Abstract
Staphylococcus aureus is a gram-positive pathogen that causes a variety of diseases, including bovine mastitis, which has severe economic consequences. Standard antibiotic treatment results in selection of resistant strains, leading to a need for alternative treatments, such as bacteriophage therapy. Forty-nine S. aureus isolates were obtained from the milk of mastitic cows for use in screening of staphylococcal phages. Fifteen isolates which were positive for both coagulase and hemolysin were assayed by PCR for variation in the X region and the immunoglobulin G-binding region of the protein A gene (spa) and in the carboxy terminus of the coagulase gene (coa) and for the presence of enterotoxin C, G, H, and I genes. The host ranges of 52 phages isolated from sewage influent were determined by performing spot tests with the 15 S. aureus isolates, and two phages were subsequently chosen for further analysis. ΦSA039 had the widest host range, producing clear plaques on 13 of the 15 isolates (87%), while ΦSA012 produced clear plaques on 8 isolates (53%) and was the only phage that produced a clear plaque on a nonmastitic S. aureus strain. Transmission electron microscopy revealed that the phages were similar sizes and belonged to the Myoviridae family. Measurement of optical densities during coculture with S. aureus isolates confirmed the breadth of the ΦSA039 host range and showed that ΦSA012 had potent lytic capability. ΦSA012-resistant bacteria did not appear for three of seven isolates tested (43%) after 65 h of incubation. These two phages are proposed as candidates for phage therapy of bovine mastitis.
In the dairy industry, mastitis is a widespread problem responsible for important decreases in milk production. Economic losses of $100 million per year have been estimated for farms in Hokkaido, one of the largest milk-producing areas in Japan (28). Mastitis can be caused by over 150 different microorganisms, and one of the most important of these organisms is Staphylococcus aureus (22). After diagnosis of mastitis, the standard treatment regimen consists of isolating the diseased cow and treating it with antibiotics. However, this approach has drawbacks, such as its high cost and the eradication of harmless or beneficial organisms due to the lack of specificity of antibiotics. Additionally, the incidence of antibiotic-resistant bacteria has increased in recent years (4). As a result, there has been renewed interest in the use of other natural or engineered antimicrobial agents as an alternative or supplementary treatment for staphylococcal diseases such as mastitis (11, 21, 26). One group of alternatives with great potential involves bacteriophages (phages) and their derivatives, and a number of promising candidates have been described (2, 5, 7, 13, 17, 18, 27), notably bacteriophage K.
One of the main obstacles to successful treatment of mastitis using phages is the fact that most phages are able to infect only a very narrow range of hosts. Given the plural etiology of many mastitis cases, it is desirable to find a phage-based treatment that targets a wide range of pathogens. One proposed method is a combination of two or more phages with different and broad host ranges in a cocktail (25, 29). This method has been shown to delay considerably the appearance of phage-resistant cells in Escherichia coli cultures, providing hope that similar success can be achieved using S. aureus phages.
In this paper we describe the isolation of two novel phages, designated ΦSA012 and ΦSA039, that were found to have a lytic effect on a broad range of S. aureus isolates obtained from mastitic milk. ΦSA012 and ΦSA039 were selected after host range analysis from a total of 52 phages screened from sewage influent. These two phages were characterized morphologically by using electron microscopy, and their effects on representative strains of S. aureus were examined by coculture in milk.
MATERIALS AND METHODS
S. aureus strains.
Forty-five distinct raw milk samples from cows with mastitis were provided by Rakuno Gakuen University in Hokkaido. Staphylococcus was identified as the main causative agent by standard culture-based and biochemical procedures on site. All samples were collected in sterile tubes, kept on ice, and then stored at −20°C during transport to the laboratory where an analysis was performed. The milk was diluted when necessary, and 100 μl was plated on brain heart agar. Colonies were assayed for coagulase activity on mannitol salt agar plates and for hemolysin activity on sheep blood agar plates. Fifteen isolates with a double-positive phenotype were characterized genetically. JCM 2151 (= ATCC 6538) was used as an S. aureus reference strain.
S. aureus characterization.
The isolates were characterized by PCR amplification of the immunoglobulin G (IgG)-binding region and the hypervariable X region of the protein A gene (spa) and the carboxy terminus of the coagulase gene (coa). Additionally, strains were checked for possession of the genes for enterotoxins C, G, H, and I. The primers used were obtained from previous studies, as shown in Table 1 (10, 19, 20). The genomic DNA used as a template was obtained from overnight cultures by phenol-chloroform extraction. When necessary, an additional homogenization step was performed using a glass plunger and tube.
TABLE 1.
