Abstract
A sulfate-reducing phenol-degrading bacterium, strain AK1, was isolated from a 2-bromophenol-utilizing sulfidogenic estuarine sediment enrichment culture. On the basis of phylogenetic analysis of the 16S rRNA gene and DNA homology, strain AK1 is most closely related to Desulfobacterium anilini strain Ani1 (= DSM 4660T). In addition to phenol, this organism degrades a variety of other aromatic compounds, including benzoate, 2-hydroxybenzoate, 4-hydroxybenzoate, 4-hydroxyphenylacetate, 2-aminobenzoate, 2-fluorophenol, and 2-fluorobenzoate, but it does not degrade aniline, 3-hydroxybenzoate, 4-cyanophenol, 2,4-dihydroxybenzoate, monohalogenated phenols, or monohalogenated benzoates. Growth with sulfate as an electron acceptor occurred with acetate and pyruvate but not with citrate, propionate, butyrate, lactate, glucose, or succinate. Strain AK1 is able to use sulfate, sulfite, and thiosulfate as electron acceptors. A putative phenylphosphate synthase gene responsible for anaerobic phenol degradation was identified in strain AK1. In phenol-grown cultures inducible expression of the ppsA gene was verified by reverse transcriptase PCR, and 4-hydroxybenzoate was detected as an intermediate. These results suggest that the pathway for anaerobic degradation of phenol in D. anilini strain AK1 proceeds via phosphorylation of phenol to phenylphosphate, followed by carboxylation to 4-hydroxybenzoate. The details concerning such reaction pathways in sulfidogenic bacteria have not been characterized previously.
Phenol is a key aromatic intermediate in the anaerobic degradation of natural and industrial aromatic compounds. Phenol is highly water soluble, which leads to widespread contamination of river, lake, estuarine, and other aquatic environments. Estuarine sediments serve as important sinks for such contaminants irrespective of the point of origin of the pollutants. Phenol and p-cresol were found in 10 and 38%, respectively, of streambed sediments assessed at 536 sites in 20 major river basins across the United States from 1992 to 1995 (22). The p-cresol and phenol concentrations were up to 210 and 4,800 mg/kg, respectively (22).
Phenolic compounds are readily degraded by aerobic bacteria, but they are more recalcitrant under anaerobic conditions. Although anaerobic degradation of phenol has been known to occur for a long time (11, 12), biochemical and genetic investigations have been limited. To date, anaerobic biodegradation and remediation studies have focused on consortia, but recently anaerobic phenol-degrading bacteria have been isolated under denitrifying (4, 37, 39, 41), iron-reducing (23), sulfidogenic (3, 5, 18), and fermentative-methanogenic conditions (10, 15, 21, 28, 42). Biodegradation of phenolic compounds under denitrifying conditions is well documented, and several bacteria have been characterized, including Azoarcus evansii, “Aromatoleum aromaticum” strain EbN1, Azoarcus sp. strain BH72, and several Thauera aromatica strains (13, 29, 30, 31). In these organisms, anaerobic degradation of phenol is initiated by conversion to phenylphosphate by phenylphosphate synthase (20, 33). Phenylphosphate is further metabolized by a second enzyme, phenylphosphate carboxylase, which catalyzes the carboxylation of phenylphosphate to 4-hydroxybenzoate (36). This reaction pathway in denitrifying bacteria has been characterized, but little is known about the biodegradation of phenolics coupled to sulfate reduction, and details concerning the metabolic pathways are poorly characterized.
Sulfate reduction is the dominant process in carbon metabolism in marine sediments where sulfate is present at high concentrations (20 to 30 mM) and thus is readily available as an electron acceptor. A sulfidogenic culture was previously enriched from estuarine sediment (from the Arthur Kill intertidal strait in New Jersey) with 2-bromophenol (2BP) as the sole carbon source (9, 16, 25). 2BP was reductively debrominated by a Desulfovibrio sp. (9), and phenol was then degraded further. Through clone library and terminal restriction fragment length polymorphism (T-RFLP) analysis the enrichment was found to consist of several types of organisms, including dehalogenating and phenol-degrading organisms that in combination were capable of complete degradation of 2BP coupled to sulfate reduction. In the present study, we report the isolation and detailed characterization of the phenol-degrading bacterium Desulfobacterium anilini strain AK1 from this enrichment culture.
MATERIALS AND METHODS
Source of inoculum and composition of medium.
