Abstract
Chemotaxis is the migration of cells in gradients of chemoeffector molecules. Although multiple, competing gradients must often coexist in nature, conventional approaches for investigating bacterial chemotaxis are suboptimal for quantifying migration in response to gradients of multiple signals. In this work, we developed a microfluidic device for generating precise and stable gradients of signaling molecules. We used the device to investigate the effects of individual and combined chemoeffector gradients on Escherichia coli chemotaxis. Laminar flow-based diffusive mixing was used to generate gradients, and the chemotactic responses of cells expressing green fluorescent protein were determined using fluorescence microscopy. Quantification of the migration profiles indicated that E. coli was attracted to the quorum-sensing molecule autoinducer-2 (AI-2) but was repelled from the stationary-phase signal indole. Cells also migrated toward higher concentrations of isatin (indole-2,3-dione), an oxidized derivative of indole. Attraction to AI-2 overcame repulsion by indole in equal, competing gradients. Our data suggest that concentration-dependent interactions between attractant and repellent signals may be important determinants of bacterial colonization of the gut.
Bacteria sense chemoeffectors using cell surface receptors (13, 29). Cells constantly monitor the concentration of specific molecules, comparing the current concentration to the concentration detected a few seconds earlier. This comparison determines the net direction of movement (6, 22). Chemotaxis allows bacteria to approach sources of attractant chemicals or to avoid sources of repellent chemicals. Natural habitats for Escherichia coli, such as the gastrointestinal (GI) tract, are typically heterogeneous and contain multiple chemoeffectors with potentially opposing effects. The integrated chemotactic response in such environments is thus likely to be an important factor in bacterial colonization.
Conventional approaches for investigating bacterial chemotaxis, such as the swim plate and capillary (1) assays, are not ideal for quantifying bacterial migration. Chemotactic-ring formation in semisolid agar requires metabolizable attractants and is subject to multiple factors, and both it and the traditional capillary assay are poorly designed to investigate repellent taxis. Mao et al. (23) were the first to investigate bacterial taxis in a microfluidic flow cell. In their device, a concentration gradient is formed by the diffusive mixing of two inlet streams. However, the exposure to a fully developed gradient in this device is limited because it takes time for the gradient to develop.
Variations of this technique, such as three-channel microfluidic devices (7, 8) in which a linear gradient is generated in the absence of flow or a T-channel device that monitors chemotaxis perpendicular to the direction of fluid flow (18), were developed subsequently. The T-channel system has many of the limitations of the device developed by Mao et al. (23), and nonflow systems, like the capillary assay (1), suffer from a lack of temporal stability of the gradients.
Here, we report a flow-based microfluidic chemotaxis device that is coupled to a gradient generator. Bacteria are exposed to precise and temporally stable concentration gradients of chemoeffectors over the length of the microfluidic channel. This device was used to quantify E. coli chemotaxis in response to the canonical chemoeffectors l-aspartate and Ni2+. The device was also used to investigate chemotaxis toward cell-cell communication signals such as autoinducer-2 (AI-2), indole, and isatin that are likely to be present in the in vivo microenvironment in which E. coli is present (e.g., the human GI tract). The data obtained reinforce the idea that concentration-dependent interactions between different chemical signals could be important determinants of bacterial colonization in natural environments.
MATERIALS AND METHODS
Bacterial strains, materials, and growth media.
E. coli RP437 (23) is wild type for chemotaxis, and strain RP437 eda+ Δtar (9) is blind to aspartate and Ni2+. E. coli TG1 was obtained from Stratagene (La Jolla, CA). Plasmids pCM18 (10) and pDS-RedExpress (Clontech, CA) were used to express green fluorescent protein (GFP) and red fluorescent protein (RFP), respectively, at high, constitutive levels. Tryptone broth (TB) (10 g liter−1 tryptone and 8 g liter−1 NaCl) was used for liquid cultures. Erythromycin (200 μg ml−1) and ampicillin (100 μg ml−1) were used for plasmid selection and retention. Chemically synthesized AI-2 [(S)-4,5-dihydroxy-2,3-pentanedione] was purchased from Omm Scientific (Dallas, TX) and exists as an equilibrium mixture of different isomers; the sample contains dimethyl sulfoxide (DMSO) and methanol as solvents in about a 1:1 ratio with AI-2 (information from the manufacturer). Indole (99% pure) and isatin (indole-2,3-dione) (98% pure) were purchased from Fisher Scientific (Pittsburgh, PA). All other chemicals were purchased from Sigma Chemical.
Fabrication of the microfluidic device.
Microfluidic devices were fabricated, as previously described (16, 27, 31), in the Materials Characterization Facility at Texas A&M University. Briefly, device designs were drawn in AutoCAD and used to create a high-resolution (>3,000 dots per inch) photolithography mask produced with a laser printer (Advanced Reproductions, North Andover, MA). Standard photolithography techniques with an SU-8 2015 apparatus (Microchem Corp., MA) were used to generate imprints of the microfluidic devices on silicon wafers. The silicon wafer templates served as negative molds to generate the chemotaxis device in poly(dimethyl)siloxane (PDMS) using standard soft-lithography protocols (24). Channel dimensions were measured using a profilometer. Devices were fabricated by bonding the patterned PDMS slab to clean glass slides using oxygen-plasma bonding in a plasma etcher (150 mTorr and 50 W for 20 s) to create optically transparent devices. Access ports were punched into PDMS using a blunt 20-gauge needle.
Growth of bacteria for the chemotaxis assay.