Oligonucleotide primers used for PCR amplification of S. aureus-specific genes or gene segments
| Primer | Direction | Sequence (5′-3′) | Reference |
|---|---|---|---|
| Coagulase C terminus primers | |||
| coa-F1 | Forward | ATAGAGATGCTGGTACAGG | 15 |
| coa-R1 | Reverse | GCTTCCGATTGTTCGATGC | 15 |
| Protein A segment primers | |||
| SpaX-F1 | Forward | GACGATCCTTCGGTGAGC | 13 |
| SpaX-R1 | Reverse | CAGCAGTAGTGCCGTTTGC | 13 |
| SpaIgG-F1 | Forward | CACCTGCTGCAAATGCTGCG | 15 |
| SpaIgG-R1 | Reverse | GGCTTGTTGTTGTCTTCCTC | 15 |
| Enterotoxin gene primers | |||
| SEC-3 | Forward | CTCAAGAACTAGACATAAAAGCTAGG | |
| SEC-4 | Reverse | TCAAAATCGGATTAACATTATCC | 14 |
| SEG-1 | Forward | AAGTAGACATTTTTGGCGTTCC | 14 |
| SEG-2 | Reverse | AGAACCATCAAACTCGTATAGC | 14 |
| SEH-1 | Forward | GTCTATATGGAGGTACAACACT | 14 |
| SEH-2 | Reverse | GACCTTTACTTATTTCGCTGTC | 14 |
| SEI-1 | Forward | GGTGATATTGGTGTAGGTAAC | 14 |
| SEI-2 | Reverse | ATCCATATTCTTTGCCTTTACCAG | 14 |
All PCR programs were begun with a 5-min denaturation step at 94°C and ended with a 20-min extension step at 72°C. The programs were adjusted according to the target sequence, as follows: for coa, 30 cycles of 40 s at 94°C, 60 s at 58 or 62°C, and 60 s at 72°C; for the spa IgG-binding region, 30 cycles of 40 s at 94°C, 40 s at 60°C, and 40 s at 72°C; for the spa X region, 35 cycles of 60 s at 94°C, 60 s at 60°C, and 60 s at 72°C; and for the staphylococcal enterotoxin genes, 30 cycles of 30 s at 94°C, 60 s at 58°C, and 30 s at 72°C. Amplicons were visualized on a 2% agarose gel and sequenced using a BigDye Terminator version 3.1 kit (ABI, Japan) and an ABI Prism 3130 genetic analyzer (Applied Biosystems, Japan).
Bacteriophage isolation and purification.
Influent collected from a municipal wastewater treatment plant in Tokyo, Japan, was the source for screening phages capable of lysing S. aureus by the method described previously for isolating E. coli phages (24). Phage strains were purified by repeatedly plating and picking individual plaques and were concentrated by plate lysate and polyethylene glycol 6000-NaCl centrifugation.
Determination of phage host range (spot test).
The host range of each phage strain was determined using individual strains of S. aureus isolated from mastitic milk. Two microliters of concentrated phage lysate (>108 PFU/ml) was dropped onto an Luria-Bertani (LB) medium plate overlaid with S. aureus mixed with 0.5% top agar and incubated overnight. The lytic ability of phage isolates was assessed using the clarity of plaques, which was scored as clear, turbid or faint.
TEM imaging of phages.
Phages were purified from polyethylene glycol 6000-NaCl-concentrated lysate by CsCl step centrifugation (step densities, 1.13, 1.22, 1.29, 1.38, 1.46, and 1.55 g/ml). Six microliters of a concentrated phage suspension (minimum concentration, 1010 PFU/ml) in SM buffer (100 mM NaCl, 8 mM MgSO4, 50 mM Tris-HCl [pH 7.5], 0.01% gelatin) was spotted on top of either a hydrophilic Formvar-carbon-coated copper grid or a collodion-carbon-coated copper grid (Nissin EM Corporation), and the phages were allowed to adsorb for 2 min. Excess sample was removed by carefully touching the side of a grid with filter paper; then 6 μl of distilled water was spotted on the grid and removed after a short time. Phages were stained by addition of 6 μl of 1.6% uranyl acetate. After 2 min, excess stain was removed, and the grid was allowed to air dry for 30 min. The grids were observed with an Hitachi H7500 transmission electron microscope (TEM) operating at 80 kV.
Average phage dimensions were calculated by measuring the head diameter, head length, tail diameter, and tail length of 25 phages of each type, and the results were compared to measurements for a T4 phage whose dimensions are known.
Coculture of phage and S. aureus.
The effects of phages on representative strains of S. aureus in LB medium were observed by measuring the optical density at 660 nm (OD660) with a TVS062CA biophotorecorder (Advantec, Tokyo, Japan). Four milliliters of LB broth in an L-shaped glass test tube was inoculated (1%) with an overnight S. aureus culture and incubated at 37°C with shaking at 40 rpm until early logarithmic phase (OD660, 0.1) was reached. Phages were added at a multiplicity of infection of 10 in single-phage experiments and at an multiplicity of infection of 5 for each phage in experiments in which both phages were added. The OD660 of each culture was monitored at 15-min intervals for a minimum of 65 h following phage addition.