A 2BP-utilizing sediment enrichment culture from the Arthur Kill intertidal strait in the New York-New Jersey Harbor estuary (9, 16) was used as the starting culture for this study. The enrichment culture was maintained in anaerobic minimal salts medium under a 70% N2-30% CO2 headspace as described by Fennell et al. (9). For enrichment of phenol-degrading bacteria, we prepared sulfidogenic medium (20 mM Na2SO4) with 1 mM phenol as the electron donor. After three transfers (total dilution from the original sediment, 10−9), a portion of the culture was serially diluted in agar shake culture tubes (7), solidified with Noble agar (10 g/liter; Difco), and incubated in the dark at 28°C. After 10 days, six distinct colonies were picked using a strict anaerobic technique, transferred to liquid medium, and cultivated to check for phenol degradation activity. One of the cultures was selected for further study, and colonies were reisolated in agar shake tube dilution cultures.
Microscopy.
The isolate grown in anaerobic sulfate medium was harvested for microscopic observation. Gram staining was performed using the protocols of Murray et al. (26). The morphology of the isolate was observed using a phase-contrast microscope and an epifluorescence microscope (Olympus BX 60).
T-RFLP.
Total DNA was extracted from the cultures, and 16S rRNA genes were PCR amplified using universal primers (fluorescently labeled 27F and 1525R) (1). Fluorescently labeled PCR products were purified with a Geneclean kit II (Qbiogene, Inc., Carlsbad, CA) and digested for 6 h at 37°C with MnlI (New England Biolabs, Beverly, MA). Twenty-five nanograms of labeled PCR product was analyzed on an ABI 310 genetic analyzer with Genescan software and internal standards (1).
16S rRNA gene analysis, DNA-DNA hybridization, and determination of the G+C content of genomic DNA.
DNA was extracted as described previously (1) and used for PCR amplification, cloning, and sequencing of the 16S rRNA genes. DNA-DNA hybridization was performed at the Identification Service of the DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany) according to its protocol. The G+C content of DNA was determined using high-performance liquid chromatography (HPLC), as described by Mesbah et al. (24). The nucleosides were analyzed using a model 1100 series HPLC (Agilent Technologies, California) with a reverse-phase C18 column (Partisil ODS-3; 4.6 by 250 mm; particle size, 5 μm; Whatman, New Jersey) and phosphate-methanol (90:10) as the solvent, and the absorbance at 254 nm was monitored. Phosphate buffer (pH 4.0) was prepared by dissolving KH2PO4 (0.049 M) in distilled water and adjusting the pH with 85% H3PO4.
Fatty acid composition.
Cellular lipids from strain AK1 and D. anilini strain Ani1 (= DSM 4660T) grown on 1 mM phenol at 28°C for 4 days in sulfate medium were saponified, methylated, and extracted into hexane-methyl tertiary butyl ether as described previously (38). Fatty acid methyl esters were analyzed by gas chromatography (GC) using the SHERLOCK microbial identification system (MIDI), and identities were verified by gas chromatography-mass spectrometry (MS) with an Agilent series 6890 GC system and a 5973 mass-selective detector equipped with an HP5 MS capillary column (30 m by 0.25 mm; film thickness, 0.25 μm) using helium as the carrier gas.
Analytical methods.
Phenol and other halogenated aromatic compounds were analyzed by HPLC (Agilent 1100 series) with a C18 column (Spherisorb; 4.6 by 250 mm; particle size, 5 μm; Phenomenex, California) using a flow rate of 1 ml/min, an eluent consisting of methanol, water, and acetic acid (40:58:2), and a UV detector set to 280 nm (1). Ion chromatography (Dionex DX-120) with an Ion Pac AS9 column was used for measurement of sulfate, sulfite, and thiosulfate using conductivity detection (25).
Metabolites of phenol were analyzed by GC-MS using an HP 5890 gas chromatograph with an HP 5971 mass-selective detector (Agilent Technologies, Wilmington, DE) and a DB-5MS fused silica column (length, 30 m; inside diameter, 0.25 mm; film thickness, 0.2 μm; J&W Scientific, Folsom, CA). One milliliter of a culture sample was first acidified by adding 0.1 ml 6.0 N HCl and saturated with NaCl, and then it was extracted with 3 ml ethyl ether. The ether extract was evaporated to dryness under a vacuum and then derivatized with 0.5 ml bis(trimethylsilyl)trifluoroacetamide containing 1% trimethylchlorosilane (Sigma) with heating at 70°C for 20 min.
PCR amplification of the phenylphosphate synthase subunit A gene.