Bacteria were prepared for chemotaxis assays as described previously by Mao et al. (23). Briefly, cultures of bacteria grown overnight were used to inoculate 10-ml samples of TB containing erythromycin to a turbidity of ∼0.05 at 600 nm. Cultures were grown to late exponential phase (turbidity of 0.45 at 600 nm). The appropriate chemoeffectors at the midpoint concentrations were added to the cells during growth in case an induction of receptors was necessary. The cells were harvested by filtering 1 to 3 ml of culture through a 0.4-μm filter, and the cells on the filter were washed twice with 15 ml of chemotaxis buffer (CB) (1× phosphate-buffered saline, 0.1 mM EDTA, 0.01 mM l-methionine, and 10 mM dl-lactate). The filter paper was placed at the bottom of a 50-ml Falcon tube, and the bacteria were resuspended by gentle shaking in 1 to 2 ml of CB containing the midpoint concentration of chemoeffector to a turbidity of ∼0.3 at 600 nm (∼1.5 × 108 cells/ml). For example, for a 0 to 100 μM gradient of l-aspartate, CB contained 50 μM aspartate. TG1 cells expressing RFP were killed by exposure to 1 mM kanamycin for 1 h (complete killing was verified based on a lack of growth on LB agar plates) and mixed with GFP-expressing RP437 cells at approximately equal densities. Assays were performed within 60 min of resuspension in CB.
Microfluidic chemotaxis.
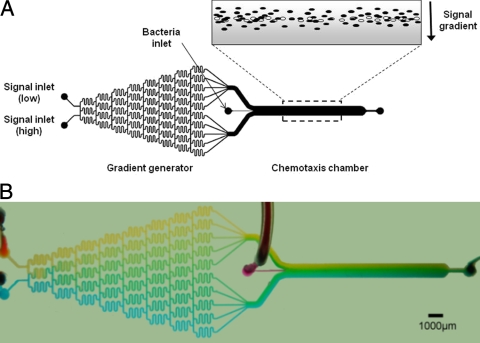
The microfluidic chemotaxis (μFlow) device (Fig. 1) consists of a gradient generator and a chemotaxis observation module. The gradient generator was inspired by the designs described previously by Jeon et al. (15) and Thompson et al. (27) and consists of a network of microfluidic channels (20 by 100 by 18,750 μm) that enables diffusive mixing from two inputs to generate a nearly linear concentration gradient (Fig. 2A). The width of each channel exiting the gradient mixer and entering the chemotaxis observation chamber is 500 μm. The observation module comprises a channel (20 by 1,050 by 11,500 μm) connected to the gradient generator module. A secondary inlet (50 μm) was used to introduce bacteria into the observation module at the midpoint of the concentration gradient. Silicone tubing was used to introduce the bacteria and the established concentration gradient into the device. All experiments were conducted at room temperature (∼ 23°C).
FIG. 1.
μFlow chemotaxis device. (A) Schematic representation of the μFlow device. The device consists of a gradient-mixing module (20 by 100 by 18,750 μm) and a chemotaxis observation module (20 by 1,050 by 11,500 μm). The inset schematically depicts a gradient of a repellent molecule (gray) and bacteria migrating in response to it. Live bacteria are depicted as solid ovals, whereas dead bacteria are shown as open ovals. (B) Food dye representation of gradient formation in the μFlow device, showing the formation of a range of greens from blue and yellow inputs. Scale bar, 1,000 μm.
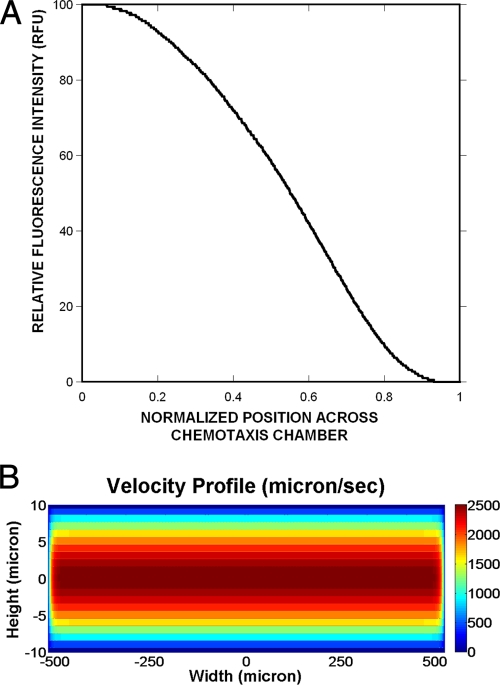
FIG. 2.
Formation of concentration gradients in the μFlow device. (A) A concentration gradient of 0 to 100 ng/ml of fluorescein isothiocyanate was established with the μFlow device. Fluorescence images were acquired after 30 min, and the fluorescence intensity was determined at 16-μm intervals, using Matlab. (B) The velocity profile in the device along the z direction was simulated using the equations described previously by Brody et al. (5).
The flow rate in the microfluidic device was controlled using a PicoPlus programmable pump (Harvard Apparatus, Holliston, MA). The assembled device was positioned on the stage of a Leica TCS SP5 resonant-scanner confocal microscope. Two 500-μl syringes containing either CB or CB with the chemoeffector molecule being tested were connected to the two inlets of the gradient generator module, with care being taken to avoid air bubbles. The bacteria, prepared as described above, were introduced into a 50-μl syringe that was connected to the bacterial inlet port. The syringes connected to the gradient generator and the bacterial inlet were operated using different pumps. The flow rate through each of the two gradient inlets was 1,000 nl/min, and the flow rate of the bacteria was 100 nl/min, such that the total flow rate through the observation module was 2,100 nl/min. For experiments in which a combined gradient of two molecules was applied, one of the inlets contained both chemicals being tested. The different concentration gradients (i.e., concentration range per 1,050 μm of chamber width) used in these studies were as follows: l-aspartate, 0 to 100 μM; NiSO4, 0 to 225 μM; AI-2, 0 to 500 μM; indole, 0 to 500 μM; and isatin, 0 to 250 μM. The velocity profile along the x, y, and z directions of the device was simulated for the channel dimensions and volumetric flow rate used in this study using equations derived previously by Brody et al. (5) with Matlab, version 7.4.0.
Green and red fluorescence images were acquired for ∼20 min after cells initially entered the observation module. For each experiment, 100 images for each fluorophore were collected approximately two-thirds down the length of the observation chamber (∼7,000 μm from the inlet) at 2.5-s intervals. The 2.5-s imaging interval was chosen based on our observation that bacteria took ∼2.5 to 3 s to traverse a 1,000-μm imaging field of view; therefore, bacteria were exposed to the gradient for ∼18 to 21 s prior to imaging. At the flow rate used, the gradient remained intact for ∼90% of the length of the chamber. The middle 90% of the width of the observation module was included in each image.
