Abstract
Enterococcus faecalis WHE 96, a strain isolated from soft cheese based on its anti-Listeria activity, produced a 5,494-Da bacteriocin that was purified to homogeneity by ultrafiltration and cation-exchange and reversed-phase chromatographies. The amino acid sequence of this bacteriocin, named enterocin 96, was determined by Edman degradation, and its structural gene was sequenced, revealing a double-glycine leader peptide. After a comparison with other bacteriocins, it was shown that enterocin 96 was a new class II bacteriocin that showed very little similarity with known structures. Enterocin 96 was indeed a new bacteriocin belonging to class II bacteriocins. The activity spectrum of enterocin 96 covered a wide range of bacteria, with strong activity against most gram-positive strains but very little or no activity against gram-negative strains.
Bacteriocins are a heterogeneous group of ribosomally synthesized antibacterial peptides that inhibit strains and species that are usually, but not always, closely related to the producing bacteria (16). Enterococcal bacteriocins, often termed enterocins, have been widely investigated, mainly because they are active against gram-positive food-borne pathogens, such as Listeria monocytogenes, Staphylococcus aureus, and Bacillus cereus. The vast majority of enterocins are active only against gram-positive bacteria (10, 17); however, some exceptions with broad activity spectra described in recent years showed the ability to inhibit the growth of gram-negative microorganisms (5, 11, 13).
The increasing number of enterocins reported in the literature and the emergence of novel structures that could not be included in classical bacteriocin classifications (12, 14, 17) prompted the grouping of enterocins into a new four-class scheme by Franz et al. (8). Most enterocins known so far were included in class II (small, nonlantibiotic peptides), which was divided into three subgroups: class II.1, enterocins of the pediocin family; class II.2, enterocins synthesized without a leader peptide; and class II.3, other linear, non-pediocin-like enterocins.
The fact that numerous Enterococcus strains found in a variety of fermented and nonfermented foods produce bacteriocins, often more than one per strain, has sparked interest in their use in food preservation (4). Despite the concerns over enterococci as opportunistic pathogens and indicators of fecal contamination, they are indigenous species in the gastrointestinal tract and have long been used as human and/or animal probiotics (1, 2, 7).
In this work, we describe and characterize a new class II enterocin produced by Enterococcus faecalis WHE 96, previously isolated from Munster cheese, for its anti-Listeria properties. The amino acid sequence, the structural gene, and the spectrum of activity of this bacteriocin are reported.
MATERIALS AND METHODS
Bacterial strains and cultures.
The bacteriocin producer E. faecalis WHE 96 was isolated from Munster cheese. E. faecalis CECT 481 was chosen as the indicator strain to monitor bacteriocin activity during the purification process. The strains used to determine the activity spectrum of the purified bacteriocin are listed in Table 1. All cultures were maintained as frozen stocks held at −80°C in brain heart infusion broth with 30% glycerol (Bio-Rad, Marnes-La-Coquette, France). For short storage, the strains were subcultured on brain heart infusion agar (Bio-Rad) and kept at 4°C. Before experimental use, the strains were cultivated twice for 18 to 24 h in MRS broth (Biokar Diagnostics, Beauvais, France). Culturing was done without aeration.
TABLE 1.
