Abstract
The γ-aminobutyrate (GABA) shunt, an alternative route for the conversion of α-ketoglutarate to succinate, involves the glutamate decarboxylase Gad1p, the GABA transaminase Uga1p and the succinate semialdehyde dehydrogenase Uga2p. This pathway has been extensively described in plants and animals, but its function in yeast remains unclear. We show that the flux through Gad1p is insignificant during fermentation in rich sugar-containing medium, excluding a role for this pathway in redox homeostasis under anaerobic conditions or sugar stress. However, we found that up to 4 g of exogenous GABA/liter was efficiently consumed by yeast. We studied the fate of this consumed GABA. Most was converted into succinate, with a reaction yield of 0.7 mol/mol. We also showed that a large proportion of GABA was stored within cells, indicating a possible role for this molecule in stress tolerance mechanisms or nitrogen storage. Furthermore, based on enzymatic and metabolic evidence, we identified an alternative route for GABA catabolism, involving the reduction of succinate-semialdehyde into γ-hydroxybutyric acid and the polymerization of γ-hydroxybutyric acid to form poly-(3-hydroxybutyric acid-co-4-hydroxybutyric acid). This study provides the first demonstration of a native route for the formation of this polymer in yeast. Our findings shed new light on the GABA pathway and open up new opportunities for industrial applications.
The γ-aminobutyric acid (GABA) shunt bypasses two steps of the tricarboxylic acid (TCA) cycle—the α-ketoglutarate (α-KG) dehydrogenase complex and the succinyl coenzyme A (CoA) synthase—for the conversion of α-KG into succinate (Fig. 1). It involves three enzymes: a glutamate decarboxylase (GAD; EC 4.1.1.15), which catalyzes the decarboxylation of glutamate to GABA, a GABA transaminase (EC 2.6.1.19), which converts GABA to succinate semialdehyde (SSA), and an SSA dehydrogenase (EC 1.2.1.16), which catalyzes the oxidation of SSA to succinate (36). This metabolic route, also called the GAD/GABA pathway, is conserved from bacteria, through yeasts and plants, to vertebrates. In higher eukaryotes, SSA can be reduced to γ-hydroxybutyric acid (GHB) by an alternative reaction catalyzed by a GHB dehydrogenase (4, 7, 44).
FIG. 1.
GABA uptake and catabolism as a function of yeast central carbon metabolism. The following processes are as denoted: GABA uptake (arrow 1); endogenous formation of GABA from α-KG, involving a glutamate dehydrogenase (arrow 2) and a GAD (arrow 3); conversion of GABA into succinate through a GABA transaminase (arrow 4) and an SSA dehydrogenase (arrow 5); formation of succinate by oxidative decarboxylation of α-KG via the α-KG dehydrogenase complex (arrow 6); formation of succinate through the reductive branch of the TCA cycle, involving fumarate reductase (arrow 7); SSA reduction by the GHB dehydrogenase (arrow 8); polymerization of GHB and 3-HB by P(3HB-co-4HB) synthetase (arrow 9); 3HB formation from acetyl-CoA involving an acetoacetyl-CoA thiolase (arrow 10) and a 3HB-CoA dehydrogenase (arrow 11); and vacuolar import of GABA (arrow 12). Metabolic routes and gene names previously identified in yeast are indicated by black lines. Metabolic routes identified in the present study are indicated by gray lines.
The role of the GAD/GABA pathway has been extensively investigated in plants and animals. In mammals, GABA is a key neurotransmitter in the central nervous system (32) and also serves as a hormone or trophic factor in nonneuronal peripheral and endocrine systems (19). The regulation of intracellular GABA levels is critical in cellular physiology and a deficiency in the metabolism of this signaling molecule results in severe clinical manifestations (22, 35). In plants, there is experimental evidence to suggest that GABA is synthesized as a metabolite or signal molecule, in response to a number of biotic and abiotic stresses, including pathogen infection, thermal shock, hypoxia, water stress (12, 37, 39). The GABA shunt is thus thought to contribute to a wide range of cellular functions, including the regulation of cytosolic pH, nitrogen storage and metabolism, plant defense, and osmoprotection (6, 46). It has also recently been suggested that GABA may affect gene expression in plants (28).
The yeast GAD/GABA pathway has received less attention. Efforts have focused mostly on identification of the key components of the pathway and its transcriptional regulation by nitrogen. The Saccharomyces cerevisiae enzymes GAD, GABA transaminase and SSA dehydrogenase are encoded by GAD1, UGA1, and UGA2, respectively (3, 15, 36). GABA uptake is facilitated by a specific transporter encoded by UGA4 (2) but may also be mediated by the general amino acid permease Gap1p and the proline-specific permease Put4p (20, 25). The genes involved in GABA transport and degradation are subject to nitrogen catabolite repression. In the presence of preferred nitrogen sources, their expression is repressed to low levels (16, 17, 48). In contrast, UGA1, UGA2, and UGA4 are upregulated by GABA (51). The induction of these genes requires at least two positive regulatory proteins: the specific Uga3p factor (2) and the pleiotropic Dal81p factor (8). Whereas GAP1 is not regulated by GABA (20), PUT4 expression increases in the presence of this amino acid (13).
The UGA regulon has clearly been implicated in the use of GABA as a source of nitrogen (3, 36), but little is known about the physiological function of the GAD/GABA pathway in yeast. GAD1 expression is probably controlled by the Tor signaling pathway, which mediates nutrient-sensitive regulation, particularly in response to carbon and nitrogen signals (27). Moreover, it has been suggested that Gad1p regulation is linked to calcium levels, since this protein binds calmodulin (15). A role of the GAD/GABA pathway in oxidative stress tolerance has also been proposed, as GAD1 and UGA2 are upregulated under oxidative stress conditions and S. cerevisiae cells in which the GABA shunt has been blocked appear to be more sensitive to oxidants than cells in which this pathway is functional (15). Recent analysis of the transcriptome of a wine yeast strain during wine fermentation showed that all genes of the GAD/GABA pathway are upregulated during the stationary phase, in conditions of nitrogen starvation (42). GAD1, UGA1, and UGA2 have also been reported to be activated by high levels of sugar stress, a finding consistent with the involvement of this pathway in the production of succinate from glutamate in the presence of large amounts of sugar (18). Cakir et al. (9) found that environmental disturbances (i.e., aerobic versus anaerobic or low versus high sugar content) considerably modified the levels of metabolites associated with the GAD/GABA regulon. These findings raise questions about the possible involvement of this route in yeast metabolism under anaerobiosis or in the presence of high glucose concentration.