Quantification of chemotaxis using image analysis.
The migration and distribution of bacteria in each image were quantified using a Matlab (Mathworks, Natick, MA) image analysis subroutine developed in-house. The analysis consisted of the following steps: (i) removal of background pixels in the image based on pixel size and intensities; (ii) determination of the center of the image (i.e., where bacteria enter the observation chamber) using the dead cells (red fluorescence) as the reference; (iii) location of green cells (i.e., live bacteria expressing GFP) in the images relative to the center, determined by calculating the centroid; and (iv) quantification of the number of live cells in 16-μm-wide intervals (channels). There are a total of 64 channels across the width of the chemotaxis chamber. These steps were repeated for each image, and the total counts of cells in each image were summed for analysis. The quantified live- and dead-cell counts in each channel were scaled to facilitate plotting and comparison.
Chemotaxis partition and migration coefficients.
The migration profiles were used to calculate parameters for bacterial responses to chemoeffectors. The chemotaxis partition coefficient (CPC) and the chemotaxis migration coefficient (CMC) were calculated based on the locations of green bacteria in 100 images, as previously described (23). The CPC value represents the direction of migration (i.e., toward or away from a gradient) and quantifies the number of bacteria on either side of the bacterial inlet. A CPC value of 0.20 indicates that 20% more of the total bacteria (e.g., 60% moved to the right, and 40% moved to the left) moved to the higher-concentration side than the lower-concentration side for an attractant, whereas a CPC value of −0.20 indicates that 20% fewer of the total bacteria moved to the lower-concentration side than the higher-concentration side for a repellent.
The CPC value considers only the direction of bacterial migration. The CMC weights the migration of cells by the distance they move. For example, a cell that moves to the right to the farthest high-concentration position (channel 64) would be given a weighting factor of 1, whereas one that moves halfway into the higher-concentration side (channel 48) would be given a weighting factor of 0.5. Thus, CMC values will always be less than or equal to CPC values.
The center of each image (i.e., the region where bacteria move due to flow and not motility, corresponding to the place where signal noise is highest) was not included in the CPC/CMC calculations. The average location (i.e., the channel interval) of the red (dead) cells was calculated based on all of the images generated (100 images per experiment over 45 experiments). Any cell that was located within three channels of the mean location of the dead cells (∼ 48 μm on either side) was excluded from the analysis. Less than 1% of the dead bacteria were detected outside of this limit, whereas live bacteria were detected at all locations in the channel. This analysis allowed us to identify bacteria that exhibited significant movement from the center of the channel and eliminated biases due to bulk flow.
RESULTS
Chemotaxis toward a canonical attractant and away from a canonical repellent.
The formation of a concentration gradient in the device was first demonstrated using a gradient of 0 to 100 ng/ml of fluorescein. Figure 2A shows that the gradient formed in the μFlow device channel is linear for most of its 1,050-μm width. The velocity profile in the channel was simulated in order to determine the time for which bacteria were exposed to the different gradients. The velocity profile along the length and width of the channel (y and x directions, respectively) was uniform at all planes (not shown), whereas the velocity profile in the z direction varied along the height of the channel (Fig. 2B). At the midplane along the z direction, the velocity was ∼2,500 μm/s but decreased sharply beyond a distance of 5 μm on either side of the midplane. Since the bacteria were present at all z planes, our simulation results suggest that bacteria in the midplane could move at a much higher velocity than those present near the top or bottom surface of the channel. In addition, since bacterial migration is also likely to be retarded by the flagella colliding with the top or bottom surface of the channel, the average velocity of bacteria in the device is likely to be lower than the theoretical value of ∼2,500 μm/s, and the minimum residence time in the device was greater than ∼7 s. Indeed, our observations over the ∼45 experiments indicate that the average time taken for a bacterium to cover a 1,000-μm imaging field of view was ∼2.5 to 3 s. Since the imaging point was ∼7,000 μm from the inlet, the average time for which bacteria were exposed to the gradient before imaging was calculated to be ∼18 to 21 s.
The migration of E. coli RP437 was initially determined for the known attractant l-aspartate and the known repellent Ni2+(23). Gradients of 0 to 100 μM l-asparate/1,050 μm and 0 to 225 μM NiSO4/1,050 μm were established in the μFlow device. A mixture of GFP-labeled E. coli RP437 cells and RFP-labeled dead E. coli TG1 cells were introduced into the device such that the entering bacteria immediately encountered the midpoint of the established gradient (i.e., 50 μM l-aspartate and 122.5 μM NiSO4, respectively). Figures 3A and B show representative pseudocolored fluorescent micrographs of bacteria after ∼18 to 21 s of exposure to the gradient. Cells responded to l-aspartate by migrating toward the higher concentration, since more cells moved to the right. In contrast, Ni2+ repelled RP437 cells, since almost all movement was toward the left. It should be noted that 300 μM NiSO4 had little effect on the aerobic growth rate of RP437 cells in TB, nor did it have any significant effect on motility in TB (S. Meghani, unpublished data).
FIG. 3.
