Abstract
We have developed web-based software for the rapid identification of protein biomarkers of bacterial microorganisms. Proteins from bacterial cell lysates were ionized by matrix-assisted laser desorption ionization (MALDI), mass isolated, and fragmented using a tandem time of flight (TOF-TOF) mass spectrometer. The sequence-specific fragment ions generated were compared to a database of in silico fragment ions derived from bacterial protein sequences whose molecular weights are the same as the nominal molecular weights of the protein biomarkers. A simple peak-matching and scoring algorithm was developed to compare tandem mass spectrometry (MS-MS) fragment ions to in silico fragment ions. In addition, a probability-based significance-testing algorithm (P value), developed previously by other researchers, was incorporated into the software for the purpose of comparison. The speed and accuracy of the software were tested by identification of 10 protein biomarkers from three Campylobacter strains that had been identified previously by bottom-up proteomics techniques. Protein biomarkers were identified using (i) their peak-matching scores and/or P values from a comparison of MS-MS fragment ions with all possible in silico N and C terminus fragment ions (i.e., ions a, b, b-18, y, y-17, and y-18), (ii) their peak-matching scores and/or P values from a comparison of MS-MS fragment ions to residue-specific in silico fragment ions (i.e., in silico fragment ions resulting from polypeptide backbone fragmentation adjacent to specific residues [aspartic acid, glutamic acid, proline, etc.]), and (iii) fragment ion error analysis, which distinguished the systematic fragment ion error of a correct identification (caused by calibration drift of the second TOF mass analyzer) from the random fragment ion error of an incorrect identification.
Food-borne illness is a serious and continuing problem, with an estimated 76 million cases in the United States per year (http://www.cdc.gov). It is often caused by bacteria and viruses that are often ubiquitous in the environment and are difficult to eliminate due to their ability to adapt. In addition to the resulting morbidity, food-borne illness also has enormous societal costs, including losses in worker productivity due to illness, recall of food products determined (or suspected) to be contaminated, etc. Consequently, there is a critical need to develop rapid and sensitive methods for detection and accurate identification of food-borne pathogens.
A number of techniques have been developed for detection and identification of food-borne pathogens. A relatively recent technique for bacterial identification involves the use of mass spectrometry (MS). Because of its sensitivity and high specificity, MS has become a popular technique for chemicotaxonomic classification of microorganisms (16, 27). The use of MS in the analysis of microorganisms is a relatively recent application that was dramatically accelerated by the development of two ionization techniques in the late 1980s and early 1990s: electrospray ionization (15) and matrix-assisted laser desorption ionization (MALDI) (24, 37). When coupled with time of flight (TOF) MS, MALDI has been demonstrated to be a powerful tool for “fingerprinting” microorganisms by ionization and detection of proteins from intact bacterial cells or extracts resulting from bacterial cell lysis (1, 2, 3, 8-12, 19, 21, 25, 26, 29, 34, 40, 41, 42). Typically, MALDI-TOF MS “fingerprinting” of microorganisms involves analysis using either pattern recognition or bioinformatic algorithms.
Pattern recognition analysis compares MALDI-TOF MS spectra of samples of unknown microorganisms to spectra of known microorganisms. A high degree of similarity between the MS spectrum of an unknown microorganism and an MS spectrum of a known microorganism strongly suggests the identity of the unknown microorganism (22, 39, 43). It should be noted that pattern recognition analysis does not rely on actual identification of the biomarker ion peaks in an MS spectrum. It is the pattern generated by multiple ion peaks that constitutes a microorganism's “fingerprint.” The actual identities of individual ion peaks are not specified, and the peaks could be peaks for any of a number of possible biological molecules generated by a microorganism, including proteins, nucleic acids, lipids, etc.
Microorganism identification by bioinformatic analysis of MALDI-TOF MS data involves using the protein molecular weights (MWs) in bacterial genomic databases to assign biomarker ion peaks in a mass spectrum to specific proteins (4, 5, 32, 33, 45). If a significant number of biomarker ion peaks in a mass spectrum correspond to protein MWs for the open reading frames of a microorganism's genome, then the microorganism is considered identified. Such an analysis has also incorporated the simplest and most common posttranslational modification (PTM) observed for bacterial proteins, N-terminal methionine cleavage (5). It should be noted, however, that “identification” of a microorganism relies solely on a sufficient number of protein MWs derived from open reading frames of its genome corresponding to the m/z of biomarker ions in a MALDI-TOF MS spectrum. However, the protein MW alone is not sufficient to definitively identify a biomarker ion as a specific protein. Protein biomarkers are considered to be tentatively assigned instead of definitively identified.
Analysis of samples containing multiple bacterial organisms presents increased challenges for MALDI-TOF MS when protein MW is the sole criterion for protein biomarker identification. Clearly, it would be advantageous if researchers could obtain more information about a biomarker in addition to its MW. In the case of protein biomarkers, this can be accomplished by enzymatically digesting a protein in solution and analyzing its tryptic peptides by MS (peptide mass mapping) or by tandem MS (MS-MS) (sequence tags) (45). Alternatively, it is possible to fragment mature, intact proteins (without digestion) in the gas phase to obtain sequence-specific and PTM information. This approach is referred to as top-down proteomics. Until recently, top-down proteomics was possible only if Fourier transform ion cyclotron resonance MS involving complicated gas phase ion dissociation techniques was used (6, 23).
Although not originally designed for top-down proteomics, recently developed MALDI-tandem TOF (MALDI-TOF-TOF) MS was shown to fragment small or modest-size proteins (5 kDa > molecular mass < 15 kDa) without prior digestion (28). Demirev and coworkers (7) identified Bacillus atrophaeus and Bacillus cereus spores by fragmenting their protein biomarkers using a MALDI tandem mass spectrometer and analyzing the sequence-specific fragment ions generated by comparison to in silico fragment ions derived from protein amino acid sequences from genomic databases. Protein and microorganism identities were determined using a probability-based significance-testing algorithm (P value). The P value algorithm calculates the probability that a protein or microorganism identification occurred randomly. The smaller the P value, the lower the probability that an identification occurred randomly. The data analysis was performed using software developed in house (7).
In the current study, web-based software and databases, developed in house at the U.S. Department of Agriculture (USDA), were used to identify 10 protein biomarkers from three pure strains of Campylobacter by sequence-specific fragmentation using a MALDI-TOF-TOF mass spectrometer. Many of the protein biomarkers had been identified previously by bottom-up proteomics techniques (9, 11, 12), which provided an excellent data set to test the accuracy and performance of the algorithms incorporated into the software. MALDI-TOF-TOF MS-MS fragment ions were compared with a database of in silico fragment ions derived from bacterial protein sequences. The sequence-specific MS-MS fragment ions were used to identify a protein and thus the source microorganism. A simple peak-matching mathematical algorithm, incorporated into the software, was used to score and rank protein and microorganism identifications. In addition, the P value algorithm of Demirev and coworkers (7) was also incorporated into the USDA software (available with execution of appropriate control usage agreement) for comparison to the peak-matching algorithm. The peak-matching algorithm correctly identified a protein biomarker among as many as ∼1,400 possible bacterial proteins and gave rankings for protein identification comparable to the rankings obtained by more complicated and computationally intensive P value calculation. We often observed enhancement of the score for correct identification when results for MS-MS fragment ions were compared to results for residue-specific in silico fragment ions compared to non-residue-specific in silico fragment ions. In addition, the correctness of the algorithm's identification was, in certain cases, further confirmed by fragment ion error analysis which compared random error caused by false matches between MS-MS fragment ions and in silico fragment ions with the systematic error observed for correct matches due to drift in the calibration of the TOF mass analyzer (38).
(Portions of this work were presented at the 121st AOAC Conference [13] and at the 55th American Society of Mass Spectrometry Conference [14].)
MATERIALS AND METHODS
Figure 1 shows a flow chart of the process used for identification of protein biomarkers and bacteria.
FIG. 1.
Flow chart for protein and microorganism identification. TFA, trifluoroacetic acid; IDs, identifications.
Bacterial protein extraction.
Bacterial proteins were extracted from Campylobacter bacterial cells using a technique that has been previously reported (9-12, 29). Briefly, Campylobacter upsaliensis strain RM3195, C. coli strain RM2228, and C. lari strain RM2100 were each cultured on nonselective growth media for 24 to 48 h. Bacterial cells were harvested with a 1-μl loop (an amount which corresponded to 109 cells) and transferred to a microcentrifuge tube containing 0.5 ml of extraction solvents (67% water, 33% acetonitrile, and 0.1% trifluoroacetic acid) and 40 mg of 0.1-mm zirconia-silica beads (BioSpec Products Inc., Bartlesville, OK). The tube was capped and agitated for 60 s with a bead beater, resulting in cell lysis. The tube was then centrifuged at 8,161 × g for 4 to 5 min in order to pellet insoluble cellular debris.
MALDI-TOF-TOF MS and MS-MS.
