Abstract
Methanogens are of great importance in carbon cycling and alternative energy production, but quantitation with culture-based methods is time-consuming and biased against methanogen groups that are difficult to cultivate in a laboratory. For these reasons, methanogens are typically studied through culture-independent molecular techniques. We developed a SYBR green I quantitative PCR (qPCR) assay to quantify total numbers of methyl coenzyme M reductase α-subunit (mcrA) genes. TaqMan probes were also designed to target nine different phylogenetic groups of methanogens in qPCR assays. Total mcrA and mcrA levels of different methanogen phylogenetic groups were determined from six samples: four samples from anaerobic digesters used to treat either primarily cow or pig manure and two aliquots from an acidic peat sample stored at 4°C or 20°C. Only members of the Methanosaetaceae, Methanosarcina, Methanobacteriaceae, and Methanocorpusculaceae and Fen cluster were detected in the environmental samples. The three samples obtained from cow manure digesters were dominated by members of the genus Methanosarcina, whereas the sample from the pig manure digester contained detectable levels of only members of the Methanobacteriaceae. The acidic peat samples were dominated by both Methanosarcina spp. and members of the Fen cluster. In two of the manure digester samples only one methanogen group was detected, but in both of the acidic peat samples and two of the manure digester samples, multiple methanogen groups were detected. The TaqMan qPCR assays were successfully able to determine the environmental abundance of different phylogenetic groups of methanogens, including several groups with few or no cultivated members.
Methanogens are integral to carbon cycling, catalyzing the production of methane and carbon dioxide, both potent greenhouse gases, during organic matter degradation in anaerobic soils and sediment (8). Methanogens are widespread in anaerobic environments, including tundra (36), freshwater lake and wetland sediments (9, 12), estuarine and marine sediments (2), acidic peatlands (4, 14), rice field soil (10, 16), animal guts (41), landfills (30), and anaerobic digesters treating animal manure (1), food processing wastewater (27), and municipal wastewater and solid waste (37, 57). Methane produced in anaerobic digesters may be captured and used for energy production, thus offsetting some or all of the cost of operation and reducing the global warming potential of methane release to the atmosphere.
Methanogens are difficult to study through culture-based methods, and therefore many researchers have instead used culture-independent techniques to study methanogen populations. The 16S rRNA gene is the most widely used target for gene surveys, and a number of primers and probes have been developed to target methanogen groups (9, 11, 31, 36, 38, 40, 46, 48, 57). To eliminate potential problems with nonspecific amplification, some researchers have developed primers for the gene sequence of the α-subunit of the methyl coenzyme M reductase (mcrA) (17, 30, 49). The Mcr is exclusive to the methanogens with the exception of the methane-oxidizing Archaea (18) and shows mostly congruent phylogeny to the 16S rRNA gene, allowing mcrA analysis to be used in conjunction with, or independently of, that of the 16S rRNA gene (3, 30, 49). A number of researchers have examined methanogen communities with mcrA and have found uncultured clades quite different in sequence from cultured methanogen representatives (9, 10, 12, 14, 17, 22, 28, 47).
Previous studies described methanogen communities by quantitation of different clades through the use of rRNA-targeted or rRNA gene-targeted probes with techniques such as dot blot hybridization (1, 27, 37, 38, 48) and fluorescent in situ hybridization (11, 40, 44, 57). Real-time quantitative PCR (qPCR) is an alternate technique capable of determining the copy number of a particular gene present in the DNA extracted from an environmental sample. Only a few studies have used qPCR to quantitatively examine different clades within methanogen communities, and most of these studies have exclusively targeted the 16S rRNA gene (19, 41, 42, 54-56). Far fewer researchers have used qPCR to quantify methanogen clades by targeting the mcrA (21, 34, 45), and these studies were limited to only a few phylogenetic groups.
In this paper we present a methodology for determining methanogen gene copy numbers through the use of qPCR targeting the mcrA. Methanogens were quantified in total using methanogen-specific primers in SYBR green assays and also as members of nine different phylogenetic groups using TaqMan probes targeting specific subsets of methanogens.
MATERIALS AND METHODS
Design of methanogen-specific probes.
