Abstract
Adapted Pseudomonas putida strains grew in the presence of up to 6% (vol/vol) butanol, the highest reported butanol concentration tolerated by a microbe. P. putida might be an alternative host for biobutanol production, overcoming the primary limitation of currently used strains—insufficient product titers due to low butanol tolerance.
The focus of biofuel production research has recently shifted from ethanol to bioenergy carriers that are more compatible with existing infrastructure (e.g., refineries, transport, and car engines). At the forefront is n-butanol (hereafter referred to as butanol) for which large-scale production processes have been implemented (16, 35). Existing fermentations, however, are limited in energetically attractive butanol titers, because butanol inhibits microbial growth at concentrations above 16 g/liter (2, 10). As reported for other organic solvents with low logarithm of the partition coefficient in a two-phase octanol/water system (log Pow), this toxicity is due primarily to accumulation of butanol (log Pow, 0.8) in the cell membrane and subsequent impairment (4, 17, 30, 33). With the maximum aqueous solubility of 0.97 M (8.8% [vol/vol]), the maximum membrane concentration of butanol was calculated to be 1.59 M (17), spotlighting its potential toxicity. The low achievable butanol titers have necessitated large reactor volumes, resulting in high purification costs (8, 15). Recent metabolic engineering strategies for improving biobutanol fermentation have focused on maximization of butanol production rates (10, 19), reducing the levels of by-products (20), finding alternative substrates (20), or finding alternative hosts (2, 12, 21, 31). However, recently engineered microbial strains (1, 14) have not overcome butanol toxicity.
High organic solvent concentrations are tolerated by strains of the bacterial species Pseudomonas putida reported to grow in a second phase of octanol (25), toluene (13), or styrene (32). This suggests that solvent-tolerant P. putida strains withstand high butanol titers and therefore warrant exploitation as host for butanol production. Indeed, viable solvent-tolerant P. putida S12 cells were observed at butanol concentrations of up to 10% (vol/vol) by live-dead staining and fluorescence microscopy (5) (see supplemental material). We used growth as the parameter of interest, because growth in the presence of butanol directly indicates the potential of selected P. putida strains as hosts for recombinant butanol production.
Three solvent-tolerant P. putida strains, DOT-T1E (23), S12 (32), and Pseudomonas sp. strain VLB120 (18), and the solvent-sensitive P. putida reference strain KT2440 (24) were examined for their ability to grow in the presence of butanol. Toxicity assays were performed in 96-well microtiter plates (System Duetz [7]) at 30°C and 300 rpm using glucose-supplemented LB and M9 media (with 10 and 5 g/liter glucose, respectively) (26). Higher glucose concentrations in LB medium did not increase butanol tolerance (data not shown). Butanol was added in all experiments to cells in the mid-exponential phase. Cell growth was monitored by changes in optical density, and substrate and butanol concentrations were analyzed by high-pressure liquid chromatography (Trentec 308R-Gel.H; VWR Hitachi). Comparable low butanol concentrations were withstood by all P. putida strains, with butanol tolerance highly dependent on the medium composition (Table 1). Growth was observed at butanol concentrations up to 3% (vol/vol), occurring in a culture of Pseudomonas sp. strain VLB120 using glucose-supplemented LB medium.
TABLE 1.
Tolerated butanol concentrations in different growth media
| Pseudomonas strain and treatment or cell type | Maximum butanol concn [% (vol/vol)]a
|
||
|---|---|---|---|
| M9 minimal medium with glucose (5 g/liter) | LB medium | LB medium with glucose (10 g/liter) | |
| P. putida DOT-T1E | |||
| Untreated | 1.5 | 1.5-2.0 | 2.5 |
| Adapted | 1.0 (1.0) | 1.5 (2.0) | 6.0 (5.0) |
| P. putida KT2440 | |||
| Untreated | 1.0 | 1.5 | 2.0 |
| Treated | 1.0 (1.0) | 1.5 (1.0) | 1.5 (1.5) |
| P. putida S12 | |||
| Untreated | 1.5 | 2.0 | 2.5 |
| Adapted | 1.0 (1.0) | 1.5-2.0 (1.5) | 6.0 (5.0) |
| Pseudomonas sp. strain VLB120 | |||
| Untreated | 1.5 | 2.0 | 2.5-3.0 |
| Adapted | 1.0 (1.5) | 1.5-2.0 (1.5) | 6.0 (6.0) |
Values represent the maximum butanol concentration allowing growth (growth rate of ≥0.05 h−1). Data in parentheses were measured in experiments with cells that were stored at −80°C.