Samples were analyzed with a 4800 TOF-TOF proteomics analyzer (Applied Biosystems, Foster City, CA). A 0.5-μl aliquot of sample supernatant was mixed with an equal volume of a saturated solution of MALDI matrix and deposited onto a 384-spot stainless steel target. Two MALDI matrices were utilized: 3,5-dimethoxy-4-hydroxycinnamic acid (sinapinic acid), a “cold” matrix; and α-cyano-4-hydroxycinnamic acid, a “hot” matrix. Laser desorption ionization was accomplished using a pulsed solid-state YAG laser (repetition rate, 200 Hz; wavelength, 355 nm; pulse width, ∼5 ns). Spectra were acquired in positive-ion mode for both MS (linear mode) and MS-MS (reflectron mode). In MS linear mode, after laser desorption ionization, ions were accelerated from the first source by delayed ion extraction at 20 kV, separated over an effective ion path length of 1.5 m, and detected with a multichannel plate detector. In MS-MS reflectron mode, ions were accelerated from the first source by delayed ion extraction at 8.0 kV. Ions were separated spatially and temporally in the first field free region. Ions of interest were mass selected with a timed ion selector (TIS) or mass “gate” based on their arrival time at the TIS gate. The TIS was used to mass isolate specific protein ions for fragmentation based on their mass-to-charge ratio (m/z). The TIS was operated with a “window” of either ±50 Da or ±100 Da. Mass-selected ions were decelerated to 1.70 kV prior to entry into a floating collision cell at 2 kV. Ions were fragmented either by high-energy collision-induced dissociation or by postsource dissociation. The target gas for high-energy collision-induced dissociation was filtered air. Fragment ions exiting the collision cell were reaccelerated to 15 kV. A Bradbury-Neilsen ion gate after the second source could be used to suppress (and thus exclude) the precursor ion signal. MS-MS data were collected with the precursor ion suppressor gate in both the “on” and “off” modes. A two-stage reflectron mirror assembly was operated at 10.910 kV (mirror 1) and at 18.750 kV (mirror 2). Both linear and reflectron multichannel plate detectors were operated at 2.190 kV. The effective ion path length from the second source to the reflectron detector was 2.3 m. The instrument was externally calibrated in linear mode with the following calibrants: bovine insulin (MW, 5,733.58), Escherichia coli thioredoxin (MW, 11,673.47), and horse heart apomyoglobin (MW, 16,951.55). The instrument was externally calibrated in reflectron MS-MS mode using the y fragment ions of glu1-fibrino-peptide B (MW, 1570.60) at m/z 175.120 and 1441.635. MS and MS-MS data were processed using commercially available instrument software (Data Explorer software, version 4.9). The software parameters are described in the supplemental material.
Peak-matching algorithm.
The peak-matching algorithm involves counting the number of MS-MS fragment ions whose intensity is equal to or greater than a relative intensity threshold (e.g., 2%). The algorithm then counts the number of in silico fragment ions whose m/z fall within a specified m/z tolerance (e.g., ±2.5 thomson [Th]) to that of the m/z of MS-MS fragment ions; i.e., it counts the number of “matches” between MS-MS fragment ions and in silico fragment ions for the two data sets. The number of “matches” is then divided by the total number of MS-MS fragment ions whose m/z are above the specified intensity threshold. The resulting number is then multiplied by 100% to obtain the peak-matching score, as follows: score = 100 × (number of MS-MS fragment ion peaks that “matched” in silico fragment ion peaks)/(number of MS-MS fragment ion peaks).
The peak-matching score has a theoretical range of 0 to 100%. Zero percent indicates that no matches were identified, and 100% indicates that every MS-MS fragment ion matched an in silico fragment ion for identification. A nonzero fragment ion m/z tolerance indicates that it is possible for an MS-MS fragment ion m/z to “match” the m/z of two (or more) in silico fragment ions (or vice versa). Such multiple matches are counted only once by the algorithm; otherwise, a score greater than 100% could be obtained. The highest-scoring protein or microorganism identification that is significantly higher than the second-highest-scoring protein or microorganism identification is a presumptive correct identification. “Significantly” is defined here as a relative difference between the scores of the highest-scoring identification and the second-highest-scoring identification of 15 to 20% or greater. For comparison, the more mathematically complicated P value algorithm, developed by Demirev and coworkers, was also incorporated into the USDA software. In brief, the P value algorithm calculates the probability that an identification occurred randomly. The lower the P value of an identification, the less likely that the identification occurred randomly. A confident protein or microorganism identification is one in which the P value of the “top” identification is significantly (typically several orders of magnitude) lower than the P values of the “runner-up” identifications (7).
The peak-matching and P value algorithms are completely independent, and the results of each calculation are displayed in the software window. Software functionality allows selective operation of one of the algorithms or both algorithms. Algorithm computation time is provided by the software. In addition, the protein and microorganism identifications can be “ranked” by either the peak-matching scores or the P values.
Residue-specific and non-residue-specific in silico fragment ion comparisons.
The peak-matching and P value algorithms described above are used under the assumption that the polypeptide backbone has an equal probability of fragmenting at every residue of the protein to produce the a, b, b-18, y, y-17, and y-18 fragment ions. However, it has been shown experimentally that singly protonated (charged) protein ions are more likely to fragment at aspartic acid (D), glutamic acid (E), and proline (P) residues (7, 28, 31, 44, 46). As discussed below, each in silico fragment ion is identified by its m/z, ion type and number, and the two amino acid residues on either side of the backbone cleavage site that resulted in formation of the fragment ion. Software functionality allows comparison of the m/z of MS-MS fragment ions to the m/z of all in silico fragment ions of a particular protein (i.e., a non-residue-specific comparison). Alternatively, MS-MS fragment ions can be compared to residue-specific in silico fragment ions (e.g., D-, E-, and P-specific in silico fragment ions). Residue-specific and non-residue specific comparisons are discussed in greater detail in the supplemental material.
In silico bacterial protein sequence database.
Figure 1 outlines the process by which in silico bacterial protein sequences (and their associated fragment ions) were obtained. A detailed description of the construction of the in silico database (as well as software and database architecture and function) is given in the supplemental material. In brief, in silico bacterial protein sequences were downloaded using the TagIdent software at the ExPASy public website (http://ca.expasy.org/tools/tagident.html) for proteins having a pI in the range from 0.00 to 14.00 and a molecular mass that was within 5 Da of that of the singly protonated protein biomarker ion after removal of its proton charge. The searches were conducted using both the UniProtKB/Swiss-Prot (versions 55.5 to 56.0) and UniProtKB/TrEMBL (versions 38.5 to 39.0) databases. Bacterial protein sequences with possible PTM (e.g., N-terminal methionine cleavage, signal peptides, etc.) were also retrieved. Multiprotein sequence FASTA files obtained from the ExPASy website were processed using a beta version (version 8.01a5) of the commercial GPMAW software (Lighthouse Data, Denmark) to generate individual text files for each protein sequence which contain the in silico fragment ions, the protein name, the amino acid sequence, the average MW of the protein, and the taxonomic classification of the bacterium. Each in silico fragment ion is identified by its m/z, ion type and number (a, b, b-18, y, y-17, and y-18), and the two amino acid residues adjacent to the polypeptide cleavage site that resulted in formation of the fragment ion. Individual in silico text files were batch uploaded to the in silico database of the USDA software.
RESULTS
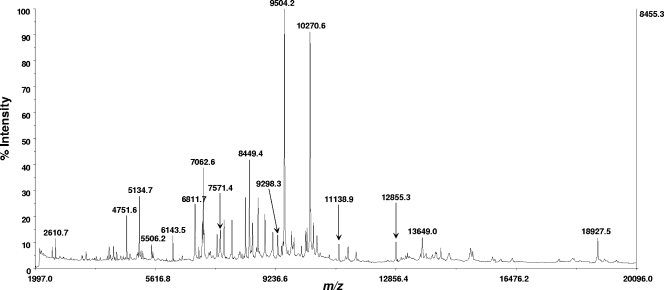
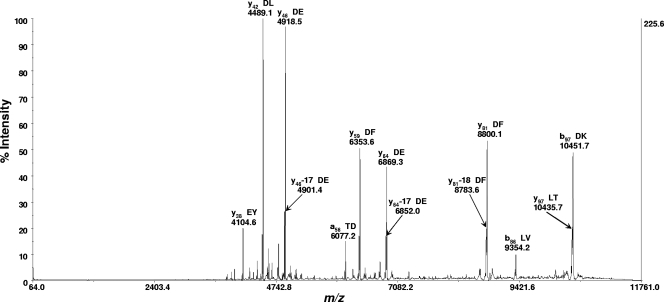
Figure 2 shows a typical MS spectrum of a bacterial cell lysate of C. upsaliensis strain RM3195 analyzed by MALDI-TOF-TOF MS in linear mode using the sinapinic acid matrix. Figure 3 shows a typical MS-MS spectrum of the protein biomarker ion at m/z 11138.9 shown in Fig. 2. This protein biomarker had been previously identified by bottom-up proteomics as thioredoxin (12). Prominent fragment ions are identified by their m/z, ion type and number, and amino acid residues adjacent to the site of polypeptide cleavage that resulted in the fragment ion. As the spectrum shows, many of the fragment ions are the result of polypeptide cleavage adjacent to an aspartic acid or glutamic acid residue.