A fragment of the ppsA (phenylphosphate synthase subunit A) gene was PCR amplified using primers 5′-GTCGAGCACTGGTTCCAC-3′ (forward) and 5′-ACCGCCGGCATGCCGTATTC-3′ (reverse), which were designed on the basis of conserved regions of known and putative ppsA nucleotide sequences of T. aromatica (accession number CAC12685) (6), “A. aromaticum” EbN1 (accession number CAI07888) (29), and Geobacter metallireducens GS-15 (accession number ABB32329). The PCR product was obtained using a 25-μl reaction mixture containing 1.6 pmol of each primer, 12.5 μl of ReadyMix Taq PCRmix (Sigma, St. Louis, MO), and approximately 100 ng of genomic DNA as the template. The reactions were initiated by incubation at 94°C for 2 min, which was followed by 40 cycles of denaturation at 94°C for 30 s, annealing at 50°C for 30 s, and extension at 72°C for 30 s and then by incubation at 72°C for 10 min. The PCR products were directly cloned into the pGEM-T vector (Promega, Madison, WI) according to the manufacturer's instructions and then sequenced.
IPCR and sequence analysis.
Total genomic DNA was extracted from strain AK using an UltraClean microbial DNA isolation kit (MO BIO, Carlsbad, CA). One microgram of total genomic DNA was digested with several endonucleases and purified with a NucleoTrap purification kit (BD Biosciences, Palo Alto, CA) used according to the manufacturer's protocol. The eluted DNA was self-ligated with T4 DNA ligase (Invitrogen, Carlsbad, CA) for 3 h at room temperature and then used as a template for inverse PCR (IPCR). The sequences of the IPCR products were obtained by using a primer-walking strategy.
Nucleotide sequences were determined with an ABI PRISM 3130 genetic analyzer (Applied Biosystems, Foster City, CA). The sequence data were compared with sequences in the GenBank database by performing a BLAST search (2) and were further analyzed with the Lasergene software (DNASTAR Inc., Madison, WI) and the Molecular Evolutionary Genetics Analysis software (19).
Isolation of total RNA and RT-PCR.
Strain AK1 was cultivated in sulfidogenic medium containing either 1 mM phenol, benzoate, 4-hydroxybenzoate, or acetate and harvested in exponential phase by centrifugation. The cell pellet was resuspended in a lysozyme solution (3 mg/ml), and total RNA was extracted with Trizol reagent (Invitrogen, Carlsbad, CA). The extracted total RNA was purified by spin column and DNase I treatment according to the manufacturer's instructions (Qiagen, Valencia, CA). The reverse transcriptase PCRs (RT-PCRs) were performed using 25-μl mixtures containing 0.5 μg of total RNA and 3.2 pmol of each primer with the Qiagen OneStep RT-PCR enzyme mixture (Qiagen, Valencia, CA). The thermocycler program used for the RT-PCRs was as follows: 50°C for 30 min, 95°C for 15 min, 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s, and then 72°C for 10 min. The following primers were used to amplify 517 bp of the ppsA gene: 5′-TGAACGGATCAACCAG-3′ (forward) and 5′-GATATCCGTTACGGCG-3′ (reverse).
Nucleotide sequence accession numbers.
The 16S rRNA gene sequence of strain AK1 has been deposited in the GenBank database under accession number EU020016. The putative ppsA gene sequences obtained in this study have been deposited in the GenBank database under accession number EF127527.
RESULTS AND DISCUSSION
Isolation of a phenol-degrading bacterium.
A phenol-degrading sulfidogenic consortium was enriched from a 2BP-utilizing enrichment culture (9) from estuarine sediment (from the Arthur Kill intertidal strait in New Jersey) by repeated feeding with phenol and serial dilution into fresh medium over a 6-year period. T-RFLP analysis and DGGE were used to analyze the composition of the culture during enrichment. From a final dilution series in soft agar, six colonies were picked and transferred to liquid medium to verify the phenol degradation activity. One of these colonies was selected for further study, and a culture of it was serially diluted in agar shake tubes until a pure culture was obtained. 16S rRNA gene T-RFLP analysis of the purified colony revealed a peak with a 220-bp terminal restriction fragment (see Fig. S1 in the supplemental material). DGGE analysis also showed that there was only one band, whose sequence exactly matched that of the isolate (data not shown). This bacterium most likely corresponds to the phylotype previously implicated in phenol degradation in the original consortium (9).
Phylogeny.