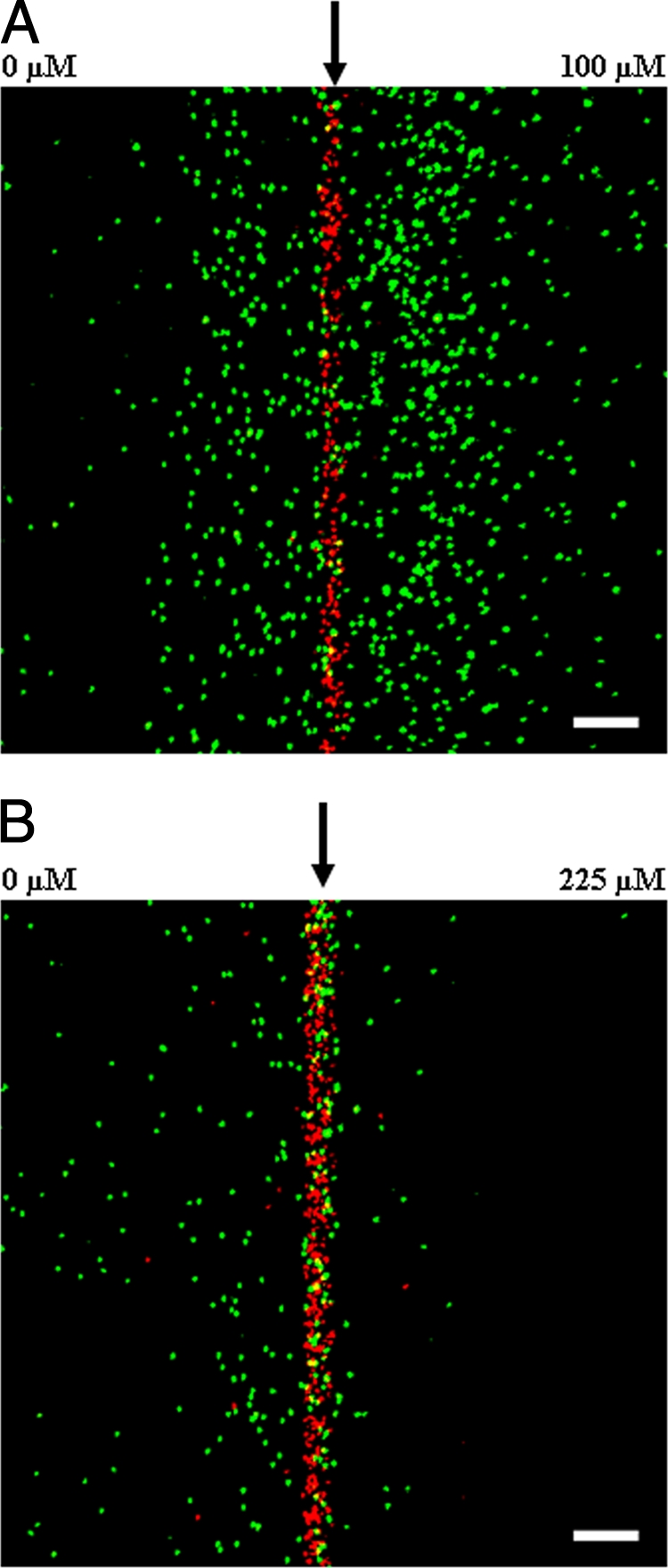
Chemotaxis in response to gradients of l-aspartate and NiSO4. RP437 cells were exposed to gradients of 0 to 100 μM l-aspartate and 0 to 225 μM NiSO4. Representative fluorescence images of movement toward l-aspartate (A) and away from Ni2+ (B) are shown. Live cells are green, and dead cells are red. An arrow indicates the position along the width of the chamber where cells enter. The low concentration is to the left, and the high concentration is to the right. Scale bar, 100 μm.
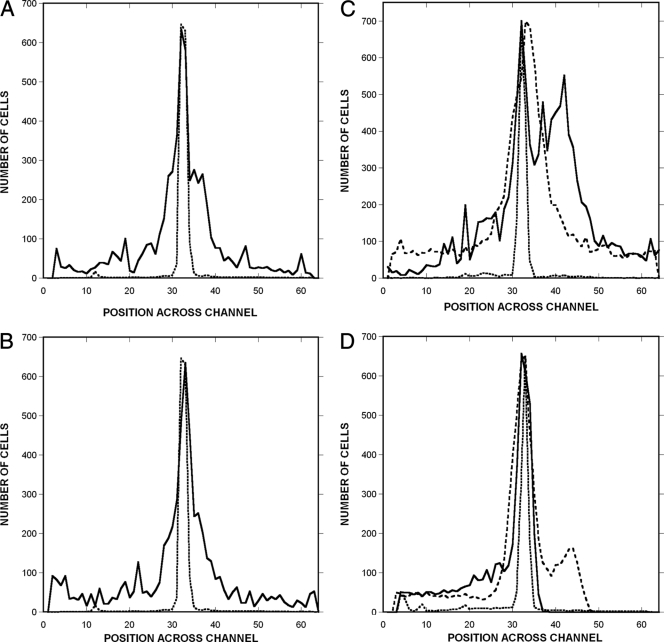
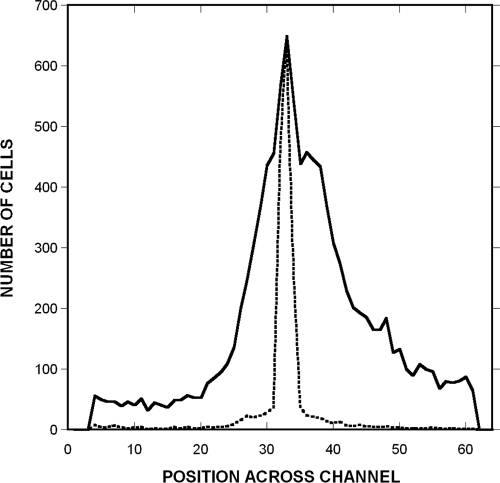
Chemotaxis was quantified by determining the spatial distribution of GFP-expressing RP437 cells and RFP-expressing dead TG1 cells. Dead bacteria were tightly distributed and were located predominantly in a ∼50-μm region at the center of the channel (Fig. 4). Thus, the effect of bulk flow on the distribution of these cells was minimal. In contrast, live bacteria were detected at all positions across the channel. In the absence of any signal (chemotaxis buffer in both signal inlets) or with a uniform signal concentration across the channel (100 μM to 100 μM l-aspartate), the mean position of the live cells was indistinguishable from that of the dead cells (Fig. 4A and B), although their distribution was much broader.
FIG. 4.