FIG. 2.
MS spectrum (as displayed in commercial instrument software) of extracted cell lysate of C. upsaliensis strain RM3195 analyzed with the MALDI-TOF-TOF MS instrument (in linear mode) using sinapinic acid as the MALDI matrix.
FIG. 3.
MS-MS spectrum (as displayed in commercial instrument software) of the protein biomarker ion at m/z 11138.9 (see Fig. 2) analyzed with the MALDI-TOF-TOF MS instrument (in reflectron mode) using sinapinic acid as the MALDI matrix. Fragmentation was by postsource dissociation. The precursor ion suppressor was “on.” This protein biomarker had been previously identified by bottom-up proteomics as thioredoxin (12). Prominent fragment ions are identified by their m/z, ion type and number, and amino acid residues adjacent to the site of polypeptide backbone cleavage resulting in the fragment ion. Many of the fragment ions are the result of polypeptide cleavage adjacent to an aspartic acid or glutamic acid residue. The MS-MS data were centroided before they were exported as an ASCII-formatted file for upload into the USDA software database.
The protein biomarkers of the following three strains of Campylobacter were analyzed by top-down proteomics: C. upsaliensis strain RM3195, C. lari strain RM2100, and C. coli strain RM2228. Many of the protein biomarkers had been identified previously by bottom-up proteomics techniques (9, 11, 12).
C. upsaliensis strain RM3195.
Table 1 shows the top five identifications for a protein biomarker of C. upsaliensis strain RM3195 at m/z 11138.9 (Fig. 2) analyzed by MS-MS using MALDI-TOF-TOF MS (Fig. 3; see Fig. S1 in the supplemental material) and compared to all in silico fragment ions of bacterial protein sequences having the same molecular mass as the biomarker (within 5 Da). This corresponded to 1,409 in silico bacterial protein sequences. The protein biomarker had been identified previously by bottom-up proteomics techniques as thioredoxin (12). The rankings for the top five identifications based on the USDA scores and the P value calculations are identical, and both algorithms correctly identify the protein as thioredoxin with an N-terminal methionine cleavage PTM and its source microorganism as C. upsaliensis strain RM3195. The computation time of the USDA peak-matching algorithm is ∼35% shorter than the computation time of the P value calculation. Table 1 also shows the top five identifications for an analysis that was the same as that described above above except that only D-, E-, and P-specific in silico fragment ions were used for comparison. The numbers of in silico bacterial protein sequences are identical. The top identification for both algorithms correctly identifies the protein and its source microorganism. There also is significant relative enhancement of the top identification score compared to the “runner-up” identification scores when a comparison of D-, E-, and P-specific in silico fragment ions is used instead of a comparison of all in silico fragment ions (Table 1). In addition, the “runner-up” identification is different for the non-residue-specific comparison (all in silico fragment ions) than for the residue-specific comparison. The computation time of both algorithms for the residue-specific analysis is lower than that for the non-residue-specific analysis. The USDA peak-matching algorithm is ∼116% faster than the P value calculation for the D-, E-, and P-specific in silico fragment ion comparison. Fragment ion error analysis for this protein biomarker identification is described and discussed in the supplemental material.
TABLE 1.
Top five identifications of a protein biomarker of C. upsaliensis strain RM3195 at m/z 11138.9 based on all in silico fragment ions and on D-, E-, and P-specific in silico fragment ionsa
| Fragment ions | In silico identification | Identifier | Sample name | Protein (MW) | USDA score | P value |
|---|---|---|---|---|---|---|
| All in silico fragment ions | 1262 | >Q4HNM5 Q4HNM5_CAMUP | Campylobacter upsaliensis RM3195 | Thioredoxin PTM N-Met (11,136.76) | 82.69 | 7.7E−19 |
| 323 | >B1WRD3 B1WRD3_CYAA5 | Cyanothece sp. strain ATCC 51142 | Carbon dioxide-concentrating mechanism protein (11,133.85) | 53.85 | 3.6E−6 | |
| 71 | >A4T659 A4T659_MYCGI | Mycobacterium gilvum strain PYR-GCK (Mycobacterium flavescens strain ATCC 700033 [=PYR-GCK]) | Putative uncharacterized protein (11,142.24) | 46.15 | 3.1E−4 | |
| 697 | >Q1LMQ8 Q1LMQ8_RALME | Ralstonia metallidurans strain CH34 (=ATCC 43123 = DSM 2839) | Putative uncharacterized protein PTM 23SigPep (11,128.81) | 44.23 | 1.1E−3 | |
| 1154 | >Q18XI2 Q18XI2_DESHD | Desulfitobacterium hafniense strain DCB-2 | Small multidrug resistance protein PTM N-Met (11,138.67) | 44.23 | 1.6E−3 | |
| D-, E-, and P-specific in silico fragment ions | 1262 | >Q4HNM5 Q4HNM5_CAMUP | Campylobacter upsaliensis strain RM3195 | Thioredoxin PTM-N-Met (11,136.76) | 61.54 | 3.0E−21 |
| 143 | >A7A905 A7A905_BIFAD | Bifidobacterium adolescentis L2-32 | Putative uncharacterized protein (11,134.94) | 25.00 | ||
| 942 | >A8LX75 A8LX75_SALAI | Salinira arenicola strain CNS-205 | Putative uncharacterized protein PTM N-Met (11,140.56) | 9.0E−5 | ||
| 136 | >A6QD71 A6QD71_STAAE | Staphylococcus aureus strain Newman | Putative uncharacterized protein (11,138.98) | 23.08 | ||
| 143 | >A7A905 A7A905_BIFAD | Bifidobacterium adolescentis L2-32 | Putative uncharacterized protein (11,134.94) | 9.4E−5 | ||
| 145 | A7BQP4 A7BQP4_9GAMM | Beggiatoa sp. strain PS | Putative uncharacterized protein (11,132.06) | 23.08 | ||
| 136 | >A6QD71 A6QD71_STAAE | Staphylococcus aureus strain Newman | Putative uncharacterized protein (11,138.98) | 1.5E−4 | ||
| 271 | >B1BI09 B1BI09_CLOPE | Clostridium perfringens C strain JGS1495 | Putative uncharacterized protein (11,140.05) | 23.08 | ||
| 449 | >Q2G1R1 Q2G1R1_STAA8 | Staphylococcus aureus strain NCTC 8325 | Putative uncharacterized protein (11,138.98) | 1.5E−4 |
The protein marker at m/z 11138.9 (Fig. 2) was analyzed by MS-MS using MALDI-TOF-TOF MS (Fig. 3; see Fig. S1 in the supplemental material) and compared to all in silico fragment ions and D-, E-, and P-specific in silico fragment ions of bacterial protein sequences having the same molecular mass as the biomarker (within 5 Da). This corresponded to 1,409 in silico bacterial protein sequences. The protein biomarker had been identified previously by bottom-up proteomics techniques as thioredoxin (12). The top identification of both algorithms was the correct identification of the protein and its source microorganism. In addition, the top identification scores of the two algorithms are relatively enhanced compared to the “runner-up” identification scores. The parameters for comparison of MS-MS data and in silico data were as follows: intensity threshold, 2%; number of MS-MS peaks with an intensity of ≥2%, 52; m/z range for comparison, 0 to 14,000 Th; fragment ion tolerance, 2.5 Th; and protein MW, 11,138 ± 10. “PTM N-Met” indicates that the in silico protein sequence was modified to remove the N-terminal methionine.“PTM 23SigPep” indicates that the in silico protein sequence was modified to remove a signal peptide. The algorithm computation times for all in silico fragment ions and for the D-, E-, and P-specific in silico fragment ions were as follows: USDA peak-matching algorithm, 35.7 and 18.4 s, respectively; and P value calculation, 48.8 and 39.7 s, respectively.
