Abstract
Botulinum neurotoxin type A (BoNT/A), the most poisonous substance known to humans, is a potential bioterrorism agent. The light-chain protein induces a flaccid paralysis through cleavage of the 25-kDa synaptosome-associated protein (SNAP-25), involved in acetylcholine release at the neuromuscular junction. BoNT/A is widely used as a therapeutic agent and to reduce wrinkles. The toxin is used at very low doses, which have to be accurately quantified. With this aim, internally quenched fluorescent substrates containing the fluorophore/repressor pair pyrenylalanine (Pya)/4-nitrophenylalanine (Nop) were developed. Nop and Pya were, respectively, introduced at positions 197 and 200 of the cleavable fragment (amino acids 187 to 203) of SNAP-25 (with norleucine at position 202 [Nle202]), which is acetylated at its N terminus and amidated at its C terminus. Cleavage of this peptide occurred between positions 197 and 198, as in SNAP-25, and was easily quantified by the strong fluorescence emission of the metabolite. To increase the assay sensitivity, the peptide sequence of the previous substrate was lengthened to account for exosite binding to BoNT/A. We synthesized the peptide PL50 (SNAP-25-NH2 acetylated at positions 156 to 203 [Nop197, Pya200, Nle202]) and its analogue PL51, in which all methionines were replaced by nonoxidizable Nle. Consistent with a large increase in affinity for BoNT/A, PL50 and PL51 exhibit catalytic efficiencies of 2.6 × 106 M−1 s−1 and 8.85 × 106 M−1 s−1, respectively, and behave as the best fluorigenic substrates of BoNT/A reported to date. Under optimized assay conditions, they allow simple quantification of as little as 100 and 60 pg of BoNT/A, respectively, within 2 h with a classical fluorimeter. Calibration of the method against the mouse 50% lethal dose assay unequivocally validates the enzymatic assay.
The botulinum neurotoxin (BoNT) family consists of seven antigenically distinct serotypes, BoNT/A to BoNT/G, which act on the peripheral nervous system (19). Of these toxins, serotypes A, B, E, and F cause botulism in humans, a disease characterized by flaccid muscular paralysis. The neurotoxins are produced as single inactive polypeptides of 150 kDa, which are subsequently processed by proteolytic cleavage into biologically active di-chains (19). These forms consist of an approximately 50-kDa light chain (LC) linked by a disulfide bridge to a 100-kDa heavy chain (HC) that contains two domains, designated the binding and translocation domains. The neurotoxins reach their intracellular targets by translocating the LC into the cytosol after endocytosis via interaction of the HC with a high-affinity membrane-bound receptor complex (9, 20). The LC, which possesses a highly specific zinc-endopeptidase activity (29), then blocks the fusion of synaptic vesicles with the presynaptic membrane by selectively cleaving one of the three polypeptides involved in neuroexocytosis. BoNT/A, for instance, cleaves the 206-amino-acid, 25-kDa synaptosome-associated protein (SNAP-25) exclusively between the Q197 and R198 residues, thus inhibiting neurotransmitter release at the neuromuscular junction (37, 38).
BoNT/A is recognized as the most toxic serotype; its oral 50% lethal dose (LD50) for humans is estimated at 1 μg/kg of body weight (2). Because of this extreme toxicity and prolonged effect, BoNTs are classified by the Centers for Disease Control and Prevention (CDC) as one of the six highest-risk threat agents for bioterrorism in “category A” (27). In spite of this, BoNT/A and -B are widely used as therapeutic agents for the treatment of muscular and nerve disorders, as well as in the treatment of neurological diseases (14, 15, 28). There is also an increasing use of BoNT/A in esthetics for wrinkle reduction (4). Because of their high toxicity, BoNTs are used at very low concentrations, and procedures to be used for their detection and quantification in toxin preparations for medical applications or in the event of malevolent bioterrorist acts have to be highly sensitive, rapid, and easy to use; the use of all lengthy in vivo assays is excluded (2, 11). The advantage of the currently used pharmacotoxicological mouse LD50 (MLD50) assay, considered the gold standard assay, is that it provides the in vivo toxicity of a given botulinum toxin sample, whatever the nature of the infected medium. However, this assay is time-consuming, requires the use of a large number of animals, and has poor repeatability due to many fluctuant parameters involved in this method (22). Several in vitro assays have been reported for the detection of BoNT/A, relying either on mass spectrometry (3, 16), immunological detection (10, 25), or BoNT/A's endopeptidase activity (12, 30). The advantage of the endopeptidase assay is that it measures and quantifies the “active” part of the toxin, which is directly responsible for neurotransmission inhibition. Various methods have been developed to quantify the BoNT/A proteolytic activity (12, 23, 32-33). Although some of these assays are very sensitive (11), they cannot be used for the field detection of BoNT/A, as they require a multistep procedure, and they are also not easily amenable to quantification of toxin preparations used for medical applications.
In this paper, we have designed novel, specific, high-affinity, mimetic peptide substrates for BoNT/A using the internal-collision-induced fluorescence-quenching technique (13). This technique, the use of which has previously been successful in the design of peptide substrates for other Zn-metallopeptidases, e.g., ECE-1 (18) and BoNT/B (1, 26), involves the introduction of a fluorophore/repressor pair, here the highly fluorescent pyrenylalanine (Pya) along with a nitro-phenylalanine (Nop) repressor residue on each side of the cleavage site. Once the better positions of the fluorophore/repressor pair Pya/Nop were determined using a fragment of the SNAP-25 sequence from amino acids 187 to 203 [(187-203) SNAP-25] (30), the kinetic parameters of the peptide substrate were optimized and the stability of the final substrate, acetylated SNAP-25 from positions 156 to 203 [(Ac-156-203) SNAP-25] (Nop197, Pya200, Nle202), also called PL50, was finally improved in PL51 by replacing the oxidizable methionine residues within the sequence with norleucines. Thus, the specificity constants (catalytic constant [kcat]/Michaelis constant [Km]) of PL50 and of its analogue PL51 were 2.6 × 106 M−1 s−1 and 8.85 × 106 M−1 s−1, respectively. The use of these novel high-affinity substrates provides a simple, one-step, specific, robust, and rapid enzymatic assay, thus fulfilling all the requirements for BoNT/A field detection and for BoNT/A's quantification in preparations for medical applications.
MATERIALS AND METHODS
Peptide synthesis.
9-Fluorenylmethoxy carbonyl (Fmoc) amino acids and coupling reagents were purchased from Applied Biosystems (Courtaboeuf, France). Fmoc-l-(para nitro)-phenylalanine was from Bachem Distribution (Germany), and Fmoc-l-pyrenylalanine was from Polypeptides (Strasbourg, France). HMP (4-hydroxymethylphenoxyacetyl-4′-methylbenzyhydrylamine) and MBHA (methylbenzhydrylamine) resins were from Novabiochem (VWR, Fontenay sous Bois, France), and all the solvents were from Carlo Erba-SDS (Vitry, France).
Peptides were synthesized by the solid-phase method, with an ABI 433A Applied Biosystems automated synthesizer, coupled to a model 785A programmable absorbance UV detector. The synthesis was carried out using the classical Fmoc protocol with HBTU [O-(benzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate], HOBt (1-hydroxybenzotriazole), and diisopropylethylamine as coupling reagents. By this strategy, the amino acid side chains were protected by a t-butyl group, except Asn, Cys, Gln, and His, which were protected by a trityl group, Trp, which was protected by a t-butoxy group, and Arg, which was protected by a Pmc (2,2,5,7,8-pentamethylchroman-6-sulfonyl) group.
The synthesis of the various SNAP-25 fragments was performed at a 100 μM scale on an MBHA resin for the peptides bearing a carboxamide at their C termini and on an HMP resin for those having a free carboxylate group, using a precharged resin for the first amino acid.
