutrient removal from simulated wastewater using Schoenoplectus validus in wetland microcosm. Chemosphere 72:1823-1982. [DOI] [PubMed] [Google Scholar]
- 48.Zintl, A., A. F. Proctor, C. Read, T. Dewaal, N. Shanaghy, S. Fanning, and G. Mulcahy. 2009. The prevalence of Cryptosporidium species and subtypes in human faecal samples in Ireland. Epidemiol. Infect. 137:270-277. [DOI] [PubMed] [Google Scholar]