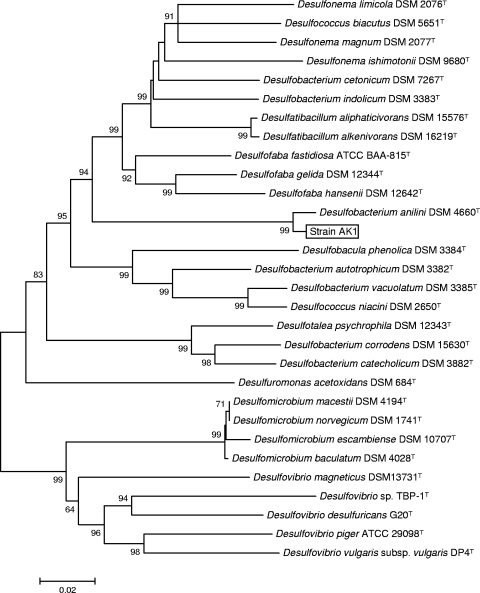
The 16S rRNA gene sequence analysis showed that strain AK1 (= ATCC BAA1486) was a member of the genus Desulfobacterium, with 98% sequence similarity to D. anilini strain Ani1 (= DSM 4660T; accession number AJ237601) (Fig. 1). Strain AK1 shared 70% DNA-DNA hybridization similarity with D. anilini Ani1 and thus can be identified as a D. anilini strain. The G+C content of the DNA of strain AK1 was 57.7 mol%, compared to 59.1 mol% for strain Ani1.
FIG. 1.
Neighbor-joining phylogenetic tree of D. anilini strain AK1 and related species based on 16S rRNA gene sequences. Bootstrap values less than 50% are not indicated at nodes. Scale bar = 0.02 substitution per nucleotide position.
Physiological and morphological characterization.
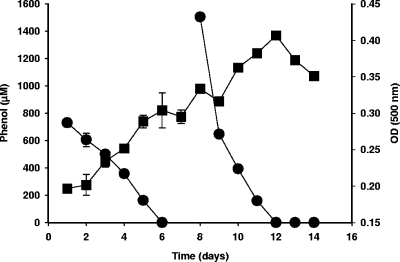
The key physiological and biochemical characteristics of strain D. anilini AK1 are shown in Table 1. Cells were gram-negative rods that were approximately 1 by 2 to 4 μm. Rod-shaped cells were observed in early growth phases but were observed less frequently in older cultures (see Fig. S2 in the supplemental material). Spores were not detected in any old cultures, and strain AK1 could not grow in oxidized medium. The effects of various phenol concentrations on the growth of strain AK1 were determined using anaerobic minimal salts medium with sulfate (Fig. 2). The first spike, 800 μM phenol, was degraded within 6 days, and an additional 1,500 μM was utilized in only 4 days. Strain AK1 grew at temperatures up to 45°C, and optimum utilization of phenol occurred at 37°C. Although strain AK1 was isolated from an estuarine sediment and was initially enriched in a saline medium, it utilizes phenol in medium without NaCl. The optimum NaCl concentration for growth of strain AK1 was between 0 and 12.5 g NaCl per liter, and it grew very slowly in medium containing more than 32.5 g NaCl per liter. Phenol was utilized when the initial concentration was up to 10 mM, but higher concentrations were inhibitory to growth. D. anilini strain AK1 was able to degrade various aromatic compounds (Table 1). In addition to phenol, it utilized benzoate, 2-hydroxybenzoate, 4-hydroxybenzoate, 4-hydroxyphenylacetate, 2-aminobenzoate, 2-fluorophenol, and 2-fluorobenzoate, but it did not utilize aniline, 3-hydroxybenzoate, 4-cyanophenol, 2,4-dihydroxybenzoate, monohalogenated phenols, or monohalogenated benzoates. Thiosulfate and sulfite were used as electron acceptors, but elemental sulfur was not used (Table 1). In general, these physiological characteristics are similar to those of D. anilini strain Ani1 (34, 35) and Desulfobacterium phenolicum (3).
TABLE 1.
Comparison of morphological and phenotypic characteristics of D. anilini strains AK1 and Ani1
| Characteristic | Strain AK1 | Strain Ani1a |
|---|---|---|
| Gram stain | − | − |
| Cell morphology | Short rod | Short rod |
| Cell width (μm) | 1 | 1.25 |
| Cell length (μm) | 2-4 | 1.5-3.0 |
| Motility | − | − |
| Optimum NaCl concn (%) | 0.9 | 0.9 |
| Optimum temp (°C) | 37 | 35 |
| Use of compounds (2.5 mM) as electron donors | ||
| Acetate | + | + |
| Lactate | − | −b |
| Pyruvate | + | + |
| Phenol | + | + |
| Benzoate | + | + |
| Aniline | − | + |
| 2-Hydroxybenzoate | + | + |
| 3-Hydroxybenzoate | − | + |
| 4-Hydroxybenzoate | + | + |
| 2-Aminobenzoate | + | + |
| 4-Hydroxyphenylacetate | + | + |
| 2-Fluorophenol | + | −b |
| 2-Fluorobenzoate | + | −b |
| Use of sulfate (20 mM) as electron acceptor | + | + |
| G+C content (mol%) | 57.8 | 59.1 |
Data from reference 34.