Table 2 shows the top five identifications for a protein biomarker of C. upsaliensis strain RM3195 at m/z 12855.3 (Fig. 2) analyzed by MS-MS using MALDI-TOF-TOF MS and compared to all in silico fragment ions of bacterial protein sequences having the same molecular mass (within 5 Da) as the biomarker. This corresponded to 1,315 in silico protein sequences. This protein biomarker had been identified previously by bottom-up proteomics techniques as the 50S L7/L12 ribosomal protein (12). The top identification of both algorithms correctly identifies the protein as ribosomal protein 50S L7/L12 (with an N-terminal methionine cleavage PTM) and its source microorganism as C. upsaliensis strain RM3195. The “runner-up” identification is the 50S L7/L12 ribosomal protein of C. coli strain RM2228, whose molecular mass differs from that of the ribosomal protein of C. coli strain RM3195 by only ∼2 Da. The primary amino acid sequences of the C. upsaliensis strain RM3195 and C. coli strain RM2228 50S L7/L12 ribosomal proteins are shown in Fig. 4. The homologies of these two sequences with respect to aspartic acid, glutamic acid, and proline residues are identical, which results in fragmentation channels with high levels of similarity (13, 14). However, variations in non-D, non-E, non-P amino acid residues between these sequences result in “shifts” in the m/z of some in silico fragment ions, allowing differentiation of the these two proteins in a comparison of MS-MS data to in silico data. It is also interesting that there was incorrect identification of the 50S L7/L12 protein of Prosthecochloris aestuarii strain DSM 271 (ranked fourth by the USDA score and fifth by the P value), which highlights the fact that, although these high-copy-number housekeeping proteins have nearly identical MWs, their amino acid sequences are significantly different for protein and source identification by MS-MS analysis. Table 2 also shows the top five identifications for an analysis that was the same as that described above except that only D-, E-, and P-specific in silico fragment ions were used for comparison. The top identification for both algorithms correctly identifies the protein and its source microorganism. Again, there is enhancement of the top (and correct) identification compared to the “runner-up” identification when a comparison of D-, E-, and P-specific in silico fragment ions is used instead of a comparison of all in silico fragment ions. The computation time of both algorithms is lower for a residue-specific in silico comparison than for a non-residue-specific in silico comparison. However, the analysis time for the USDA peak-matching algorithm was reduced by ∼60%, whereas the analysis time for the P value calculation was reduced by only ∼20%.
TABLE 2.
Top five identifications of a protein biomarker of C. upsaliensis strain RM3195 at m/z 12855.3 based on all in silico fragment ions and on D-, E-, and P-specific in silico fragment ionsa
| Fragment ions | In silico identification | Identifier | Sample name | Protein (MW) | USDA score | P value |
|---|---|---|---|---|---|---|
| All in silico fragment ions | 2551 | >Q4HS60 Q4HS60_CAMUP | Campylobacter upsaliensis RM3195 | Ribosomal protein L7/L12 PTM N-Met (12,855.65) | 66.07 | 1.1E−10 |
| 2550 | >Q4HDZ9 Q4HDZ9_CAMCO | Campylobacter coli RM2228 | Ribosomal protein L7/L12 PTM N-Met (12,853.74) | 51.79 | 1.5E−5 | |
| 2152 | >A5ZFY2 A5ZFY2_9BACE | Bacteroides caccae ATCC 43185 | Putative uncharacterized protein PTM N-Met (12,852.15) | 42.86 | 8.7E−4 | |
| 2532 | >Q3VUB8 Q3VUB8_PROAE | Prosthecochloris aestuarii DSM 271 | Ribosomal protein L7/L12 PTM N-Met (12,855.76) | 42.86 | ||
| 2368 | >B2HXJ0 B2HXJ0_ACIBA | Acinetobacter baumannii ACICU | Putative transcriptional regulator, TetR family PTM N-Met (12,853.04) | 2.8E−3 | ||
| 1427 | >A7NKT8 A7NKT8_ROSCS | Roseiflexus castenholzii strain DSM 13941 (=HLO8) | Putative uncharacterized protein PTM 41SigPep (12,858.11) | 41.07 | ||
| 2532 | >Q3VUB8 Q3VUB8_PROAE | Prosthecochloris aestuarii DSM 271 | Ribosomal protein L7/L12 PTM N-Met (12,855.76) | 2.8E−3 | ||
| D-, E-, and P-specific in silico fragment ions | 2551 | >Q4HS60 Q4HS60_CAMUP | Campylobacter upsaliensis RM3195 | Ribosomal protein L7/L12 PTM N-Met (12,855.65) | 53.57 | 1.3E−15 |
| 2550 | >Q4HDZ9 Q4HDZ9_CAMCO | Campylobacter coli RM2228 | Ribosomal protein L7/L12 PTM N-Met (12,853.74) | 39.29 | 1.4E−8 | |
| 2254 | >A9IY33 A9IY33_BART1 | Bartonella tribocorum strain CIP 105476 (=IBS 506) | Putative uncharacterized protein PTM N-Met (12,848.37) | 23.21 | ||
| 1676 | >B0ACQ4 B0ACQ4_9CLOT | Clostridium bartlettii DSM 16795 | Putative uncharacterized protein (12,849.21) | 1.5E−3 | ||
| 1462 | >A3WE41 A3WE41_9SPHN | Erythrobacter sp. strain NAP1 | Putative uncharacterized protein (12,859.01) | 21.43 | ||
| 2592 | >Q63UK3 Q63UK3_BURPS | Burkholderia pseudomallei (Pseudomonas pseudomallei) | Putative membrane protein PTM N-Met (12,847.95) | 2.8E−3 | ||
| 2373 | >B2JBB3 B2JBB3_NOSPU | Nostoc punctiforme PCC 73102 | Excalibur domatin protein PTM N-Met (12,846.38) | 21.43 | ||
| 1617 | >A0HKV3 A0HKV3_COMTE | Comamonas testosteroni KF-1 | Iron-sulfur cluster assembly accessory protein (12,856.3) | 4.6E−3 |
The protein marker at m/z 12855.3 (Fig. 2) was analyzed by MS-MS using MALDI-TOF-TOF MS and compared to all in silico fragment ions and D-, E-, and P-specific in silico fragment ions of bacterial protein sequences having the same molecular mass as the biomarker (within 5 Da). This corresponded to 1,315 in silico bacterial protein sequences. The protein biomarker had been identified previously by bottom-up proteomics techniques as ribosomal protein 50S L7/L12 (12). The top identification of both algorithms was the correct identification of the protein and its source microorganism. In addition, the top identification scores of the two algorithms are enhanced compared with the “runner-up” identification scores. The parameters for comparison of MS-MS data and in silico data were as follows: intensity threshold, 2%; number of MS-MS peaks with an intensity of ≥2%, 56; m/z range for comparison, 0 to 14,000 Th; fragment ion tolerance, 2.5 Th; and protein MW, 12,854 ± 10. “PTM N-Met” indicates that the in silico protein sequence was modified to remove the N-terminal methionine. “PTM 41SigPep” indicates that the in silico protein sequence was modified to remove a signal peptide. The algorithm computation times for all in silico fragment ions and for the D-, E-, and P-specific in silico fragment ions were as follows: USDA peak-matching algorithm, 48.0 and 19.7 s, respectively; and P value calculation, 57.5 and 44.4 s, respectively.
FIG. 4.
Amino acid sequences of the 50S L7/L12 ribosomal proteins of C. upsaliensis strain RM3195 and C. coli strain RM2228. The MWs of the two proteins are nearly identical, and the sequences are also identical with respect to aspartic acid, glutamic acid, and proline residues (bold type), but amino acid sequence variations (boxes) between the two proteins result in “shifts” in fragment ion m/z for this protein biomarker between the two strains (13, 14). These differences in fragment ion m/z allow differentiation of the two sequences which have a high level of homology. Both proteins undergo posttranslational N-terminal methionine cleavage.
Identifications of other protein biomarkers of C. upsaliensis strain RM3195 are shown in the supplemental material.
C. lari strain RM2100.
Table 3 shows the top five identifications for a protein biomarker of C. lari strain RM2100 observed at m/z 11253.3 obtained by MALDI-TOF MS and analyzed by MS-MS using MALDI-TOF-TOF MS. The MS-MS fragment ions were compared to all in silico fragment ions of bacterial protein sequences having the same molecular mass (within 5 Da) as the biomarker. This corresponded to 1,548 in silico bacterial protein sequences. The protein biomarker had been identified previously by bottom-up proteomics techniques as thioredoxin (11). The top identification of both algorithms correctly identifies the protein and its source microorganism. Table 3 also shows the results of an analysis that was the same as that described above except that only D-, E-, and P-specific in silico fragment ions were compared to MS-MS fragment ions. Again, the top identification of both algorithms correctly identifies the protein and its source microorganism. There is also enhancement of the top identification score compared to the “runner-up” scores when the residue-specific analysis results are compared to the non-residue-specific analysis results. The peak-matching algorithm is ∼40% and ∼70% faster than the P value calculation for the comparisons of all in silico ions and residue-specific ions, respectively. In addition, the computation time for the peak-matching algorithm is cut in half for the residue-specific comparison, whereas the P value computation time is slightly increased compared to the non-residue-specific analysis time.
TABLE 3.