We previously constructed clone libraries for the mcrA and mrtA genes from two distinct methanogenic environments: sediment of an acidic transitional fen known as Bear Meadows Bog, and the primary digester of a municipal wastewater treatment plant treating combined primary and secondary sludge (50). The alignment of these clone sequences along with sequences from cultured methanogens were used to design TaqMan probes for groups of methanogens dominant in both environments (Fig. 1; Table 1). Originally we attempted to design primers to differentiate these phylogenetic groups in a SYBR green-based qPCR, but we were unable to achieve the necessary specificity without the inclusion of the group-specific TaqMan probes. Targeted groups that were observed in the municipal wastewater sludge digester included members of the Methanosaetaceae, Methanobacteriaceae mcrA and the isoenzyme mrtA, Methanospirillaceae, MCR-7, and two subgroups of MCR-2 which were labeled MCR-2a and MCR-2b. Uncultured clades MCR-7 and MCR-2 derive from a naming scheme used by Castro et al. (9). Targeted groups observed in the clone library from the acidic fen include Methanosarcina spp., Methanocorpusculaceae, and the Fen cluster, which was named in a study by Galand et al. (14). Probes were designed for a melting temperature of 68 to 72°C, a G+C content of 40 to 60%, and a length of 23 to 30 bp. The melting temperatures of probes were calculated with Integrated DNA Technologies’ Oligo Analyzer program (http://www.idtdna.com/analyzer/Applications/OligoAnalyzer/). Probe specificity was checked with a BLAST search of GenBank sequences (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and confirmed experimentally as described below. Probes were ordered from Biosearch Technologies (Novato, CA) and labeled with 6-carboxyfluorescein, Cal Fluor Orange, or Cal Fluor Red as the reporter dye and Black Hole 1 or Black Hole 2 as the quencher dye.
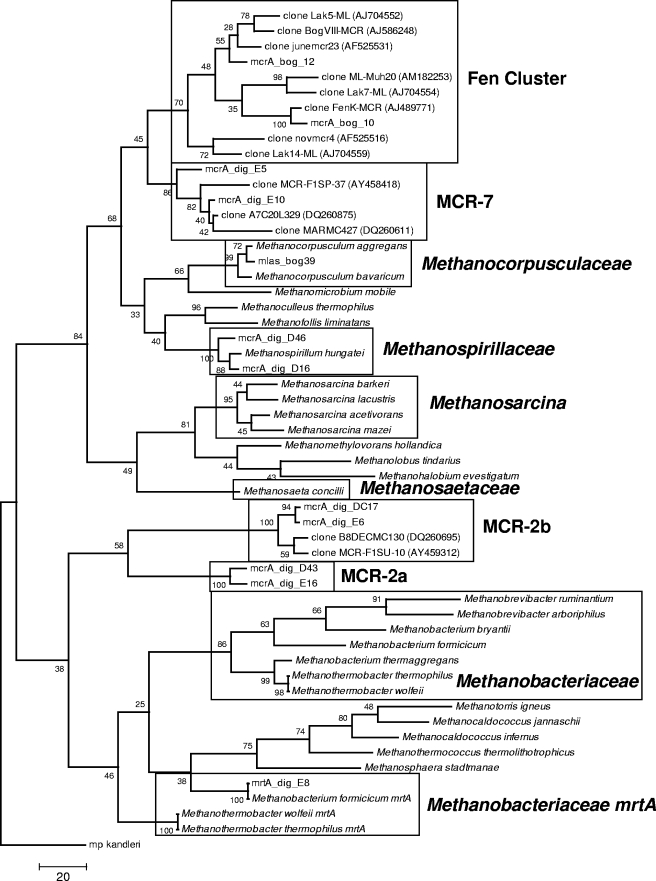
FIG. 1.
Phylogenetic tree of representative mcrA sequences and standards used for real-time quantitative PCR. Boxes denote sequences targeted by the respective TaqMan probe listed in Table 1. The tree was constructed as a maximum parsimony tree using close-neighbor interchange level 1 and bootstrapped with 1,000 trials. All positions containing gaps and missing data were eliminated from the data set. The scale bar represents the number of changes over the whole sequence. Classification of clusters is based on that reported by Castro et al. (8) for MCR-2, MCR-5, and MCR-7 and that of Juottonen et al. (24) for the Fen cluster.
TABLE 1.
TaqMan probes designed for this study and used to quantify different methanogen groups from environmental DNA
| Probe name | Target group | Probe sequence (5′-3′)a | Positionb | Clone(s) used as standard(s) (GenBank accession no.) |
|---|---|---|---|---|
| mbac-mcrA | Methanobacteriaceae mcrA | ARGCACCKAACAMCATGGACACWGT | 1154 | M. wolfeii (AB300780), M. arboriphilus (AF 414035) |
| mrtA | Methanobacteriaceae mrtA | CCAACTCYCTCTCMATCAGRAGCG | 1433 | M. thermoauto-trophicum (NC 000916), mrtA_dig_E8 (EU980398) |
| mcp | Methanocorpusculaceae | AGCCGAAGAAACCAAGTCTGGACC | 1319 | mlas_bog39 (DQ680603) |
| msp | Methanospirillaceae | TGGTWCMACCAACTCACTCTCTGTC | 1419 | mcrA_dig_D16 (EU980421), mcrA_dig_D46 (EU980419) |
| MCR-7 | Uncultured MCR-7 group | TGSCTTGACCTTRTCCWTCTCGYTS | 1134 | mcrA_dig_E5 (EU980422), mcrA_dig_E10 (EU980423) |
| MCR-2a | Uncultured MCR-2 group | CCACTCTACTGCCGGTATCAACG | 1317 | mcrA_dig_D43 (EU980402), mcrA_dig_E16 (EU980418) |
| MCR-2b | Uncultured MCR-2 group | ATGTATCTCTGCAGCAGCCGGTACA | 1269 | mcrA_dig_DC17 (EU980412), mcrA_dig_E6 (EU980407) |
| Fen | Uncultured Fen Cluster group | AAVCACGGYGGYMTCGGMAAG | 1071 | mcrA_bog_10 (EU980434), mcrA_bog_12 (EU980424) |
| msar | Methanosarcina | TCTCTCWGGCTGGTAYCTCTCCATGTAC | 1272 | M. barkeri (Y00158), M. acetivorans (NC 003552) |
| msa | Methanosaetaceae | CCTTGGCRAATCCKCCGWACTTG | 1107 | M. concilli (AF414037) |
R = A or G; K = G or T; M = A or C; W = A or T; Y = C or T; S = G or C; V = A, C, or G.