The aim of the present study was to gain further insight into the role of the GABA shunt during glucose fermentation. In particular, we sought to assess the role of GAD1 in the formation of GABA and the relative importance of the GABA degradation route in succinate formation. Functional characterization of the genes of this pathway demonstrated that Gad1p was barely operational under the experimental conditions studied. We also showed that GABA was efficiently converted into succinate through the reactions catalyzed by Uga1p and Uga2p. However, a substantial part of this amino acid also accumulates in yeast cells, pointing out a possible role as in nitrogen storage. We also demonstrated the existence, in yeast, of an alternative route of GABA degradation involving the reduction of SSA to GHB by a GHB dehydrogenase, as previously described in higher eukaryotes. Analysis of the monomer composition of yeast polyhydroxybutyrates (PHBs) also showed that yeast cells were able to further incorporate GHB monomers into PHB.
MATERIALS AND METHODS
Strains and growth conditions.
In the present study, deletion mutants were generated in the S. cerevisiae strain V5 (MATa ura3), a haploid derivative of wine yeast. Strains were propagated on rich YPD medium (1% Bacto yeast extract, 2% Bacto peptone, 2% glucose). Initial cultures in YPD medium were grown in 50-ml flasks at 28°C, with shaking (150 rpm) for 12 h. They were used to inoculate cultures at a density of 106 cells/ml. Fermentation was carried out in synthetic MS medium, which contains 200 g of glucose/liter, 6 g of malic acid/liter, 6 g of citric acid/liter, and 460 mg of nitrogen/liter in the form of amino acids (340 mg of N/liter) and NH4Cl (120 N of mg/liter), at pH 3.5 (5). Synthetic MG medium was prepared and consisted of MS medium with the nitrogen source replaced by glutamate at a final concentration of 460 mg of N/liter. Ergosterol (7.5 mg/liter), oleic acid (2.5 mg/liter), and Tween 80 (0.21 g/liter) were provided as anaerobic growth factors. When necessary, GABA was added to the culture medium. Fermentation was carried out in 1.1-liter fermentors equipped with fermentation locks, at 28°C, with continuous magnetic stirring (500 rpm). The CO2 released was followed by automatic measurements of the weight lost by the fermentor every 20 min, and the rate of CO2 production was calculated by polynomial smoothing of the curve for CO2 release. Each fermentation was carried out in triplicate.
Construction of deletion mutants.
GAD1 and UGA2 were deleted with the short flanking homology method, using the kanMX4 deletion cassette (21). The primers used for amplification of the kanMX4 cassette from pUG6 are listed in Table 1 (21). These primers have 40-nucleotide extensions corresponding to the regions immediately upstream from the target gene start codon (forward primer) or downstream from the stop codon (reverse primer). The PCR fragments were used to transform V5 by the lithium acetate procedure (45). The deletions were verified by PCR on genomic DNA, using primers complementary to regions upstream and downstream from the GAD1 and UGA2 open reading frames.
TABLE 1.
Oligonucleotides used in this study
| Gene deletion or hybridization | Orientation | Primer or probe sequencea | Position/ATG |
|---|---|---|---|
| Gene deletions | |||
| GAD1 | F | GCT GAA CAA AAC AAG GAA TAA TGT TAC ACA GGC ACG GTT CTT CGT ACG CTG CAG GTC GAC (primer) | -19 to +21 |
| R | GGA TGA CCT TTT CAA CTC AAC ATG TTC CTC TAT AGT TGC ATA GGC CAC TAG TGG ATC TG (primer) | +1738 to +1797 | |
| UGA2 | F | CTA CTA TTT CAA CAT GAC TTT GAG TAA GTA TTC TAA ACC ATT CGT ACG CTG CAG GTC GAC C (primer) | -12 to +27 |
| R | GTA TCG TCC ACG ATT ACT ATT GCT TTA AAT GCT GTT TGG CGC ATA GGC CAC TAG TGG ATC TG (primer) | +1479 to +1518 | |
| Hybridizations | |||
| GAD1 | AGAATGCTGGTAAAGATGCT (probe) | +531 to +540 | |
| UGA2 | TACAACACCAAGAGAGAGG (probe) | +218 to +237 |
Letters in boldface type indicate the plasmid sequence; normal typeface indicate the gene sequence.
RNA extraction and Northern blot analysis.
RNA samples (15 μg/lane) isolated from 2 × 109 cells with Trizol reagent (Gibco-BRL, Life Technology) were separated by electrophoresis and blotted by capillary transfer onto Hybond-N membranes (Amersham) as previously described (38). Membranes were exposed to low-wavelength UV radiation for 1 min and prehybridized for 2 h at 65°C in 1% (wt/vol) bovine serum albumin, 1 mM EDTA, 0.5 M Na2HPO4, and 7% sodium dodecyl sulfate (pH 7.2). Hybridization was carried out in the same solution, at 50°C for 18 h, using specific oligonucleotides (Table 1) labeled with [γ-32P]ATP by T4 polynucleotide kinase (Promega) (2 × 106 cpm/ml) and purified on a silica gel membrane (QIAquick Nucleotide Removal; Qiagen) as probes. The membranes were washed twice in 2.5 mM phosphate buffer (pH 7) containing 0.9 M NaCl, 90 mM sodium citrate, and 0.25% sodium dodecyl sulfate for 5 min at room temperature and twice for 5 min at the hybridization temperature. They were then analyzed with a PhosphorImager (Molecular Dynamics). S25 rRNA gene was used as a loading control and was detected by probing the membrane with the labeled oligonucleotide 5′-CCTCCGCTTATTGATATGCTTAAG-3′.
Cell extracts and enzyme assays.
Cell extracts were obtained as previously described (38). Protein concentrations were determined with a BC assay kit (Uptima; Interchim). γ-Hydroxybutyrate dehydrogenase (GHB-DH) activity was determined as the rate of enzymatic oxidation of NADPH. Spectrophotometric assays were performed at 340 nm, immediately after the preparation of cell extracts, with a reaction mixture containing 0.1 M sodium phosphate buffer (pH 7.0), 0.05 mM NADPH, and 0.1 mM SSA in a final volume of 2 ml. The reaction was started by adding cell extract.
Extraction of intracellular GABA.
Samples (50 ml) of cultures were harvested by centrifugation at 2,000 × g and washed twice in 100 mM phosphate buffer (pH 7.5). The cells were suspended in 5% trichloroacetic acid (TCA) in 50 mM phosphate buffer and broken with 0.5-mm glass beads. Cell debris was removed by centrifugation (10,000 × g) at 4°C, and the supernatant was stored at −20°C for further high-pressure liquid chromatography (HPLC) analysis of GABA content. Intracellular GABA concentrations (in mM) were calculated from these data and by determining the number and volume of cells in culture samples with an electronic particle counter (model ZB2; Beckman-Coulter) fitted with a probe with a 100-μm pore size.
Extraction and determination of the monomer composition of PHB.