All peptides, except those corresponding to the C-terminal metabolites, were acetylated at the end of the synthesis at their N-terminal amino group by action of acetic anhydride in N-methylpyrrolidone for 30 min. The peptidyl resin was treated with a solution of trifluoroacetic acid (TFA), phenol, triisopropylsilane (TIPS), and H2O (90:5:2.5:2.5) for the cleavage of both the resin and side chain protections. After filtration of the resin, the solution was evaporated under vacuum and the residue precipitated in cold diethyl ether. Peptides were purified by semipreparative reversed-phase chromatography (Waters) on a Kromasil C18 column (10 by 250 mm, 5-mm inside diameter,100 Å) with different gradients of CH3CN-H2O (0.05% TFA) at a flow rate of 10 ml/min with UV-spectrophotometric monitoring at wavelengths of 210 and 343 nm. The fractions were analyzed by reversed-phase high-performance liquid chromatography (HPLC) (Shimadzu) on a Kromasil C18 column (4.6 by 250 mm, 5-μm inside diameter, 100 Å) at a flow rate of 1 ml/min with monitoring at both 210 and 343 nm. Unless indicated, HPLC analysis were performed using a Kromasil C18 column with a 10% CH3CN-90% H2O (0.1% TFA) to 90% CH3CN-10% H2O (0.1% TFA) elution gradient in 30 min. The purity of the peptides determined by HPLC was found to be greater than 98.5%, and their molecular weights (MWs) were confirmed by deconvolution of the mass spectra obtained with an electrospray mass spectrometer (Thermo Finnigan) using Xcalibur software.
The sequence of peptide 3 is Ac-SNKTRIDEAN-Pya-Nop-ATK-Nle-L-NH2 (MW, 2,179.11). Positive mass electrospray ionization [mass ESI(+)] results were as follows: (M + 2H)+/2 = 1,090.6 and (M + 3H)/3 = 727.5 (retention time [Rt] = 15.4 min).
The sequence of peptide 4 is Ac-SNKTRIDEAN-Pya-Nop-ATK-Nle-L-NH2 (MW, 2,179.11). Mass ESI(+) results were as follows: (M + 2H)+/2 = 1,090.6 and (M + 3H)+/3 = 727.5 (Rt = 15.6 min).
The sequence of peptide 5 is Ac-SNKTRIDEAN-Pya-R-Nop-TK-Nle-L-NH2 (MW, 2,264.15). Mass ESI(+) results were as follows: (M + 2H)+/2 = 1,133.1 and (M + 3H)+/3 = 755.7 (Rt = 13.6 min).
The sequence of peptide 6 is Ac-SNKTRIDEAN-Nop-R-Pya-TK-Nle-L-NH2 (MW, 2,264.15). Mass ESI(+) results were as follows: (M + 2H)+/2 = 1,133.5 and (M + 3H)+/3 = 755.8 (Rt = 13.8 min).
The sequence of peptide 7 is Ac-SNKTRIDEAN-Pya-RA-Nop-K-Nle-L-NH2 (MW, 2,234.16). Mass ESI(+) results were as follows: (M + 2H)+/2 = 1,118.6 and (M + 3H)+/3 = 746.1 (Rt = 14.7 min).
The sequence of peptide 8 is Ac-SNKTRIDEAN-Nop-RA-Pya-K-Nle-L-NH2 (MW, 2,234.16). Mass ESI(+) results were as follows: (M + 2H)+/2 = 1,118.1 and (M + 3H)+/3 = 745.3. For this peptide, HPLC was performed using the same Kromasil C18 column, but the protein was eluted isocratically in 34% CH3CN-66% H2O (0.1% TFA) for 30 min (Rt = 9.81 min).
The sequence of peptide 9 is Ac-KSDSNKTRIDEAN-Nop-RA-Pya-K-Nle-LGSG-NH2 (MW, 2,764.8). Mass ESI(+) results were as follows: (M + 2H)+/2 = 1,383.0 and (M + 3H)+/3 = 922.58. For this peptide, HPLC analysis was performed using the same column, but the peptide was eluted with 20% CH3CN-90% H2O (0.1% TFA) to 40% CH3CN-10% H2O (0.1% TFA) for 30 min (Rt = 19.31 min).
The sequence of peptide 10 is Ac-IIGNLRHMALDMGNEIDTQNRQIDRIMEKADSNKTRIDEN-Nop-RA-Pya-K-Nle-L-NH2 (PL50) (MW, 5,827.7). Mass ESI(+) results were as follows: (M + 4H)+/4 = 1,457.2, (M + 5H)+/5 = 1,166.2, (M + 6H)+/6 = 972.2, and (M + 7H)+/7 = 833.8 (Rt = 15.96 min).
The sequence of peptide 11 is Ac-IIGNLRH-Nle-ALD-Nle-GNEIDTQNRQIDRI-Nle-EKADSNKTRIDEAN-Nop-RA-Pya-K-Nle-L-NH2 (PL51) (MW, 5,773.58). Mass ESI(+) results were as follows: (M + 5H)+/5 = 1,157.3, (M + 6H)+/6 = 964.6, (M + 7H)+/7 = 826.4, and (M + 8H)+/8 = 723.3.
Metabolites.
The sequence of peptide 12 is RA-Pya-K-Nle-L-NH2 (MW, 869.53). Mass ESI(+) results were as follows: (M + H)+ = 870.61 (Rt = 13.03 min).
The sequence of peptide 13 is RA-Nop-K-Nle-L-NH2 (MW, 791.49). Mass ESI(+) results were as follows: (M + H) + = 792.6 (Rt = 10.8 min).
The sequence of peptide 14 is Ac-SNKTRIDEAN-Pya (MW, 1,460.68). Mass ESI(+) results were as follows: (M + H)+ = 1,462.0 and (M + 2H)+/2 = 731.4 (Rt = 15.6 min).
The sequence of peptide 15 is Ac-SNKTRIDEAN-Nop (MW, 1,381.63). Mass ESI(+) results were as follows: (M + H)+ = 1,382.80 and (M + 2H)+/2 = 691.8 (Rt = 14.6 min).
The sequence of peptide 16 is Ac-IIGNLRHMALDMGNEIDTQNRQIDRIMEKADSNKTRIDEAN-Nop (MW, 4,957.59). Mass ESI(+) results were as follows: (M + 4H)+/4 = 1,240.4, (M + 5H)+/5 = 992.6, and (M + 6H)+/6 = 827.8 (Rt = 12.8 min).
The sequence of peptide 17 is Ac-IIGNLRH-Nle-ALD-Nle-GNEIDTQNRQIDRI-Nle-EKADSNKTRIDEAN-Nop (MW, 4,923.48). Mass ESI(+) results were as follows: (M + 4H)+/4 = 1,231.9, (M + 5H)+/5 = 985.9, and (M + 6H)+/6 = 821.4 (Rt = 12.2 min).
BoNT/A purification.
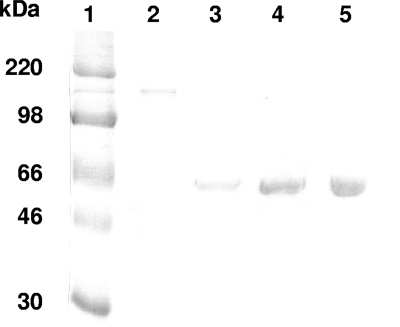
BoNT/A was produced and purified from Clostridium botulinum (A/B) strain NCTC2916 as previously described (34). Briefly, C. botulinum was grown in TGY (Trypticase, 30 g/liter; yeast extract, 20 g/liter; glucose, 5 g/liter; cysteine chlorhydrate, 0.05 g/liter; pH 7.5) for 4 days at 37°C under anaerobic conditions. The culture was precipitated at pH 3.5 with sulfuric acid. The precipitate was collected by centrifugation and extracted with 0.2 M sodium phosphate buffer (pH 6). After centrifugation, the supernatant was precipitated with ammonium sulfate (39 g/100 ml). The precipitate was suspended in distilled water, dialyzed against 50 mM sodium citrate, pH 5.4, and loaded onto a QAE-Sepharose column (Amersham) equilibrated with the same buffer. The flowthrough and the 0.15 M NaCl eluate were precipitated with ammonium sulfate (39 g/100 ml), dialyzed against Tris-HCl 10 mM, pH 8.2, and loaded onto a QAE-Sepharose column equilibrated with the same buffer. The column was eluted using a 0-to-0.15 M NaCl gradient in the same buffer. The fractions containing BoNT/A as evidenced by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) were pooled, concentrated, and stored in aliquots at −80°C.
Toxin quantification.