Because reported adaptation approaches (3, 17, 18, 32) were not successful (see supplemental material), a modified adaptation protocol was developed. Cells were incubated at 30°C on LB agar plates in an airtight desiccator with a butanol saturated gas phase. Colonies were repeatedly transferred every 2 days to new plates for at least 15 times. Cells that underwent this procedure, referred to as treated cells, were harvested and either stored at −80°C prior to testing or assessed immediately for tolerance to butanol (Fig. 1).
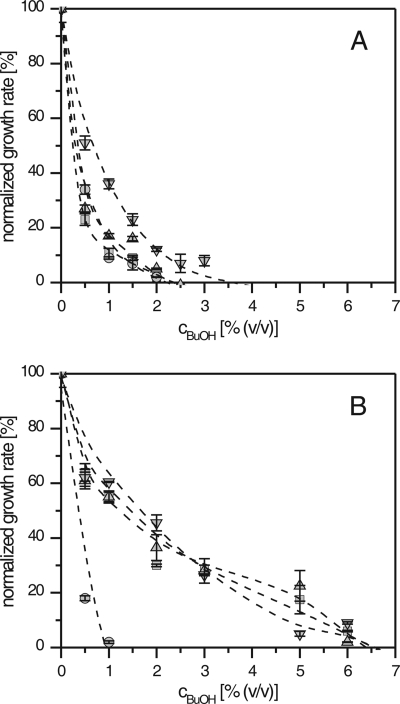
FIG. 1.
Butanol tolerance of P. putida. Growth rates of untreated (A) and adapted (B) cells in LB medium with 10 g/liter glucose as an additional energy and carbon source. The concentration of butanol (cBuOH) is shown on the x axis. The growth rates are normalized to the growth rate in the respective control experiments without butanol. Lines are drawn for better visualization. Error bars present standard deviations of independent experiments (n = 3 to 6). Symbols: ▪, P. putida DOT-T1E; •, P. putida KT2440; ▴, P. putida S12; ▾, Pseudomonas sp. strain VLB120.
The treated solvent-tolerant cells grew at rates above 0.05 h−1 (approximately 5% of the maximum growth rate without butanol) in the presence of up to 6% (vol/vol) butanol. Butanol concentrations in the medium decreased during the experiments due to evaporation (i.e., at a rate of 0.76 ± 0.03 mmol l−1 h−1) from an initial concentration of 5% (vol/vol) and, more significantly, due to consumption. Similar butanol uptake rates were observed for all four strains at 5% (vol/vol) initial butanol, ranging from 5.2 to 6.6 mmol l−1 h−1. Therefore, the butanol concentration decreased to only 3.5% (vol/vol) and 4% (vol/vol) after 9 h of cultivation in experiments at initial butanol concentrations of 5% (vol/vol) and 6% (vol/vol), respectively. This decrease resulted in an average butanol concentration of 4.5% (vol/vol) tolerated by the DOT-T1E, S12, and VLB120 cells. Notably, the time course of butanol concentration did not differ significantly with solvent-sensitive P. putida KT2440 that did not grow above 1.5% (vol/vol) butanol.