We determined the monomer composition of PHB by gas chromatography, following the depolymerization of PHB and its derivatization to yield propyl esters, as previously described (41). Cells were harvested by centrifugation, washed twice, and lyophilized. PHB from 100 mg of dried yeast was subjected to propanolysis in a tightly sealed vial (10 ml) to which 2 ml of dichloroethane and 2 ml of a solution of propanol-hydrochloric acid (4:1 [vol/vol]) had been added, which was then incubated for 2 h at 100°C. The mixture was cooled to room temperature, 4 ml of water was added, and the mixture was shaken for 20 to 30 s. The organic phase (dichloroethane-propanol) was used for the gas chromatography-mass spectrometry identification and quantification of esterified monomers. The propyl esters of 3-hydroxy acid and of 4-hydroxy acid were separated with a Varian 3800 gas chromatograph equipped with a 1-m fused silica column (0.25-mm internal diameter and 0.53-μm film thickness), coated with DB-WAX (J&W Scientific). Compounds were detected with a Varian Saturn 2000 mass spectrometer (full scan detection mode). Identification was achieved by comparing the retention times and mass spectra of commercial propyl ester of 3-hydroxy acid and of synthesized propyl ester of 4-hydroxy acid, used as standards, to those of the sample.
Analytical methods.
Growth was monitored by determining the optical density at 600 nm. Glucose and fermentation products (acetate, succinate, hydroxyglutarate, α-KG, glycerol, and ethanol) were analyzed by HPLC (HPLC 1100; Agilent Technologies) on an HPX-87H Aminex column (Bio-Rad). Dual detection was performed with a refractometer and a UV detector (Hewlett-Packard).
GABA and other amino acids were quantified with a specific amino acid analyzer (Biochrom 20; Pharmacia). Amino acids were separated by liquid chromatography on an ion-exchange column (Ultrapac-8 Lithium form; Amersham Pharmacia Biotech), with subsequent detection by the ninhydrin reaction, followed by absorbance measurement at 570 nm, except for proline, which was detected by measuring the absorbance at 440 nm. Norleucine (0.5 mM) was added to the samples and used as an internal standard.
Liquid chromatography-mass spectrometry was used to assay GHB in a Waters 2690 system equipped with a 0.25-m reversed-phase Lichrospher 100-RP18 column (2-mm internal diameter, 5-μm film thickness; Merck) (1). After 1 min of elution with 10 mM ammonium formate and 1% methanol in distilled water, separation was achieved with a linear gradient from 0 to 100% methanol in 4 min. Elution with 100% methanol was continued for 8 min. The flow rate was 0.25 ml/min. GHB was identified and quantified by mass spectrometry (negative ion mode, 50 to 300 arbitrary mass units) by using a ThermoFinnigan LCQ Advantage MS detector equipped with an electrospray ion source and an ion mass trap analyzer.
RESULTS
Expression of GAD1, UGA1, and UGA2 during fermentation.
We investigated whether the GAD/GABA pathway was operational during glucose fermentation by first analyzing the expression of GAD1, UGA1, and UGA2. Changes in mRNA levels were determined by Northern blotting during fermentation on synthetic high-sugar MS in the presence or absence of GABA. Under these conditions, nitrogen was depleted after 30 h, resulting in growth arrest (Fig. 2A). Most of the sugar was therefore consumed by resting cells, during the stationary phase.
FIG. 2.
Expression of GAD1, UGA1, and UGA2 during fermentation of synthetic MS medium by the V5 strain. (A) Growth (circles), assimilable nitrogen (triangles), and GABA (squares) consumption curves for fermentation by V5 in the absence (closed symbols) or presence of 500 mg of GABA/liter (open symbols). The arrow indicates the point at which growth is arrested. The data presented here are representative of three independent experiments. (B) Northern blot analysis of GAD1, UGA1, and UGA2 mRNAs during the course of fermentation on MS in the presence or absence of GABA.
In the absence of GABA, the expression of GAD1 and, to a lesser extent, of UGA2, was induced when the cells reached stationary phase (Fig. 2B). This is consistent with the upregulation of GAD/GABA genes in response to nitrogen exhaustion previously observed in transcriptional analyses of yeast during wine fermentation (42). When GABA (4.8 mM) was provided in the growth medium, it was consumed only during growth, with an assimilation profile similar to that of other nitrogen sources (Fig. 2A). However, this consumption was incomplete, and 1.1 mM residual GABA was present throughout the stationary phase. The addition of GABA to the growth medium did not affect GAD1 mRNA levels. In contrast, the expression of UGA1 and UGA2 was significantly induced by GABA (Fig. 2B) during the growth phase, a finding consistent with the findings of previous studies (14, 15). The decrease in UGA1 and UGA2 mRNA levels observed at the beginning of the stationary phase may account for the residual GABA present at the end of fermentation.
Analysis of GAD1 and UGA2 deletion mutants.
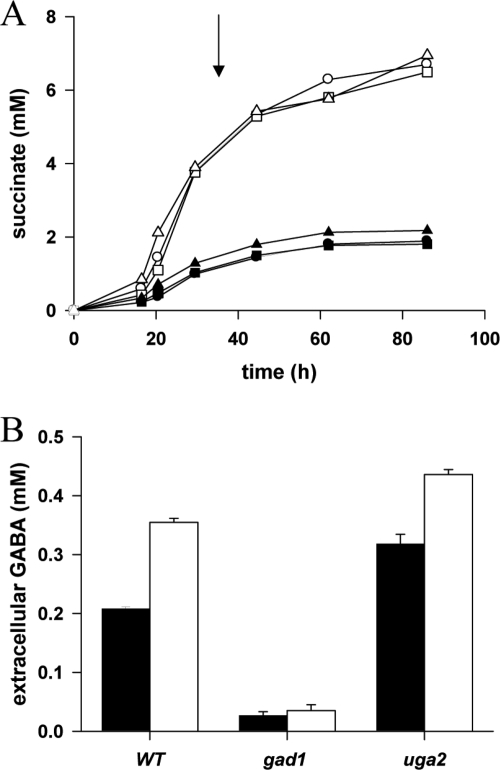
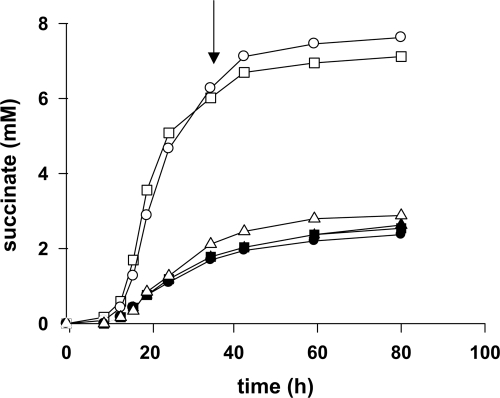
We assessed the potential role of the GAD/GABA pathway in glutamate degradation and GABA utilization, by constructing V5 deletion mutant strains lacking either GAD1 or UGA2. We studied the ability of the deletion mutants to produce GABA and succinate during batch fermentation in MS medium, which contains ammonium and amino acids as nitrogen sources, or in the same medium but with glutamate as sole nitrogen source (MG medium). We have previously demonstrated by using nuclear magnetic resonance (10) that, on MS medium, 75 and 25% of the succinate present arises from the reductive and oxidative branches of the TCA cycle, respectively (Fig. 1). In cells grown on MG medium, additional succinate is generated by oxidative decarboxylation of glutamate via glutamate dehydrogenase and the α-KG dehydrogenase complex. Consistent with this, levels of succinate production were much higher in MG medium than in MS medium (Fig. 3A). Strains lacking GAD or SSA dehydrogenase produced normal amounts of succinate on MS or MG medium, indicating that the GAD/GABA pathway plays only a minor role in the production of succinate from α-KG. We then studied the capacity of the mutants to produce GABA from α-KG (Fig. 3B). In MS medium, the wild-type strain excreted GABA, albeit at very low levels (0.22 mM). The amount of extracellular GABA produced was 63% higher in the presence of glutamate (0.35 mM), and GABA production was almost entirely abolished in the gad1 mutant, suggesting a minor contribution of GAD to fermentative metabolism. However, levels of extracellular GABA produced by the uga2 mutant were 57 and 27% higher than those produced by the control in MS and MG medium, respectively, demonstrating that GABA could be converted into succinate by SSA dehydrogenase. These data demonstrate that the oxidation of glutamate to succinate through the GABA pathway is possible but only poorly efficient under the conditions studied, consistent with the low levels of expression of GAD1, UGA1, and UGA2 during the growth phase, when succinate is produced (Fig. 2). The capacity of S. cerevisiae to synthesize succinate from GABA was confirmed by studying the ability of the various strains to assimilate exogenous GABA during fermentation in MS medium. The presence of 9.7 mM GABA significantly increased succinate production by the wild-type and gad1 strains (up to 5 mM) (Fig. 4). In contrast, the uga2 mutant produced similar amounts of succinate in the presence or absence of GABA. Thus, during wine fermentation, the major function of the GAD/GABA pathway is the assimilation of GABA and its conversion into succinate, with Gad1p playing a very limited role.