Data from this study.
FIG. 2.
Growth of strain AK1 on phenol. Phenol was added twice (800 μM and 1,500 μM). ▪, optical density at 500 nm [OD (500)]; •, phenol concentration.
Strain AK1 had a cellular fatty acid profile that included large amounts of unsaturated, straight-chain, and hydroxy fatty acids; the major components were C14:0 (2.4%), iso-C14:0 (3.1%), anteiso-C15:0 (7.3%), C16:0 (10.5%), C18:0 (6.8%), and C18:1ω7c (61.8%). This fatty acid profile was similar to that of D. anilini strain Ani1, although there were differences in the proportions of some fatty acids (see Table S1 in the supplemental material).
Metabolism of phenolic compounds.
Anaerobic phenol degradation typically proceeds via a pathway involving carboxylation in the para position, yielding 4-hydroxybenzoate (36). Production of 4-hydroxybenzoate has been detected in methanogenic consortia (17, 21, 42), an iron-reducing organism (23), and the denitrifying organisms Azoarcus sp. strain PH002 (41) and T. aromatica K172 (20, 39). However, little is known about this degradation pathway in sulfate-reducing bacteria. In phenol-fed cultures of strain AK1 a metabolite was detected by GC-MS. The mass spectrum of the trimethylsilyl (TMS) derivative eluting at 11.60 min had a fragmentation profile identical to that of a 4-hydroxybenzoate TMS standard, with a molecular ion (M) at m/z 282 and characteristic fragmentation ions at m/z 267 (M-15), 223 (M-59), 193 (M-89), 126 (M-TMS), and 73 (M-TMS) (data not shown).
Identification of the phenylphosphate synthase gene and sequence analysis.
The anaerobic phenol degradation pathway has been studied in great detail using the denitrifying betaproteobacterium T. aromatica (6, 14, 20, 36, 39, 40). Anaerobic metabolism of phenol in T. aromatica proceeds via sequential conversion to 4-hydroxybenzoate by phenylphosphate synthase and phenylphosphate decarboxylase (6, 27, 33, 36). In T. aromatica (6, 27, 33) the genes encoding phenylphosphate synthase are inducible by phenol. The enzyme is homologous to phosphoenolpyruvate synthase and is composed of three polypeptides (27, 33). Subunits A and B play an essential role in phosphorylation of phenol, which is the first step in anaerobic phenol degradation (33), while phosphoenolpyruvate synthase is homomultimeric (8). The corresponding ppsA and ppsB genes are located together in the genomes of “A. aromaticum” EbN1, G. metallireducens, and T. aromatica (6, 29, 32).
Using PCR primers specific for a portion of the phenylphosphate synthase gene, a fragment of DNA that was the predicted size (602 bp) was PCR amplified from genomic DNA extracted from strain AK1. The nucleotide sequence of the PCR product from strain AK1 shows 50% identity with the putative ppsA gene of “A. aromaticum” EbN1. This sequence was used as the starting material for IPCR and primer walking to determine the nucleotide sequence of the flanking regions. The final 6.6 kbp of sequence (accession number EF127527) contains the putative ppsA and ppsB genes encoding phenylphosphate synthase subunits A and B. We were not able to amplify a putative ppsA product from strain Ani1.
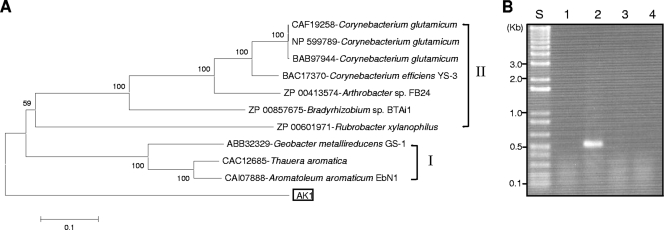
The deduced amino acid sequence of the putative PpsA protein shows the highest identity (42%) to that of T. aromatica (6) and has a relatively distant relationship with a group of phosphoenolpyruvate synthases (group II in Fig. 3A) which have molecular and biochemical similarities (27, 33). His569 encoded by the ppsA gene in T. aromatica (6) is necessary for phosphoryl group transfer onto phenol by a catalytic ping-pong mechanism (27). The analogous His571 amino acid of PpsA of strain AK1 aligns well with residues in phenylphosphate synthases of T. aromatica (6) and “A. aromaticum” EbN1 (29, 32) in group I (Fig. 3A) (data not shown). Despite the identity of these sequences, phylogenetic tree reconstruction shows that the phenylphosphate synthase gene of D. anilini strain AK1 forms a distinct group separate from previously described phenylphosphate synthase genes (Fig. 3A).