Nucleotide sequence accession numbers.
The sequences of the spa X regions have been deposited in the GenBank database under accession numbers FJ463840 to FJ463854.
RESULTS
Variation of S. aureus in mastitic milk samples.
In order to find suitable hosts for screening staphylococcal phages, 49 isolates of S. aureus were obtained from 45 distinct samples of milk taken from cows with mastitis. The concentration of bacteria in the milk varied greatly, from 101 CFU per ml to as much as 108 CFU per ml. Fifteen isolates which were positive for both coagulase and hemolysin were characterized genetically by PCR using primers (Table 1) for the carboxy terminus of the coagulase gene (coa), the X region of the protein A gene (spa), the IgG-binding domain of the spa gene, and the genes for staphylococcal enterotoxins C, G, E, and I (Table 2).
TABLE 2.
Characterization of S. aureus isolates obtained from mastitic cow's milk
| S. aureus isolate | Concn in original milk sample (CFU/ml) |
spa X region
|
Size of coa (bp)a | Enterotoxin genesb | |
|---|---|---|---|---|---|
| Size (bp) | No. of repeat motifs | ||||
| SA001 | NDc | 295 | 6 | 1,079 | None |
| SA002 | ND | 295 | 6 | 1,079 | None |
| SA003 | 104 | 295 | 6 | 836 | None |
| SA009 | 103 | 295 | 6 | 836 | None |
| SA019 | 4.6 × 108 | 199 | 2 | 593 | SEC, SEG, SEI |
| SA020 | 3.1 × 102 | 199 | 2 | 593 | SEG, SEI |
| SA021 | 4.8 × 105 | 199 | 2 | 836 | SEG, SEI |
| SA026 | 1.72 × 103 | 271 | 5 | 674 | SEC, SEG, SEI |
| SA028 | 2 × 105 | 199 | 2 | 593 | None |
| SA029 | 4.3 × 104 | 199 | 2 | 674 | None |
| SA031 | 2 × 105 | 343 | 8 | 593 | None |
| SA033 | 3 × 105 | 391 | 10 | 593 | None |
| SA047 | >103 | 391 | 10 | 593 | None |
| SA048 | >106 | 271 | 5 | 836 | None |
| SA049 | 101 | 367 | 9 | 350 | None |
| ATCC 6538 | ND | 367 | 9 | 925 | None |
The sizes of amplicons were determined using the spaX-F1/spaX-R1 and coa-F1/coa-R1 primer pairs.
SEC, SEG, and SEI, staphylococcal enterotoxins C, G, and I, respectively.
ND, no data obtained.
Amplification of the IgG-binding domain of the spa gene produced amplicons that were the same size (approximately 900 bp [data not shown]) for all isolates. In contrast, six spa X region amplicon sizes were observed: 199 bp, 271 bp, 295 bp, 343 bp, 367 bp, and 391 bp. Correspondingly, the number of 24-bp (8-amino-acid) repeats varied from 2 to 10; these repeats had either or both of two repeats with the amino acid sequence KPGKEDNK or KPGKEDGN. Five sizes of coa amplicons were detected: 350 bp, 593 bp, 674 bp, 836 bp, and 1079 bp. As the C terminus of coa contains a variable number of 81-bp repeats, it was possible to estimate the sizes relatively reliably. Four isolates (SA019, SA020, SA021, and SA026) were found to be positive for the enterotoxin G and I genes, whereas only SA019 and SA026 possessed enterotoxin C genes.
Isolation of lytic phages.
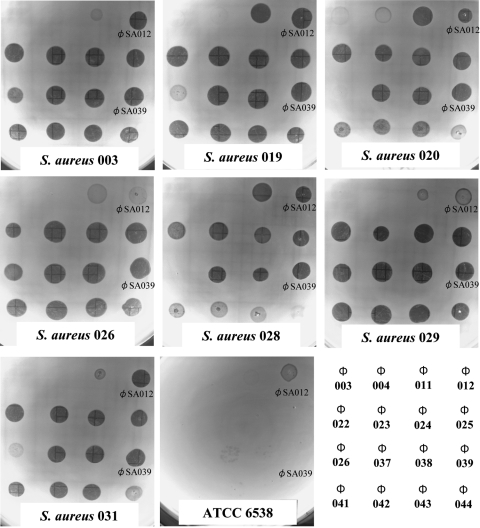
Screening of the influent of a municipal wastewater treatment plant yielded phages with various degrees of lytic activity against S. aureus. In an effort to determine the most promising candidates for lysis of the widest range of mastitis-related S. aureus strains, each of the 52 phage types was spotted onto lawns of the 15 S. aureus isolates described above and a reference strain, ATCC 6538. Sixteen phages were capable of forming some plaques on more than four isolates. Figure 1 shows the results of representative spot tests with seven isolates and ATCC 6538, and Fig. 2 summarizes the host ranges of these phages. Clear, turbid, and faint plaques indicated complete lysis, partial lysis, and minimal lysis, respectively. For 14 mastitis-related S. aureus isolates clear plaques were produced by more than one kind of phage. None of the phages were able to produce clear plaques on mastitis-related isolate SA001, although some phages did produce turbid plaques. One phage, designated ΦSA012, produced the clearest plaque on ATCC 6538.