Quantification of profiles of migration to a canonical attractant and repellent. Shown is the spatial distribution of RP437 and RP437 eda+ Δtar cells in response to gradients of l-aspartate and NiSO4. (A and B) Controls with strain RP437 were run with no chemoeffector (A) or uniform 100 μM l-aspartate (B). (C and D) Responses to a 0 to 100 μM gradient of aspartate (C) and a 0 to 225 μM gradient of NiSO4 (D). The distribution of TG1 (dead) cells is shown as a dotted line, the distribution of RP437 cells is shown as a solid line, and the distribution of RP437 eda+ Δtar cells is shown as a dashed line. The number of live and dead bacteria in 16-μm-wide channels across the width of the chemotaxis chamber was calculated using image analysis. The midpoint of the chamber (where bacteria enter) corresponds to channels 32 and 33. Data shown are averages from three independent experiments.
Chemotaxis toward l-aspartate and away from Ni2+ is evident from the markedly different profiles obtained with RP437 and RP437 Δtar cells. For example, RP437 cells moved preferentially to the right (high concentration) in the aspartate gradient (Fig. 4C), whereas no such bias was seen with the Δtar mutant. Similarly, virtually no RP437 cells were detected on the right (high-concentration) side of the device in the NiSO4 gradient (Fig. 4D), whereas the Δtar cells moved to both sides of the cell inlet, although not as far to the right. The device thus yields the expected results for a canonical attractant and a canonical repellent sensed by Tar.
Table 1 shows the CPC and CMC values for RP437 cells (see Materials and Methods). For the 0 to 100 μM l-aspartate gradient, the CPC value was 0.33 ± 0.05, and the CPC value for the 0 to 225 μM NiSO4 gradient was −0.34 ± 0.04. The corresponding CMC values were 0.13 ± 0.03 and −0.14 ± 0.03. The CPC and CMC values for the Δtar strain were significantly smaller (CPC and CMC values for l-aspartate of 0.02 ± 0.02 and 0.01 ± 0.01, and CPC and CMC values for NiSO4 of −0.07 ± 0.03 and −0.05 ± 0.02), a result that confirms the specificity of the responses.
TABLE 1.
CPC and CMC values of RP437 and RP437 Δtar cells for concentration gradients of different chemoeffectorsb
| Signal(s) | Concn gradient (μM) | Mean CPC ± SD | Mean CMC ± SD |
|---|---|---|---|
| No signal (blank) | 0-0 | 0.03 ± 0.01 | −0.01 ± 0.01 |
| Null gradient (l-aspartic acid) | 100-100 | −0.01 ± 0.01 | −0.01 ± 0.01 |
| l-Aspartic acid | 0-100 | 0.33 ± 0.05 | 0.13 ± 0.03 |
| l-Aspartic acida | 0-100 | 0.02 ± 0.02 | 0.01 ± 0.01 |
| Nickel sulfate | 0-225 | −0.34 ± 0.04 | −0.14 ± 0.03 |
| Nickel sulfatea | 0-225 | −0.07 ± 0.03 | −0.05 ± 0.02 |
| AI-2 | 0-500 | 0.28 ± 0.05 | 0.11 ± 0.02 |
| DMSO solvent | 0-500 | −0.01 ± 0.01 | 0.01 ± 0.01 |
| Indole | 0-500 | −0.35 ± 0.03 | −0.15 ± 0.02 |
| Isatin | 0-250 | 0.21 ± 0.05 | 0.09 ± 0.01 |
| Indole + AI-2 | 0-500/0-500 | 0.19 ± 0.03 | 0.08 ± 0.02 |
RP437 eda+ Δtar.
Mean values and standard deviations for data from 100 images per experiment over three independent experiments are shown.
Chemotaxis toward or away from bacterial cell-cell communication signals.
Next, chemotaxis toward or away from different potential chemoeffectors generated by bacterial metabolism was investigated. Specifically, we focused on the general quorum-sensing signal AI-2 (14) and on the stationary-phase signal indole (21). The responses of RP437 cells to gradients of 0 to 500 μM AI-2/1,050 μm and 0 to 500 μM indole/1,050 μm were measured. The migration profiles in each case were significantly different from the blank and null-gradient controls (Fig. 5A and B). The DMSO solvent present in the AI-2 sample was shown to be inactive (Fig. 5A). The 0 to 500 μM gradient of AI-2 (CPC and CMC values of 0.28 ± 0.05 and 0.11 ± 0.02, respectively) was also only slightly less effective than a 0 to 100 μM gradient of l-aspartate as an attractant (Table 1). Similarly, the distribution of cells within a gradient of 0 to 500 μM indole (CPC and CMC values of −0.35 ± 0.03 and −0.15 ± 0.02, respectively) was comparable to that observed with a 0 to 225 μM gradient of NiSO4 (Fig. 5B).
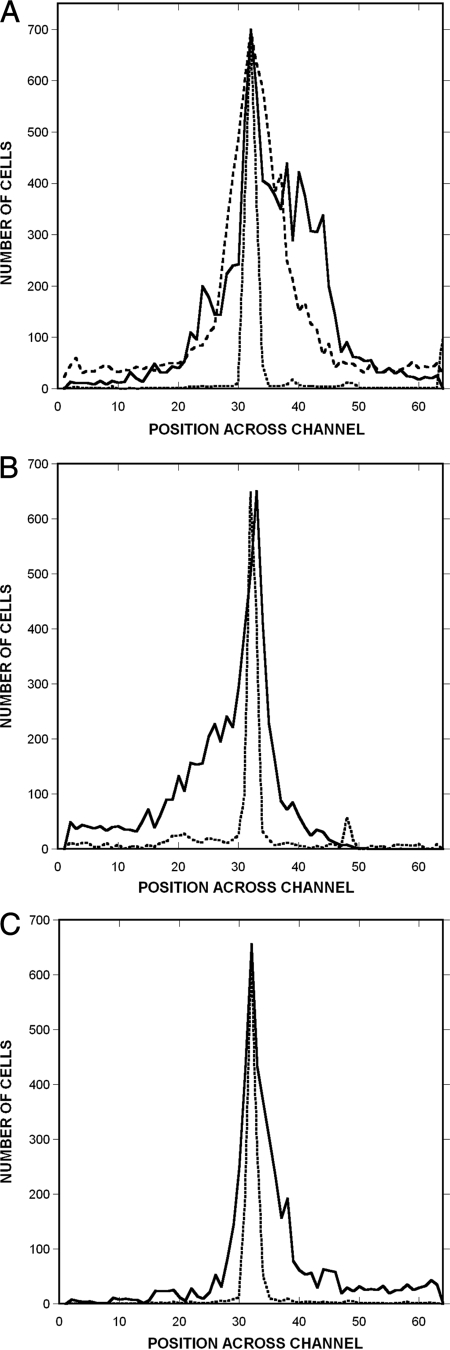
FIG. 5.