The 5′ end of the sense strand, based on M. thermoautotrophicum mcrA delta H (U10036) numbering.
Collection of environmental DNA.
Methanogen communities were analyzed from four anaerobic digesters and two incubations of acidic peat. Case studies for the design and operation of the digesters examined in this study are available through the Pennsylvania State University Department of Agricultural and Biological Engineering website (www.biogas.psu.edu). The Penn England dairy farm began operating a two-compartment reactor with a flexible cover in August 2006. Each compartment is heated to 37°C and mixed for approximately 1 hour twice daily. The design retention time in each compartment of the reactor is 20 days with 8 to 9% solids, and the system receives dairy manure, bedding, and milk parlor wash water. Bedding used in the dairy barns is composed entirely of the dried digested solids from the reactor. Grease trap waste from local restaurants is added to increase methane production. Brookside Farms began operating a plug flow reactor in April 2006 with a retention time of 30 to 33 days and 8 to 10% solids. The reactor is heated to 37°C and receives manure, sawdust bedding, and milk parlor wash water. Brewery waste and cheese whey are also added to the influent of the reactor. The Schrack dairy farm began operating a plug flow reactor in August 2006 with a retention time of 30 days and 10 to 12% solids. The reactor is heated to 37°C and receives manure, sawdust bedding, and milk parlor wash water. Crone Farms operates a plug flow digester with a retention time of 30 days and 8 to 10% solids. The reactor is heated to 37°C and treats only raw pig manure with no bedding or other amendments.
In addition to the anaerobic digesters, samples were also taken from acidic peat that had been stored for nearly 2 years at either room temperature (about 20°C) or 4°C. The peat incubations were included because, based on the previous cloning results, we expected that some of the groups targeted by these TaqMan probes would not be present in manure digesters but would be present in incubations of peat. The peat was collected in September 2006 from Bear Meadows Bog, an acidic transitional fen. Peat was transferred to sterile Pyrex medium bottles under aerobic conditions, and bottles were capped and stored with no amendments for nearly 2 years before withdrawing samples for DNA analysis. DNA from all samples was extracted using a PowerSoil DNA extraction kit (MoBio, Carlsbad, CA) according to the manufacturer's instructions. Contaminants carried over during DNA extraction inhibited PCR, but inhibition was eliminated by diluting extracts fivefold in PCR-grade water.
Preparation of templates for qPCR standard curves.
Cultures of Methanobacterium thermoautotrophicum, Methanobacterium wolfeii, Methanosarcina acetivorans, and Methanosarcina barkeri MS were generously provided by the laboratory of Christopher House (Penn State University, University Park, PA). DNA from these methanogen cultures was extracted with a DNeasy DNA extraction kit (Qiagen, Valencia, CA) following the manufacturer's instructions for DNA extraction from gram-positive cells. The mcrA or mrtA gene was amplified with primers mlas and mcrA-rev by using a previously described method (50). PCR products were ligated into a pCR 2.1 vector and used to transform Escherichia coli Top10 cells according to the manufacturer's instructions (TA cloning kit; Invitrogen, Carlsbad, CA). Blue-white screening was used to identify transformants, and positive clones were grown overnight at 37°C in LB broth containing ampicillin. Plasmids were purified with a PrepEase plasmid purification kit (USB, Cleveland, OH) and quantified based on absorbance at 260 nm. To confirm the presence of the correct insert, plasmids were sequenced at the Nucleic Acid Facility at Penn State University. As we did not have methanogen cultures representing Methanospirillaceae, Methanocorpusculaceae, or the Fen cluster, we used environmental clones previously obtained from a municipal sludge anaerobic digester and Bear Meadows Bog (50). Supercoiled plasmid DNA containing each of the 18 cloned fragments was used as templates for standard curves (Table 1). Plasmid DNA from each clone was diluted 10-fold in PCR-grade distilled water to create a dilution series ranging from 2.1 × 109 to approximately 2 copies per μl. From this original dilution series, it was determined experimentally that the consistent linear range for obtaining quantitative data was 2.1 × 107 to approximately 207 copies per μl, which translated to 4.15 × 107 to 415 copies per reaction mixture.
SYBR green I assays for total mcrA gene copies.