To rationalize the metabolic responses of untreated and treated strains to butanol, we performed 13C-labeled tracer-based flux analysis (3, 18, 27, 34), using minimal medium with 20% U-13C-labeled and 80% naturally labeled glucose, as reported recently (3, 6, 9). During growth without butanol, the four Pseudomonas strains had similar intracellular carbon flux distributions, independent of any prior adaptation to butanol (data not shown). In the presence of butanol, all untreated cells revealed significantly higher specific glucose uptake rates while growth rates decreased (Fig. 2). The reduced biomass yield was not caused by by-product formation (data not shown) but by changes in intracellular flux distribution: the carbon flux was rerouted from biomass synthesis to the tricarboxylic acid (TCA) cycle, which was fueled by pyruvate via pyruvate dehydrogenase and citrate synthase activity. The anaplerotic and gluconeogenic reactions were unaffected. The overall redox cofactor regeneration rates (approximately fourfold higher) resulting from this rerouting suggest that larger amounts of energy are demanded for cell maintenance during butanol stress, similar to the response of P. putida during growth in the presence of other organic solvents with low log Pow (22, 23, 28).
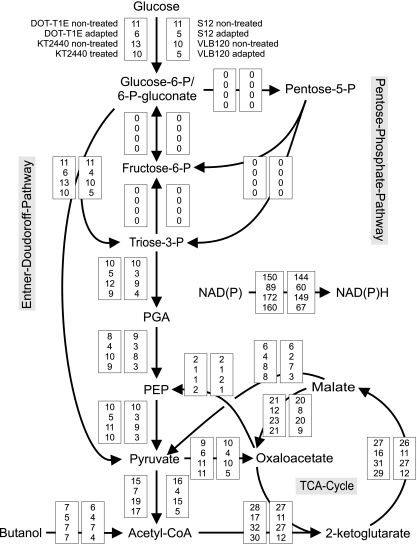
FIG. 2.
Flux distributions in P. putida under butanol stress conditions. The flux distributions in the P. putida strains DOT-T1E, KT2440, and S12 and Pseudomonas sp. strain VLB120 (from top to bottom) were determined during growth in glucose-containing M9 medium supplemented with 1% (vol/vol) butanol using untreated and adapted cells. Butanol catabolism was traced by the fractional labeling of central carbon metabolites (see text for details). The errors for all fluxes were below 10% with the exception of highly active or negligibly fluxes including PEP carboxykinase, pentose-phosphate-pathway (PPP), and phosphoglycoisomerase. The upper bound of the NAD(P)H regeneration rate is presented. Glucose-6-P, glucose-6-phosphate; PGA, 3-phosphoglycerate; PEP, phosphoenolpyruvate.
In contrast, physiology and flux distributions differed for adapted DOT-T1E, S12, and VLB120 cells, but not treated KT2440 cells. These strains, coping with high butanol concentrations, had low net glucose consumptions, resulting in comparably lower TCA cycle fluxes and consequently lower redox cofactor regeneration rates (Fig. 2). As indicated above (Fig. 1), P. putida KT2440 did not adapt to butanol, and no metabolic changes were observed compared with the untreated strain.
Coconsumption of butanol was considered in calculating the absolute intracellular fluxes by correcting the fractional labeling [FL = n13C/(n12C + n13C)] of the affected amino acids—aspartate, glutamine, isoleucine, leucine, and threonine. The dilution of the fractional isotope label due to butanol coconsumption decreased from acetyl coenzyme A (acetyl-CoA) (FL = 8%) to 2-ketoglutarate (FL = 13%) and oxaloacetate (FL = 15%), suggesting that butanol is cometabolized via β-oxidation to acetyl-CoA, followed by oxidation in the TCA cycle.