FIG. 3.
Succinate formation during fermentation on the synthetic media MS and MG. (A) Production of succinate by V5 (circles), V5 gad1 (squares), and V5 uga2 (triangles) strains during fermentation in MS (closed symbols) or MG (open symbols). The arrow indicates the time at which growth was arrested. The data presented here are representative of three experiments. (B) Formation of GABA by V5, V5 gad1, and V5 uga2 strains at the end of fermentation in MS (▪) or MG (□). The data are means ± the standard deviation of three independent experiments.
FIG. 4.
Impact of the presence of extracellular GABA on the formation of succinate during fermentation. V5 (circles), V5 gad1 (squares), and V5 uga2 (triangles) strains were grown in MS with (open symbols) or without (closed symbols) 1 g of GABA/liter; succinate production was measured during the course of fermentation. The data presented here are representative of three independent experiments.
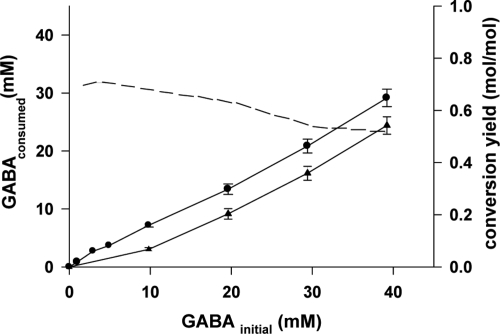
We assessed the efficiency of GABA conversion into succinate further, by culturing the wild-type and uga2 strains in MS medium with initial GABA concentrations of 0 to 40 mM and measuring the residual amount of GABA and the amount of succinate produced. The conversion yield for the generation of succinate from GABA was constant (0.7 mol/mol) for GABA concentrations up to 10 mM but decreased to ca. 0.5 mol/mol for higher concentrations of GABA (Fig. 5). In all cases, a substantial proportion of the GABA present (e.g., 5 mM for an initial GABA concentration of 20 mM) was consumed by yeast cells but not transformed into succinate. Interestingly, uga2 cells, in which the last step of the GABA pathway was inactivated, were able to consume up to 24 mM GABA, despite being unable to convert this compound into succinate (Fig. 5). Therefore, in both wild-type and uga2 strains, a substantial proportion of the GABA present can be used by yeast cells without being channeled toward succinate production.
FIG. 5.
GABA consumption and conversion to succinate. GABA consumption by the wild-type strain V5 (circles) and the uga2 mutant (triangles) was determined by measuring residual levels of GABA in the culture medium at the end of fermentation, with an initial GABA concentration of 0 to 40 mM. All values are means ± the standard deviations of three independent experiments. For V5 fermentations, the molar yield for the conversion of GABA into succinate by the strain was calculated (dashed line).
Alternative routes of GABA utilization.
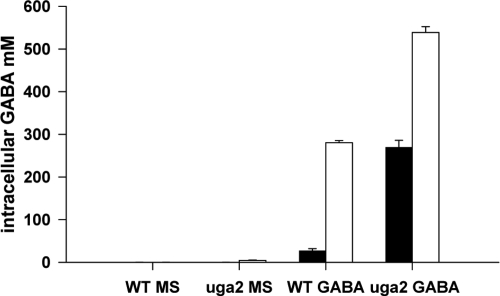
We investigated the fate of GABA further by first considering the possibility of intracellular storage. The accumulation of GABA within yeast cells, at concentrations several thousand times higher than those in the external medium, has been reported in the presence of low (0.4 mM) external GABA concentration (31). We determined intracellular GABA concentration in V5 and V5 uga2 strains, during growth on MS medium and MS medium containing 20 mM GABA (Fig. 6). During growth on MS medium, small amounts of GABA, generated by GAD, were detected in V5 and V5 uga2 strains. Larger amounts of GABA accumulated in uga2 than in V5 (4 and 1 mM, respectively), a finding consistent with the levels of GABA excreted in the medium by these two strains under these conditions (Fig. 3B). In the presence of GABA, the wild type accumulated this amino acid to a concentration of 27 mM within its cells during the growth phase. A large proportion of the GABA present was efficiently metabolized by Uga1p and Uga2p, and concentrations reached 280 mM during the stationary phase. In the uga2 mutant, which cannot convert GABA into succinate, GABA was stored more efficiently, with intracellular concentrations exceeding 270 mM throughout the course of fermentation. From these data, we estimate that, during fermentation in the presence of 20 mM extracellular GABA, a considerable proportion of this amino acid accumulates within the cells without being metabolized (20 and 64% of GABA consumed for V5 and V5 uga2 strains, respectively). Also, the uga1 deletion strain was studied and as expected, GABA consumed was mainly accumulated intracellularly (60% of GABA consumed) and not catabolized further (data not shown).
FIG. 6.
Intracellular accumulation of GABA. V5 and uga2 cells were harvested after 17 h (growth phase, □) or 44 h (stationary phase, ▪) of fermentation in MS in the presence of 20 mM GABA for determination of the GABA content.