For densitometric quantification of the purified BoNT/A, the stock solution (5 μl) was diluted in sample loading buffer (0.1 M Tris-HCl, pH 6.8, 15% glycerol, 3% SDS, and 0.02% bromophenol blue), loaded onto a 10% polyacrylamide gel, and subjected to SDS-PAGE in parallel with at least three concentrations of bovine serum albumin (BSA). Proteins were stained with Coomassie brilliant blue R-250 (Bio-Rad). The stained gel was subjected to densitometric analysis using the Quantity One software (Bio-Rad), and BoNT/A was quantified using BSA standards.
MLD50 determinations.
Male Swiss mice (18 to 24 g; Charles River) were used in these experiments. The BoNT/A stock solution was diluted appropriately in sterile phosphate buffer (50 mM, pH 6.3, 0.2% gelatin). The toxin solutions (0.5 ml) were administered intraperitoneally and the mice observed for 4 days.
In a preliminary experiment, six dilutions of BoNT/A ranging from 10−4 to 10−9 of the stock solution were administered to two mice per dose and the mortality rate was recorded after 4 days. Based on the results of this preliminary experiment, a second experiment was performed with dilutions ranging from 5.0 × 10−7 to 7.8 × 10−9, using groups of four mice. The death rate was again recorded 4 days after injection. Based on the obtained results, the toxicity of the injected solution was quantified according to the following formula: toxicity = (1/dilution)·(LD50/V), where V is the injected volume.
Substrate solubilization.
Because of their low solubility in water, peptide stock solutions (100 or 500 μM) were prepared in 50:50 (vol/vol) dimethylformamide (DMF) and H2O. The subsequent working dilutions were then made in the appropriate assay buffer. Under optimized conditions, 100 μM peptide stock solutions were made in 15:85 (vol/vol) DMF-H2O.
Spectrophotometric measurements.
UV absorption spectra of the various substrates and metabolites (0.25 μM in assay buffer with 5% DMF) were recorded on a Shimadzu UV mini 1240 spectrophotometer. Spectrofluorimetric measurements were performed on a PerkinElmer LS50B spectrometer equipped with a thermostated cell holder.
HPLC analysis of peptide cleavage.
The hydrolysis of PL50 (15 μM) by 10 ng/ml of toxin was studied by HPLC after a 5-h incubation at 37°C in standard assay buffer (20 mM HEPES buffer, pH 7.4, containing 5 mM dithiothreitol [DTT], 0.2 mM ZnSO4, and 1 mg/ml BSA). At the end of the incubation, the hydrolysis products were loaded onto a Kromasil C18 column (4.6 by 250 mm, 5-μm inside diameter, 100 Å) and separated using a 10-to-90% CH3CN (0.1% TFA) gradient in 30 min with a flow rate of 1 ml/min. The detection was performed with UV at 343 nm. The same experiment was also performed using the PL51 substrate. The nature of the observed metabolites was confirmed by coinjections with pure synthetic compounds as well as by mass spectrometry analysis.
Fluorimetric enzymatic assays for BoNT/A quantification.
The various peptide substrates were tested in enzymatic assays using purified BoNT/A based on a protocol previously described by Schmidt and Bostian (30, 31). Briefly, the purified 150-kDa toxin was preincubated in 20 mM HEPES buffer, pH 7.4, containing 5 mM DTT, 0.2 mM ZnSO4, and 1 mg/ml BSA for 30 min at 37°C to separate the LC from the HC. The enzymatic reaction was then initiated by the addition of the peptide substrate, and the incubation was performed at 37°C for 1 to 5 h. The experiments were performed using a multiwell microplate reader fluorimeter (Twinkle LB 970; Berthold Technologies) in a total volume of 100 μl. The fluorescence was measured every 15 min at an excitation wavelength of 340 nm and an emission wavelength (λem) of 405 nm (lamp energy, 10,000). Negative controls without toxin were also included in every experiment. All measurements were performed in duplicate in at least two independent experiments.
For the peptides derived from the (187-203) SNAP-25 fragment (peptides 8 and 9), kinetic parameters were determined using 100 ng/ml of purified BoNT/A in assay buffer, with peptide concentrations ranging from 1 to 50 μM and with incubation for 1 h at 37°C. Determination of the kinetic parameters of the longer peptides derived from the (156-203) SNAP-25 fragment (peptides 10 and 11 or PL50 and -51) were performed using 10 ng/ml of BoNT/A and peptide concentrations ranging from 1 to 5 or 10 μM, with incubation for 30 min at 37°C. Calibration curves connecting the increase in fluorescence intensity to changes in the molar ratio of the substrate to the metabolite were established in assay buffer by mixing increasing concentrations of the fluorescent metabolite and related decreasing concentrations of the substrate.
Optimization of the assay conditions involved testing the effects of various reducing agents, i.e., dithioerythritol or Tris(2-carboxyethyl)phosphine hydrochloride (TCEP), at different concentrations between 1 and 10 mM on the fluorescent signal produced by the toxin. The effect of removing BSA from the assay buffer was also investigated.
RESULTS
Design of “quenched fluorescent substrates” for BoNT/A based on the (187-203) SNAP-25 peptide: optimization of the Pya and Nop positions.
Previous studies have identified the (187-203) SNAP-25 fragment (30) as the “minimal” substrate of BoNT/A (Table 1, peptide 1). To increase its stability in the incubation medium, this 17-mer peptide was acetylated at its N terminus (position 187) and protected by a carboxamide at its C terminus (position 203). Moreover, the rapidly oxidized Met202 residue was replaced by its isosteric analogue, Nle (35) (Table 1, peptide 2). Thus, starting from the peptide 2 sequence, the fluorophore/repressor pair Pya and Nop were introduced in various positions replacing or flanking the Q197↓R198 scissile bond of SNAP-25 (Table 1, peptides 3 to 8).
TABLE 1.
BoNT/A cleavage of various peptides containing the fluorophore/repressor pair Pya/Nop or Nop/Pya introduced at different positions in the (187-203) SNAP-25 substrate
| Peptide | Sequence | BoNT/A cleavagea |
|---|---|---|
| 1 | 187S-N-K-T-R-I-D-E-A-N-Q↓R-A-T-K-M-L203 | + |
| 2 | Ac-S-N-K-T-R-I-D-E-A-N-Q↓R-A-T-K-Nle-L-NH2 | + |
| 3 | Ac-S-N-K-T-R-I-D-E-A-N-Pya-Nop-A-T-K-Nle-L-NH2 | 0 |
| 4 | Ac-S-N-K-T-R-I-D-E-A-N-Nop-Pya-A-T-K-Nle-L-NH2 | 0 |
| 5 | Ac-S-N-K-T-R-I-D-E-A-N-Pya-R-Nop-T-K-Nle-L-NH2 | 0 |
| 6 | Ac-S-N-K-T-R-I-D-E-A-N-Nop-R-Pya-T-K-Nle-L-NH2 | 0 |
| 7 | Ac-S-N-K-T-R-I-D-E-A-N-Pya↓R-A-Nop-K-Nle-L-NH2 | + |
| 8 | Ac-S-N-K-T-R-I-D-E-A-N-Nop↓R-A-Pya-K-Nle-L-NH2 | + |
+, present (at ↓); 0, absent.
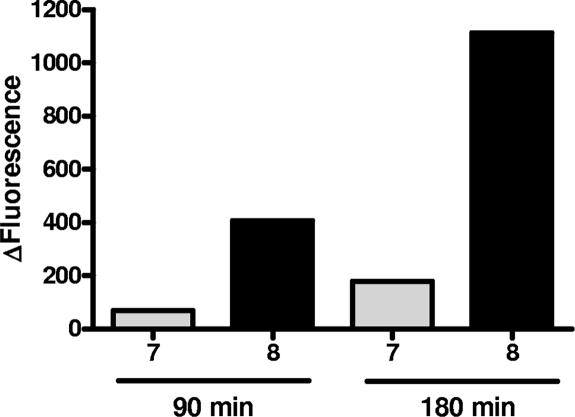
Incubation of peptides 3 to 8 with purified BoNT/A (Fig. 1) in reaction buffer with 1 mg/ml BSA showed that peptides 3 and 4 (Table 1) were not cleaved by BoNT/A. Introducing the SNAP-25 R198 residue between the fluorophore and repressor pair in either order (Table 1, peptides 5 and 6) did not restore BoNT/A cleavage. Spacing the fluorophore/repressor pair by 2 amino acids by introducing both R198 and Ala199 residues, however, restored BoNT/A cleavage (Table 1, peptides 7 and 8; Fig. 2), and there was a marked preference for the peptide containing the Nop197-R-A-Pya200 sequence (Fig. 2). The basal levels of fluorescence of peptides 7 and 8 (20 μM) in assay buffer were not significantly different from one another (110 arbitrary units [AU] and 75 AU, respectively) showing equivalent quenching efficiencies. After a 3-h incubation with BoNT/A (50 ng/ml), the cleavage of peptide 7 produced a fluorescence increase of 176 AU, while cleavage of peptide 8 led to a much greater increase of 1,100 AU (Fig. 2).