As calculated from the fractional label of the m-15 isotopomer of leucine (FL = 14%), approximately 60% of the acetyl-CoA originated from butanol. For example, in P. putida KT2440, butanol contributed to the synthesis of acetyl-CoA about 7.22 ± 0.23 mmol g−1 h−1, corresponding to the measured glucose uptake rate of 11.22 ± 0.74 mmol g−1 h−1 [(7.22/11.22) × 100 = 64%]. The untreated solvent-tolerant strains had slightly lower consumption rates of approximately 6.5 mmol g−1 h−1 for butanol and 10.2 mmol g−1 h−1 for glucose. Compared with the untreated strains, adapted DOT-T1E, S12, and VLB120 cells had lower uptake rates of 3.8 to 5.2 mmol g−1 h−1 for butanol and 4.9 to 6.6 mmol g−1 h−1 for glucose. Butanol did not contribute significantly to the synthesis of pyruvate (FL = 19%) and PEP (FL = 20%, or the contribution was below the FL detection limit of 0.5%), suggesting that malic enzyme and phosphoenolpyruvate (PEP) carboxykinase are marginally active under these conditions. This suggests that a synthetic pathway for butanol synthesis from glucose can be implemented in P. putida using native genes for butanol dehydrogenase and aldehyde dehydrogenase with a concomitant decrease of ß-oxidation activity.
Butanol degradation of P. putida KT2440 was comparable with the rates of solvent-tolerant cells, but butanol tolerance was not induced, suggesting activity of additional mechanisms of adaptation or tolerance, such as solvent removal by efflux pumps and physiochemical changes of membrane lipids (11, 22). These mechanisms reduce cellular growth rates and biomass yields by imposing higher energy demands. Additionally, energy loss can be caused by swelling and alteration of the lipid layer due to increased proton permeability of the membrane (4) and by reduced efficiency of the electron transport chain (30). In butanol-tolerant cells, the observed reduction in TCA cycle use and energy production in the presence of butanol suggests cell membrane adaptation by lowering its energy demands for maintenance.
The observed higher tolerance to butanol in LB medium compared with minimal medium can also be explained by decreased metabolic costs for sustaining biomass synthesis due to direct supply of biomass precursors like amino acids (29). Additional supplementation of LB medium with glucose enhanced butanol tolerance, most likely due to increased energy supplies. For P. putida S12, we calculated glucose uptake rates of 8.01 ± 0.21 mmol g−1 h−1 and 13.53 ± 0.34 mmol g−1 h−1 at initial butanol concentrations of 1% (vol/vol) and 3% (vol/vol), respectively, translating into an increased ATP regeneration rate at 3% (vol/vol) butanol of minimally 13.5 mmol g−1 h−1 (substrate phosphorylation via the Entner-Doudoroff pathway) and up to approximately 350 mmol g−1 h−1 (oxidative phosphorylation). The additional energy demand in the presence of butanol necessitates particular attention during strain and medium engineering.
We report solvent-tolerant P. putida strains growing at butanol concentrations as high as 6% (vol/vol). Metabolic flux analysis suggests that this is not based on glucose-butanol coconsumption but rather effected by lowered cell maintenance costs.
In conclusion, butanol-tolerant P. putida strains are promising candidates as production hosts, overcoming the principal limitation of biobutanol production—product inhibition at low concentrations.
Supplementary Material
Acknowledgments
We acknowledge financial support from Deutsche Bundesstiftung Umwelt (DBU).