We then considered the possibility that SSA, the metabolic intermediate in the conversion of GABA into succinate, might be stored or excreted. We tried to detect SSA in crude extracts and culture supernatants of V5 and V5 uga2 strains grown in the presence of GABA. We were unable to detect even trace amounts of this compound, ruling out the possibility of significant intra- or extracellular SSA accumulation.
In plants, mammals and bacterial cells, SSA may be oxidized into succinate or reduced into GHB by a GHB-DH (7, 44). If such an activity, which has never been described in yeast, exists in S. cerevisiae, it may be involved in the catabolism of GABA. We explored this hypothesis by trying to detect GHB-DH activity in yeast crude cell extracts. This experiment was conducted on strain V5 uga2, for two main reasons. First, V5 uga2 is able to assimilate GABA without converting it into succinate. We therefore anticipated that flux through this alternative metabolic route might be favored in this strain. Second, this mutant has no SSA dehydrogenase, which, due to its high affinity for SSA, might interfere with the enzymatic assay. In this genetic background, we detected GHB-DH activity of 5.2 ± 0.2 U/g or 0.7 ± 0.2 U/g in extracts from V5 uga2 cells grown in MS medium with or without 40 mM GABA, respectively (means ± standard deviations from atleast three independent experiments). This demonstrates that S. cerevisiae has the enzymatic equipment to convert SSA into GHB.
Moreover, the levels of GHB-DH activity were seven times higher in the presence of GABA than during fermentation on MS medium (P = 6.9 × 10−7, Student t test). We then tried to demonstrate the formation of GHB under these conditions, by trying to detect this compound in the culture supernatant by liquid chromatography-mass spectrometry. We showed that the V5 uga2 strain produced large amounts of GHB when grown in the presence of GABA (2.3 ± 0.1 mM [mean ± standard deviation from three independent experiments]), whereas no significant GHB excretion was detected (detection threshold of 0.1 mM) in MS medium (see above).
Thus, yeast displays GHB-DH activity, leading to the generation of GHB from GABA in the absence of SSA dehydrogenase activity. Conversely, the amount of GHB produced by V5 uga2 (2.3 mM) was much lower than the amount of GABA consumed (24 mM). We were also unable to detect GHB in supernatants from V5 cultures in MS medium supplemented with 40 mM GABA. These observations suggest that GHB synthesis plays only a minor role in GABA degradation. However, it is also possible that yeast cells metabolize this compound, which would then be an intermediate of a catabolic pathway. Little is known about the route of GHB catabolism. Some bacteria, such as Alcaligenes eutrophus and Ralstonia eutropha, have been reported to polymerize GHB with 3-hydroxybutyrate (3-HB) to form a heteropolymeric PHB, poly(3-hydroxybutyrate-co-4-hydroxybutyrate) [P(3HB-co-4HB)] (50). The extent of GHB incorporation into PHB depends mostly on the availability of the different precursors in cells. In S. cerevisiae, PHB has been found in membranes (40), but the monomer composition of this class of polyesters and the metabolic routes involved in their synthesis have not been elucidated in full. It has been shown that a yeast strain expressing a bacterial PHB synthase can form PHB from 3-HB, demonstrating the presence in yeast of the enzymatic equipment required to produce 3-HB from acetyl-CoA (29). We investigated whether yeast could use GHB as amonomeric precursor for P(3HB-co-4HB) and assessed the possible contribution of this pathway to GABA utilization by carrying out a qualitative analysis of the monomer composition of PHB from V5 and V5 uga2 cells grown in MS medium with or without supplementation with 40 mM GABA. An analysis of samples from the V5 culture in MS medium revealed two distinct peaks, with retention times (26.5 and 37 min, respectively) corresponding to those observed in the mass spectra of synthetic propyl 3-HB and propyl GHB (data not shown). Thus, both 3-HB and GHB are used as monomers for PHB synthesis in yeast. We detected no significant GHB excretion into the medium. These observations show that S. cerevisiae can synthesize GHB during alcoholic fermentation and are consistent with the hypothesis that this metabolic intermediate is one of the principal components incorporated into PHB. GHB was also detected in extracts from cells deleted for uga2 or growing in the presence of GABA. The extraction yield for cellular PHB and the recovery yield for propyl esters were not determined, so it was not possible to compare the intensities of GHB signals on mass spectra directly. We investigated the impact of uga2 deletion or the assimilation of GABA on the incorporation of GHB into PHB by assuming that extraction yields were similar for 3-HB and GHB and evaluating and analyzing the relative proportions of GHB to 3-HB in PHB by calculating the ratio between the areas of the GHB and 3-HB peaks from the mass spectra of each sample (values expressed as means ± standard deviations of three independent experiments).
In wild-type cells grown on MS medium, the conditions in which we expected PHB to be synthesized essentially from 3-HB, the GHB/3-HB ratio was low (∼0.4 ± 0.04). This ratio was significantly increased to 2.37 ± 0.03 by the addition of GABA to the culture medium (six times higher in the presence of GABA [P = 3.1 × 10−7, Student t test]). In addition, if the SSA dehydrogenase was inactivated, the relative proportion of GHB with respect to 3-HB (6.91 ± 0.05 or 0.71 ± 0.01 during cultures with or without 4 g/liter GABA, respectively; the culture conditions are significantly different [P = 9.4 × 10−6, Student t test]) during culture in the presence of GABA is ∼17 times higher than that in reference conditions (V5 MS medium). These data suggest that the pathway through which PHB is generated from SSA via GHB (i) is used as an alternative route for SSA catabolism and (ii) may contribute to GABA utilization, despite the low level of flux through this metabolic pathway in normal conditions.
DISCUSSION
The GABA shunt is a ubiquitous metabolic pathway, but its function widely differs between organisms. In S. cerevisiae, the coordinated response of the genes or of other metabolites of the GAD/GABA pathway to changes in oxygen availability (9, 42) or an osmotic (18) or oxidative (15) environment suggested the possible involvement of this entire pathway in the formation of succinate from α-KG. Reduced dinucleotides are synthesized during the oxidation of SSA into succinate. It has therefore been suggested that the GAD/GABA pathway may help to buffer the redox changes likely to occur in stressful conditions (9, 15, 18). Our findings highlight the important and different roles of Gad1p and of the UGA regulon in yeast physiology.
Functional analysis of the GABA shunt established the virtual absence of α-KG oxidation into succinate via the alternative GAD/GABA route in the experimental conditions studied (fermentation under oxygen limitation and in the presence of high sugar content). This is due to the very limited flux through Gad1p, which catalyzes the first step of the pathway. We therefore suggest that this enzyme is not involved in redox homeostasis during anaerobic or osmotic (sugar) stress. Consistent with this hypothesis, GAD1 inactivation had no effect on succinate formation under sugar stress conditions (B. Cambon, C. Camarasa, and S. Dequin, unpublished data). The GAD1 mRNA levels determined during fermentation—under hyperosmotic conditions and anaerobiosis—were markedly low, but this gene has previously been reported to display significant induction by oxidants (47). This previous study also reported that increasing the number of copies of the GAD1 gene increases tolerance to oxidative challenge (15). These observations suggest that Gad1p plays a physiological role in the response to oxidative stress. This enzyme may allow the formation, through the GABA shunt, of the reduced cofactors required for the regeneration of reduced glutathione and thioredoxin, key components for resistance to oxidative stress (24).