Migration profiles in response to gradients of potential chemoeffectors. (A) Spatial distribution of RP437 cells from one of three representative experiments in gradients of 0 to 500 μM AI-2 and 0 to 500 μM DMSO. The distribution of dead cells is shown with a dotted line, the distribution of living cells in the AI-2 gradient is shown with a solid line, and the distribution of living cells in the DMSO gradient is shown with a dashed line. (B and C) Distribution of cells in gradients of 0 to 500 μM indole (B) and 0 to 250 μM isatin (C). The distribution of dead cells is shown with a dotted line, and the distribution of living cells is shown with a solid line. Data were collected and analyzed as described in the legend of Fig. 4.
The migration of RP437 cells in response to a 0 to 250 μM/1,050 μm gradient of isatin (indole-2,3-dione) (20) was also investigated (Fig. 5C). Although E. coli lacks the enzymes required to convert indole to isatin, the monooxygenase activity required to generate isatin is present in other bacteria that coexist with E. coli (e.g., Pseudomonas). Therefore, isatin is likely to be present in mixed bacterial communities. The CPC and CMC values for isatin were 0.21 ± 0.05 and 0.09 ± 0.01, respectively (Table 1), indicating that E. coli senses isatin as an attractant, although these cells move away from indole, a precursor of isatin.
Chemotaxis in mixed gradients.
Bacteria are likely to encounter gradients of different chemoeffectors simultaneously in their local microenvironments; for example, both AI-2 and indole are produced by commensal E. coli. Since indole is a repellent and AI-2 is an attractant, the migration of E. coli RP437 in the presence of competing gradients of both chemicals was investigated. Cells were exposed to combined gradients of 0 to 500 μM indole and 0 to 500 μM AI-2/1,050 μm, and the resulting migration profiles were determined (Fig. 6). The CPC value for the combined gradient was 0.19 ± 0.03 (Table 1), and the corresponding CMC value was 0.08 ± 0.02. These values are about 70% of those found in the absence of a competing indole gradient, indicating that at these concentrations, attraction to AI-2 dominates over repulsion by indole.
FIG. 6.
Migration profiles in response to combined gradients of potential chemoeffectors. Shown is the spatial distribution of RP437 cells from one of three representative experiments for a combined gradient of 0 to 500 μM AI-2 and 0 to 500 μM indole. The distribution of dead cells is shown with a dotted line, and the distribution of living cells is shown with a solid line. Data were collected and analyzed as described in the legend of Fig. 4.
DISCUSSION
Bacteria are constantly exposed to competing gradients of different chemicals in their local microenvironments. However, conventional methods for investigating bacterial chemotaxis have focused primarily on responses to single chemoeffectors (1). In this study, we report the development of a flow-based microfluidic system capable of investigating chemotaxis in response to stable concentration gradients of single and multiple chemoeffectors.
Different microfluidic devices, ranging from static (no-flow) systems to simple flow devices, have been developed. Static chemotaxis systems, such as hydrogel and agarose-based devices (7, 8), are useful because they facilitate the rapid screening of different signals with minimal equipment needs. The μFlow device described here generates gradients that are stable in space and time, and it can be used to generate multiple gradients simultaneously. It also represents a physiologically relevant model for environments such as the human GI tract, since it creates an environment in which chemotaxis occurs transverse to bulk flow. It should be noted that Lanning and coworkers (18) also recently demonstrated bacterial chemotaxis transverse to fluid flow.
In our μFlow device, bacteria entering the chemotaxis chamber immediately encounter established gradients of chemoeffectors, and the cells experience a stable gradient throughout the length of the channel. This situation contrasts with those of the systems described previously by Mao et al. (23) and Lanning et al. (18), in which gradients are established in the observation chamber such that the bacteria flowing through the chamber are exposed to a changing gradient. This difference in design leads to a longer time available for cells to respond to the gradient in our device, making it more appropriate for investigating signals that elicit weaker responses.
Lamanna et al. (17) previously suggested that sudden, large increases in attractant concentrations may disrupt the localization of chemoreceptors and inhibit signal transduction. Thus, bacteria suddenly exposed to a high signal concentration, as in the device described previously by Mao et al. (23), could show an atypical, nonphysiological response. However, by establishing the final gradient at the chamber inlet, the μFlow device enables an investigation of responses in the absence of perturbing “edge” or “step” effects. Furthermore, defined gradients can be generated at any absolute concentration and steepness desired.
Since chemotaxis occurs transverse to bulk fluid flow in the μFlow device, it could be argued that hydrodynamic effects might have an impact on the chemotaxis response. We addressed this problem by differentially labeling dead bacteria so that motility could be distinguished from movement due to bulk flow. Data from nearly 50 independent experiments show that dead bacteria (labeled red) always migrate near the center of the observation chamber, whereas live bacteria (labeled green) are present in all positions across the width of the channel. Thus, the analysis presented here accurately represents chemotaxis, since the position of dead cells that move only through bulk flow is accounted for in each experiment. The increased dispersal observed with RP437 in the absence of chemoeffector or in a null gradient (i.e., a uniform concentration of chemoeffector) also indicates that the migration observed is due to the intrinsic motility of the cells (Fig. 4A and B).