FIG. 1.
Spot tests for the 16 phages with the widest host ranges and representative S. aureus lawns in 0.5% agar LB medium. The host bacterial strain is indicated at the bottom of each plate. A key for the phage spots is shown in the bottom right panel.
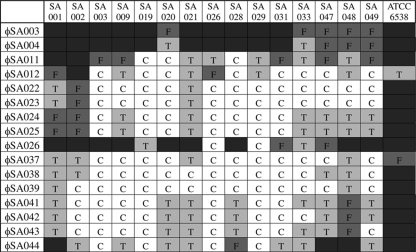
FIG. 2.
Host ranges of S. aureus phages. Host bacterial strains are indicated at the top, and phage isolates are indicated on the left. The extent of lysis is indicated by both letters and colors, as follows: C and white, clear plaques; T and light gray, turbid plaques; F and darker gray, faint plaques; dark gray with no letter, no plaques.
The two most important criteria for further narrowing down the candidates for phage therapy were the host range and the lytic ability, as judged by the relative clarity of plaques. Two of the 52 phages, ΦSA012 and ΦSA039, were found to be the most promising candidates. ΦSA012 produced clear plaques on 8 of 15 mastitis-related S. aureus isolates (53%) and less clear plaques on another 6 isolates and on ATCC 6538, but it produced no plaques on SA002. ΦSA039 produced clear plaques on 13 of the 15 mastitic isolates (87%) and turbid plaques on the 2 remaining isolates, but it did not produce plaques on ATCC 6538. These two phages were used for further analysis.
Morphological characterization and classification of phages.
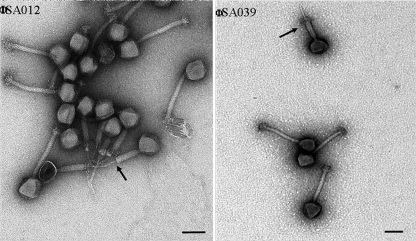
Concentrated and purified solutions of phages ΦSA012 and ΦSA039 were examined with a TEM (Fig. 3). A number of shared features were observed: an isometric capsid whose diameter slightly exceeded its length and a long, narrow tail approximately 2.5 times longer than the capsid (Table 3), as well as the presence of both a sheath and a baseplate. No long tail fibers were observed for either type. Instances of sheath contraction were observed for both ΦSA012 and ΦSA039.
FIG. 3.
TEM images of bacteriophages. Bar, 100 nm. The arrows indicate contracted sheaths.
TABLE 3.
Dimensions of bacteriophagesa
| Bacteriophage | Head diam (nm) (avg ± SD) | Head length (nm) (avg ± SD) | Tail diam (nm) (avg ± SD) | Tail length (nm) (avg ± SD) | Family |
|---|---|---|---|---|---|
| ΦSA012 | 97 ± 7 | 93 ± 9 | 23 ± 3 | 237 ± 19 | Myoviridae |
| ΦSA039 | 95 ± 14 | 93 ± 13 | 22 ± 4 | 240 ± 31 | Myoviridae |
The length of a T4 phage measured under the same conditions was found to exceed the real length by 1%.
Coculture of phages and S. aureus.
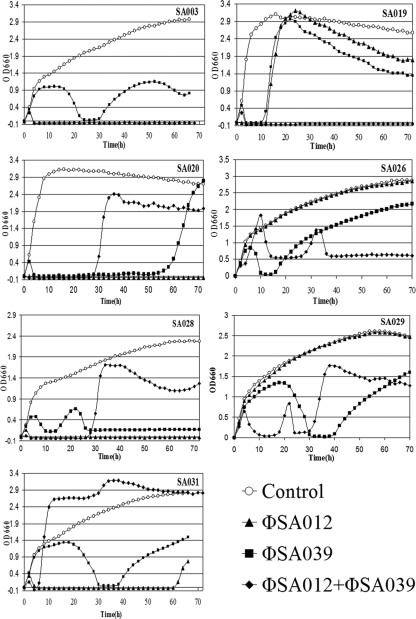
The abilities of ΦSA012 and ΦSA039 to lyse 7 of the 15 coagulase- and hemolysin-posititve S. aureus isolates obtained from mastitic cow milk were tested using liquid LB medium cultures (Fig. 4). The two phages were cultured both separately and together with each bacterial isolate. Three isolates that tested positive for enterotoxin genes, SA019, SA020, and SA026, and four additional isolates, SA003, SA028, SA029, and SA031, were used as host bacteria. The turbidity of each culture was measured by determining the OD660.