However, we found that the UGA regulon was involved in yeast fermentative metabolism in the presence of exogenous GABA, which is consistent with the considerable induction of UGA1, UGA2 (15), and UGA4 (2) expression by GABA. This pathway plays a key role in the use of GABA as a nitrogen source (36), allowing its efficient conversion into succinate. During fermentation, this organic acid is mostly synthesized by the reductive branch of the TCA cycle, which is independent of the nitrogen source. Additional succinate is also generated by the oxidative decarboxylation of α-KG, this process being enhanced by the use of glutamate as the nitrogen source (10). As a result, V5 generally produces ∼2.9 mM succinate from sugars under fermentative conditions (MS medium, 20% glucose). As we observed the synthesis of 2.6 mM succinate from 4.8 mM exogenous GABA via Uga1p and Uga2p, this metabolic route may be an important source of succinate, depending on the amount of GABA on the medium.
Nevertheless, the conversion yield for the generation of succinate from the consumed GABA did not exceed 0.7 mol/mol during fermentation. In addition, we unexpectedly found that the UGA2 mutant consumed a substantial proportion of the GABA present, despite being unable to transform this amino acid into succinate. These observations are consistent with the presence of alternative routes of GABA utilization in yeast (Table 2). A large proportion of the GABA consumed (20%) accumulates within cells and undergoes no further catabolism, despite the expression of UGA1 and UGA2, encoding the cytoplasmic enzymes (23) involved in the main pathway of GABA degradation. This finding suggests that this amino acid is stored in a cellular compartment other than the cytoplasm—probably the vacuole, as previously proposed (31). GABA is not metabolized during the stationary phase, when the other available nitrogen sources are depleted. A role for GABA accumulation as a nitrogen reserve for cells, similar to that of the intracellular stock of other basic amino acids (26), seems therefore unlikely. GABA storage may play a physiological function in stress tolerance mechanisms, similar to that of vacuolar sequestration of arginine and proline (30, 34). Finally, the import of GABA in the vacuole, mediated by the H+/GABA symporter Uga4p (49), may also help to control cytosolic pH. Consistent with this hypothesis, UGA4 is a pH-responsive gene induced in acidic conditions (33).
TABLE 2.
Distribution of fluxes through the various metabolic routes involved in 20 mM GABA utilization by the V5 and V5 uga2 strains
| Metabolic routea | Value observed for strain:
|
|
|---|---|---|
| V5 | V5 uga2 | |
| Initial GABA | 100 | 100 |
| GABA uptake* | 66.0 ± 1.4 | 45.5 ± 1.9 |
| GABA → storage* | 13.5 ± 0.7 | 29 ± 0.8 |
| GABA → excreted succinate* | 41 ± 1.5 | |
| GABA → excreted GHB* | NDc | 10.5 ± 0.5 |
| GABA → PHB*b | 11.5 ± 3.6 | 6 ± 3.2 |
*, values are expressed as a percentage of the initial GABA concentration (20 mM).
Calculated from the other experimental values.
ND, not detected.
In yeast, several enzymes are generally involved in aldehyde catabolism, to ensure that these highly toxic molecules are broken down. For example, seven alcohol dehydrogenases and five acetaldehyde dehydrogenases may catalyze the reduction and oxidation of acetaldehyde, respectively. However, to date, oxidation by Uga2p is the only catabolic pathway identified as involved in SSA detoxification through GABA transamination. The data presented here provide evidence that an alternative route exists in S. cerevisiae, involving GHB dehydrogenase activity catalyzing the reduction of SSA to GHB, which is used as a monomeric precursor for the synthesis of P(3HB-co-4HB), rather than being excreted into the culture medium. PHB has been detected in the yeast plasma membrane, in which it associates with polyphosphates to form complexes involved in transport processes (40). It has also been suggested that these compounds are synthesized principally through the homopolymerization of 3-HB-CoA, a monomeric unit produced from acetyl-CoA by an acetoacetyl-CoA thiolase and a 3-HB-CoA dehydrogenase (29). Our findings demonstrate that, in addition to making use of this metabolic pathway, S. cerevisiae can synthesize P(3HB-co-4HB) from endogenously produced GHB.
Both the induction of GHB dehydrogenase activity and the greater proportion of GHB incorporated into PHB in the presence of GABA suggest that the flux through the alternative GHB pathway contributes to GABA catabolism. Comparisons of GABA consumption with the amounts of this amino acid stored in the cell or converted into succinate during fermentation in the presence of 20 mM GABA suggested that ca. 17% of the GABA consumed may be used for PHB synthesis (Table 2). This value is consistent with the PHB content of up to 59 mg/g cell dry weight reported for yeast cells grown on YPD (43) and with the significantly larger amount of GHB found in PHB in the presence of GABA. A very different pattern of GABA utilization was observed in the UGA2 mutant, with (i) substantial GABA accumulation within cells, (ii) a much lower proportion of GABA effectively catabolized (3.3 mM versus 10.5 mM GABA metabolized by V5 under the same conditions), and (iii) greater flux through GHB dehydrogenase, the only available pathway for removing SSA, an aldehyde compound toxic for yeast (36). The main consequence of this metabolic remodeling is the excretion of some of the GHB synthesized by the mutant, probably reflecting the limited capacity of the metabolic route by which GHB is polymerized into P(3HB-co-4HB) in S. cerevisiae.
Overall, the present study provides new insight into the functioning of the GAD/GABA pathway, identifies alternative metabolic routes for GABA assimilation and highlights the relevance of these pathways for biotechnological purposes. First, when present in significant amounts in the raw materials used in bioindustries (e.g., brewing, winemaking), GABA may play an important role as a source of succinate. Efforts are currently made to favor the production of this organic acid, which contributes to acidity, in industrial bioprocesses. Another field of interest is the bioindustry based around the production of PHBs, which are attracting interest as possible biosynthetic alternatives to petroleum-based thermoplastics. These copolyesters are both biodegradable and biocompatible and P(3HB-co-4HB), in particular, has considerable potential for medical and pharmaceutical applications, due to its mechanical properties and high degree of flexibility. The main disadvantage of the bacterial synthesis method used in its production is its low yields (50). However, it has been shown that the use of S. cerevisiae as a biotechnological platform for poly-3HB production is feasible (11). In this context, the evidence for an endogenous route of P(3HB-co-4HB) formation from GABA, via GHB, opens up new prospects for improving the development of strategies (genetic engineering, design of fermentation) for P(3HB-co-4HB) production in yeast.