FIG. 3.
(A) Diversity of phenylphosphate synthase subunit A (PpsA) of strain AK1. The bar indicates 10% estimated sequence divergence. The deduced amino acid sequence of the ppsA gene was aligned using ClustalW with homologues imported from the GenBank database. The phylogenetic tree was constructed by neighbor-joining analysis using Molecular Evolutionary Genetics Analysis software (19) with 1,000 bootstrap replicates. (B) RT-PCR products of phenylphosphate synthase gene (ppsA) following growth on different substrates. Lane 1, acetate; lane 2, phenol; lane 3, benzoate; lane 4, hydroxybenzoate; lane S, 1-kb ladder.
In order to investigate whether the identified ppsA gene in strain AK1 is involved in phenol degradation, RT-PCR was performed with mRNA extracted following growth on different substrates (Fig. 3B). No RT-PCR product was generated from mRNA extracted from cells grown on acetate, benzoate, or 4-hydroxybenzoate. In contrast, an RT-PCR product for ppsA was obtained from strain AK1 grown on phenol. This suggests that the ppsA gene is inducibly expressed by phenol and might be involved in phenol degradation in strain AK1.
Conclusions.
D. anilini strain AK1 was identified as the phenol-degrading bacterium in a sulfidogenic 2BP-utilizing enrichment culture from an estuarine sediment. In addition to phenol, this strain utilizes a variety of other phenolic compounds. The detection of a gene encoding phenylphosphate synthase in strain AK1 suggests that phenol is metabolized through phenylphosphate to 4-hydroxybenzoate. This hypothesis was further supported by the detection of 4-hydroxybenzoate in the culture medium of phenol-grown strain AK1.
Supplementary Material
Acknowledgments
This research was funded in part by DoD grant SERDP ER-1492 and NSF grants OCE-451708 and CHE-0221978.
We thank Lee J. Kerkhof and Lora McGuinness for assisting with the microbial community analyses.
Footnotes
Published ahead of print on 1 May 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ahn, Y.-B., S.-K. Rhee, D. E. Fennell, L. J. Kerkhof, U. Hentschel, and M. M. Häggblom. 2003. Reductive dehalogenation of haloaromatics by microorganisms associated with the marine sponge Aplysina aerophoba. Appl. Environ. Microbiol. 69:4159-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bak, F., and F. Widdel. 1986. Anaerobic degradation of phenol and phenol derivatives by Desulfobacterium phenolicum sp. nov. Arch. Microbiol. 146:177-180. [Google Scholar]
- 4.Bakker, G. 1977. Anaerobic degradation of aromatic compounds in the presence of nitrate. FEMS Microbiol. Lett. 1:103-108. [Google Scholar]
- 5.Boopathy, R. 1995. Isolation and characterization of a phenol-degrading, sulfate-reducing bacterium from swine manure. Bioresour. Biotechnol. 54:29-33. [Google Scholar]
- 6.Breinig, S., E. Schiltz, and G. Fuchs. 2000. Genes involved in anaerobic metabolism of phenol in the bacterium Thauera aromatica. J. Bacteriol. 182:5849-5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breznak, J. A., and R. N. Costilow. 1994. Physicochemical factors in growth, p. 137-154. In P. Gerhardt, R. G. E. Murray, W. A Wood, and N. R. Krieg (ed.), Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC.