Antimicrobial activity of purified enterocin 96
| Indicator species | Straina | Bacteriocin activityb |
|---|---|---|
| Enterococcus faecalis | CECT 481 | 0.41 ± 0.05 |
| LC E5 | 0.16 ± 0.02 | |
| LC E3 | 0.44 ± 0.04 | |
| LC E4 | 0.41 ± 0.08 | |
| CECT 795 | 0.45 ± 0.02 | |
| Enterococcus faecium | WHE 81 | 0.14 ± 0.02 |
| LC E14 | 0.24 ± 0.02 | |
| Enterococcus hirae | CIP 5855 | 0.37 ± 0.09 |
| Enterococcus pseudoavium | LC E18 | 0.18 ± 0.01 |
| Enterococcus sulfureus | LC E22 | 0.40 ± 0.04 |
| Enterococcus saccharolyticus | ATCC 43076 | 0.10 ± 0.04 |
| Enterococcus columbae | LC E2 | 0.33 ± 0.02 |
| Lactobacillus plantarum | CIP A159 | 0.15 ± 0.03 |
| CECT 749 | 0.25 ± 0.03 | |
| 299V | 0.24 ± 0.02 | |
| Lactobacillus acidophilus | LC 660 | 0 |
| Lactobacillus sakei subsp. sakei | ATCC 15521 | 0 |
| Lactobacillus paracasei subsp. paracasei | LC 94 | 0.49 ± 0.08 |
| Lactococcus lactis | LC RO50 | 0 |
| LC 72 | 0.17 ± 0.01 | |
| Lactococcus lactis subsp. cremoris | LC 657 | 0.35 ± 0.02 |
| Leuconostoc mesenteroides | LC258 | 0.14 ± 0.03 |
| Bacillus subtilis | CIP 7718 | 0.17 ± 0.04 |
| CIP 5262 | 0.23 ± 0.08 | |
| Bacillus cereus | CIP 78-3 | 0.21 ± 0.01 |
| LC 447 | 0.17 ± 0.03 | |
| Listeria innocua | LC14 | 0.13 ± 0.03 |
| Listeria monocytogenes | CIP 7838 | 0.35 ± 0.10 |
| LC 12 | 0.20 ± 0.02 | |
| LC 40 | 0.19 ± 0.01 | |
| Staphylococcus xylosus | LC 57 | 0.23 ± 0.03 |
| Staphylococcus aureus | ATCC 6538 | 0.44 ± 0.02 |
| Salmonella enterica serovar Typhimurium | LC 443 | 0.13 ± 0.03 |
| Salmonella enterica serovar Infantis | LC 8 | 0 |
| Klebsiella pneumoniae | LC 261 | 0.081 ± 0.005 |
| Serratia liquefaciens | 384 | 0.076 ± 0.01 |
| Proteus vulgaris | 243 | 0.068 ± 0.008 |
| Enterobacter cloacae | 407 | 0 |
| Escherichia coli | LC 22 | 0.063 ± 0.005 |
| CIP 7624 | 0 | |
| ATCC 10536 | 0.093 ± 0.008 |
LC, laboratory collection; CIP, Collection de L'Institut Pasteur; ATCC, American Type Culture Collection; CECT, Coleccion Española de Cultivos Tipo.
Expressed as the highest difference in the absorbance at 600 nm between the indicator strain and the control observed during incubation. Means ± standard deviations for four independent experiments are given.
Antibacterial activity determination.
Bacteriocin activity in the cell-free culture supernatant and in fractions obtained during the purification process was tested by the spot-on-a-lawn test (6), by aliquoting a 10-μl sample onto MRS agar that was overlaid with Lactobacilli Agar AOAC (Difco, Le Pont-de-Claix, France) inoculated with approximately 106 CFU/ml of indicator cells. An overnight culture of E. faecalis CECT 481 in MRS broth at 37°C was used for inoculation. To monitor antibacterial activity during the purification process, the diameter of the inhibition zone (in millimeters) was measured. The activities of fractions collected after each purification step were quantified by serial twofold dilutions and expressed in activity units (AU) per milliliter as previously described (6).
The antimicrobial spectrum of enterocin 96 was determined using a Bioscreen 200C automated turbidometer (Labsystems, Helsinki, Finland) and a wide range of target bacteria (Table 1). Strains were grown for 18 h in MRS broth at 37°C. Cultures were then diluted in fresh broth to reach a final concentration of ca. 105 CFU/ml, and 190 μl of each was loaded into microplate wells. Ten microliters of the purified bacteriocin solution was then added to these wells. Controls consisted of 190 μl of diluted cultures supplemented with 10 μl of sterile deionized water. All assays were repeated four times, and duplicates were run for each repeat. The growth of indicator strains was monitored at 600 nm for 24 h at 37°C. Bacteriocin activity was expressed as the maximal difference in absorbance between controls and assays, which was reached at the end of the exponential-growth phase (between 7 and 10 h depending on the strain) (9).
Bacteriocin purification.
E. faecalis WHE 96 was inoculated in 5 liters of MRS broth to yield an initial count of ca. 105 CFU/ml and was then incubated for 15 h at 37°C. Cells were removed by centrifugation at 4,000 × g for 15 min, and the supernatant was filter sterilized (pore size, 0.45 μm; Durapore mixed cellulose ester filter; Millipore, Carrigtwohill, Ireland).
The culture extract obtained was ultrafiltrated using a Centramate tangential flow system (Pall, East Hills, NY) through a membrane with a molecular weight cutoff (MWCO) of 10,000 (Centramate cassette; Pall). The permeate obtained was concentrated to a final volume of 220 ml using a membrane with an MWCO of 5,000.