Acknowledgments
We thank Raymond Baumes for useful advice on the PHB analyses and helpful comments on the manuscript.
Footnotes
Published ahead of print on 1 May 2009.
REFERENCES
- 1.Allan, W. L., R. W. Smith, and B. J. Shelp. 2003. Direct measurement of gamma-hydroxybutyrate (GBH) in crude plant extracts by liquid chromatography/negative ion electrospray: mass spectrometry. Application bulletin no. AB-0015:4. Agilent Technologies, Inc., Mississauga, Ontario, Canada.
- 2.Andre, B., C. Hein, M. Grenson, and J. C. Jauniaux. 1993. Cloning and expression of the UGA4 gene coding for the inducible GABA-specific transport protein of Saccharomyces cerevisiae. Mol. Gen. Genet. 237:17-25. [DOI] [PubMed] [Google Scholar]
- 3.Andre, B., and J. C. Jauniaux. 1990. Nucleotide sequence of the yeast UGA1 gene encoding GABA transaminase. Nucleic Acids Res. 18:3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andriamampandry, C., J. C. Siffert, M. Schmitt, J. M. Garnier, A. Staub, C. Muller, S. Gobaille, J. Mark, and M. Maitre. 1998. Cloning of a rat brain succinic semialdehyde reductase involved in the synthesis of the neuromodulator gamma-hydroxybutyrate. Biochem. J. 334:43-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bely, M., J. M. Sablayrolles, and P. Barre. 1990. Description of alcoholic fermentation kinetics: its variability and significance. Am. J. Enol. Vitic. 41:319-324. [Google Scholar]
- 6.Bouche, N., and H. Fromm. 2004. GABA in plants: just a metabolite? Trends Plant Sci. 9:110-115. [DOI] [PubMed] [Google Scholar]
- 7.Breitkreuz, K. E., W. L. Allan, O. R. Van Cauwenberghe, C. Jakobs, D. Talibi, B. Andre, and B. J. Shelp. 2003. A novel gamma-hydroxybutyrate dehydrogenase: identification and expression of an Arabidopsis cDNA and potential role under oxygen deficiency. J. Biol. Chem. 278:41552-41556. [DOI] [PubMed] [Google Scholar]
- 8.Bricmont, P. A., J. R. Daugherty, and T. G. Cooper. 1991. The DAL81 gene product is required for induced expression of two differently regulated nitrogen catabolic genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 11:1161-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cakir, T., K. R. Patil, Z. Onsan, K. O. Ulgen, B. Kirdar, and J. Nielsen. 2006. Integration of metabolome data with metabolic networks reveals reporter reactions. Mol. Syst. Biol. 2:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Camarasa, C., J. P. Grivet, and S. Dequin. 2003. Investigation by 13C-NMR and tricarboxylic acid (TCA) deletion mutant analysis of pathways for succinate formation in Saccharomyces cerevisiae during anaerobic fermentation. Microbiology 149:2669-2678. [DOI] [PubMed] [Google Scholar]
- 11.Carlson, R., and F. Srienc. 2006. Effects of recombinant precursor pathway variations on poly[(R)-3-hydroxybutyrate] synthesis in Saccharomyces cerevisiae. J. Biotechnol. 124:561-573. [DOI] [PubMed] [Google Scholar]
- 12.Cholewa, E., and C. A. Peterson. 2004. Evidence for symplastic involvement in the radial movement of calcium in onion roots. Plant Physiol. 134:1793-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coffman, J. A., H. M. el Berry, and T. G. Cooper. 1994. The URE2 protein regulates nitrogen catabolic gene expression through the GATAA-containing UASNTR element in Saccharomyces cerevisiae. J. Bacteriol. 176:7476-7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coffman, J. A., R. Rai, and T. G. Cooper. 1995. Genetic evidence for Gln3p-independent, nitrogen catabolite repression-sensitive gene expression in Saccharomyces cerevisiae. J. Bacteriol. 177:6910-6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coleman, S. T., T. K. Fang, S. A. Rovinsky, F. J. Turano, and W. S. Moye-Rowley. 2001. Expression of a glutamate decarboxylase homologue is required for normal oxidative stress tolerance in Saccharomyces cerevisiae. J. Biol. Chem. 276:244-250. [DOI] [PubMed] [Google Scholar]
- 16.Cunningham, T. S., and T. G. Cooper. 1991. Expression of the DAL80 gene, whose product is homologous to the GATA factors and is a negative regulator of multiple nitrogen catabolic genes in Saccharomyces cerevisiae, is sensitive to nitrogen catabolite repression. Mol. Cell. Biol. 11:6205-6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daugherty, J. R., R. Rai, H. M. el Berry, and T. G. Cooper. 1993. Regulatory circuit for responses of nitrogen catabolic gene expression to the GLN3 and DAL80 proteins and nitrogen catabolite repression in Saccharomyces cerevisiae. J. Bacteriol. 175:64-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erasmus, D. J., G. K. van der Merwe, and H. J. van Vuuren. 2003. Genome-wide expression analyses: metabolic adaptation of Saccharomyces cerevisiae to high sugar stress. FEMS Yeast Res. 3:375-399. [DOI] [PubMed] [Google Scholar]
- 19.Gladkevich, A., J. Korf, V. P. Hakobyan, and K. V. Melkonyan. 2005. The peripheral GABAergic system as a target in endocrine disorders. Auton. Neurosci. 124:1-8. [DOI] [PubMed] [Google Scholar]
- 20.Grenson, M., C. Hou, and M. Crabeel. 1970. Multiplicity of the amino acid permeases in Saccharomyces cerevisiae. IV. Evidence for a general amino acid permease. J. Bacteriol. 103:770-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guldener, U., S. Heck, T. Fielder, J. Beinhauer, and J. H. Hegemann. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24:2519-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hogema, B. M., M. Gupta, H. Senephansiri, T. G. Burlingame, M. Taylor, C. Jakobs, R. B. Schutgens, W. Froestl, O. C. Snead, R. Diaz-Arrastia, T. Bottiglieri, M. Grompe, and K. M. Gibson. 2001. Pharmacologic rescue of lethal seizures in mice deficient in succinate semialdehyde dehydrogenase. Nat. Genet. 29:212-216. [DOI] [PubMed] [Google Scholar]
- 23.Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson, J. S. Weissman, and E. K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature 425:686-691. [DOI] [PubMed] [Google Scholar]
- 24.Jamieson, D. J. 1998. Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast 14:1511-1527. [DOI] [PubMed] [Google Scholar]