Our data provide several insights into chemotaxis responses to inter- and intraspecies cell-cell communication signals. First, the CPC and CMC values observed with AI-2 and isatin as attractants and with indole as a repellent, respectively, were comparable to those seen with the canonical attractant and repellent sensed by Tar: l-aspartate and Ni2+, respectively. Indole is known to be sensed by Tsr (28), but there is very limited information about how AI-2 and isatin are perceived by E. coli. Since AI-2 can be taken into cells via the lsr genes (30, 33), it is possible that this operon is involved in the chemotactic response to AI-2. The attraction observed with isatin is intriguing, as isatin is a derivative of the repellent indole. The basis for the diametrically opposed effects of these two structurally similar compounds remains to be determined, as is the attraction observed with a combined gradient of AI-2 and indole.
The generation of countervailing gradients of different attractants or repellents can help determine the relative strengths of the responses to the different chemoeffectors. Adler and Tso (2) previously suggested that bacteria receiving opposing (i.e., attractant and repellent) signals sum up the signals to decide the direction of movement. In other words, the signal that is present at the most effective concentration will be the one that dominates the response. This suggests that the 0 to 500 μM gradient of AI-2 was more effective than the 0 to 500 μM gradient of indole. It should be noted that any synergism or additivity would presumably be seen only for suboptimal gradients, since the intrinsic properties of the biased random walk are likely to set an upper limit on the drift rate in single or combined gradients. It will be interesting to investigate whether all cells respond the same averaged way or if they fall into two populations that respond more strongly to different chemoeffectors. If two populations are observed, it will also be instructive to determine if this is true only for chemoeffectors sensed by different receptors and, if so, whether differences in the abundances of different receptor types define the two populations.
Although we used the well-characterized, nonpathogenic E. coli laboratory strain RP437 in these studies, the results here potentially describe mechanisms underlying the colonization of the human GI tract by pathogenic strains such as enterohemorrhagic E. coli (EHEC) (D. L. Englert and A. Jayaraman, unpublished data). The four classical E. coli chemotaxis receptors Tar, Tsr, Tap, and Trg as well as the aerotaxis transducer Aer (26) are also present in EHEC. Thus, chemosensing capabilities are expected to be conserved between these strains. Indeed, unpublished data from our laboratory show that E. coli RP437 and EHEC strains demonstrate similar chemotaxis toward a broad range of signals, ranging from classical chemoeffectors (l-aspartate and Ni2+) to eukaryotic hormones (norepinephrine) to bacterial molecules (AI-2, indole, and isatin).
The initial migration of pathogenic bacteria to epithelial cell surfaces is a key step in GI tract infections, and recent studies suggest that chemotaxis is important for this migration. Williams et al. (32) and McGee et al. (25) previously demonstrated, using an in vivo model, that the adherence of the ulcer-causing bacterium Helicobacter pylori is attenuated in nonmotile chemotaxis mutants, suggesting that this pathogen uses chemotaxis to guide itself to the stomach epithelium.
Our data showing that the chemotactic attraction toward AI-2 is stronger than the repulsion by indole at the concentrations tested could also be a significant factor in EHEC colonization, since both of these molecules are likely to be present in the same local microenvironment. For example, nonpathogenic E. coli strains, which constitute ∼1% of the commensal microflora in the human GI tract (12, 19), secrete ∼500 μM indole (21) and ∼100 μM AI-2 (4) per 109 cells. Prior work demonstrated that AI-2 increases EHEC motility and attachment to epithelial cells, whereas indole decreases these phenotypes (3, 4). Although many other combinations of competing concentration gradients need to be tested, it is intriguing to speculate that colonization occurs primarily in regions in which the effects of AI-2 dominate over those of indole.
In summary, we have developed a microfluidic flow-based device for investigating interactions between different bacterial chemoeffectors. We have demonstrated that the device allows E. coli chemotaxis in response to individual and combined signals to be quantified. Our data suggest that interactions between signaling pairs, such as AI-2 and indole, could be important for determining the extent of pathogen colonization in the GI tract.
Acknowledgments
This work was supported by funds from the Texas Engineering Experiment Station (to A.J.) and by the Bartoszek Fund for Basic Biological Science (to M.D.M.).
Footnotes
Published ahead of print on 1 May 2009.
REFERENCES
- 1.Adler, J. 1973. A method for measuring chemotaxis and use of the method to determine optimum conditions for chemotaxis by Escherichia coli. J. Gen. Microbiol. 74:77-91. [DOI] [PubMed] [Google Scholar]
- 2.Adler, J., and W.-W. Tso. 1974. “Decision”-making in bacteria: chemotactic response of Escherichia coli to conflicting stimuli. Science 184:1292-1294. [DOI] [PubMed] [Google Scholar]
- 3.Bansal, T., D. Englert, J. Lee, M. Hegde, T. K. Wood, and A. Jayaraman. 2007. Differential effects of epinephrine, norpeinpehrine, and indole on Escherichia coli O157:H7 chemotaxis, colonization, and gene expression. Infect. Immun. 75:4597-4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bansal, T., P. Jesudhasan, S. Pillai, T. K. Wood, and A. Jayaraman. 2008. Temporal regulation of enterohemorrhagic Escherichia coli virulence mediated by autoinducer-2. Appl. Microbiol. Biotechnol. 78:811-819. [DOI] [PubMed] [Google Scholar]
- 5.Brody, J. P., P. Yager, R. E. Goldstein, and R. H. Austin. 1996. Biotechnology at low Reynolds numbers. Biophys. J. 71:3430-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, D. A., and H. C. Berg. 1974. Temporal stimulation of chemotaxis in Escherichia coli. Proc. Natl. Acad. Sci. USA 71:1388-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng, S. Y., S. Heilman, M. Wasserman, S. Archer, M. L. Shuler, and M. Wu. 2007. A hydrogel-based microfluidic device for the studies of directed cell migration. Lab Chip 7:763-769. [DOI] [PubMed] [Google Scholar]
- 8.Diao, J., L. Young, S. Kim, E. A. Fogarty, S. M. Heilman, P. Zhou, M. L. Shuler, M. Wu, and M. P. DeLisa. 2006. A three-channel microfluidic device for generating static linear gradients and its application to the quantitative analysis of bacterial chemotaxis. Lab Chip 6:381-388. [DOI] [PubMed] [Google Scholar]
- 9.Gardina, P., C. Conway, M. Kossman, and M. D. Manson. 1992. Aspartate and maltose-binding protein interact with adjacent sites in the Tar chemotactic signal transducer of Escherichia coli. J. Bacteriol. 174:1528-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansen, M. C., R. J. Palmer, Jr., C. Udsen, D. C. White, and S. Molin. 2001. Assessment of GFP fluorescence in cells of Streptococcus gordonii under conditions of low pH and low oxygen concentration. Microbiology 147:1383-1391. [DOI] [PubMed] [Google Scholar]
- 11.Reference deleted.