Quantitation of total mcrA gene copies was performed with primers mlas and mcrA-rev (50) using the nonspecific fluorophore SYBR green I (Molecular Probes, Invitrogen). The full genomes from 19 methanogens submitted to GenBank (http://www.ncbi.nlm.nih.gov/GenBank/index.html) were examined for the presence of multiple copies of mcrA. All the genomes contained only one copy of mcrA, but members of the Methanococcales and Methanobacteriales also contained a copy of the gene for the isoenzyme Mrt. The primers mlas and mcrA-rev also amplify mrtA, so methanogen numbers inferred by gene copy numbers may be overestimated by as much as a factor of 2 when these two orders are prevalent.
The SYBR green qPCR conditions included 1× Taq polymerase buffer and 0.03 U/μl Taq (USB), 2.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates containing uracil in place of thymidine, 0.01 U/μl of heat-labile uracil-DNA glycosylase, a 0.25 μM concentration of each primer, 0.3 M betaine, 250 μg/ml of bovine serum albumin, 10 nM fluorescein (Bio-Rad, Hercules, CA) as a reference dye, and a 75,000× dilution of SYBR green I dye (Molecular Probes). Quantitative PCR was run on a Bio-Rad iCycler with the following protocol: 20 min at 37°C to degrade contaminating PCR products, then 3.5 min at 95°C, followed by 40 cycles of denaturation at 95°C for 30 s, annealing at 55° for 45 s, extension at 72°C for 30 s, and image capture at 83°C, followed by a final extension at 72°C for 7 min. Melt curve analysis to detect the presence of primer dimers was performed after the final extension by increasing the temperature from 50 to 95°C in 0.5°C increments every 10 s. Image capture at 83°C was necessary to exclude fluorescence from the amplification of primer dimers.
SYBR green qPCR was first performed in triplicate on a dilution series of each of 18 clones used as a standard for the group-specific methanogen TaqMan probes (Table 1). This was done to ensure similar amplification efficiencies when using these primers with all phylogenetic groups, as the primers used for the SYBR green assays were also used for the TaqMan probe assays. Three separate 96-well plates were necessary to quantify the dilution series for all clones. The means, standard deviations, and coefficients of variance were calculated for the dilution series within each plate and among all three plates. After testing the assay with these 18 standards, total mcrA copies from environmental DNA were determined in triplicate, with dilution series of clones containing the M. thermoautotrophicum mrtA and Methanosaeta concilli mcrA genes run in duplicate as controls.
TaqMan qPCR for mcrA copies of individual methanogen groups.
TaqMan assays for mcrA copies from methanogen groups were verified before quantitation of environmental DNA. Each probe was checked for specificity in TaqMan assays by running all of the nontarget clone standards as negative controls. A dilution series of clones used as standards for each probe was run in three separate reactions to test the reproducibility of the TaqMan probe quantitation. The master mix for TaqMan qPCR was identical to that used for SYBR green I assays, including the primers, except that it contained 3.5 mM MgCl2 instead of 2.5 mM MgCl2 and 150 nM of TaqMan probe in place of SYBR green I and fluorescein dyes. Quantitative PCR was performed with the following protocol: 20 min at 37°C to degrade any contaminating product and 3.5 min at 95°C, followed by 40 cycles of denaturation at 95°C for 30 s and annealing/extension with image capture at 55° for 1 min. For quantitation of environmental DNA, one or two clones were used as standards for each TaqMan assay. One dilution series was constructed for each standard, and standards were run in duplicate while environmental samples were run in triplicate. The standard deviation of mcrA concentration for each environmental sample was determined from the standard deviation of the three threshold cycle values, which was then log transformed to gene copy number.
RESULTS
Quantitative PCR assays.
Standard curves for the SYBR green I qPCR were run with dilution series of all clones used as standards for methanogen group-specific TaqMan assays. A total of 18 curves were collected from three separate qPCR 96-well plates, and the means and standard deviations were calculated for assays within each plate and for all the data combined (Table 2). Similar results were obtained for each standard curve within and among plates, with an average slope of −3.451, average efficiency of 94.9%, average y intercept of 39.40, average R2 of 0.9962, and a lower limit of detection of approximately 415 copies/reaction mixture. This limit of detection corresponds to approximately 2.6 × 104 gene copies/ml sludge, which takes into account the fivefold dilution of the original DNA extractions to eliminate inhibition. A slope of −3.32 represents 100% efficiency, or a doubling of the DNA products in each cycle of PCR.
TABLE 2.
Reproducibility of the SYBR green I assay for total mcrA copies conducted for 18 standard curves in three separate qPCRsa
| Plate no. | Slope | Efficiency (%) | y intercept | R2 |
|---|---|---|---|---|
| 1 | −3.416 ± 0.054 | 96.2 ± 2.1 | 38.78 ± 0.72 | 0.9954 ± 0.0037 |
| 2 | −3.470 ± 0.097 | 94.2 ± 3.7 | 39.48 ± 0.49 | 0.9964 ± 0.0030 |
| 3 | −3.464 ± 0.099 | 94.4 ± 3.9 | 40.24 ± 0.36 | 0.9974 ± 0.0069 |
| Combined | −3.451 ± 0.079 | 94.9 ± 3.1 | 39.40 ± 0.69 | 0.9962 ± 0.0030 |
Values are means ± standard deviations.