FIG. 1.
SDS-PAGE analysis of purified BoNT/A. A BoNT/A sample (5 μl) was electrophoresed under nonreducing conditions on a 10% polyacrylamide gel in parallel with BSA standards. After Coomassie brilliant blue staining, purified BoNT/A migrating as a single 150-kDa band was quantified by densitometric analysis against the BSA standards. Lane 1 shows the molecular mass standards, while lane 2 contains BoNT/A. Lanes 3 to 5 were loaded with the BSA standards (2, 4, and 6 μg, respectively).
FIG. 2.
Comparative hydrolysis of peptides 7 and 8. BoNT/A (150 kDa; 50 ng/ml) was added to peptide substrates 7 and 8 (20 μM) in 100 μl of reaction buffer (HEPES, 20 mM, pH 7.4; ZnSO4, 200 μM; DTT, 5 mM; BSA 1 mg/ml) and incubated at 37°C for 90 and 180 min, times at which the fluorescent signal was monitored. The bar graph shows the mean deltas of fluorescence (total fluorescence − fluorescence of the peptide substrate alone in buffer) measured with peptide 7 (gray bars) and peptide 8 (black bars).
This result was confirmed by HPLC analysis of the incubation medium performed with UV detection at a λ of 343 nm, a wavelength corresponding to the absorption of Pya. In all these experiments, the formed peaks were characterized by coinjection with pure synthetic peptides. Thus, after a 3-h incubation with BoNT/A at 50 ng/ml, peptide 7 yielded 6.1% of the fluorescent metabolite (peptide 15, Ac-S-N-K-T-R-I-D-E-A-N-Pya [Rt = 13.2 min]), while peptide 8 yielded 51% of its own fluorescent metabolite (peptide 12, R-A-Pya-K-Nle-L-NH2 [Rt = 9.38 min]). These metabolites allow the identification of the amide bond cleaved by BoNT/A as that holding residues 197 and 198, i.e., between Nop197 and R198, revealing that the presence of the nonnatural residues in the synthetic peptides did not change the cleavage site observed in native SNAP-25. Moreover, the fact that only two metabolites were formed indicated that the high specificity of BoNT/A was preserved in this minimal sequence (see Fig. S1 in the supplemental material).
Based on these results, peptide 8 was selected as the first “lead” in the search for an efficient fluorigenic substrate for BoNT/A, and its kinetic parameters were determined. The affinity (Km) of the peptide toward BoNT/A was 13 ± 3 μM, whereas its kcat was 0.79 ± 0.06 s−1, leading to a catalytic efficiency (kcat/Km) of (6.1 ± 0.5) × 104 M−1 s−1 (Table 2; see also Fig. S2 in the supplemental material). Peptide 8 possessed a slightly better catalytic efficiency for BoNT/A than the natural (187-203) SNAP-25 peptide (2.75 × 104 M−1 s−1 [30]), essentially due to a significant improvement of its Michaelis constant.
TABLE 2.
Kinetic parameters of peptides 8 to 11a
| Peptide | Km (μM) | kcat (s−1) | kcat/Km (M−1 s−1) |
|---|---|---|---|
| 8 | 13 ± 3 | 0.79 ± 0.06 | 6.1 (±0.5) × 104 |
| 9 | 11 ± 1 | 1.01 ± 0.04 | 9.2 (±0.4) × 104 |
| 10 | 0.79 ± 0.20 | 2.08 ± 0.12 | 2.63 (±0.15) × 106 |
| 11 | 0.13 ± 0.02 | 1.15 ± 0.05 | 8.85 (±0.38) × 106 |
Kinetic parameters were obtained under the conditions described in Materials and Methods. The plotted Km results for peptides 9, 10 (PL50), and 11 (PL51) are represented in Fig. S2 in the supplemental material.
Lengthening of the substrate sequence of peptide 8.
To improve the efficiency of peptide 8 as a BoNT/A substrate, its size was increased by adding at its N terminus a K-S-D sequence mimicking the 184K-A-D186 sequence of SNAP-25, as well as the three (G-S-G) amino acids corresponding to the sequence from positions 203 to 206 of SNAP-25 at its C terminus. The resulting peptide, peptide 9 (Ac-K-S-D-S-N-K-T-R-I-D-E-A-N-Nop-R-A-Pya-K-Nle-L-G-S-G-NH2) displayed better kinetic constants than those of the lead peptide, peptide 8. Indeed, the Km of peptide 9 was estimated at 11 ± 1 μM and its kcat at 1.01 ± 0.04 s−1, leading to an improved catalytic efficiency (kcat/Km) of (9.2 ± 0.4) × 104 M−1 s−1 (Table 2; Fig. S2 in the supplemental material).
Design and properties of fluorescently quenched substrates based on the sequence of (156-203) SNAP-25.
To further improve the sensitivity and velocity of the BoNT/A fluorescence assay, we took into account the demonstrated presence of BoNT/A exosites able to bind BoNT/A's substrate at a more or less great distance from the scissile bond, thus positively impacting its cleavage (5, 8). Consequently, the fluorophore/repressor pair Pya/Nop was introduced into the (156-203) SNAP-25 fragment at the previously determined optimal positions in peptides 8 and 9, i.e., at positions 197 for Nop and 200 for Pya, to yield peptide 10.
Finally, the sequence of peptide 10, or PL50 (Ac-IIGNLRHMALDMGNEIDTQNRQIDRIMEKADSNKTRIDEAN- Nop-RA-Pya-K-Nle-L-NH2) was modified to increase its stability. To prevent their possible oxidation, the three Met residues at positions 153, 157, and 172 of peptide 10 were replaced by their isosteric analogue, Nle, leading to peptide 11, or PL51 (Ac-IIGNLRH-Nle-ALD-Nle-GNEIDTQNRQIDRI-Nle- EKADSNKTRIDEAN-Nop-RA-Pya-K-Nle-L-NH2).
Spectral properties of PL50 and PL51.
The UV spectra of peptide 10 (PL50) and of its fluorescent metabolite R-A-Pya-K-Nle-L-NH2 (peptide 12) reveal wavelengths of maximum absorption at 343.5 nm (ɛmol = 29,750 liters·mol−1 cm−1) and at 342.5 nm (ɛmol = 27,500 liters·mol−1 cm−1), respectively, showing that the chemical environment of the fluorophore has only a slight effect on the absorption maximum but a significant effect on the molar absorption.
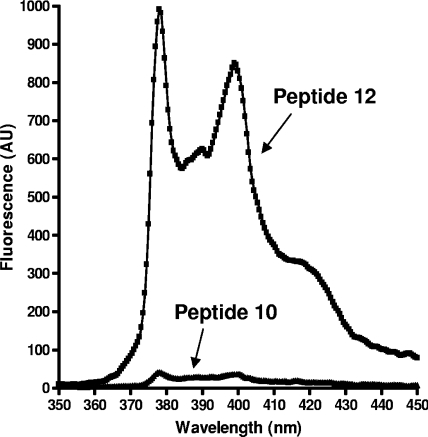
The fluorescence spectra obtained for these compounds after excitation at 343 nm display two maxima at λems of 378 and 398 nm (Fig. 3). The superimposition of the emission spectra of peptide 10 and of its fluorescent metabolite (peptide 12) at 0.25 μM in assay buffer with 5% DMF shows that the intense fluorescence of the pyrenylalanine moiety observed in peptide 10 is almost completely quenched by the p-nitrophenylalanyl residue. To determine the fluorescence ratio between peptide 11 (PL51) and its fluorescent metabolite (peptide 12), the former compound was used at 10 μM and the latter at 0.25 μM (2.5% degradation). The fluorescence intensities measured at 378 nm for both peptides, for which identical concentrations were extrapolated, gave a quenching factor of 150 (not shown).