- 25.Jauniaux, J. C., M. Vandenbol, S. Vissers, K. Broman, and M. Grenson. 1987. Nitrogen catabolite regulation of proline permease in Saccharomyces cerevisiae: cloning of the PUT4 gene and study of PUT4 RNA levels in wild-type and mutant strains. Eur. J. Biochem. 164:601-606. [DOI] [PubMed] [Google Scholar]
- 26.Klionsky, D. J., P. K. Herman, and S. D. Emr. 1990. The fungal vacuole: composition, function, and biogenesis. Microbiol. Rev. 54:266-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuruvilla, F. G., A. F. Shamji, and S. L. Schreiber. 2001. Carbon- and nitrogen-quality signaling to translation are mediated by distinct GATA-type transcription factors. Proc. Natl. Acad. Sci. USA 98:7283-7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lancien, M., and M. R. Roberts. 2006. Regulation of Arabidopsis thaliana 14-3-3 gene expression by gamma-aminobutyric acid. Plant Cell Environ. 29:1430-1436. [DOI] [PubMed] [Google Scholar]
- 29.Leaf, T. A., M. S. Peterson, S. K. Stoup, D. Somers, and F. Srienc. 1996. Saccharomyces cerevisiae expressing bacterial polyhydroxybutyrate synthase produces poly-3-hydroxybutyrate. Microbiology 142:1169-1180. [DOI] [PubMed] [Google Scholar]
- 30.Matsuura, K., and H. Takagi. 2005. Vacuolar functions are involved in stress-protective effect of intracellular proline in Saccharomyces cerevisiae. J. Biosci. Bioeng. 100:538-544. [DOI] [PubMed] [Google Scholar]
- 31.McKelvey, J., R. Rai, and T. G. Cooper. 1990. GABA transport in Saccharomyces cerevisiae. Yeast 6:263-270. [DOI] [PubMed] [Google Scholar]
- 32.Mody, I., Y. De Koninck, T. S. Otis, and I. Soltesz. 1994. Bridging the cleft at GABA synapses in the brain. Trends Neurosci. 17:517-525. [DOI] [PubMed] [Google Scholar]
- 33.Moretti, M. B., A. Batlle, and S. C. Garcia. 2001. UGA4 gene encoding the gamma-aminobutyric acid permease in Saccharomyces cerevisiae is an acid-expressed gene. Int. J. Biochem. Cell Biol. 33:1202-1207. [DOI] [PubMed] [Google Scholar]
- 34.Morita, Y., S. Nakamori, and H. Takagi. 2002. Effect of proline and arginine metabolism on freezing stress of Saccharomyces cerevisiae. J. Biosci. Bioeng. 94:390-394. [DOI] [PubMed] [Google Scholar]
- 35.Nisbet, A. P., D. J. Eve, A. E. Kingsbury, S. E. Daniel, C. D. Marsden, A. J. Lees, and O. J. Foster. 1996. Glutamate decarboxylase-67 messenger RNA expression in normal human basal ganglia and in Parkinson's disease. Neuroscience 75:389-406. [DOI] [PubMed] [Google Scholar]
- 36.Ramos, F., M. el Guezzar, M. Grenson, and J. M. Wiame. 1985. Mutations affecting the enzymes involved in the utilization of 4-aminobutyric acid as nitrogen source by the yeast Saccharomyces cerevisiae. Eur. J. Biochem. 149:401-404. [DOI] [PubMed] [Google Scholar]
- 37.Ramputh, A. I., and A. W. Bown. 1996. Rapid γ-aminobutyric acid synthesis and the inhibition of the growth and development of oblique-banded leaf-roller larvae. Plant Physiol. 111:1349-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Remize, F., L. Barnavon, and S. Dequin. 2001. Glycerol export and glycerol-3-phosphate dehydrogenase, but not glycerol phosphatase, are rate limiting for glycerol production in Saccharomyces cerevisiae. Metab. Eng. 3:301-312. [DOI] [PubMed] [Google Scholar]
- 39.Rentsch, D., B. Hirner, E. Schmelzer, and W. B. Frommer. 1996. Salt stress-induced proline transporters and salt stress-repressed broad specificity amino acid permeases identified by suppression of a yeast amino acid permease-targeting mutant. Plant Cell 8:1437-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reusch, R. N. 1989. Poly-β-hydroxybutyrate/calcium polyphosphate complexes in eukaryotic membranes. Proc. Soc. Exp. Biol. Med. 191:377-381. [DOI] [PubMed] [Google Scholar]
- 41.Riis, V., and W. Mai. 1988. Gas chromatographic determination of poly-[β]-hydroxybutyric acid in microbial biomass after hydrochloric acid propanolysis. J. Chromatogr. A 445:285-289. [Google Scholar]
- 42.Rossignol, T., L. Dulau, A. Julien, and B. Blondin. 2003. Genome-wide monitoring of wine yeast gene expression during alcoholic fermentation. Yeast 20:1369-1385. [DOI] [PubMed] [Google Scholar]
- 43.Şafak, S., M. Mercan, B. Aslim, and Y. Beyatli. 2002. A study on the production of poly-β-hydroxybutyrate by some eukaryotic microorganisms. Turk. Electr. J. Biotechnol. 1:11-17. [Google Scholar]
- 44.Schaller, M., M. Schaffhauser, N. Sans, and B. Wermuth. 1999. Cloning and expression of succinic semialdehyde reductase from human brain: identity with aflatoxin B1 aldehyde reductase. Eur. J. Biochem. 265:1056-1060. [DOI] [PubMed] [Google Scholar]
- 45.Schiestl, R. H., and R. D. Gietz. 1989. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 16:339-346. [DOI] [PubMed] [Google Scholar]
- 46.Shelp, B. J., A. W. Bown, and M. D. McLean. 1999. Metabolism and functions of gamma-aminobutyric acid. Trends Plant Sci. 4:446-452. [DOI] [PubMed] [Google Scholar]
- 47.Sulahian, R., D. Sikder, S. Johnston, T. Kodadek, S. Coleman, T. Fang, S. Rovinsky, F. J. Turano, and W. S. Moye-Rowley. 2006. The proteasomal ATPase complex is required for stress-induced transcription in yeast. Nucleic Acids Res. 34:1351-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Talibi, D., M. Grenson, and B. Andre. 1995. Cis- and trans-acting elements determining induction of the genes of the gamma-aminobutyrate (GABA) utilization pathway in Saccharomyces cerevisiae. Nucleic Acids Res. 23:550-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uemura, T., Y. Tomonari, K. Kashiwagi, and K. Igarashi. 2004. Uptake of GABA and putrescine by UGA4 on the vacuolar membrane in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 315:1082-1087. [DOI] [PubMed] [Google Scholar]
- 50.Valentin, H. E., and D. Dennis. 1997. Production of poly(3-hydroxybutyrate-co-4-hydroxybutyrate) in recombinant Escherichia coli grown on glucose. J. Biotechnol. 58:33-38. [DOI] [PubMed] [Google Scholar]
- 51.Vissers, S., B. Andre, F. Muyldermans, and M. Grenson. 1989. Positive and negative regulatory elements control the expression of the UGA4 gene coding for the inducible 4-aminobutyric-acid-specific permease in Saccharomyces cerevisiae. Eur. J. Biochem. 181:357-361. [DOI] [PubMed] [Google Scholar]