TaqMan probes were tested in triplicate for specificity and reproducible amplicon detection with dilution series of clones used as standards for each probe (Table 1). Amplification and detection varied from probe to probe, with efficiencies ranging from 91 to 99% (Table 3). The lower limit of detection for each TaqMan assay was approximately 415 copies/reaction mixture, and similar y intercepts were obtained with each probe, suggesting all probes had about the same sensitivity for their respective targets. Quantitative PCR assays with the same probe were also similar for all three assays, which demonstrated the reproducibility of these methods. No negative controls or controls lacking template were amplified in any of the TaqMan qPCR assays, which confirmed the specificity of the probes to their targets. Several probes had two clones that were used as standards, and amplification of both clones was nearly identical for these probes (data not shown).
TABLE 3.
Reproducibility of TaqMan assays targeting different methanogen groups from three standard curves from three separate qPCRsa
| Probe | Slope | Efficiency (%) | y intercept | R2 |
|---|---|---|---|---|
| mbac-mcrA | −3.518 ± 0.141 | 92.4 ± 5.3 | 42.20 ± 0.89 | 0.9949 ± 0.0004 |
| mrtA | −3.538 ± 0.064 | 91.7 ± 2.3 | 41.06 ± 0.32 | 0.9937 ± 0.0035 |
| mcp | −3.487 ± 0.040 | 93.6 ± 1.5 | 42.75 ± 1.44 | 0.9912 ± 0.0041 |
| msp | −3.570 ± 0.104 | 90.6 ± 3.7 | 41.66 ± 0.43 | 0.9930 ± 0.0040 |
| MCR-7 | −3.505 ± 0.040 | 92.9 ± 1.5 | 41.41 ± 0.26 | 0.9964 ± 0.0014 |
| MCR-2a | −3.513 ± 0.077 | 92.6 ± 2.8 | 40.67 ± 0.97 | 0.9953 ± 0.0057 |
| MCR-2b | −3.427 ± 0.355 | 95.8 ± 15.8 | 42.73 ± 1.47 | 0.9864 ± 0.0130 |
| Fen | −3.345 ± 0.121 | 99.4 ± 5.2 | 41.24 ± 1.46 | 0.9930 ± 0.0043 |
| msar | −3.477 ± 0.094 | 93.9 ± 3.6 | 43.64 ± 1.24 | 0.9953 ± 0.0034 |
| msa | −3.481 ± 0.074 | 93.8 ± 2.8 | 42.45 ± 1.45 | 0.9977 ± 0.0016 |
Values are means ± standard deviations.
Quantitation of environmental DNA.
DNA was extracted from six environmental samples, including four anaerobic digesters used to treat primarily animal waste and two incubations of an acidic peat maintained at either 20°C or 4°C for nearly 2 years. Total mcrA was measured using a SYBR green I method, and mcrA from different methanogen groups was measured using the TaqMan probe method. Total mcrA was also calculated as the sum of the methanogen numbers obtained with each TaqMan probe. Total mcrA numbers obtained with both the SYBR green I and TaqMan probe methods were similar, although not identical (Table 4). The structure of the methanogen communities differed in the six environments both in terms of total mcrA numbers and in the numbers of different methanogen groups that were represented. All three cattle manure digesters were dominated by members of the Methanosarcina genus, whereas the two incubations of peat were dominated by both members of Methanosarcina and the Fen cluster. The Brookside digester sample showed detectable levels of only Methanosarcina, and the Crone digester sample showed only Methanobacteriaceae. The Penn England digester sample contained Methanosaetacae, Methanosarcina, Methanobacteriaceae, and Methanocorpusculaceae, and the Schrack digester sample contained Methanosarcina and Methanocorpusculaceae. The two peat incubations were the only environmental samples to contain members of the Fen cluster, but although both incubations were inoculated from the same sample, the methanogen communities were different in the two incubations. Besides members of the Methanosarcina genus and Fen cluster, the 20°C incubation mixture also contained Methanosaetaceae, whereas the 4°C incubation mixture contained members of Methanobacteriaceae.
TABLE 4.