FIG. 4.
Coculture of bacteriophages and S. aureus in LB medium. The bacterial strain is indicated in each graph. ○, bacterium-only controls; ▴, ΦSA012 added to bacteria; ▪, ΦSA039 added to bacteria; ⧫, both ΦSA012 and ΦSA039 added to bacteria.
Coculture of ΦSA012 with five of the seven S. aureus isolates resulted in a decrease in the absorbance, whereas the OD660 plots for the SA026 and SA029 cocultures were nearly identical to those for the bacterium-only controls. Coculture with SA019 resulted in an immediate decline followed by a rise in the OD660 after 12 h. The OD660 started to decrease gradually again after 25 h and continued to decrease until the experiment was stopped. Coculture with SA031 resulted in a decrease in the OD660 after 3 h to nearly zero, and then it started to recover after 60 h. During incubation with SA003, SA020, and SA028 the OD660 decreased to nearly zero very soon after inoculation and remained constant with no recovery until the experiments were stopped after more than 65 h.
Coculture of ΦSA039 with the seven S. aureus strains resulted in decreases in absorbance in all cases which exhibited three distinct patterns. The first and most common pattern (SA003, SA026, SA029, and SA031) was an initial increase in the OD660 for 5 to 20 h depending on the strain before the OD660 started to decrease until it was nearly zero. After 10 h or less, the absorbance started to increase gradually again until it reached or surpassed the previous peak, but the level was never as high as the level for the bacterium-only control. In the case of SA003, there was another slight decrease after 52 h. The second pattern, observed for cocultures with SA019 and SA020, was a nearly immediate decrease in the OD660 to almost zero, followed by recovery to the same levels as those of the bacterium-only control. The recovery occurred early for SA019, starting after approximately 11 h, and was followed by a gradual decline after the OD660 peaked at around 22 h. For SA020 the recovery was delayed until after 54 h, and no subsequent decrease was observed. The last pattern, observed with SA028, consisted of a decline in the OD660 starting after 6 h until recovery began at 14 h. This was followed by another decrease before the value stabilized at an average of 0.17 absorbance unit.
Simultaneous coculture of both phages with each S. aureus isolate resulted in three broad patterns. The first pattern, observed with SA003 and SA019, included a decrease to a value below zero immediately after phage addition, from which the bacteria did not recover in the time span of the experiment. The second pattern, observed with SA020, SA028, and SA031, was characterized by an initial decrease in absorbance, followed by an eventual recovery, within 30 h in the case of SA020 and SA028 and within 5 h in the case of SA031. The third pattern, observed with SA026 and SA029, had a double peak and culminated in an absorbance value lower than the value that either phage produced individually by the end of the experiment.
DISCUSSION
This paper describes the isolation from sewage influent of bacteriophages which exhibited lytic capabilities with a range of S. aureus isolates from mastitic cow milk. In previous studies workers have isolated phages from sewage with the ultimate aim of controlling S. aureus-caused disease (27), and other workers have tested phage therapy candidates with S. aureus isolated from mastitic milk (7). However, the renowned specificity of the interaction of a phage with its bacterial host means that the chances of successful treatment can be greatly increased by having multiple phages with strong lytic capabilities and wide host ranges that complement those of other phages. Isolation of novel phages by screening various sources with hosts different than those used previously is thus essential.
Selection of phage hosts among S. aureus isolates.
In our effort to isolate novel staphylococcal phages, the first step was to isolate a variety of hosts that were both virulent and genetically diverse. The source of the bacteria was logically determined based on the target of the eventual phage therapy, i.e., cows with staphylococcal mastitis. Forty-nine isolates were narrowed down to 15 isolates and finally to 7 isolates with the aim of testing phages with a practical number of S. aureus hosts that represented the most virulent and varied isolates available. The 15 isolates used to test the host ranges of phages were chosen because they produced both coagulase and hemolysin and were thus thought to have the greatest potential virulence. Genetic characterization was performed in order to confirm that the bacteria were indeed S. aureus and to ascertain their diversity. PCR-based assays of the IgG-binding region and the hypervariable X region of the protein A gene spa and the carboxy terminus of the coagulase gene coa are established methods for molecular typing of S. aureus isolates (1, 6, 8, 10, 19, 23). All three of these regions contain tandem repeats that differ in sequence and number.