The retentate obtained was adjusted to pH 5.5 with NaOH and was further purified by passage on a cation exchanger (SP-Sepharose HP; length, 100 mm; internal diameter, 26 mm; Amersham Biosciences, Orsay, France). Equilibration and washing were done with 20 mM sodium acetate at pH 5.5 (buffer A) prior to elution with an NaCl gradient in 60 min from 0 to 1 M NaCl in buffer A at a flow rate of 5.0 ml/min. Active fractions were collected and applied to C8 reversed-phase (Polaris C8-A; length, 250 mm; internal diameter, 10 mm; pore size, 180 Å; particle size, 5 μm; Varian, Les Ulis, France) high-performance liquid chromatography. After a 20-min wash with 20% acetonitrile in water containing 0.1% trifluoroacetic acid, elution was done with a gradient from 20% to 60% acetonitrile in water containing 0.1% trifluoroacetic acid in 60 min at a flow rate of 2 ml/min. Detection was performed at 280 nm using a UV-900 photodiode array detector (Amersham Biosciences). The sample containing the purified bacteriocin was vacuum dried using a DD-4X centrifugal evaporator (Genevac, Ipswich, United Kingdom) and stored at −20°C. Prior to activity testing and structural determination, the powder obtained was redissolved in deionized water.
Mass analysis and amino acid sequencing.
The molecular mass of the purified bacteriocin was determined by a Voyager DE-PRO matrix-assisted laser desorption ionization-time-of-flight (MALDI-TOF) mass spectrometer (Applied Biosystems, Foster City, CA).
The amino acid sequence of the reduced and alkylated bacteriocin was determined by Edman degradation on an Applied Biosystems Procise 494 automatic sequencer. The sample was loaded onto a Biobrene (Applied Biosystems) pretreated fiber glass disc and placed in the reaction chamber.
PCR amplification and sequencing of the bacteriocin gene.
Plasmid DNA was isolated from cells from 5-ml overnight cultures with a GeneJET plasmid miniprep kit (Fermentas, Hanover, MD) as described by the manufacturer. The oligonucleotide primers used in this study were obtained from Millegen (Labège, France). PCR was carried out using Taq (Invitrogen, Cergy Pontoise, France) and KOD (Novagen, Darmstadt, Germany) DNA polymerases under the following amplification conditions: an initial cycle of denaturation (97°C for 2 min), followed by 35 cycles of denaturation (94°C for 45 s), annealing (50 to 62°C for 30 s), and elongation (72°C for 15 s to 3 min), and a final extension step at 72°C for 7 min in a 2720 DNA thermal cycler (Applied Biosystems, Foster City, CA). The resulting PCR products were cleaned with a QIAquick PCR purification kit (Qiagen, Courtaboeuf, France) and analyzed by electrophoresis in 1% agarose gels with a Gel Doc 1000 documentation system (Bio-Rad). They were then purified from agarose gels using an UltraClean Gel Spin kit (MoBio, Solana Beach, CA) before they were used for nucleotide sequencing. The primers used for PCR amplification were Ent96S1 (5′-TTGAAAGATTTTGATAATTT-3′) and Ent96R2 (5′-TCACAATATATTAATCTATA-3′), yielding PCR products of 348 and 170 bp, respectively. DNA sequencing was performed with a BigDye Terminator cycle sequencing ready reaction kit (version 3.1) and a model ABI3130XL genetic analyzer (Applied Biosystems) at a DNA sequencing facility (Millegen). All products were used according to the manufacturers' instructions. The nucleotide sequence was analyzed with the DNA Strider (version 1.2) program. A search for homology of the predicted amino acid sequences was done with the BLAST network service at the National Center for Biotechnology Information. Homology comparisons and calculations were done with the DNAStar program.
Nucleotide sequence accession number.
The sequence of the enterocin 96 gene determined in this study has been submitted to GenBank under accession number FJ769024.
RESULTS AND DISCUSSION
Purification and characterization of enterocin 96.