Total methanogen mcrA copies and mcrA from different methanogen groups from six different environments
| Targeted group | Copies/ml of original samplea
|
|||||
|---|---|---|---|---|---|---|
| Penn England | Brookside | Schrack | Crone | 20°C incubation | 4°C incubation | |
| Methanosaetaceae | 7.07 × 105 ± 1.91 × 105 | 0 | 0 | 0 | 3.45 × 104 ± 1.88 × 104 | 0 |
| Methanosarcina | 3.20 × 106 ± 9.18 × 105 | 1.54 × 106 ± 2.42 × 105 | 4.49 × 106 ± 1.54 × 106 | 0 | 1.24 × 105 ± 1.19 × 105 | 1.27 × 106 ± 1.12 × 105 |
| Methanobacteriaceae | 6.81 × 105 ± 3.96 × 105 | 0 | 0 | 1.40 × 106 ± 2.29 × 106 | 0 | 2.98 × 106 ± 1.50 × 106 |
| Methanocorpusculaceae | 5.95 × 105 ± 9.78 × 104 | 0 | 3.90 × 105 ± 5.23 × 104 | 0 | 0 | 0 |
| Fen cluster | 0 | 0 | 0 | 0 | 6.36 × 105 ± 1.62 × 105 | 6.94 × 105 ± 1.25 × 105 |
| Total of TaqMan assays | 5.18 × 106 ± 1.60 × 106 | 1.54 × 106 ± 2.42 × 105 | 4.88 × 106 ± 1.59 × 106 | 1.40 × 106 ± 2.29 × 106 | 7.95 × 105 ± 3.00 × 105 | 4.94 × 106 ± 1.74 × 106 |
| SYBR green I | 2.53 × 106 ± 3.99 × 105 | 1.04 × 106 ± 4.36 × 105 | 3.13 × 106 ± 6.68 × 105 | 3.63 × 106 ± 2.37 × 105 | 3.55 × 106 ± 3.61 × 105 | 3.95 × 106 ± 5.57 × 105 |
Values represent copies per milliliter of original sample, which was digester effluent for the Penn England, Brookside, Schrack, and Crone environments and peat slurry for the 20°C and 4°C incubations. Only those methanogen groups detected in at least one environment are shown.
DISCUSSION
A number of studies have quantified methanogens belonging to different phylogenetic groups with techniques such as dot blot hybridization and fluorescent in situ hybridization (27, 38, 57), as well as qPCR (19, 41, 42, 54-56). Quantitation in these studies has targeted 16S rRNA or the rRNA gene, but studies by Springer et al. (49) and Bapteste et al. (3) suggest that the mcrA gene demonstrates the same phylogenetic relationships as the 16S rRNA gene between organisms. A major advantage to the use of mcrA as a target is that this gene is exclusive to methanogens, with the exception of the closely related anaerobic methane-oxidizing Archaea. To the best of our knowledge, the only other study besides ours that quantified phylogenetically different groups of methanogens by targeting mcrA is by Shigematsu et al., who designed TaqMan probes to target Methanosaeta, Methanosarcina, and Methanoculleus in acetate-fed anaerobic reactors (45). Our study is the first to quantify methanogens of other phylogenetic groups using mcrA as the target, including methanogens of the uncultured group MCR-2, and also members of the previously uncultured Fen cluster and MCR-7 groups.
Methanosarcina was the most commonly detected methanogen clade in the six samples and was the only methanogen group detected in the Brookside digester. Both Methanosarcina and Methanosaetaceae were detected in the Penn England digester sample and the acidic peat incubation at 20°C, and in both samples there were approximately four times more Methanosarcina than Methanosaetaceae detected. Although Methanosarcina and Methanosaetaceae are the only aceticlastic methanogens, Methanosaetaceae are obligately aceticlastic, whereas Methanosarcina are also able to produce methane from H2/CO2 and methylated compounds (52). Methanosaetaceae have a lower Ks for acetate than Methanosarcina, but as cleavage of acetate yields the least energy for Methanosarcina among its metabolic options (52), members of these two genera may not always directly compete for acetate, and Methanosarcina has been reported to dominate the methanogenic community of anaerobic digesters even when acetate levels are low (26, 55). Although both Methanosarcina and Methanosaetaceae were detected in the 20°C acidic peat incubation, Methanosarcina was the only detected aceticlastic methanogen in the 4°C incubation (Table 4). At colder temperatures homoacetogenic bacteria, which produce acetate from H2 and CO2, outcompete hydrogenotrophic methanogens for H2 (33, 43) and more methane is produced from acetate than H2/CO2 (36). Thus, in tundra, deep lake sediments, and other permanently cold environments methanogenesis proceeds predominantly from cleavage of acetate (32, 33, 43). It is possible that in the 20°C incubation, acetate was sufficiently low to allow Methanosaetaceae to outcompete Methanosarcina, but at 4°C homoacetogenesis may have led to the accumulation of acetate and dominance of the faster-growing Methanosarcina. Similar to our findings, some studies of permanently cold environments have found Methanosarcina to be the only aceticlastic methanogen present (23, 32).