The sizes of the amplicons produced by cloning the spa IgG-binding domain were the same (900 bp) for all 15 isolates tested, and this was consistent with previous reports (1, 15). The homogeneous size indicated that there was no variation in the number of repeats, which was not unexpected as relatively high levels of conservation are common in regions that are functionally important. However, it is possible there was some variation in the sequence of the 58-amino-acid repeat.
On the other hand, substantially different sizes were observed for the coa and spa X regions, as determined by PCR, which is also consistent with previous reports. However, further examination of the spa X repeats revealed little diversity in the sequences, although, as expected, the number of copies varied greatly in proportion to the amplicon length. Although it is not clear whether the differences in the spa and coa repeats truly reflect the overall diversity of S. aureus strains, we used them as the best available indicators to decrease the number of isolates used in coculture experiments.
Enterotoxin genes were detected in 4 of the 15 isolates examined (27%), indicating that some bacteria that cause mastitis could also cause food poisoning. The detection of enterotoxin C in two isolates (13%) was of particular interest, as this toxin has been linked to the severity of mastitis symptoms in other studies (9).
Host ranges and morphological characterization of phages.
Once suitable S. aureus hosts had been isolated, the next step was to screen bacteriophages and assess their lytic abilities and host ranges. Sewage influent was chosen as the phage source because it has yielded a wide diversity of phages in the past (14). Fifty-two staphylococcal phages were isolated from samples over the course of 1 year. The phages were tested using lawns of the 15 S. aureus isolates, and the clarity of plaques was observed. ΦSA039 exhibited a surprising ability to produce plaques on all mastitis-related hosts tested, although the plaques were turbid in the case of SA001 and SA048. However, no phage produced a clear plaque on SA001 or ATCC 6538. The latter strain was isolated from a human lesion and may therefore be considerably genetically and morphologically different than the mastitic strains. ΦSA012 was the phage which had the greatest lytic effect on ATCC 6538. For this reason, and because preliminary coculture experiments showed that this phage lysed certain mastitis-related isolates, it was chosen to complement ΦSA039 as a candidate for phage therapy.
TEM observation of the two phages revealed that they had similar tail lengths (more than twice the length of the T4 tail [12]), similar tail diameters, and heads that were similar sizes and whose diameter and length were nearly the same. Observation of sheath contraction led us to classify both ΦSA012 and ΦSA039 as members of the Myoviridae family according to the guidelines of the International Committee on Taxonomy of Viruses. Further TEM observation supplemented by genome sequencing will be undertaken in order to better characterize both phages.
Effects of phages on bacteria and prospects for phage therapy.
In order to assess the effects of ΦSA012 and ΦSA039 on S. aureus in a liquid medium over time, each phage was cocultured with 7 of the 15 coagulase- and hemolysin-positive S. aureus isolates. SA019, SA020, and SA026 were chosen because they possessed enterotoxin genes and may therefore be relevant to applications other than bovine mastitis-related applications. SA003, SA028, SA029, and SA031 were selected by taking into account their host ranges and the differences in their coa and spa X region amplicon sizes and repeat patterns. A decline in the OD660 was considered to be due to the lytic activity of a phage on its host.
The patterns of lysis observed in liquid culture confirmed the results obtained using solid LB medium since ΦSA039 successfully lysed all of the host isolates and ΦSA012 lysed all of the host isolates except the two isolates on which it had produced only turbid plaques in solid medium, SA026 and SA029. Monitoring of the OD660 of S. aureus isolates inoculated with ΦSA039 resulted in the expected pattern, an initial increase followed by a steady decrease as the phage began to lyse its host and then by eventual recovery as the number of phage-resistant bacteria increased. A subsequent decline in the OD660 was considered to be due to the reduction in the nutrients available as the OD660 of the bacterium-only control also decreased. These results confirmed the ability of ΦSA039 to infect a wide range of mastitis-associated S. aureus isolates in solid medium. Given the fact that a narrow host range, which is typical of most phages, is one of the biggest obstacles to therapy, these findings underlined the considerable potential for in vivo use of ΦSA039 with S. aureus.
The dynamic lytic effect of ΦSA012 on four of the five bacteria that it infected successfully was remarkable. In all cases the bacterial levels plummeted almost immediately, and three of the isolates (SA003, SA020, and SA028) never recovered, while SA031 recovered only slightly and belatedly. This ability to delay and perhaps even prevent the appearance of resistant cells was a demonstration of a potency that is very promising for the eventual use of ΦSA012 in bovine mastitis therapy and possibly other applications.