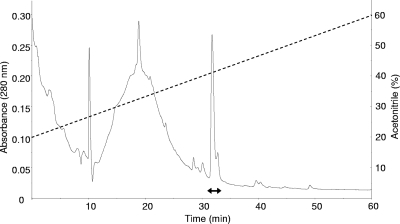
E. faecalis WHE 96, a strain isolated from Munster cheese, produces an antimicrobial peptide that was purified and biochemically characterized. The changes in the sample's biological activity and overall purity following each purification step are summarized in Table 2. As a first purification step, ultrafiltration was preferred, since it resulted in much higher bacteriocin yields than the classical ammonium sulfate precipitation (data not shown). After tangential flow filtration through a 10,000 MWCO membrane, the bacteriocin activity was detected in the permeate fraction. However, a membrane with a 5,000 MWCO retained the bacteriocin, and no activity was detected in the permeate fraction, indicating that the bacteriocin produced by E. faecalis WHE 96 had a molecular mass between 5 and 10 kDa. The crude bacteriocin sample was further purified successively by cation-exchange and reversed-phase chromatography. After the last step, the bacteriocin was detected as a single peak of activity corresponding to a chromatographic retention time of 31.5 min (Fig. 1). Active fractions were rechromatographed and purified to homogeneity (until a single peak was obtained). The final bacteriocin yield corresponded to 10% of the original activity in the culture extract, and specific activity was increased 30,000 times (Table 2).
TABLE 2.
Changes in the antibacterial activity and overall purity of enterocin 96 following successive purification steps
| Purification stage | Vol (ml) | Total A280a | Activity (AU/ml) | Total activity (AU)b | Sp actc | Yield (%)d |
|---|---|---|---|---|---|---|
| Culture extract | 5,000 | 140,250 | 100 | 500,000 | 7.13 | 100 |
| Tangential filtration | 220 | 19,712 | 1,600 | 352,000 | 17.85 | 70 |
| Cation exchange | 25 | 51 | 3,200 | 80,000 | 1,564 | 16 |
| Reversed-phase chromatography | 4 | 0.25 | 12,800 | 51,200 | 206,451 | 10 |
Calculated as A280 multiplied by the volume (in milliliters).
Calculated as the bacteriocin activity (in activity units per milliliter) multiplied by the volume (in milliliters).
Calculated as the bacteriocin activity (in activity units per milliliter) divided by the A280.
One hundred percent yield is defined as the total activity in the culture extract (in activity units).
FIG. 1.
Profile of high-performance liquid chromatography elution from a C8 column at 280 nm. The dashed line indicates the percentage of acetonitrile. The double-headed arrow indicates the active fractions.
The molecular mass of the purified bacteriocin obtained by MALDI-TOF was 5,493.92 Da, which did not correspond to that of any previously described bacteriocin. The bacteriocin amino acid sequence, determined by Edman degradation, revealed the following relatively large peptide made up of 48 unmodified amino acids: MSKRDCNLMKACCAGQAVTYAIHSLLNRLGGDSSDPAGCNDIVRKYCK. The estimated isoelectric point of this bacteriocin is 8.7, and the overall charge at pH 7.0 is 3. The calculated mass of this sequence is 5,178.98 Da, which is 314.94 Da smaller than the experimental mass obtained for the purified bacteriocin. This discrepancy indicates that the bacteriocin structure may include chemical modifications. However, since Edman degradation sequencing was not hampered, it can be concluded that the bacteriocin is a linear, nonlantibiotic peptide. This new bacteriocin, which was termed enterocin 96, belongs to the nonpediocin-like class II bacteriocins.
Comparison of enterocin 96 with other bacteriocins showed no significant amino acid sequence homologies and revealed that its structure was in fact new. Otherwise, a search in protein databases revealed that enterocin 96 was identical to the C-terminal part of a putative uncharacterized protein from E. faecalis V583 (GenBank accession number AA083145) identified after complete genome sequencing of this strain (15). The amino acid sequence of this protein contains an N-terminal extension of 26 amino acids with two C-terminal glycyl residues. The likely presence of such an extension for enterocin 96 was investigated by sequencing its structural gene.
Analysis of the gene encoding enterocin 96.
Primers Ent96S1 and Ent96R2, based on the amino acid sequence of enterocin 96, but also on the previously published nucleotide sequence of plasmid pTEF2 (GenBank accession number AE016831), which encodes the putative uncharacterized protein from E. faecalis V583, were constructed, and PCR was used to obtain the DNA sequence of the region encoding enterocin 96. This gene could be amplified using plasmid DNA isolated from E. faecalis WHE 96, showing that it was plasmid encoded. Figure 2 shows the 348-bp DNA sequence obtained (GenBank accession number FJ769024), which was identical to the 49368-to-49715 region of plasmid pTEF2 (15). The DNA sequencing confirmed the results of the amino acid sequencing and revealed that the bacteriocin is translated as a prebacteriocin of 74 amino acids, which is then processed to give the mature enterocin 96 of 48 amino acids. Enterocin 96 is synthesized with a double-glycine-type leader peptide, which is very common among enterocins. This N-terminal extension of the class II bacteriocins is important in the recognition of the prebacteriocin by the transport and maturation machinery of bacteriocin-dependent secretion systems.