- 8.Cicicopol, C., J. Peters, A. Lupas, Z. Cejka, S. A. Müller, R. Golbik, G. Pfeifer, H. Lilie, A. Engel, and W. Baumeister. 1999. Novel molecular architecture of the multimeric archaeal PEP-synthase homologue (MAPS) from Staphylothermus marinus. J. Mol. Biol. 290:347-361. [DOI] [PubMed] [Google Scholar]
- 9.Fennell, D. E., S.-K. Rhee, Y.-B. Ahn, M. M. Häggblom, and L. J. Kerkhof. 2004. Detecting the dehalogenating microorganism in a sulfidogenic, 2-bromophenol-utilizing enrichment by T-RFLP fingerprinting of ribosomes. Appl. Environ. Microbiol. 70:1169-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia, M. T., V. Gallego, A. Ventosa, and E. Mellado. 2005. Thalassobacillus devorans gen. nov., sp. nov., a moderately halophilic, phenol-degrading, Gram-positive bacterium. Int. J. Syst. Evol. Microbiol. 55:1789-1795. [DOI] [PubMed] [Google Scholar]
- 11.Healy, J. B., and L. Y. Young. 1978. Catechol and phenol degradation by a methanogenic population of bacteria. Appl. Environ. Microbiol. 35:216-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Healy, J. B., and L. Y. Young. 1979. Anaerobic biodegradation of eleven aromatic compounds to methane. Appl. Environ. Microbiol. 38:84-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heider, J., and G. Fuchs. 1997. Microbial anaerobic aromatic metabolism. Anaerobe 3:1-22. [DOI] [PubMed] [Google Scholar]
- 14.Heider, J., M. Boll, K. Breese, S. Breinig, C. Ebenau-Jehle, U. Feil, N. Gad'on, B. Laempe, B. Leuthner, M. E. Mohamed, S. Schneider, G. Burchhardt, and G. Fuchs. 1998. Differential induction of enzymes involved in anaerobic metabolism of aromatic compounds in the denitrifying bacterium Thauera aromatica. Arch. Microbiol. 170:120-131. [DOI] [PubMed] [Google Scholar]
- 15.Juteau, P., V. Côté, M.-F. Duckett, R. Beaudet, F. Lépine, R. Villemur, and J.-G. Bisaillon. 2005. Cryptanaerobacter phenolicus gen. nov., sp. nov., an anaerobe that transforms phenol into benzoate via 4-hydroxybenzoate. Int. J. Syst. Evol. Microbiol. 55:245-250. [DOI] [PubMed] [Google Scholar]
- 16.Knight, V. K., L. J. Kerkhof, and M. M. Häggblom. 1999. Community analyses of sulfidogenic 2-BP-dehalogenating and phenol-degrading microbial consortia. FEMS Microbiol. Ecol. 29:137-147. [Google Scholar]
- 17.Knoll, G., and J. Winter. 1989. Degradation of phenol via carboxylation to benzoate by a defined, obligately syntrophic consortium of anaerobic bacteria. Appl. Microbiol. Biotechnol. 30:318-324. [Google Scholar]
- 18.Kuever, J., J. Kulmer, S. Janssen, U. Fischer, and K.-H. Blotevogel. 1993. Isolation and characterization of a new spore-forming sulfate-reducing bacterium growing by complete oxidation of catechol. Arch. Microbiol. 159:282-288. [DOI] [PubMed] [Google Scholar]
- 19.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 20.Lack, A., and G. Fuchs. 1994. Evidence that phenol phosphorylation to phenylphosphate is the first step in anaerobic phenol metabolism in a denitrifying Pseudomonas sp. Arch. Microbiol. 161:132-139. [DOI] [PubMed] [Google Scholar]
- 21.Li, T., J.-G. Bisaillon, R. Villemur, L. Létourneau, K. Bernard, F. Lépine, and R. Beaudet. 1996. Isolation and characterization of a new bacterium carboxylating phenol to benzoic acid under anaerobic conditions. J. Bacteriol. 178:2551-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopes, T. J., and E. T. Furlong. 2001. Occurrence and potential adverse effects of semivolatile organic compounds in streambed sediment, United States, 1992-1995. Environ. Toxicol. Chem. 20:727-737. [PubMed] [Google Scholar]
- 23.Lovley, D. R., and D. J. Lonergan. 1990. Anaerobic oxidation of toluene, phenol, and p-cresol by the dissimilatory iron-reducing organism, GS-15. Appl. Environ. Microbiol. 56:1858-1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mesbah, M., U. Premachandran, and W. B. Whitman. 1989. Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int. J. Syst. Bacteriol. 39:159-167. [Google Scholar]
- 25.Monserrate, E., and M. M. Häggblom. 1997. Dehalogenation and biodegradation of brominated phenols and benzoic acids under iron-reducing, sulfidogenic, and methanogenic conditions. Appl. Environ. Microbiol. 63:3911-3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murray, R. G. E., R. N. Doetsch, and C. F. Robinow. 1994. Determinative and cytological light microscopy, p. 21-41. In P. Gerhardt, R. G. E. Murray, W. A Wood, and N. R. Krieg (ed.), Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC.