It was hoped that adding ΦSA012 and ΦSA039 simultaneously to S. aureus cultures would lead to a combination of the two phages' effects which suppressed bacterial growth, as determined by the turbidity of the culture. However, in three of seven cases (SA020, SA028, and SA030), the turbidity was greater after 30 h than it was when either phage was added individually. In three other cases (SA019, SA026, and SA029) the combined phages suppressed bacterial growth better than the individual phages. A further important observation was that the reproducibility of this experiment was low, even when identical starting conditions were used. This phenomenon was ascribed to the selection of different types of cells possessing different resistance mechanisms. Investigation of this possibility is a research topic in its own right, and the mechanisms will hopefully be elucidated in the near future. The creation of a broad-host-range, potently lytic phage cocktail remains a tangible objective but requires more detailed analysis of phage-S. aureus host interactions.
This study was limited to an in vitro analysis of the lytic effects of ΦSA012 and ΦSA039 on representative S. aureus strains associated with bovine mastitis. As milk is known to have detrimental effects on the interaction between a phage and its host (3, 16), it is desirable to examine the effects of these phages when they are cocultured with host bacteria in milk and ultimately to demonstrate their effectiveness in controlling bovine mastitis in vivo. It would be interesting to test them with S. aureus isolates from different sources and particularly with antibiotic-resistant strains. This study provides a solid basis on which to continue the exploration of the potential of ΦSA012 and ΦSA039 for treatment of staphylococcal diseases.
Acknowledgments
We are grateful to Fumio Arisaka for advice concerning TEM imaging and to Damien Synnott for critical reading of the manuscript.
Footnotes
Published ahead of print on 1 May 2009.
REFERENCES
- 1.Akineden, O., C. Annemuller, A. A. Hassan, C. Lammler, W. Wolter, and M. Zschock. 2001. Toxin genes and other characteristics of Staphylococcus aureus from milk of cows with mastitis. Clin. Diagn. Lab. Immunol. 8:959-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Capparelli, R., M. Parlato, G. Borriello, P. Salvatore, and D. Iannelli. 2007. Experimental phage therapy against Staphylococcus aureus in mice. Antimicrob. Agents Chemother. 51:2765-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Das, N. K., and R. T. Marshall. 1967. Adsorption of staphylococcal bacteriophage by milk proteins. Appl. Microbiol. 15:1095-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Oliveira, A. P., J. L. Watts, S. A. Salmon, and F. M. Aarestrup. 2000. Antimicrobial susceptibility of Staphylococcus aureus isolated from bovine mastitis in Europe and the United States. J. Dairy Sci. 83:855-862. [DOI] [PubMed] [Google Scholar]
- 5.Donovan, D. M., M. Lardeo, and J. Foster-Frey. 2006. Lysis of staphylococcal mastitis pathogens by bacteriophage phi11 endolysin. FEMS Microbiol. Lett. 265:133-139. [DOI] [PubMed] [Google Scholar]
- 6.Fournier, C., P. Kuhnert, J. Frey, R. Miserez, M. Kirchhofer, T. Kaufmann, A. Steiner, and H. U. Graber. 2008. Bovine Staphylococcus aureus: association of virulence genes, genotypes and clinical outcome. Res. Vet. Sci. 85:439-448. [DOI] [PubMed] [Google Scholar]
- 7.Gill, J. J., J. C. Pacan, M. E. Carson, K. E. Leslie, M. W. Griffiths, and P. M. Sabour. 2006. Efficacy and pharmacokinetics of bacteriophage therapy in treatment of subclinical Staphylococcus aureus mastitis in lactating dairy cattle. Antimicrob. Agents Chemother. 50:2912-2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hookey, J. V., J. F. Richardson, and B. D. Cookson. 1998. Molecular typing of Staphylococcus aureus based on PCR restriction length polymorphism and DNA sequence analysis of the coagulase gene. J. Clin. Microbiol. 36:1083-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones, T. O. 1998. Toxin production by Staphylococcus aureus from cases of bovine mastitis. Proc. Br. Mastitis Conf., p. 94.