- 12.Hartl, D. L., and D. E. Dykhuizen. 1984. The population genetics of Escherichia coli. Annu. Rev. Genet. 18:31-68. [DOI] [PubMed] [Google Scholar]
- 13.Hazelbauer, G. L., R. E. Mesibov, and J. Adler. 1969. Escherichia coli mutants defective in chemotaxis towards specific chemicals. Proc. Natl. Acad. Sci. USA 64:1300-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jayaraman, A., and T. K. Wood. 2008. Bacterial quorum sensing: signals, circuits, and implications for biofilms and disease. Annu. Rev. Biomed. Eng. 10:146-157. [DOI] [PubMed] [Google Scholar]
- 15.Jeon, N. L., H. Baskaran, S. K. W. Dertinger, G. M. Whitesides, L. V. D. Water, and M. Toner. 2002. Neutrophil chemotaxis in linear and complex gradients of interleukin-8 formed in a microfabricated device. Nat. Biotechnol. 20:826-830. [DOI] [PubMed] [Google Scholar]
- 16.King, K. R., S. Wang, A. Jayaraman, M. Toner, and M. L. Yarmush. 2007. A high-throughput microfluidic real-time gene expression living cell array. Lab Chip 7:77-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamanna, A. C., G. W. Ordal, and L. L. Kiessling. 2005. Large increases in attractant concentration disrupt the polar localization of bacterial chemoreceptors. Mol. Microbiol. 57:774-785. [DOI] [PubMed] [Google Scholar]
- 18.Lanning, L. M., R. M. Ford, and T. Long. 2008. Bacterial chemotaxis transverse to axial flow in a microfluidic channel. Biotechnol. Bioeng. 100:653-663. [DOI] [PubMed] [Google Scholar]
- 19.Leclerc, H., D. A. A. Mossel, S. C. Edberg, and C. B. Struijk. 2001. Advances in the bacteriology of the coliform group: their suitability as markers of microbial water safety. Annu. Rev. Microbiol. 55:201-234. [DOI] [PubMed] [Google Scholar]
- 20.Lee, J., T. Bansal, A. Jayaraman, W. E. Bentley, and T. K. Wood. 2007. Enterohemorrhagic Escherichia coli biofilms are inhibited by 7-hydroxyindole and stimulated by isatin. Appl. Environ. Microbiol. 73:4100-4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, J., A. Jayaraman, and T. K. Wood. 2007. Indole is an inter-species biofilm signal mediated by SdiA. BMC Microbiol. 7:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macnab, R. M., and D. E. Koshland. 1972. The gradient-sensing mechanism in bacterial chemotaxis. Proc. Natl. Acad. Sci. USA 69:2509-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mao, H., P. S. Cremer, and M. D. Manson. 2003. A sensitive, versatile microfluidic assay for bacterial chemotaxis. Proc. Natl. Acad. Sci. USA 100:5449-5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDonald, J. C., M. L. Chabinyc, S. J. Metallo, J. R. Anderson, A. D. Stroock, and G. M. Whitesides. 2002. Prototyping of microfluidic devices in poly(dimethylsiloxane) using solid-object printing. Anal. Chem. 74:1537-1545. [DOI] [PubMed] [Google Scholar]
- 25.McGee, D. J., M. L. Langford, E. L. Watson, J. E. Carter, Y. T. Chen, and K. M. Ottemann. 2005. Colonization and inflammation deficiencies in Mongolian gerbils infected by Helicobacter pylori chemotaxis mutants. Infect. Immun. 73:1820-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parkinson, J. S., P. Ames, and C. A. Studdert. 2005. Collaborative signaling by bacterial chemoreceptors. Curr. Opin. Microbiol. 8:116-121. [DOI] [PubMed] [Google Scholar]
- 27.Thompson, D. M., K. R. King, K. J. Wieder, M. Toner, M. L. Yarmush, and A. Jayaraman. 2004. Dynamic gene expression profiling using a microfabricated living cell array. Anal. Chem. 76:4098-4103. [DOI] [PubMed] [Google Scholar]
- 28.Tso, W.-W., and J. Adler. 1974. Negative chemotaxis in Escherichia coli. J. Bacteriol. 118:560-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wadhams, G. H., and J. P. Armitage. 2004. Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell Biol. 5:1024-1037. [DOI] [PubMed] [Google Scholar]
- 30.Wang, L., Y. Hashimoto, C. Y. Tsao, J. J. Valdes, and W. E. Bentley. 2005. Cyclic AMP (cAMP) and cAMP receptor protein influence both synthesis and uptake of extracellular autoinducer 2 in Escherichia coli. J. Bacteriol. 187:2066-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wieder, K. J., K. R. King, D. M. Thompson, C. Zia, M. L. Yarmush, and A. Jayaraman. 2005. Optimization of reporter cells for expression profiling in a microfluidic device. Biomed. Microdevices 7:213-222. [DOI] [PubMed] [Google Scholar]
- 32.Williams, S. M., Y. T. Chen, T. M. Andermann, J. E. Carter, D. J. McGee, and K. M. Ottemann. 2007. Helicobacter pylori chemotaxis modulates inflammation and bacterium-gastric epithelium interactions in infected mice. Infect. Immun. 75:3747-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xavier, K. B., and B. L. Bassler. 2005. Regulation of uptake and processing of the quorum-sensing autoinducer AI-2 in Escherichia coli. J. Bacteriol. 187:238-248. [DOI] [PMC free article] [PubMed] [Google Scholar]