FIG. 3.
Comparison of the emission fluorescent spectra (λex = 343 nm) of peptide substrate 10 (Ac-IIGNLRHMALDMGNEIDTQNRQIDRIMEKADSNKTRIDEAN-Nop-RA-Pya-K-Nle-L-NH2) and of its fluorescent metabolite RA-Pya-K-Nle-L-NH2 (peptide 12) at the same concentration (0.25 μM).
Cleavage site determination and kinetic parameters of PL50 and PL51.
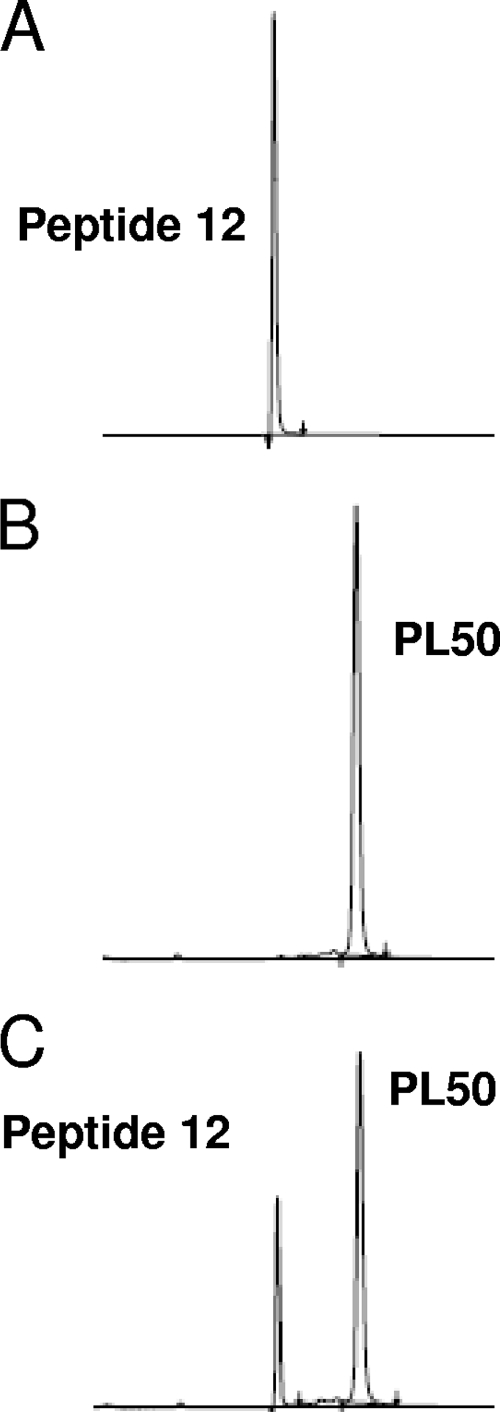
HPLC analysis of the cleavage products of PL50 by BoNT/A shows the appearance of two chemical entities at 210 nm (not shown), only one of which was detected at 343 nm (UV absorption of Pya) or with a fluorimetric detector (λem = 377 nm) (Fig. 4). The Rt of this fluorescent metabolite (13.03 min) corresponded to that of peptide 12 (RA-Pya-K-Nle-L-NH2). The second nonfluorescent metabolite had an Rt of 14.6 min and was shown by coinjection to correspond to the N-terminal synthetic peptide (156-197) SNAP-25 (Nop197), or peptide 16. The same results were observed with PL51, whose cleavage by BoNT/A also led to only two metabolites, one corresponding to peptide 17 and the other to fluorescent peptide 12 (not shown).
FIG. 4.
HPLC analysis of the PL50 cleavage products. BoNT/A (10 ng/ml) was incubated for 5 h at 37°C with PL50 (15 μM) in standard assay buffer (HEPES, 20 mM, pH 7.4; ZnSO4, 200 μM; DTT, 5 mM; BSA, 1 mg/ml) in a final reaction volume of 100 μl. The reaction products were then separated by HPLC on a Kromasil C18 column using a gradient of 10 to 90% CH3CN (0.1% TFA) in 30 min, and the resulting chromatograms are shown. (A) Peptide 12 metabolite in assay buffer (1 μM), with an Rt of 13.03 min; (B) PL50 (15 μM) incubated in assay buffer without toxin (Rt = 15.96 min); (C) result of the hydrolysis of PL50 (15 μM) by BoNT/A. At 343 nm, the wavelength at which Pya emits, the only observed peaks are those corresponding to PL50 and to peptide 12.
The kinetic parameters of PL50 were determined and are reported in Table 2 (see also Fig. S2 in the supplemental material). The Km value was 0.79 ± 0.20 μM, and the kcat value was 2.08 ± 0.12 s−1, leading to a kcat/Km of 2.63 × 106 ± 0.15 × 106 M−1 s−1. Thus, the kcat value of PL50 was increased by a factor 2 compared to that of peptide 9, while its apparent affinity was improved by a factor 10. Finally, while the kcat value of PL51 was not improved compared to that of PL50, its apparent affinity was increased sixfold, yielding a better kcat/Km value for this last peptide substrate (Km = 0.13 ± 0.02 μM, kcat = 1.15 ± 0.05 s−1, kcat/Km = 8.85 × 106 ± 0.38 × 106 M−1 s−1) (Table 2; Fig. S2 in the supplemental material).
Characterization and calibration of the BoNT/A-PL50 enzymatic assay against the MLD50 assay.
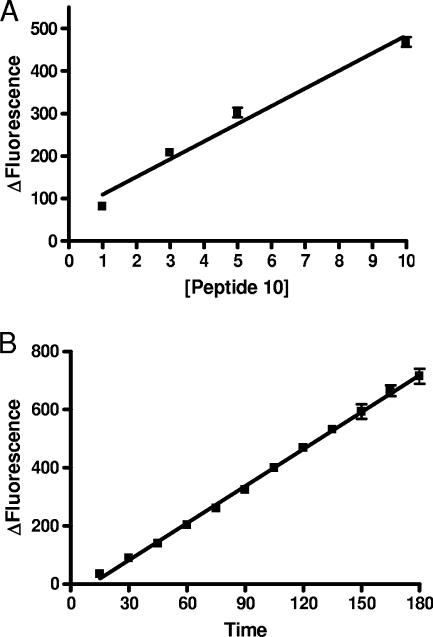
Using PL50, the novel BoNT/A enzymatic assay was characterized. There was a linear correlation between the fluorescent signal and PL50 substrate concentration used in the 1 μM-to-10 μM range (R2 = 0.994) (Fig. 5A). There was also a linear correlation between the fluorescent signal and time over 150 min using a 10 μM PL50 substrate concentration and 10 ng/ml of BoNT/A (R2 = 0.994) (Fig. 5B). Based on these data, PL50 was used in the assay at a concentration of 10 μM. Under these conditions, a detection of 10 ng/ml of the toxin could be obtained in less than 1 h.
FIG. 5.
Characterization of the BoNT/A enzymatic assay using PL50. (A) There was a linear correlation (R2 = 0.97) between the fluorescent response (Δfluorescence = endpoint fluorescence − fluorescence of PL50 in assay buffer) produced by 10 ng/ml of the 150-kDa BoNT/A toxin incubated for 60 min at 37°C in assay buffer (HEPES, 20 mM, pH 7.4; ZnSO4, 200 μM; DTT, 5 mM; BSA, 1 mg/ml) and the PL50 concentration between 1 and 10 μM. (B) With the highest concentration of PL50 (10 μM), the BoNT/A (10 ng/ml) fluorescent response was linear (R2 = 0.99) over 180 min. Under these conditions, 10 ng/ml of BoNT/A toxin was detected in about 30 min.
In order to calibrate the assay not only versus protein weight but also versus the commonly accepted unit concentration, it was necessary to be able to convert one scale into another. As units, in the case of toxins, correspond to MLD50 values, we performed an MLD50 assay using our stock solution of purified toxin. For this purpose, the stock solution was serially diluted, and on the same day, dilutions were either injected into mice intraperitoneally (5 × 10−7 to 7.81 × 10−9) to determine the MLD50 or used in the fluorescent PL50 enzymatic assay (0.5 × 10−3 to 10−6).