- 10.Kalorey, D. R., Y. Shanmugam, N. V. Kurkure, K. K. Chousalkar, and S. B. Barbuddhe. 2007. PCR-based detection of genes encoding virulence determinants in Staphylococcus aureus from bovine subclinical mastitis cases. J. Vet. Sci. 8:151-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knight, C. H., J. L. Fitzpatrick, D. N. Logue, and D. J. Platt. 2000. Efficacy of two non-antibiotic therapies, oxytocin and topical liniment, against bovine staphylococcal mastitis. Vet. Rec. 146:311-316. [DOI] [PubMed] [Google Scholar]
- 12.Leiman, P. G., S. Kanamaru, V. V. Mesyanzhinov, F. Arisaka, and M. G. Rossmann. 2003. Structure and morphogenesis of bacteriophage T4. Cell. Mol. Life Sci. 60:2356-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez, B., J. M. Obeso, A. Rodriguez, and P. Garcia. 2008. Nisin-bacteriophage crossresistance in Staphylococcus aureus. Int. J. Food Microbiol. 122:253-258. [DOI] [PubMed] [Google Scholar]
- 14.Namura, M., T. Hijikata, K. Miyanaga, and Y. Tanji. 2008. Detection of Escherichia coli with fluorescent labeled phages that have a broad host range to E. coli in sewage water. Biotechnol. Prog. 24:481-486. [DOI] [PubMed] [Google Scholar]
- 15.Nashev, D., K. Toshkova, S. O. Salasia, A. A. Hassan, C. Lämmler, and M. Zschöck. 2004. Distribution of virulence genes of Staphylococcus aureus isolated from stable nasal carriers. FEMS Microbiol. Lett. 233:45-52. [DOI] [PubMed] [Google Scholar]
- 16.O'Flaherty, S., A. Coffey, W. J. Meaney, G. F. Fitzgerald, and R. P. Ross. 2005. Inhibition of bacteriophage K proliferation on Staphylococcus aureus in raw bovine milk. Lett. Appl. Microbiol. 41:274-279. [DOI] [PubMed] [Google Scholar]
- 17.O'Flaherty, S., A. Coffey, W. Meaney, G. F. Fitzgerald, and R. P. Ross. 2005. The recombinant phage lysin LysK has a broad spectrum of lytic activity against clinically relevant staphylococci, including methicillin-resistant Staphylococcus aureus. J. Bacteriol. 187:7161-7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Flaherty, S., R. P. Ross, R. Edwards, W. Meaney, G. F. Fitzgerald, M. F. Elbreki, and A. Coffey. 2005. Potential of the polyvalent anti-Staphylococcus bacteriophage K for control of antibiotic-resistant staphylococci from hospitals. Appl. Environ. Microbiol. 71:1836-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oliveira, D. C., I. Crisostomo, I. Santos-Sanches, P. Major, C. R. Alves, M. Aires-De-Sousa, M. K. Thege, and H. De Lencastre. 2001. Comparison of DNA sequencing of the protein A gene polymorphic region with other molecular typing techniques for typing two epidemiologically diverse collections of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 39:574-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Omoe, K., M. Ishikawa, Y. Shimoda, D. L. Hu, S. Ueda, and K. Shinagawa. 2002. Detection of seg, seh, and sei genes in Staphylococcus aureus isolates and determination of the enterotoxin productivities of S. aureus isolates harboring seg, seh, and sei genes. J. Clin. Microbiol. 40:857-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petitclerc, D., K. Lauzon, A. Cochu, C. Ster, M. S. Diarra, and P. Lacasse. 2007. Efficacy of a lactoferrin-penicillin combination to treat β-lactam-resistant Staphylococcus aureus mastitis. J. Dairy Sci. 90:2778-2787. [DOI] [PubMed] [Google Scholar]
- 22.Rebhun, W. C. 1995. Diseases of dairy cattle. Lippincott, Williams & Wilkins, Philadelphia, PA.
- 23.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanji, Y., K. Hattori, K. Suzuki, and K. Miyanaga. 2008. Spontaneous deletion of 209-kilobase-pair gene fragment from the Escherichia coli genome occurs with acquisition of resistance to an assortment of infectious phages. Appl. Environ. Microbiol. 74:4256-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanji, Y., T. Shimada, M. Yoichi, K. Miyanaga, K. Hori, and H. Unno. 2004. Toward rational control of Escherichia coli O157:H7 by a phage cocktail. Appl. Microbiol. Biotechnol. 64:270-274. [DOI] [PubMed] [Google Scholar]
- 26.Twomey, D. P., A. I. Wheelock, J. Flynn, W. J. Meaney, C. Hill, and R. P. Ross. 2000. Protection against Staphylococcus aureus mastitis in dairy cows using a bismuth-based teat seal containing the bacteriocin, lacticin 314. J. Dairy Sci. 83:1981-1983. [DOI] [PubMed] [Google Scholar]
- 27.Wills, Q. F., C. Kerrigan, and J. S. Soothill. 2005. Experimental bacteriophage protection against Staphylococcus aureus abscesses in a rabbit model. Antimicrob. Agents Chemother. 49:1220-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamane, I. 2006. Epidemiological survey and economical evaluation of bovine mastitis in tie-stall dairy farms. J. Jpn. Vet. Med. Assoc. 59:674-678. [Google Scholar]
- 29.Yoichi, M., M. Morita, K. Mizoguchi, C. R. Fischer, H. Unno, and Y. Tanji. 2004. The criterion for selecting effective phage for Escherichia coli O157:H7 control. Biochem. Eng. J. 19:221-227. [Google Scholar]