Top five identifications of a protein biomarker of C. lari strain RM2100 at m/z 11253.3 based on all in silico fragment ions and on D-, E-, and P-specific in silico fragment ionsa
| Fragment ions | In silico identification | Identifier | Sample name | Protein (MW) | USDA score | P value |
|---|---|---|---|---|---|---|
| All in silico fragment ions | 9623 | >Q4HJN3 Q4HJN3_CAMLA | Campylobacter lari RM2100 | Thioredoxin PTM N-Met (11,246.88) | 53.62 | 6.4E−9 |
| 9224 | >A7H3X2 A7H3X2_CAMJD | Campylobacter jejuni subsp. doylei strain ATCC BAA-1458 (=RM4099 = 269.97) | Putative uncharacterized protein PTM N-Met (11,252.08) | 44.93 | 3.0E−6 | |
| 8933 | >Q8XUD3 Q8XUD3_RALSO | Ralstonia solanacearum | Putative uncharacterized protein (11,251.85) | 39.13 | ||
| 9223 | >A7H3K3 A7H3K3_CAMJD | Campylobacter jejuni subsp. doylei strain ATCC BAA-1458 (=RM4099 = 269.97) | Transcriptional regulator, Cro/CI family (11,249.82) | 2.1E−4 | ||
| 9223 | >A7H3K3 A7H3K3_CAMJD | Campylobacter jejuni subsp. doylei strain ATCC BAA-1458 (=RM4099 = 269.97) | Transcriptional regulator, Cro/CI family (11,249.82) | 39.13 | ||
| 9233 | >A7JU22 A7JU22_PASHA | Mannheimia hemolytica PHL213 | Putative uncharacterized protein PTM N-Met (11,251.98) | 4.3E−4 | ||
| 9233 | >A7JU22 A7JU22_PASHA | Mannheimia hemolytica PHL213 | Putative uncharacterized protein PTM N-Met (11,251.98) | 39.13 | ||
| 8933 | >Q8XUD3 Q8XUD3_RALSO | Ralstonia solanacearum | Putative uncharacterized protein (11,251.85) | 7.4E−4 | ||
| D-, E-, and P-specific in silico fragment ions | 9623 | >Q4HJN3 Q4HJN3_CAMLA | Campylobacter lari RM2100 | Thioredoxin PTM N-Met (11,246.88) | 37.68 | 2.2E−11 |
| 9224 | >A7H3X2 A7H3X2_CAMJD | Campylobacter jejuni subsp. doylei strain ATCC BAA-1458 (=RM4099 = 269.97) | Putative uncharacterized protein PTM N-Met (11,252.08) | 23.19 | 6.6E−5 | |
| 8398 | >A6E0L1 A6E0L1_9RHOB | Roseovarius sp. strain TM1035 | Putative uncharacterized protein (11,253.5) | 21.74 | ||
| 9294 | >A9HBE8 A9HBE8_N-METNO | Methylobacterium nodulans ORS 2060 | Thioredoxin PTM N-Met (11,253.06) | 1.2E−4 | ||
| 9294 | >A9HBE8 A9HBE8_N-METNO | Methylobacterium nodulans ORS 2060 | Thioredoxin PTM N-Met (11,253.06) | 21.74 | ||
| 9223 | >A7H3K3 A7H3K3_CAMJD | Campylobacter jejuni subsp. doylei strain ATCC BAA-1458 (=RM4099 = 269.97) | Transcription regulator, Cro/CI family, PTM N-Met (11,249.82) | 1.7E−4 | ||
| 9540 | >Q2CJW4 Q2CJW4_9RHO | Oceanicola granulosus HTCC2516 | Thioredoxin PTM N-Met (11,252.78) | 21.74 | ||
| 8819 | >Q4ZTY4 Q4ZTY4_PSEU2 | Pseudomonas syringae pv. syringae strain B728a | Putative uncharacterized protein (11,245.79) | 2.9E−4 |
A protein marker at m/z 11253.3 was analyzed by MS-MS using MALDI-TOF-TOF MS and compared to all in silico fragment ions and D-, E-, and P-specific in silico fragment ions of bacterial protein sequences having the same molecular mass as the biomarker (within 5 Da). This corresponded to 1,548 in silico bacterial protein sequences. The protein biomarker had been identified previously by bottom-up proteomics techniques as thioredoxin (12). The top identification of both algorithms was the correct identification of the protein and its source microorganism. In addition, the top identification scores of the two algorithms are enhanced compared with the “runner-up” identification scores. The parameters for comparison of MS-MS data and in silico data were as follows: intensity threshold, 4%; number of MS-MS peaks with an intensity of ≥4%, 69; m/z range for comparison, 0 to 10,000 Th; fragment ion tolerance, 2.5 Th; and protein MW, 11,252 ± 10. “PTM N-Met” indicates that the in silico protein sequence was modified to remove the N-terminal methionine. The algorithm computation times for all in silico fragment ions and for the D-, E-, and P-specific in silico fragment ions were as follows: USDA peak-matching algorithm, 44.7 and 22.0 s, respectively; and P value algorithm, 73.6 and 75.3 s, respectively.
Identifications of other protein biomarkers of C. lari strain RM2100 are shown in the supplemental material.
C. coli strain RM2228.
Table 4 shows the top five identifications for a protein biomarker of C. coli strain RM2228 at m/z 8571.4 obtained by MALDI-TOF MS and analyzed by MS-MS using MALDI-TOF-TOF MS. The MS-MS fragment ions were compared to all in silico fragment ions of bacterial protein sequences having the same molecular mass (within 5 Da) as the biomarker. This corresponded to 1,425 in silico bacterial protein sequences. This protein biomarker had been identified previously as the DUF-465 protein in another strain of C. coli by bottom-up proteomics techniques (11). The top identification of both algorithms correctly identifies the protein and its source microorganism. Table 4 shows the results of an analysis that was the same as that described above except that only D-, E,- and P-specific in silico fragment ions were compared to MS-MS fragment ions. Again, the top identification of both algorithms correctly identifies the protein and source microorganism. In addition, there is enhancement of the USDA score and P value of the top identification compared to the “runner-up” identification when the residue-specific analysis results are compared to the non-residue-specific analysis results.
TABLE 4.
Top five identifications of a protein biomarker of C. coli strain RM2228 at m/z 8571.4 based on all in silico fragment ions and on D-, E-, and P-specific in silico fragment ionsa
| Fragment ions | In silico identification | Identifier | Sample name | Protein (MW) | USDA score | P value |
|---|---|---|---|---|---|---|
| All in silico fragment ions | 4675 | >Q4HFY1 Q4HFY1_CAMCO | Campylobacter coli RM2228 | Putative uncharacterized protein or DUF-465 (8,571.7) | 62.50 | 3.2E−8 |
| 4352 | >A9E284 A9E284_9FLAO | Kordia algicida OT-1 | Putative uncharacterized protein (8,568.96) | 52.50 | 8.7E−5 | |
| 5084 | >A9VG47 A9VG47_BACWK | Bacillus weihenstephanensis strain KBAB4 | Zinc finger CDGSH-type domain protein, PTM N-Met (8,569.86) | 52.50 | 8.7E−5 | |
| 5510 | >Q92PU4 Q92PU4_RHIME | Rhizobium meliloti (Sinorhizobium meliloti) | Putative uncharacterized protein PTM N-Met (8,570.17) | 52.50 | 8.7E−5 | |
| 4252 | >A6EXJ7 A6EXJ7_9ALTE | Marinobacter algicola DG893 | Putative uncharacterized protein (8,570.13) | 47.50 | 7.1E−4 | |
| D-, E-, and P-specific in silico fragment ions | 4675 | >Q4HFY1 Q4HFY1_CAMCO | Campylobacter coli RM2228 | Putative uncharacterized protein or DUF-465 (8,571.7) | 60.00 | 1.2E−14 |
| 5064 | >A9H9E8 A9H9E8_GLUDA | Gluconacetobacter diazotrophicus strain ATCC 49037 (=DSM 5601 = PAl5). | Putative uncharacterized protein, PTM N-Met (8,569.65) | 37.50 | ||
| 5021 | >A7ZIB1 A7ZIB1_ECO24 | Escherichia coli O139:H28 strain E24377A/ETEC | Putative uncharacterized protein, PTM N-Met (8,566.90) | 2.3E−7 | ||
| 4506 | >B2JIT5 B2JIT5_9BURK | Burkholderia phymatum STM815 | Putative uncharacterized protein (8,566.86) | 32.50 | ||
| 5023 | >A7ZX11 A7ZX11_ECOHS | Escherichia coli O9:H4 strain HS | Putative uncharterized protein, PTM N-Met (8,566.90) | 2.3E−7 | ||
| 5393 | >Q3XZT5 Q3XZT5_ENTFC | Enterococcus faecium DO | d-Alanyl carrier protein, PTM N-Met (8,569.53) | 32.50 | ||
| 5472 | >Q7CN31 Q7CN31_STRP8 | Streptococcus pyogenes serotype M18 | Conserved hypothetical phage protein, PTM N-Met (8,568.00) | 4.3E−7 | ||
| 4202 | >A4Y5S5 A4Y5S5_SHEPC | Shewanella putrefaciens strain CN-32 (=ATCC BAA-453) | Acyl carrier protein, PTM N-Met (8,570.55) | 30.00 | ||
| 5513 | >Q99Z11 Q99Z11_STRP1 | Streptococcus pyogenes serotype M1 | Putative uncharacterized protein, PTM N-Met (8,568.00) | 4.3E−7 |
A protein marker at m/z 8571.4 was analyzed by MS-MS using MALDI-TOF-TOF MS and compared to all in silico fragment ions and D-, E-, and P-specific in silico fragment ions of bacterial protein sequences having the same molecular mass as the biomarker (within 5 Da). This corresponded to 1,425 in silico bacterial protein sequences. The protein biomarker had been identified previously as DUF-465 in another strain of C. coli by bottom-up proteomics techniques (11). The top identification of both algorithms was the correct identification of the protein and its source microorganism. In addition, the top identification scores of the two algorithms are enhanced compared with the “runner-up” identification scores. The parameters for comparison of MS-MS data and in silico data were as follows: intensity threshold, 4%; number of MS-MS peaks with an intensity of ≥4%, 40; m/z range for comparison, 0 to 14,000 Th; fragment ion tolerance, 2.5 Th; and protein MW, 8,570 ± 10. “PTM N-Met” indicates that the in silico protein sequence was modified to remove the N-terminal methionine. The algorithm computation times for all in silico fragment ions and for the D-, E-, and P-specific in silico fragment ions were as follows: USDA peak-matching algorithm, 28.6 and 15.4 s, respectively; and P value algorithm, 32.4 and 26.0 s, respectively.