The results of a typical MLD50 assay are presented in Table 3. Based on these results, the concentration of the purified toxin stock solution was determined to be 1.7 × 108 MLD50s/ml or U/ml. The same results, combined with the estimated protein concentrations, also allow us to estimate the MLD50 of the toxin as 1.9 pg (Table 3).
TABLE 3.
MLD50 assay resultsa
| Toxin concn (pg/ml) | Toxin dilution | Mouse mortality at 96 h (no. of mice that died/total) |
|---|---|---|
| 162.5 | 5 × 10−7 | 3/4 |
| 81.25 | 2.5 × 10−7 | 4/4 |
| 40.62 | 1.23 × 10−7 | 3/4 |
| 20.31 | 6.25 × 10−8 | 4/4 |
| 10.16 | 3.125 × 10−8 | 4/4 |
| 5.08 | 1.56 × 10−8 | 3/4 |
| 2.54 | 7.81 × 10−9 | 0/4 |
Evaluation of mouse mortality 4 days postinjection after intraperitoneal injection of successive dilutions of a BoNT/A (150 kDa) stock solution. Based on the toxin concentration evaluated by gel electrophoresis, the toxin concentration corresponding to each toxin dilution is also indicated. From the mouse mortality observations, the potency of the stock solution can be calculated using the formula 1/LD50 dilution/V (injected volume). If the LD50 is located at a dilution intermediary between 1.56×10−8 and 7.81×10−9, i.e., 1.17×10−8, the potency of the stock solution is 1.7×108 MLD50s/ml. Moreover, the protein concentration corresponding to the MLD50 dilution can be deduced from the data in the first column, i.e., 3.80 pg/ml, which yields an MLD50 value of 1.9 pg (injected volume = 0.5 ml).
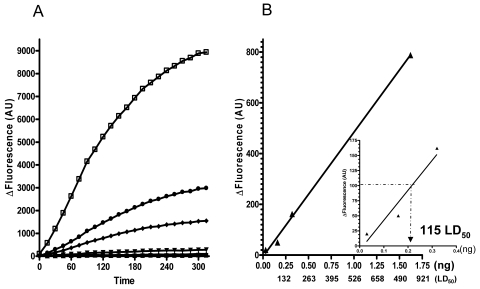
Using the 0.5 × 10−3, 10−4, 0.5 × 10−4, 10−5, 0.5 × 10−5, and 10−6 stock solution dilutions, the BoNT/A enzymatic assay was performed using 10 μM PL50. Results of the fluorescent assay reveals that, considering a 100 AU increase in fluorescence as the limit of detection, the lowest concentration of BoNT/A detected using the PL50 enzymatic assay is 2 ng/ml in 120 min or 4 ng/ml in 1 hour. These values correspond to 200 and 400 pg of purified 150-kDa toxin per assay, respectively. Using the results of the MLD50 assay performed in parallel with the same toxin solution, the detected toxin concentrations can be converted to MLD50 values. Thus, using 1.9 pg as 1 MLD50, the PL50 assay can detect 115 MLD50s in 120 min (Fig. 6).
FIG. 6.
Sensitivity of the BoNT/A-PL50 enzymatic assay and MLD50 calibration. Serial dilutions of a BoNT/A (150 kDa) stock solution (325 μg/ml) were used either in an MLD50 assay (Table 3) or in a BoNT/A-PL50 enzymatic assay. Decreasing concentrations of BoNT/A (150 kDa) were incubated for 5 h at 37°C with 10 μM PL50 in 100 μl of reaction buffer (HEPES, 20 mM, pH 7.4; ZnSO4, 200 μM; DTT, 5 mM; BSA, 1 mg/ml), and the fluorescence was read every 15 min. (A) Deltas of fluorescence (total fluorescence − fluorescence of the peptide in buffer) were measured over 5 h for the six studied dilutions (ranging from 0.5 × 10−3 to 10−6). Based on the toxin concentration of the starting stock solution, toxin concentrations used were 162.5 (□), 32.5 (•), 16.2 (⧫), 3.2 (▾), 1.6 (▴), and 0.3 (▪) ng/ml. (B) Linearity (R2 = 0.99) of the detection at 120 min (▴) as a function of BoNT/A concentration in the assay in nanograms or in the corresponding MLD50 (using 1 MLD50 or 1.9 pg). The inset graph zooms in on the lower BoNT/A concentrations, which allowed us to determine the assay detection limit to be 115 MLD50s.
Sensitivity optimization.
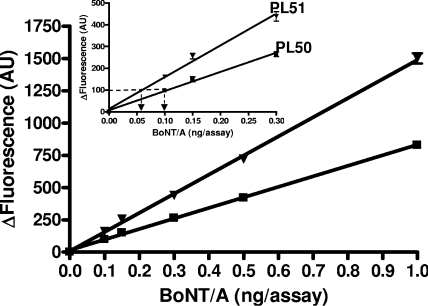
One of the aims of this study was to use the developed assay for the detection and quantification of BoNT/A in medical samples which contain between 100 and 500 MLD50s. The MLD50 being estimated at about 2 pg, it was important to detect less than 200 pg in a given sample in the shortest incubation time possible. To increase the sensitivity of the BoNT/A enzymatic assay, the assay conditions were optimized on one hand, and on the other hand, the PL50 substrate was substituted for PL51 in a comparative assay with toxin concentrations of either 0.5, 1, or 2 ng/assay and incubated for 120 min at 37°C. The results depicted in Fig. 7 show that under these optimized conditions, a detection limit of 100 pg of toxin in 120 min, corresponding to 53 MLD50s, was obtained. Under the same conditions, the use of PL51 also significantly increased the sensitivity of the assay. Thus, in 120 min using PL51, one can detect around 30 MLD50s (Fig. 7).
FIG. 7.
BoNT/A enzymatic assay optimization using PL50 and PL51. BoNT/A was incubated at increasing concentrations at 37°C with either 10 μM PL50 (▪) or 10 μM PL51 (▾) under optimized conditions. Substrate stock solutions were prepared so as to obtain a final DMF concentration of 1.5% in 100 μl of the optimized reaction buffer (HEPES, 20 mM, pH 7.4; ZnSO4, 200 μM; TCEP, 2.5 mM). The inset represents a zoom of the lower BoNT/A concentrations, which allowed us to determine the detection limits, namely, 100 pg (53 MLD50s) or 60 pg (32 MLD50s) in 120 min using PL50 or PL51, respectively. Mean values of endpoint measurements (120 min) from two independent experiments performed in duplicate are represented.
DISCUSSION
Intramolecularly quenched fluorescent peptides containing the fluorophore/repressor pair Pya/Nop have successfully been developed for the detection and quantification of various specific enzymatic activities (1, 18). With these substrates, the observed quenching is attributed to collisional interactions between the side chains of the fluorophore/repressor pair members. The Pya/Nop pair is particularly well adapted due to the high quantum yield and long half-life of the fluorescence of the pyrenyl moiety (Pya) and the low absorption of the quencher residue, (p-NO2)-phenylalanine (Nop), at the excitation wavelength of Pya (1). These properties have led to the design of the most-efficient fluorescent substrate of BoNT/B described to date, by the introduction of the Pya and Nop moieties at positions 74 and 77 of synaptobrevin (positions 60 to 94), respectively, i.e., on each side of the neurotoxin scissile bond (26).
In order to develop a specific and high-affinity substrate for BoNT/A, we first introduced the fluorigenic pair at and around the 197Q-R198 cleavage site of SNAP-25 using the previously described 17-mer peptide (187-203) SNAP-25 (30) to optimize the relative positions of the fluorophore/repressor pair. As shown in Table 1, the presence of the 198R-A199 amino acids was absolutely necessary for BoNT/A recognition, and the positions of the Pya and Nop moieties around these amino acids were not interchangeable. These results are consistent with the three-dimensional structure of the BoNT/A LC cocrystallized with (141-204) SNAP-25 (5), in which the threonine amino acid at position 200 is located within a large pocket formed by hydrophobic residues (Val68, Val242, Val258, Leu256, Phe366, and Phe369), allowing its easy replacement by Pya. On the other hand, Q197 of SNAP-25, which is hydrogen bonded with Gln162 of BoNT/A, is located in a more hydrophilic environment and occupies a more restricted space, which could not accommodate the large Pya residue but allowed binding of the polar p-nitro Phe (Nop) residue. Thus, as for BoNT/B recognition, the integrity of the P′1 and P′2 amino acids of the peptide substrate were necessary, but unlike BoNT/B, BoNT/A could not accommodate the Pya residue at S1 (1).