Footnotes
Published ahead of print on 1 May 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Atsumi, S., A. F. Cann, M. R. Connor, C. R. Shen, K. M. Smith, M. P. Brynildsen, K. J. Y. Chou, T. Hanai, and J. C. Liao. 2008. Metabolic engineering of Escherichia coli for 1-butanol production. Metab. Eng. 10:305-311. [DOI] [PubMed] [Google Scholar]
- 2.Atsumi, S., T. Hanai, and J. C. Liao. 2008. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451:86-90. [DOI] [PubMed] [Google Scholar]
- 3.Blank, L. M., G. Ionidis, B. E. Ebert, B. Bühler, and A. Schmid. 2008. Metabolic response of Pseudomonas putida during redox biocatalysis in the presence of a second octanol phase. FEBS J. 275:5173-5190. [DOI] [PubMed] [Google Scholar]
- 4.de Bont, J. A. M. 1998. Solvent-tolerant bacteria in biocatalysis. Trends Biotechnol. 16:493-499. [Google Scholar]
- 5.de Carvalho, C. C. C. R., A. A. R. L. da Cruz, M.-N. Pons, H. M. R. V. Pinheiro, J. M. S. Cabral, M. M. R. da Fonseca, B. S. Ferreira, and P. Fernandes. 2004. Mycobacterium sp., Rhodococcus erythropolis, and Pseudomonas putida behavior in the presence of organic solvents. Microsc. Res. Tech. 64:215-222. [DOI] [PubMed] [Google Scholar]
- 6.del Castillo, T., J. L. Ramos, J. J. Rodríguez-Herva, T. Fuhrer, U. Sauer, and E. Duque. 2007. Convergent peripheral pathways catalyze initial glucose catabolism in Pseudomonas putida: genomic and flux analysis. J. Bacteriol. 189:5142-5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duetz, W. A. 2007. Microtiter plates as mini-bioreactors: miniaturization of fermentation methods. Trends Microbiol. 15:469-475. [DOI] [PubMed] [Google Scholar]
- 8.Ezeji, T. C., N. Qureshi, and H. P. Blaschek. 2004. Butanol fermentation research: upstream and downstream manipulations. Chem. Rec. 4:305-314. [DOI] [PubMed] [Google Scholar]
- 9.Fuhrer, T., E. Fischer, and U. Sauer. 2005. Experimental identification and quantification of glucose metabolism in seven bacterial species. J. Bacteriol. 187:1581-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris, L. M., L. M. Blank, R. P. Desai, N. E. Welker, and E. T. Papoutsakis. 2001. Fermentation characterization and flux analysis of recombinant strains of Clostridium acetobutylicum with an inactivated solR gene. J. Ind. Microbiol. Biotechnol. 27:322-328. [DOI] [PubMed] [Google Scholar]
- 11.Heipieper, H. J., B. Loffeld, H. Keweloh, and J. A. M. de Bont. 1995. The cis/trans isomerization of unsaturated fatty acids in Pseudomonas putida S12—an indicator for environmental stress due to organic compounds. Chemosphere 30:1041-1051. [Google Scholar]
- 12.Husted, G. R., J. D. Santangelo, and D. W. Bostwick. October 1988. Method for producing butanol by fermentation. U.S. patent 4777135.
- 13.Inoue, A., and K. Horikoshi. 1989. A Pseudomonas thrives in high concentrations of toluene. Nature 338:264-266. [Google Scholar]
- 14.Inui, M., M. Suda, S. Kimura, K. Yasuda, H. Suzuki, H. Toda, S. Yamamoto, S. Okino, N. Suzuki, and H. Yukawa. 2008. Expression of Clostridium acetobutylicum butanol synthetic genes in Escherichia coli. Appl. Microbiol. Biotechnol. 77:1305-1316. [DOI] [PubMed] [Google Scholar]
- 15.Jones, D. T., and D. R. Woods. 1986. Acetone-butanol fermentation revisited. Microbiol. Rev. 50:484-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, S. Y., J. H. Park, S. H. Jang, L. K. Nielsen, J. Kim, and K. S. Jung. 2008. Fermentative butanol production by clostridia. Biotechnol. Bioeng. 101:209-228. [DOI] [PubMed] [Google Scholar]
- 17.Neumann, G., N. Kabelitz, A. Zehnsdorf, A. Miltner, H. Lippold, D. Meyer, A. Schmid, and H. J. Heipieper. 2005. Prediction of the adaptability of Pseudomonas putida DOT-T1E to a second phase of a solvent for economically sound two-phase biotransformations. Appl. Environ. Microbiol. 71:6606-6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park, J. B., B. Buhler, S. Panke, B. Witholt, and A. Schmid. 2007. Carbon metabolism and product inhibition determine the epoxidation efficiency of solvent-tolerant Pseudomonas sp. strain VLB120 Delta C. Biotechnol. Bioeng. 98:1219-1229. [DOI] [PubMed] [Google Scholar]
- 19.Qureshi, N., and H. P. Blaschek. 1999. Production of acetone butanol ethanol (ABE) by a hyper-producing mutant strain of Clostridium beijerinckii BA101 and recovery by pervaporation. Biotechnol. Prog. 15:594-602. [DOI] [PubMed] [Google Scholar]
- 20.Qureshi, N., and H. P. Blaschek. 2001. Recent advances in ABE fermentation: hyper-butanol producing Clostridium beijerinckii BA101. J. Ind. Microbiol. Biotechnol. 27:287-291. [DOI] [PubMed] [Google Scholar]
- 21.Raamsdonk, L. M., W. T. A. M. de Laat, and M. A. van de Berg. October 2007. Butanol production in a eukaryotic cell. The Netherlands patent WO/2008/052991.