Identifications of other protein biomarkers of C. coli strain RM2228 are shown in the supplemental material.
DISCUSSION
Quality of MS-MS data, multiple biomarkers, and in silico identification.
The algorithms and software were tested using MS-MS data whose quality was variable. This reflected, in part, a gradual increase in our skill at acquiring MS-MS data for intact proteins using the MALDI-TOF-TOF instrument (an application for which this instrument was not originally designed). Our initial MS-MS data were not as good as the data collected in our later MS-MS experiments. However, it seemed useful to test the software with both high-quality and lower-quality MS-MS data. This approach was facilitated by the fact that most of the protein biomarkers identified by MS-MS had been identified previously by bottom-up proteomics (9, 11, 12) and so provided an excellent data set to test the limits of algorithm and software identification. Table 5 summarizes the quality of MS-MS spectra analyzed in this study. Two criteria were used to evaluate the MS-MS spectra qualitatively: (i) the number of prominent fragment ion peaks observed in the MS-MS spectrum (which is proportional to the fragmentation efficiency of the protein) and (ii) the noise background of the MS-MS spectrum. Typically, a higher-intensity-threshold cutoff was used for MS-MS spectra that exhibited a noisier baseline. The noise background was not necessarily uniform over the entire m/z range, which contributed to the problem of selecting the optimum intensity threshold to apply over the entire spectrum. Although increasing the intensity threshold cutoff can reduce chemical noise contributions from a noisy baseline, it may also eliminate genuine low-intensity fragment ions that are prominent in a less noisy part of the MS-MS spectrum. Not surprisingly, higher-quality MS-MS data resulted in higher-scoring correct identifications, whereas lower-quality MS-MS data resulted in lower-scoring correct identifications (or incorrect identifications). For the lower-quality MS-MS data, in some cases it was necessary to restrict the in silico comparison to residue-specific in silico fragment ions (e.g., D,- E,- and P-specific or D-specific in silico fragment ions) in order to obtain a top-scoring correct identification. Presumably, a non-residue-specific comparison (i.e., a comparison of all in silico ions) is likely to have an increased probability of random in silico matches to chemical noise peaks, which may contribute to the greater difficulty of correctly identifying the protein from poorer-quality MS-MS data. By narrowing the in silico comparison to only the in silico fragment ions that have the highest probability for formation (D-, E-, and P-specific or D-specific in silico fragment ions), many random (false) in silico matches are eliminated, resulting in a more prominent score for the correct identification.
TABLE 5.
Quality of MS-MS spectra analyzed in this study
| Species | Strain | Protein(s) | Table | Intensity threshold (%) | No. of MS-MS ionsa | MS-MS quality |
|---|---|---|---|---|---|---|
| C. upsaliensis | RM3195 | Thioredoxin | 1 | 2 | 52 | Excellent |
| C. upsaliensis | RM3195 | 50S L7/L12 ribosomal protein | 2 | 2 | 56 | Excellent |
| C. upsaliensis | RM3195 | 10-kDa chaperonin | S1b | 2 | 79 | Excellent |
| C. upsaliensis | RM3195 | 4-Oxalocrotonate tautomerase (DmpI)-related protein | S2b | 6 | 25 | Poor |
| C. upsaliensis | RM3195 | DUF-465 | S3b | 6 | 27 | Fair |
| C. lari | RM2100 | Thioredoxin | 3 | 4 | 69 | Fair |
| C. lari | RM2100 | DNA-binding protein HU | S4b | 10 | 60 | Poor |
| C. lari | RM2100 | 10-kDa chaperonin and cytochrome c | S5b | 4 | 52 | Fair |
| C. coli | RM2228 | DUF-465 | 4 | 4 | 40 | Good |
| C. coli | RM2228 | 4-Oxalocrotonate tautomerase (DmpI)-related protein | S6b | 4 | 57 | Poor |
Number of MS-MS ions whose relative intensity exceeds the intensity threshold.
See the supplemental material.
It should be noted that MS-MS fragment ion intensity per se is not used (by either algorithm) as a criterion for comparing MS-MS fragment ions to in silico fragment ions. Only m/z are compared. However, a minimum intensity threshold is applied to the relative intensities of MS-MS fragment ions. This intensity threshold was determined ad hoc based on the amount of baseline noise of the MS-MS spectrum after processing (but prior to centroiding). Although the absolute (or relative) intensities of fragment ions are not directly involved in algorithm calculations, one would expect that a “correct” identification by an algorithm should match a greater number of prominent fragment ions than the top incorrect identification by the algorithm. This is shown in Fig. S9 in the supplemental material. MS-MS fragment ion peaks whose relative intensities only slightly exceed the intensity threshold may or may not be caused by chemical noise. “Matches” of in silico fragment ions to the lowest-intensity MS-MS fragment ions are less significant from an MS standpoint than matches to other more prominent fragment ions. However, neither algorithm discriminates on the basis of fragment ion intensity as long as the ion peak intensity is above the preset threshold.
In addition to the problem of random in silico matches to chemical noise peaks, fragment ions from multiple protein biomarkers can increase the difficulty of identifying individual protein biomarkers. As noted previously, the nearly identical MWs of the 10,000-MW chaperonin (average MW, 9,617.3) and cytochrome c (average MW, 9,617.0) of C. lari strain RM2100 means that it is not possible to isolate these ions on the basis of m/z; i.e., fragment ions from both proteins are detected (13, 14). Consequently, fragment ions from cytochrome c probably contributed to the difficulty of identifying the 10-kDa chaperonin using a comparison of all in silico fragment ions (see Table S5A in the supplemental material). As mentioned previously, the protein sequence for cytochrome c was not included in the in silico database because the mature protein polypeptide is covalently linked with a heme group (MW, 616.5), making in silico identification complicated. Consequently, the MS-MS fragment ions of cytochrome c could not be correctly matched to their in silico sequence; however, they could be incorrectly matched to in silico fragment ions of other protein sequences in the database (i.e., false or random matches). Consequently, use of D-, E-, and P-specific in silico comparison and then D-specific in silico comparison narrowed the MS-MS in silico comparison to only the in silico fragment ions that have the greatest probability for formation. This may significantly reduce the number of random matches and result in a top score correctly identifying one of the protein biomarkers (i.e., the 10-kDa chaperonin) (see Tables S5B and S5C in the supplemental material). However, although the 10-kDa chaperonin was correctly identified with the highest USDA and P value scores in a D-specific in silico comparison, the top score is still “grouped” with the “runner-up” scores (see Table 5C in the supplemental material). This suggests the importance of using ion isolation for restricting MS-MS analysis to a single protein whenever possible.
Residue-specific versus non-residue-specific in silico comparisons.
Our analysis in the current study indicated that the simple peak-matching algorithm and the more complicated P value algorithm of Demirev and coworkers appear to perform fairly well for either a non-residue-specific in silico comparison or a D-, E-, and P-specific or D-specific in silico comparison. In 2005, Demirev and coworkers (7) reported testing their algorithm for only non-residue-specific in silico comparisons; i.e., all possible in silico fragment ions (a, b, and y ions with up to two small neutral losses [NH3 or H2O]) were compared without regard to the residues adjacent to the sites of polypeptide cleavage responsible for the in silico fragment ions formed (7). Our analysis using both the peak-matching and P value algorithms suggests that a D-, E-, and P-specific or D-specific in silico comparison can reveal a correct identification that is not always apparent from a non-residue-specific in silico comparison. This is particularly apparent in the analysis of MS-MS data whose quality is marginal (see Table S2 in the supplemental material) or of MS-MS data for fragment ions that cannot be correctly “matched” to in silico ions because of PTM of the mature protein (see Table S5 in the supplemental material).
Algorithm complexity and computation speed.