- 27.Narmandakh, A., N. Gad'on, F. Drepper, B. Knapp, W. Haehnel, and G. Fuchs. 2006. Phosphorylation of phenol by phenylphosphate synthase: role of histidine phosphate in catalysis. J. Bacteriol. 188:7815-7822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qiu, Y. L., S. Hanada, A. Ohashi, H. Harada Y. Kamagata, and Y. Sekiguchi. 2008. Syntrophorhabdus aromaticivorans gen. nov., sp. nov., the first cultured anaerobe capable of degrading phenol to acetate in obligate syntrophic associations with a hydrogenotrophic methanogen. Appl. Environ. Microbiol. 74:2051-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rabus, R. 2005. Functional genomics of an anaerobic aromatic-degrading denitrifying bacterium, strain EbN1. Appl. Microbiol. Biotechnol. 68:580-587. [DOI] [PubMed] [Google Scholar]
- 30.Rabus, R., and F. Widdel. 1995. Anaerobic degradation of ethylbenzene and other aromatic hydrocarbons by new denitrifying bacteria. Arch. Microbiol. 163:96-103. [DOI] [PubMed] [Google Scholar]
- 31.Rabus, R., and F. Widdel. 1996. Utilization of alkylbenzenes during anaerobic growth of pure cultures of denitrifying bacteria on crude oil. Appl. Environ. Microbiol. 64:1238-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rabus, R., M. Kube, J. Heider, A. Beck, K. Heitmann, F. Widdel, and R. Reinhardt. 2005. The genome sequence of an anaerobic aromatic-degrading denitrifying bacterium, strain EbN1. Arch. Microbiol. 183:27-36. [DOI] [PubMed] [Google Scholar]
- 33.Schmeling, S., A. Narmandakh, O. Schmitt, N. Gad'on, K. Schuhle, and G. Fuchs. 2004. Phenylphosphate synthase: a new phosphotransferase catalyzing the first step in anaerobic phenol metabolism in Thauera aromatica. J. Bacteriol. 186:8044-8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schnell, S., F. Bak, and N. Pfennig. 1989. Anaerobic degradation of aniline and dihydroxybenzenes by newly isolated sulfate-reducing bacteria and description of Desulfobacterium anilini. Arch. Microbiol. 152:556-563. [DOI] [PubMed] [Google Scholar]
- 35.Schnell, S., and B. Schink. 1991. Anaerobic aniline degradation via reductive deamination of 4-aminobenzoyl-CoA in Desulfobacterium anilini. Arch. Microbiol. 155:183-190. [Google Scholar]
- 36.Schühle, K., and G. Fuchs. 2004. Phenylphosphate carboxylase: a new C-C lyase involved in anaerobic phenol metabolism in Thauera aromatica. J. Bacteriol. 186:4556-4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shinoda, Y., Y. Sakai, M. Ue, A. Hiraishi, and N. Kato. 2000. Isolation and characterization of a new denitrifying spirillum capable of anaerobic degradation of phenol. Appl. Environ. Microbiol. 66:1286-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song, B., N. J., Palleroni, L. J., Kerkhof, and M. M. Häggblom. 2001. Characterization of halobenzoate-degrading, denitrifying Azoarcus and Thauera isolates and description of Thauera chlorobenzoica sp. nov. Int. J. Syst. Evol. Microbiol. 51:589-602. [DOI] [PubMed] [Google Scholar]
- 39.Tschech, A., and G. Fuchs. 1987. Anaerobic degradation of phenol by pure cultures of newly isolated denitrifying pseudomonads. Arch. Microbiol. 148:213-217. [DOI] [PubMed] [Google Scholar]
- 40.Tschech, A., and G. Fuchs. 1989. Anaerobic degradation of phenol via carboxylation to 4-hydroxybenzoate: in vitro study of isotope exchange between 14CO2 and 4-hydroxybenzoate. Arch. Microbiol. 152:594-599. [Google Scholar]
- 41.van Schie, P. M., and L. Y. Young. 1998. Isolation and characterization of phenol-degrading denitrifying bacteria. Appl. Environ. Microbiol. 64:2432-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang, X., L. Mandelco, and J. Wiegel. 1994. Clostridium hydroxybenzoicum sp. nov., an amino acid-utilizing, hydroxybenzoate-decarboxylating bacterium isolated from methanogenic freshwater pond sediment. Int. J. Syst. Bacteriol. 44:214-222. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.