The introduction of the Pya and Nop residues, both with aromatic side chains, in the vicinity of the BoNT/A cleavage site significantly improved the affinity of the substrate toward the toxin. Indeed, the observed Km of peptide 8 (13 μM) was 400-fold better than that of its parent peptide, (187-203) SNAP-25 (Km = 5 mM) (30). However, these favorable hydrophobic interactions had an adverse effect on the reaction rate, which was significantly decreased using peptide 8 as the substrate (kcat = 0.79 s−1) (Table 2) compared to that with (187-203) SNAP-25 (reported kcat = 4.7 to 47 s−1 [30]). However, despite the relatively low kcat value of 8, its catalytic efficiency (kcat/Km = 6.1 × 104 M−1 s−1) was equivalent to that of the fluorigenic substrate {SNRTRIDEAN[dnpK]RA[daciaC]RML; where dnpK is Nɛ-(2,4-dinitrophenyl)lysine and daciaC is S-(N-[4-methyl-7-dimethylaminocoumarin-3-yl]-carboxamidomethyl)- cysteine}, also derived from the fragment from positions 187 to 203 of SNAP-25 (kcat/Km = 7.5 × 104 M−1 s−1) (33). Although the sensitivity of the assay using the peptide 8 substrate was sufficient for inhibitor screening using high-throughput-screening techniques, it was considered insufficient for the quantification of toxin formulations used for medical purposes.
Clostridial toxins such as the tetanus and botulinum toxins have been shown to possess exosites whose role is to induce an increase in the velocity of the reaction by an allosteric mechanism (5, 8). Previous analyses of SNAP-25 deletion mutants have suggested that the specific binding of SNAP-25 to BoNT/A could involve binding to an exosite(s) more or less distant from the catalytic site (35-36). The three-dimensional structure of an inactive construct of the BoNT/A LC that is cocrystallized with (141-204) SNAP-25 (5) indeed reveals the presence of two main exosites and various anchor points: the α exosite, which interacts with (152-167) SNAP-25, and the C-terminal β exosite around Met202. The first fragment was shown to play an important role in the binding affinity of the peptide toward the toxin, while the second is involved in catalytic efficiency. Moreover, it has been demonstrated that the internal (168-186) SNAP-25 sequence facilitates the binding of SNAP-25 to the toxin, thereby enhancing the cleavage rate (7, 21). Taken together, these results, strongly suggested that (156-203) SNAP-25 could be optimal for BoNT/A binding. Accordingly, the fluorigenic substrates PL50 (peptide 10) and PL51 (peptide 11), corresponding to this (156-203) SNAP-25 sequence, were designed. The specific activity of the resulting PL50 substrate was found to be 100-fold better than that of peptide 8, resulting exclusively from a significant increase of the apparent affinity of PL50 (Table 2; Fig. S2 in the supplemental material) and confirming the hypothesis that increasing peptide length would ameliorate BoNT/A recognition, probably by binding one or more exosites of the BoNT/A LC. Interestingly, PL50 also displays a 50- to 60-fold-higher apparent affinity for BoNT/A than full-length SNAP-25 (Km = 40 to 50 μM), while the cleavage rate is not significantly improved (6, 17).
In an effort to calibrate this novel enzymatic assay against the gold standard, but nonetheless controversial, MLD50 assay, parallel experiments were performed using the same toxin dilutions. The ensemble of these results shows that the purified toxin used in this study possesses an MLD50 of 1.9 pg, a value in line with those found in the literature (2). Moreover, this allows the detection limit of the assay to be expressed either in toxin weight or in MLD50s. Thus, as shown in Fig. 6 and 7, using the PL50 substrate under previously published conditions, the detection limit of the assay was found to be about 115 MLD50s. Using TCEP as the reducing agent, the detection limit was lowered by over 50% to 53 MLD50s in 120 min, reaching values as low as 15 MLD50s in 300 min, providing to date the most sensitive direct assay to measure BoNT/A activity. To protect the PL50 peptide from oxidation, the methionine residues in its sequence were replaced by norleucines, to yield PL51. Interestingly, this substrate possessed a slightly better specificity constant than its parent compound (Table 2), providing better sensitivity for shorter incubation times, i.e., 32 MLD50s in 120 min (Fig. 7).
Altogether, this study reveals that PL50 and PL51, which associate excellent catalytic efficiencies with the remarkable fluorigenic properties of Pya, are at present the most efficient and easy-to-use fluorescent substrates of BoNT/A (M. C. Fournié-Zaluski and B. P. Roques, 22 December 2005, patent application PCT WO 2005 1121354 A2). The two substrates are therefore of value for measuring the concentration of BoNT/A in medicinal preparations in which the smaller amount of BoNT/A employed corresponds to 50 MLD50s. One of the main advantages of this enzymatic assay is that it is a single-step assay which circumvents any specificity problems that can be encountered with other immunoabsorbent enzyme-linked immunosorbent assay-type assays (12, 23, 24). It can be used to quantify BoNT/A in pharmaceutical toxin preparations and could also find application in the detection of toxin in complex matrices, although its efficacy will be dependent on preliminary extraction methods. This assay also drastically reduces the time necessary to obtain a result, the toxin here being quantified, and identified, within an hour. These are important considerations, particularly in the case of crisis situations where malevolent bioterrorist acts are suspected (e.g., in water systems), and the use of the assay is thus expected to severely reduce the use of the mouse bioassay, which nevertheless remains necessary for investigating the endocytosis property of a given enzymatically active toxin.
Supplementary Material
Footnotes
Published ahead of print on 8 May 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Anne, C., F. Cornille, C. Lenoir, and B. P. Roques. 2001. High-throughput fluorigenic assay for determination of botulinum type B neurotoxin protease activity. Anal. Biochem. 291:253-261. [DOI] [PubMed] [Google Scholar]
- 2.Arnon, S. S., R. Schechter, T. V. Inglesby, D. A. Henderson, J. G. Bartlett, M. S. Ascher, E. Eitzen, A. D. Fine, J. Hauer, M. Layton, S. Lillibridge, M. T. Osterholm, T. O'Toole, G. Parker, T. M. Perl, P. K. Russell, D. L. Swerdlow, and K. Tonat. 2001. Botulinum toxin as a biological weapon: medical and public health management. JAMA 285:1059-1070. [DOI] [PubMed] [Google Scholar]
- 3.Barr, J. R., H. Moura, A. E. Boyer, A. R. Woolfitt, S. R. Kalb, A. Pavlopoulos, L. G. McWilliams, J. G. Schmidt, R. A. Martinez, and D. L. Ashley. 2005. Botulinum neurotoxin detection and differentiation by mass spectrometry. Emerg. Infect. Dis. 11:1578-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blitzer, A., M. F. Brin, M. S. Keen, and J. E. Aviv. 1993. Botulinum toxin for the treatment of hyperfunctional lines of the face. Arch. Otolaryngol. Head Neck Surg. 119:1018-1022. [DOI] [PubMed] [Google Scholar]
- 5.Breidenbach, M. A., and A. T. Brunger. 2004. Substrate recognition strategy for botulinum neurotoxin serotype A. Nature 432:925-929. [DOI] [PubMed] [Google Scholar]
- 6.Cai, S., and B. R. Singh. 2001. A correlation between differential structural features and the degree of endopeptidase activity of type A botulinum neurotoxin in aqueous solution. Biochemistry 40:4693-4702. [DOI] [PubMed] [Google Scholar]
- 7.Chen, S., and T. Barbieri. 2006. Unique substrate recognition by botulinum neurotoxins serotypes A and E. J. Biol. Chem. 281:10906-10911. [DOI] [PubMed] [Google Scholar]
- 8.Cornille, F., L. Martin, C. Lenoir, D. Cussac, B. P. Roques, and M. C. Fournié-Zaluski. 1997. Cooperative exosite-dependent cleavage of synaptobrevin by tetanus toxin light chain. J. Biol. Chem. 272:3459-3464. [DOI] [PubMed] [Google Scholar]
- 9.Dong, M., F. Yeh, W. H. Tepp, C. Dean, E. A. Johnson, R. Janz, and E. R. Chapman. 2006. SV2 is the protein receptor for botulinum neurotoxin A. Science 312:592-596. [DOI] [PubMed] [Google Scholar]
- 10.Ekong, T. A., K. McLellan, and D. Sesardic. 1995. Immunological detection of Clostridium botulinum toxin type A in therapeutic preparations. J. Immunol. Methods 180:181-191. [DOI] [PubMed] [Google Scholar]
- 11.Ferreira, J. L., S. J. Eliasberg, P. Edmonds, and M. A. Harrison. 2004. Comparison of the mouse bioassay and enzyme-linked immunosorbent assay procedures for the detection of type A botulinal toxin in food. J. Food Prot. 67:203-206. [DOI] [PubMed] [Google Scholar]
- 12.Hallis, B., B. A. James, and C. C. Shone. 1996. Development of novel assays for botulinum type A and B neurotoxins based on their endopeptidase activities. J. Clin. Microbiol. 34:1934-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hof, M., V. Fidler, and R. Hutterer (ed.). 2005. Fluorescence spectroscopy in biology: advanced methods and their applications to membranes, proteins, DNA, and cells. Springer, Berlin, Germany.