The relative computational efficiency of an algorithm may play an increasingly important role as the number of in silico bacterial proteins increases due to the increasing number of bacterial genomes in public and private databases. The USDA peak-matching algorithm is mathematically much simpler than the P value formula. Not surprisingly, P value calculation is computationally more intensive and thus requires more time than the USDA algorithm, especially as the number of MS-MS fragment ions increases. The disparity in computation time between the two algorithms becomes more apparent as the number of MS-MS fragment ions increases. In the P value formula (7), the number of MS-MS fragment ions is designated “K,” the number of “matches” is designated “k,” and the number of in silico ions is designated “n.” The unexpected increase in computation time for P value calculation for a D-, E-, and P-specific analysis (Table 3; see Table S1B in the supplemental material) compared to a non-residue-specific analysis (Table 3; see Table S1A in the supplemental material), where the values of K are 79 and 69, respectively, is probably due to the calculation of factorials and powers used in the P value formula [e.g., (K − k)!]. Although fewer in silico ions (n) are compared to MS-MS fragment ions for a D-, E-, and P-specific analysis than for a non-residue-specific analysis, the number of “matches” may also decline, resulting in an increase in computation time for calculating (K − k)!.
Identification of protein biomarkers from unknown (nongenomically sequenced) bacterial strains.
Identification of bacteria (or other microorganisms) using sequence-specific fragmentation of their protein biomarkers is dependent on the availability and accuracy of the genomic information from which the in silico protein amino acid sequences are derived. In order to test the algorithms and software, we examined genomically sequenced strains of Campylobacter whose protein biomarkers had been identified previously by bottom-up proteomics techniques. Although the software and algorithm were not specifically designed to identify unknown (nongenomically sequenced) bacterial strains, the usefulness of this technique would be enhanced if unknown bacterial strains could also be identified. The ability to identify an unknown (nongenomically sequenced) bacterial strain using this technique would be dependent on the extent of sequence homology between the unknown strain and a genomically sequenced strain. A protein sequence from an unknown strain may contain amino acid substitutions compared to the same protein sequence from a genomically sequenced strain. These substitutions may result in a protein molecular mass that is outside the range specified in the initial protein search (±5 Da) of genomic and proteomic databases. However, it may still be possible to identify such proteins by expanding the protein molecular mass range for search and retrieval (e.g., ±50 Da). This would greatly expand the number of proteins retrieved from public database and uploaded to the in silico protein database. It would also allow possible protein identification from partial sequence homology between the protein sequence of an unknown strain and the protein sequence of a genomically sequenced strain. The likelihood of identification would depend on the number and location of the amino acid substitutions. The number of amino acid variations is dependent on the phylogenetic distance between the unknown and genomically sequenced strains (10). The more closely related the two strains are, the fewer the amino acid substitutions and the greater the probability that a protein from an unknown strain could be identified based on its sequence homology to a protein from a genomically sequenced strain (10).
Identification of protein biomarkers from mixtures of bacterial strains.
In the current study, the software was tested by using identification of protein biomarkers from pure bacterial strains. However, the only limitation of this technique for its application in analysis of bacterial mixtures is the resolving power of the TIS, which is used to isolate specific protein ions on basis of their m/z. Currently, the narrowest TIS “window” obtainable with the TOF-TOF instrument is ±50 Da at 10 kDa. If two protein ions (either from a single bacterial strain or from multiple strains) are separated in m/z by 50 Th (or more), then it is possible to mass isolate (resolve) these two protein ions and identify each protein from the fragment ions generated. However, if two protein ions are separated in m/z by less than 50 Th, the TIS is not able to isolate the two protein precursor ions, and fragment ions from both precursor ions may be detected (although this also depends on the fragmentation efficiency of the two protein ions). Software analysis of fragment ions from multiple precursor ions may result in “runner-up” identifications that reflect correct MS-MS-in silico matches that are different from MS-MS-in silico matches of the top identification.
We have developed web-based software for rapid top-down proteomic identification of small proteins (and their source bacterial microorganisms) from analysis of MS-MS fragment ions of intact bacterial proteins generated using MALDI-TOF-TOF MS. A simple peak-matching algorithm was used to score and rank identifications of proteins and microorganisms by comparing MS-MS fragment ions to in silico fragment ions generated from bacterial protein sequences derived from genomic databases. The P value algorithm of Demirev and coworkers was also incorporated into the software for purposes of comparison. The algorithms and software were successfully tested with protein biomarkers of species and strains of Campylobacter that had been identified previously by bottom-up proteomics techniques. A database of in silico fragment ions was constructed for bacterial protein sequences whose calculated MWs corresponded to the m/z of a protein biomarker observed in MALDI-TOF MS spectra. In silico fragment ions were identified by m/z, type, number, and the amino acid residues adjacent to the site of polypeptide fragmentation resulting in a fragment ion. Consequently, MS-MS fragment ions could be compared to in silico fragment ions without regard to the residues adjacent to the site of fragmentation (i.e., non-residue-specific comparison), or MS-MS fragment ions could be compared only to the in silico fragment ions that were formed as a result of polypeptide fragmentation adjacent to specific residues (i.e., residue-specific comparison). A D-, E-, and P-specific or D-specific analysis often enhanced the top identification score (correct identification) relative to the scores of the “runner-up” identifications compared to the top identification score for a non-residue-specific analysis. In some cases, a protein biomarker was successfully identified by a residue-specific analysis when a non-residue-specific analysis failed to correctly identify the protein or its source microorganism. The success of D-, E-, and P-specific or D-specific in silico analysis for identification confirms the importance of these residues in the fragmentation of singly charged (protonated) proteins. Although the relative intensities of protein biomarker ions are not explicit criteria used in the algorithm, it is reasonable to expect that a correct identification should “match” many of the most prominent MS-MS fragment ions. This was found to be the case. Finally, fragment ion error analysis may be successfully used to confirm an algorithm identification by distinguishing systematic fragment ion error caused by drift in the TOF calibration from random error caused by random matches between MS-MS fragment ions and in silico fragment ions.
Supplementary Material
Acknowledgments
We thank Peter Højrup at Lighthouse for modifying his existing GPMAW software and providing it to us as a beta version. We also thank Christine Hoogland at Bioinformatics Institute of Switzerland for her assistance. We thank Linda C. Whitehand for statistical discussions and Robert E. Mandrell and Al Lastovica for providing Campylobacter strains RM3195, RM2100, and RM2228.
Mention of a brand or firm does not constitute an endorsement by the U.S. Department of Agriculture over other similar brands or firms not mentioned. This article describes U.S. Government work.
Footnotes
Published ahead of print on 1 May 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Arnold, R., and J. Reilly. 1998. Fingerprint matching of E. coli strains with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry of whole cells using a modified correlation approach. Rapid Commun. Mass Spectrom. 12:630-636. [DOI] [PubMed] [Google Scholar]
- 2.Cain, T. C., D. M. Lubman, and W. J. Weber, Jr. 1994. Differentiation of bacteria using protein profiles from matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 8:1026-1030. [Google Scholar]
- 3.Dai, Y., L. Li, D. Roser, and S. R. Long. 1999. Detection and identification of low-mass peptides and proteins from solvent suspensions of Escherichia coli by high performance liquid chromatography fractionation and matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun. Mass Spectrom. 13:73-78. [DOI] [PubMed] [Google Scholar]
- 4.Demirev, P. A., Y.-P. Ho, V. Ryzhov, and C. Fenselau. 1999. Microorganism identification by mass spectrometry and protein database searches. Anal. Chem. 71:2732-2738. [DOI] [PubMed] [Google Scholar]
- 5.Demirev, P. A., J. S. Lin, F. J. Peneda, and C. Fenselau. 2001. Bioinformatics and mass spectrometry for microorganism identification: proteome-wide post-translational modifications and database search algorithms for characterization of intact H. pylori. Anal. Chem. 73:4566-4573. [DOI] [PubMed] [Google Scholar]
- 6.Demirev, P. A., J. Ramirez, and C. Fenselau. 2001. Tandem mass spectrometry of intact proteins for characterization of biomarkers from Bacillus cereus T spores. Anal. Chem. 73:5725-5731. [DOI] [PubMed] [Google Scholar]
- 7.Demirev, P. A., A. B. Feldman, P. Kowalski, and J. S. Lin. 2005. Top-down proteomics for rapid identification of intact microorganisms. Anal. Chem. 77:7455-7461. [DOI] [PubMed] [Google Scholar]
- 8.Donohue, M. J., A. W. Smallwood, S. Pfaller, M. Rodgers, and J. A. Shoemaker. 2006. The development of a matrix-assisted laser desorption/ionization mass spectrometry-based method for the protein fingerprinting and identification of Aeromonas species using whole cells. J. Microbiol. Methods 65:380-389. [DOI] [PubMed] [Google Scholar]
- 9.Fagerquist, C. K., W. G. Miller, L. A. Harden, A. H. Bates, W. H. Vensel, G. Wang, and R. E. Mandrell. 2005. Genomic and proteomic identification of a DNA-binding protein used in the “fingerprinting” of Campylobacter species and strains by MALDI-TOF-MS protein biomarker analysis. Anal. Chem. 77:4897-4907. [DOI] [PubMed] [Google Scholar]
- 10.Fagerquist, C. K., A. H. Bates, S. Heath, B. C. King, B. R. Garbus, L. A. Harden, and W. G. Miller. 2006. Sub-speciating Campylobacter jejuni by proteomic analysis of its protein biomarkers and their post-translational modifications. J. Proteome Res. 5:2527-2538. [DOI] [PubMed] [Google Scholar]
- 11.Fagerquist, C. K., E. Yee, and W. G. Miller. 2007. Composite sequence proteomic analysis of protein biomarkers of Campylobacter coli, C. lari and C. concisus for bacterial identification. Analyst 132:1010-1023. [DOI] [PubMed] [Google Scholar]
- 12.Fagerquist, C. K. 2007. Amino acid sequence determination of protein biomarkers of Campylobacter upsaliensis and C. helveticus by “composite” sequence proteomic analysis. J. Proteome Res. 6:2539-2549. [DOI] [PubMed] [Google Scholar]
- 13.Fagerquist, C. K. 2007. Identification of foodborne bacteria by MALDI-TOF-TOF analysis of protein biomarkers, abstr. 5-1604. Abstr. 121st AOAC Conf., Anaheim, CA.