FIG. 2.
Nucleotide sequence (GenBank accession number FJ769024) of the region encoding enterocin 96 of E. faecalis WHE 96, along with the deduced amino acid sequence. The portion of the amino acid sequence of enterocin 96 obtained from Edman degradation is underlined, and the signal peptidase cleavage site is indicated by an arrow. The potential Shine-Dalgarno ribosome binding sequence is double underlined.
Activity of the purified bacteriocin.
The activity of enterocin 96 was assessed by testing the sensitivities of a wide range of bacteria in liquid medium using an automated turbidometer. Sensitive strains, due to the loss of part of their population, had their growth significantly slowed down. The data for all indicator bacteria are summarized in Table 1, which represents the highest differences in population size (given by absorbance measures) obtained during growth compared to controls.
As expected, the strongest activity of enterocin 96 was detected against Enterococcus strains, especially E. faecalis strains, although most other lactic acid bacteria tested were affected. Interestingly, the bacteriocin was also active against some gram-positive pathogens, especially two tested strains of L. monocytogenes and S. aureus. Gram-negative bacteria were either slightly sensitive or resistant, which is common for the vast majority of bacteriocins from lactic acid bacteria (17). The weak activities detected against some gram-negative strains, such as Escherichia coli and Salmonella enterica serovar Typhimurium, are most certainly due to the high sensitivity of the test used in this study, which was carried out in a liquid medium. In fact, no activity of enterocin 96 could be shown against any gram-negative bacteria when the spot-in-a-lawn method was used. The antibacterial spectrum of enterocin 96 may, however, be considered relatively wide compared with those of other bacteriocins from lactic acid bacteria (3).
Concluding remarks.
The present study showed that E. faecalis WHE 96 produces a new class II bacteriocin: enterocin 96, the amino acid sequence of which is quite atypical. A search of protein databases did not reveal any significant structural similarity to other class II bacteriocins. This enterocin is produced with a leader peptide, but it lacks the pediocin consensus motif, YGNGVXC. According to the recent classification of enterocins by Franz et al. (8), enterocin 96 belongs to class II.3 (other linear nonpediocin-like enterocins), which includes nonpediocin-like enterocins that are synthesized with a leader peptide. The activity spectrum of enterocin 96 indicates biopreservation potential, especially against food-borne gram positive pathogens.
Finally, the elucidation of new bacteriocin structures remains key to understanding their structure-function relationships and mode of action. Generally, interest is focused on same-class bacteriocins with extensive sequence similarities that facilitate an analysis of the relationships between structure and activity. However, new structures that are unique are useful because they help correct misconceptions in bacteriocin classification. They also provide insights into structure-function relationships in that they show new and interesting aspects of the general mode of action of bacteriocins. In particular, such structures reveal new structural features that challenge the established structure-function dogma and may actually contribute to the development of protein-engineering strategies that use existing structures as a blueprint to develop tailor-made bacteriocin derivatives with enhanced biological activity and target cell specificity. Such attractive approaches would undoubtedly result in new and exciting applications that are able to address the challenges faced by the food industry.
Footnotes
Published ahead of print on 1 May, 2009.