- 22.Ramos, J. L., E. Duque, M. T. Gallegos, P. Godoy, M. I. Ramos-Gonzalez, A. Rojas, W. Teran, and A. Segura. 2002. Mechanisms of solvent tolerance in gram-negative bacteria. Annu. Rev. Microbiol. 56:743-768. [DOI] [PubMed] [Google Scholar]
- 23.Ramos, J. L., E. Duque, P. Godoy, and A. Segura. 1998. Efflux pumps involved in toluene tolerance in Pseudomonas putida DOT-T1E. J. Bacteriol. 180:3323-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramos-Díaz, M. A., and J. L. Ramos. 1998. Combined physical and genetic map of the Pseudomonas putida KT2440 chromosome. J. Bacteriol. 180:6352-6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rojas, A., E. Duque, A. Schmid, A. Hurtado, J. L. Ramos, and A. Segura. 2004. Biotransformation in double-phase systems: physiological responses of Pseudomonas putida DOT-T1E to a double phase made of aliphatic alcohols and biosynthesis of substituted catechols. Appl. Environ. Microbiol. 70:3637-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 27.Sauer, U., D. R. Lasko, J. Fiaux, M. Hochuli, R. Glaser, T. Szyperski, K. Wütherich, and J. E. Bailey. 1999. Metabolic flux ratio analysis of genetic and environmental modulations of Escherichia coli central carbon metabolism. J. Bacteriol. 181:6679-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Segura, A., P. Godoy, P. van Dillewijn, A. Hurtado, N. Arroyo, S. Santacruz, and J. L. Ramos. 2005. Proteomic analysis reveals the participation of energy- and stress-related proteins in the response of Pseudomonas putida DOT-T1E to toluene. J. Bacteriol. 187:5937-5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sezonov, G., D. Joseleau-Petit, and R. D'Ari. 2007. Escherichia coli physiology in Luria-Bertani broth. J. Bacteriol. 189:8746-8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sikkema, J., J. A. M. de Bont, and B. Poolman. 1994. Interactions of cyclic hydrocarbons with biological membranes. J. Biol. Chem. 269:8022-8028. [PubMed] [Google Scholar]
- 31.Steen, E. J., R. Chan, N. Prasad, S. Myers, C. J. Petzold, A. Redding, M. Ouellet, and J. D. Keasling. 2008. Metabolic engineering of Saccharomyces cerevisiae for the production of n-butanol. Microb. Cell Fact. 7:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weber, F. J., L. P. Ooijkaas, R. M. W. Schemen, S. Hartmans, and J. A. M. de Bont. 1993. Adaptation of Pseudomonas putida S12 to high concentrations of styrene and other organic solvents. Appl. Environ. Microbiol. 59:3502-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weber, J., and J. A. M. de Bont. 1996. Adaptation mechanism of microorganisms to the toxic effects of organic solvents on membranes. Biochim. Biophys. Acta 1286:225-245. [DOI] [PubMed] [Google Scholar]
- 34.Zamboni, N., E. Fischer, and U. Sauer. 2005. FiatFlux—a software for metabolic flux analysis from 13C-glucose experiments. BMC Bioinformatics 6:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zverlov, V. V., O. Berezina, G. A. Velikodvorskaya, and W. H. Schwarz. 2006. Bacterial acetone and butanol production by industrial fermentation in the Soviet Union: use of hydrolyzed agricultural waste for biorefinery. Appl. Microbiol. Biotechnol. 71:587-597. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.