- 14.Fagerquist, C. K., K. E. Williams, and A. H. Bates. 2007. Identification of foodborne bacteria by high energy collision-induced dissociation of their protein biomarkers by MALDI tandem-time-of-flight mass spectrometry, abstr. MPT-327. Proc. 55th Am. Soc. Mass Spectrom. Conf., Indianapolis, IN.
- 15.Fenn, J. B., M. Mann, C. K. Meng, S. F. Wong, and C. M. Whitehouse. 1989. Electrospray ionization for mass spectrometry of large biomolecules. Science 246:64-71. [DOI] [PubMed] [Google Scholar]
- 16.Fenselau, C., and P. A. Demirev. 2001. Characterization of intact microorganisms by MALDI mass spectrometry. Mass Spectrom. Rev. 20:157-171. [DOI] [PubMed] [Google Scholar]
- 17.Reference deleted.
- 18.Reference deleted.
- 19.Haag, A., S. Taylor, K. Johnston, and R. Cole. 1998. Rapid identification and speciation of Haemophilus bacteria by matrix-assisted laser desorption/ ionization time-of-flight mass spectrometry. J. Mass Spectrom. 33:750-756. [DOI] [PubMed] [Google Scholar]
- 20.Reference deleted.
- 21.Holland, R. D., J. G. Wilkes, F. Rafii, J. B. Sutherland, C. C. Persons, K. J. Voorhees, and J. O. Lay, Jr. 1996. Rapid identification of intact whole bacteria based on spectral patterns using matrix-assisted laser desorption/ionization with time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 10:1227-1232. [DOI] [PubMed] [Google Scholar]
- 22.Jarman, K. H., S. T. Cebula, A. J. Saenz, C. E. Petersen, N. B. Valentine, M. T. Kingsley, and K. L. Wahl. 2000. An algorithm for automated bacterial identification using matrix-assisted laser desorption/ionization mass spectrometry. Anal. Chem. 72:1217-1223. [DOI] [PubMed] [Google Scholar]
- 23.Jones, J. J., M. J. Stump, R. C. Fleming, J. O. Lay, Jr., and C. L. Wilkins. 2003. Investigation of MALDI-TOF and FT-MS techniques for analysis of Escherichia coli whole cells. Anal. Chem. 75:1340-1347. [DOI] [PubMed] [Google Scholar]
- 24.Karas, M., D. Bachmann, U. Bahr, and F. Hillenkamp. 1987. Matrix-assisted ultraviolet laser desorption of non-volatile compounds. Int. J. Mass Spectrom, Ion Proc. 78:53-68. [Google Scholar]
- 25.Krishnamurthy, T., P. L. Ross, and U. Rajamani. 1996. Detection of pathogenic and non-pathogenic bacteria by matrix-assisted laser desorption/ ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 10:883-888. [DOI] [PubMed] [Google Scholar]
- 26.Krishnamurthy, T., and P. L. Ross. 1996. Rapid identification of bacteria by direct matrix-assisted laser desorption/ionization mass spectrometric analysis of whole cells. Rapid Commun. Mass Spectrom. 10:1992-1996. [DOI] [PubMed] [Google Scholar]
- 27.Lay, J. O., Jr. 2001. MALDI-TOF mass spectrometry of bacteria. Mass Spectrom. Rev. 20:172-194. [DOI] [PubMed] [Google Scholar]
- 28.Lin, M., J. M. Campbell, D. R. Mueller, and U. Wirth. 2003. Intact protein analysis by matrix-assisted laser desorption/ionization tandem time-of-flight mass spectrometry. Rapid Comm. Mass Spectrom. 17:1809-1814. [DOI] [PubMed] [Google Scholar]
- 29.Mandrell, R. E., L. A Harden, A. H. Bates, W. G. Miller, W. F. Haddon, and C. K. Fagerquist. 2005. Speciation of Campylobacter coli, C. jejuni, C. helveticus, C. lari, C. sputorum, and C. upsaliensis by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 71:6292-6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reference deleted.
- 31.Paizs, B., and S. Suhai. 2005. Fragmentation pathways of protonated peptides. Mass Spectrom. Rev. 24:508-548. [DOI] [PubMed] [Google Scholar]
- 32.Pineda, F. J., J. S. Lin, C. Fenselau, and P. A. Demirev. 2000. Testing the significance of microorganism identification by mass spectrometry and proteome database search. Anal. Chem. 72:3739-3744. [DOI] [PubMed] [Google Scholar]
- 33.Pineda, F. J., M. D. Antoine, P. A. Demirev, A. B. Feldman, J. Jackman, M. Longenecker, and J. S. Lin. 2003. Microorganism identification by matrix-assisted laser/desorption ionization mass spectrometry and model-derived ribosomal protein biomarkers. Anal. Chem. 75:3817-3822. [DOI] [PubMed] [Google Scholar]
- 34.Ramirez, J., and C. Fenselau. 2001. Factors contributing to peak broadening and mass accuracy in the characterization of intact spores using matrix-assisted laser desorption/ionization coupled with time-of-flight mass spectrometry. J. Mass Spectrom. 36:929-936. [DOI] [PubMed] [Google Scholar]
- 35.Reference deleted.
- 36.Reference deleted.
- 37.Tanaka, K., Y. Ido, S. Akita, Y. Yoshida, and T. Yoshida. 1987. Abstr. 2nd Japan-China Joint Symp. Mass Spectrom., Osaka, Japan, p. 185-188.
- 38.Taylor, J. A., and R. S. Johnson. 2001. Implementation and uses of automated de novo peptide sequencing by tandem mass spectrometry. Anal. Chem. 73:2594-2604. [DOI] [PubMed] [Google Scholar]
- 39.Wahl, K. L., S. C. Wunschel, K. H. Jarman, N. B. Valentine, C. E. Petersen, M. T. Kingsley, K. A. Zartolas, and A. J. Saenz. 2002. Analysis of microbial mixtures by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal. Chem. 74:6191-6199. [DOI] [PubMed] [Google Scholar]
- 40.Wang, Z., L. Russon, L. Li, D. Roser, and S. R. Long. 1998. Investigation of spectral reproducibility in direct analysis of bacteria proteins by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 12:456-464. [DOI] [PubMed] [Google Scholar]
- 41.Welham, K., M. Domin, D. Scannell, E. Cohen, and D. Ashton. 1998. The characterization of micro-organisms by matrix-assisted laser desorption/ ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 12:176-180. [DOI] [PubMed] [Google Scholar]
- 42.Whiteaker, J., J. Karns, C. Fenselau, and M. L. Perdue. 2004. Analysis of Bacillus anthracis spores in milk using mass spectrometry. Foodborne Pathog. Dis. 1:185-194. [DOI] [PubMed] [Google Scholar]
- 43.Wunschel, S. C., K. H. Jarman, C. E. Petersen, N. B. Valentine, K. L. Wahl, D. Schauki, J. Jackman, C. P. Nelson, and E. White V. 2005. Bacterial analysis by MALDI-TOF mass spectrometry: an inter-laboratory comparison. J. Am. Soc. Mass Spectrom. 16:456-462. [DOI] [PubMed] [Google Scholar]
- 44.Wysocki, V. H., G. Tsaprailis, L. L. Smith, and L. A. Breci. 2000. Mobile and localized protons: a framework for understanding peptide dissociation. J. Mass Spectrom. 35:1399-1406. [DOI] [PubMed] [Google Scholar]
- 45.Yao, Z.-P., P. A. Demirev, and C. Fenselau. 2002. Mass spectrometry-based proteolytic mapping for rapid virus identification. Anal. Chem. 74:2529-2534. [DOI] [PubMed] [Google Scholar]
- 46.Yu, W., J. E. Vath, M. C. Hurberty, and S. A. Martin. 1993. Identification of the facile gas-phase cleavage of the Asp-Pro and Asp-Xxx peptide bonds in matrix-assisted laser desorption time-of-flight mass spectrometry. Anal. Chem. 65:3015-3023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.