REFERENCES
- 1.Agerbaek, M., L. U. Gerdes, and B. Richelsen. 1995. Hypocholesterolaemic effect of a new fermented milk product in healthy middle-aged men. Eur. J. Clin. Nutr. 49:346-352. [PubMed] [Google Scholar]
- 2.Buydens, P., and S. Debeuckelaere. 1996. Efficacy of SF 68 in the treatment of acute diarrhea. A placebo-controlled trial. Scand. J. Gastroenterol. 31:887-891. [DOI] [PubMed] [Google Scholar]
- 3.Cintas, L. M., P. Casaus, M. F. Fernández, and P. E. Hernández. 1998. Comparative antimicrobial activity of enterocin L50, pediocin PA-1, nisin A and lactocin S against spoilage and foodborne pathogenic bacteria. Food Microbiol. 15:289-298. [Google Scholar]
- 4.Cleveland, J., T. J. Montville, I. F. Nes, and M. L. Chikindas. 2001. Bacteriocins: safe, natural antimicrobials for food preservation. Int. J. Food Microbiol. 71:1-20. [DOI] [PubMed] [Google Scholar]
- 5.De Kwaadsteniet, M., S. D. Todorov, H. Knoetze, and L. M. T. Dicks. 2005. Characterization of a 3944 Da bacteriocin, produced by Enterococcus mundtii ST15, with activity against Gram-positive and Gram-negative bacteria. Int. J. Food Microbiol. 105:433-444. [DOI] [PubMed] [Google Scholar]
- 6.Ennahar, S., Y. Asou, T. Zendo, K. Sonomoto, and A. Ishizaki. 2001. Biochemical and genetic evidence for production of enterocins A and B by Enterococcus faecium WHE 81. Int. J. Food Microbiol. 70:291-301. [DOI] [PubMed] [Google Scholar]
- 7.Franz, C. M. A. P., W. H. Holzapfel, and M. E. Stiles. 1999. Enterococci at the crossroads of food safety? Int. J. Food Microbiol. 47:1-24. [DOI] [PubMed] [Google Scholar]
- 8.Franz, C. M. A. P., M. J. van Belkum, W. H. Holzapfel, H. Abriouel, and A. Galvez. 2007. Diversity of enterococcal bacteriocins and their grouping in a new classification scheme. FEMS Microbiol. Rev. 31:293-310. [DOI] [PubMed] [Google Scholar]
- 9.Izquierdo, E., A. Bednarczyk, C. Schaeffer, Y. Cai, E. Marchioni, A. Van Dorsselaer, and S. Ennahar. 2008. Production of enterocins L50A, L50B, and IT, a new enterocin, by Enterococcus faecium IT62, a strain isolated from Italian ryegrass in Japan. Antimicrob. Agents Chemother. 52:1917-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jack, R. W., J. R. Tagg, and B. Ray. 1995. Bacteriocins of gram-positive bacteria. Microbiol. Rev. 59:171-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang, J. H., and M. S. Lee. 2005. Characterization of a bacteriocin produced by Enterococcus faecium GM-1 isolated from an infant. J. Appl. Microbiol. 98:1169-1176. [DOI] [PubMed] [Google Scholar]
- 12.Klaenhammer, T. R. 1993. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol. Rev. 12:39-85. [DOI] [PubMed] [Google Scholar]
- 13.Line, J. E., E. A. Svetoch, B. V. Eruslanov, V. V. Perelygin, E. V. Mitsevich, I. P. Mitsevich, V. P. Levchuk, O. E. Svetoch, B. S. Seal, G. R. Siragusa, and N. J. Stern. 2008. Isolation and purification of enterocin E-760 with broad antimicrobial activity against gram-positive and gram-negative bacteria. Antimicrob. Agents Chemother. 52:1094-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nes, I. F., D. B. Diep, L. S. Havarstein, M. B. Brurberg, V. Eijsink, and H. Holo. 1996. Biosynthesis of bacteriocins in lactic acid bacteria. Antonie van Leeuwenhoek 70:113-128. [DOI] [PubMed] [Google Scholar]
- 15.Paulsen, I. T., L. Banerjei, G. S. A. Myers, K. E. Nelson, R. Seshadri, T. D. Read, D. E. Fouts, J. A. Eisen, S. R. Gill, J. F. Heidelberg, H. Tettelin, R. J. Dodson, L. Umayam, L. Brinkac, M. Beanan, S. Daugherty, R. T. DeBoy, S. Durkin, J. Kolonay, R. Madupu, W. Nelson, J. Vamathevan, B. Tran, J. Upton, T. Hansen, J. Shetty, H. Khouri, T. Utterback, D. Radune, K. A. Ketchum, B. A. Dougherty, and C. M. Fraser. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071-2074. [DOI] [PubMed] [Google Scholar]
- 16.Tagg, J. R., A. S. Dajani, and L. W. Wannamaker. 1976. Bacteriocins of gram-positive bacteria. Bacteriol. Rev. 40:722-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Belkum, M. J., and M. E. Stiles. 2000. Nonlantibiotic antibacterial peptides from lactic acid bacteria (1995 to date). Nat. Prod. Rep. 17:323-335. [DOI] [PubMed] [Google Scholar]