Both Methanocorpusculaceae and Methanobacteriaceae are commonly detected in anaerobic digesters treating a variety of wastes. In this study, Methanocorpusculaceae was detected in the Penn England and Schrack digesters, while Methanobacteriaceae was also detected in the Penn England digester and was the only methanogen clade detected in the Crone digester. The genus Methanobrevibacter, within the Methanobacteriaceae, is often the dominant methanogen in animal guts (51, 53), so it is not surprising that this clade was the only methanogen group detected in a digester treating only pig waste. It is interesting that Methanobacteriaceae were the dominant methanogens in the 4°C peat incubation but were not detected in the 20°C peat incubation (Table 4). Although Methanobacteriaceae have been detected in northern wetlands and freshwater lake sediments which experience yearly cold seasons (20, 24, 32), they are rarely detected in permanently cold sediments, such as tundra and polar lakes. This observation, coupled with the fact that no psychrophilic members of the Methanobacteriaceae have been isolated, suggests that methanogens of this family do not compete well at cold temperatures. So, while it is not unusual that Methanobacteriaceae would be present in the original peat prior to incubation, it is unusual that they would become the predominant methanogen at 4°C but not at 20°C. Although mcrA of Methanobacteriaceae was detected, the isoenzyme mrtA of Methanobacteriaceae was not. The primers used in this study previously amplified both mcrA and mrtA from Methanobacteriaceae (50), so the failure to detect the Methanobacteriaceae mrtA is most likely due to a limitation in the coverage of the probe designed for this study. For quantitative analysis, a probe for mcrA of Methanobacteriaceae alone is sufficient. Originally, probes for both mcrA and mrtA of Methanobacteriaceae were designed to allow greater coverage of this diverse family.
The Fen cluster was detected in both of the acidic peat incubations but not in any of the digester samples. Bräuer et al. recently isolated and characterized a member of the Fen cluster, “Candidatus Methanoregula boonei,” from an acidic peatland (6). This isolate was found to be hydrogenotrophic and acidophilic, with optimum growth at pH 5. The Fen cluster, which has also been referred to as MCR-5 (9) or E1 and E2 (4), commonly dominates acidic freshwater environments (7, 13). If members of this cluster exhibit maximal growth at acidic pH, then this may indicate why they were not detected in any of the circumneutral anaerobic digester environments.
Although the Penn England, Brookside, and Schrack dairy manure digesters treat similar waste, Penn England had the most diverse methanogen community. The Brookside, Schrack, and Crone digesters are all plug flow designs, whereas the Penn England reactor is a periodically mixed tank reactor. In addition, bedding used in the Penn England barns is composed of the dried, digested solids from the reactor, which return to the reactor when the barns are cleaned. If some methanogens present in the digested solids survive desiccation and oxygen exposure, they are eventually returned to a favorable environment in the reactor. Active methanogens have been obtained from oxic environments, including aerated activated sludge (15) and the A horizon of forest, savanna, and desert soils (35). This may essentially increase the retention time for both methanogens and other bacteria in this reactor and may allow a more diverse methanogen population to emerge.
As the detection limit for the TaqMan qPCR method was approximately 2.6 × 104 copies per ml of reactor effluent, minor methanogen groups may have existed in numbers lower than this in the environmental samples but could not be detected by this qPCR method. So, although only one methanogen clade was detected in the Brookside and Crone digesters, this does not mean that only one methanogen clade was present. In addition, a TaqMan probe was not designed to capture some members of the order Methanomicrobiales, including the genus Methanoculleus, which is commonly detected in anaerobic digesters (25, 39). We designed TaqMan probes to target methanogen clades present in clone libraries obtained from a peat bog and an anaerobic digester treating municipal wastewater sludge, and Methanoculleus was not detected in these libraries (50). The inability to detect minor community members may partially explain the difference between total mcrA numbers as detected by SYBR green I assays and the total mcrA calculated by adding the numbers obtained in TaqMan assays, although it is more likely that differences in these numbers are due to the errors of the methods themselves. Even though the two calculations of total mcrA do not perfectly agree, they are close (Table 4). Sufficiently large clone libraries may be able to detect minor community members that are present in numbers too low to be detected by qPCR, but PCR bias may render abundance of a sequence in a clone library an unreliable predictor for abundance of that organism in the actual sample. Quantitative PCR may overcome some of these biases and thus serve as a complementary molecular technique for determination of relative abundance of a sequence in an environment (22, 29). Other sources of bias, such as differences in cell lysis and DNA extraction efficiencies among different phylogenetic groups or environmental samples, still affect qPCR results, but the determination by qPCR of the relative numbers of microbes of different clades provides a more in-depth examination of a microbial community than can be provided by clone libraries alone.
Acknowledgments
We thank Chris House and Zhidan Zhang, Department of Geosciences, Penn State University, for providing methanogen cultures. We also thank Robb Meinen, Dairy and Animal Sciences, and Pat and Deborah Topper, Department of Agricultural and Biological Engineering, Penn State University, for help in obtaining samples from swine and dairy manure digesters.
This study was supported by the Penn State Biogeochemical Research Initiative for Education (NSF IGERT grant DGE-9972759), the Graduate Research and Education in Advanced Transportation Technologies (NSF GK-12 grant DGE-0338240), and the Cooperative State Research, Education, and Extension Service of the U.S. Department of Agriculture (grant 2006-34437-17166).
Footnotes
Published ahead of print on 15 May 2009.