- 14.Johnson, E. A. 1999. Clostridial toxins as therapeutic agents: benefits of nature's most toxic proteins. Annu. Rev. Microbiol. 53:551-575. [DOI] [PubMed] [Google Scholar]
- 15.Johnson, E. A., G. E. Borodic, and M. A. Acquadro. 2006. Medical applications of botulinum neurotoxins, p. 959-975. In E. J. Alouf and M. R. Popoff (ed.), The comprehensive source book of bacterial protein toxins. Academic Press, Burlington, MA.
- 16.Kalb, S. R., H. Moura, A. E. Boyer, L. G. McWilliams, J. L. Pirkle, and J. R. Barr. 2006. The use of endopep-MS for the detection of botulinum toxins A, B, E and F in serum and food samples. Anal. Biochem. 351:84-92. [DOI] [PubMed] [Google Scholar]
- 17.Li, L., T. Binz, H. Niemann, and B. R. Singh. 2000. Probing the mechanistic role of glutamate residue in the zinc binding motif of type A botulinum neurotoxin light chain. Biochemistry 39:2399-2405. [DOI] [PubMed] [Google Scholar]
- 18.Luciani, N., H. de Rocquigny, S. Turcaud, A. Romieu, and B. P. Roques. 2001. Highly sensitive and selective fluorescence assays for rapid screening of endothelin-converting enzyme inhibitors. Biochem. J. 356:813-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niemann, H. 1991. Molecular biology of the clostridial neurotoxins, p. 303-348. In J. H. Alouf and J. H. Freer (ed.). A source book of bacterial protein toxins. Academic Press, London, United Kingdom.
- 20.Nishiki, T., Y. Tokuyama, Y. Kamata, Y. Nemoto, A. Yoshida, K. Sato, M. Sekiguchi, M. Takahashi, and S. Kosaki. 1996. The high-affinity binding of Clostridium botulinum type B to synaptotagmin II associated with gangliosides GT1b/GD1a. FEBS Lett. 378:253-257. [DOI] [PubMed] [Google Scholar]
- 21.Ouimet, T., H. Poras, S. V. Orng, S. Duquesnoy, M. C. Fournié-Zaluski, and B. P. Roques. 2008. Design of fluorogenic substrates for botulinum toxin A, quantification and identification of a novel cooperative exosite for substrate binding. Toxicon 51(Suppl. 1):17.
- 22.Pearce, L., E. R. First, R. D. Maccallum, and A. Gupta. 1997. Pharmacological characterization of botulinum toxin for basic science and medicine. Toxicon 35:1373-1412. [DOI] [PubMed] [Google Scholar]
- 23.Phillips, R. W., and D. Abbott. 2008. High-throughput enzyme-linked immunoabsorbent assay (ELISA) electrochemiluminescent detection of botulinum toxins in food for safety and defence purposes. Food Addit. Contam. Part A 25:1084-1088. [DOI] [PubMed] [Google Scholar]
- 24.Rasooly, R., and P. Do. 2008. Development of an in vitro activity assay as an alternative to the mouse bioassay for Clostridium botulinum neurotoxin A. Appl. Environ. Microbiol. 74:4309-4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rivera, V. R., F. J. Gamez, W. K. Keener, J. A. White, and M. A. Poli. 2006. Rapid detection of Clostridium botulinum toxins A, B, E, and F in clinical samples, selected food matrices, and buffer using paramagnetic bead-based electrochemiluminescence detection. Anal. Biochem. 353:248-256. [DOI] [PubMed] [Google Scholar]
- 26.Roques, B. P., and C. Anne. 9 January 2007. Peptide substrate identified by type B BoNT/B botulinum toxin and use thereof for assaying and/or detecting said toxin or corresponding inhibitors. U.S. patent 7,160,982 B2.
- 27.Rotz, L. D., A. S. Khan, S. R. Lillibridge, S. M. Ostroff, and J. M. Hugues. 2002. Public health assessment of potential biological terrorism agents. Emerg. Infect. Dis. 8:225-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schantz, E. J., and E. A. Johnson. 1992. Properties and use of botulinum toxin and other microbial neurotoxins in medicine. Microbiol. Rev. 56:80-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schiavo, G., O. Rossetto, A. Santucci, B. P. DasGupta, and C. Montecucco. 1992. Botulinum neurotoxins are zinc proteins. J. Biol. Chem. 267:23479-23483. [PubMed] [Google Scholar]
- 30.Schmidt, J. J., and K. A. Bostian. 1995. Proteolysis of synthetic peptides by type A botulinum neurotoxin. J. Prot. Chem. 14:703-708. [DOI] [PubMed] [Google Scholar]
- 31.Schmidt, J. J., and K. A. Bostian. 1997. Endoproteinase activity of type A botulinum neurotoxin: substrate requirements and activation by serum albumin. J. Prot. Chem. 16:19-26. [DOI] [PubMed] [Google Scholar]
- 32.Schmidt, J. J., R. G. Stafford, and C. B. Millard. 2001. High-throughput assays for botulinum neurotoxin proteolytic activity: serotypes A, B, D, and F. Anal. Biochem. 296:130-137. [DOI] [PubMed] [Google Scholar]
- 33.Schmidt, J. J., and R. G. Stafford. 2003. Fluorigenic substrates for the protease activities of botulinum neurotoxins, serotypes A, B, and F. Appl. Environ. Microbiol. 69:297-303. (Erratum, 69:3025.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shone, C. C., and H. S. Tranter. 1995. Growth of Clostridia and preparation of their neurotoxins. Curr. Top. Microbiol. Immunol. 195:143-160. [DOI] [PubMed] [Google Scholar]
- 35.Sukonpan, C., T. Oost, M. Goodnough, W. Tepp, E. A. Johnson, and D. H. Rich. 2004. Synthesis of substrates and inhibitors of botulinum neurotoxin A metalloprotease. J. Peptide Res. 63:181-193. [DOI] [PubMed] [Google Scholar]
- 36.Vaidyanathan, V. V., K. Yoshino, M. Jahnz, C. Dôrries, S. Bade, S. Nauenburg, H. Niemann, and T. Binz. 1999. Proteolysis of SNAP-25 isoforms by botulinum neurotoxin types A, C and E: domains and amino acid residues controlling the formation of enzyme-substrate complexes and cleavage. J. Neurochem. 72:327-337. [DOI] [PubMed] [Google Scholar]
- 37.Washbourne, P., R. Pellizzari, G. Baldini, M. C. Wilson, and C. Montecucco. 1997. Botulinum neurotoxin types A and B require the SNARE motif in SNAP-25 for proteolysis. FEBS Lett. 418:1-5. [DOI] [PubMed] [Google Scholar]
- 38.Wictome, M., and C. C. Shone. 1998. Botulinum neurotoxins: mode of action and detection. J. Appl. Microbiol. 84:87S-97S. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.