REFERENCES
- 1.Angenent, L. T., S. W. Sung, and L. Raskin. 2002. Methanogenic population dynamics during startup of a full-scale anaerobic sequencing batch reactor treating swine waste. Water Res. 36:4648-4654. [DOI] [PubMed] [Google Scholar]
- 2.Banning, N., F. Brock, J. C. Fry, R. J. Parkes, E. R. C. Hornibrook, and A. J. Weightman. 2005. Investigation of the methanogen population structure and activity in a brackish lake sediment. Environ. Microbiol. 7:947-960. [DOI] [PubMed] [Google Scholar]
- 3.Bapteste, E., C. Brochier, and Y. Boucher. 2005. Higher-level classification of the archaea: evolution of methanogenesis and methanogens. Archaea I:353-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basiliko, N., J. B. Yavitt, P. M. Dees, and S. M. Merkel. 2003. Methane biogeochemistry and methanogen communities in two northern peatland ecosystems, New York state. Geomicrobiol. J. 20:563-577. [Google Scholar]
- 5.Reference deleted.
- 6.Brauer, S. L., H. Cadillo-Quiroz, E. Yashiro, J. B. Yavitt, and S. H. Zinder. 2006. Isolation of a novel acidiphilic methanogen from an acidic peat bog. Nature 442:192-194. [DOI] [PubMed] [Google Scholar]
- 7.Cadillo-Quiroz, H., S. Brauer, E. Yashiro, C. Sun, J. Yavitt, and S. Zinder. 2006. Vertical profiles of methanogenesis and methanogens in two contrasting acidic peatlands in central New York state, USA. Environ. Microbiol. 8:1428-1440. [DOI] [PubMed] [Google Scholar]
- 8.Cakir, F. Y., and M. K. Stenstrom. 2005. Greenhouse gas production: a comparison between aerobic and anaerobic wastewater treatment technology. Water Res. 39:4197-4203. [DOI] [PubMed] [Google Scholar]
- 9.Castro, H., A. Ogram, and K. R. Reddy. 2004. Phylogenetic characterization of methanogenic assemblages in eutrophic and oligotrophic areas of the Florida everglades. Appl. Environ. Microbiol. 70:6559-6568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chin, K. J., T. Lueders, M. W. Friedrich, M. Klose, and R. Conrad. 2004. Archaeal community structure and pathway of methane formation on rice roots. Microb. Ecol. 47:59-67. [DOI] [PubMed] [Google Scholar]
- 11.Crocetti, G., M. Murto, and L. Bjornsson. 2006. An update and optimisation of oligonucleotide probes targeting methanogenic archaea for use in fluorescence in situ hybridisation (FISH). J. Microbiol. Methods 65:194-201. [DOI] [PubMed] [Google Scholar]
- 12.Earl, J., G. Hall, R. W. Pickup, D. A. Ritchie, and C. Edwards. 2003. Analysis of methanogen diversity in a hypereutrophic lake using PCR-RFLP analysis of mcr sequences. Microb. Ecol. 46:270-278. [DOI] [PubMed] [Google Scholar]
- 13.Galand, P. E., H. Fritze, and K. Yrjala. 2003. Microsite-dependent changes in methanogenic populations in a boreal oligotrophic fen. Environ. Microbiol. 5:1133-1143. [DOI] [PubMed] [Google Scholar]
- 14.Galand, P. E., S. Saarnio, H. Fritze, and K. Yrjala. 2002. Depth related diversity of methanogen archaea in finnish oligotrophic fen. FEMS Microbiol. Ecol. 42:441-449. [DOI] [PubMed] [Google Scholar]
- 15.Gray, N. D., I. P. Miskin, O. Kornilova, T. P. Curtis, and I. M. Head. 2002. Occurrence and activity of archaea in aerated activated sludge wastewater treatment plants. Environ. Microbiol. 4:158-168. [DOI] [PubMed] [Google Scholar]
- 16.Großkopf, R., S. Stubner, and W. Liesack. 1998. Novel euryarchaeotal lineages detected on rice roots and in the anoxic bulk soil of flooded rice microcosms. Appl. Environ. Microbiol. 64:4983-4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hales, B. A., C. Edwards, D. A. Ritchie, G. Hall, R. W. Pickup, and J. R. Saunders. 1996. Isolation and identification of methanogen-specific DNA from blanket bog peat by PCR amplification and sequence analysis. Appl. Environ. Microbiol. 62:668-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hallam, S. J., P. R. Girguis, C. M. Preston, P. M. Richardson, and E. F. DeLong. 2003. Identification of methyl coenzyme M reductase a (mcrA) genes associated with methane-oxidizing archaea. Appl. Environ. Microbiol. 69:5483-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hori, T., S. Haruta, Y. Ueno, M. Ishii, and Y. Igarashi. 2006. Dynamic transition of a methanogenic population in response to the concentration of volatile fatty acids in a thermophilic anaerobic digester. Appl. Environ. Microbiol. 72:1623-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horn, M. A., C. Matthies, K. Kusel, A. Schramm, and H. L. Drake. 2003. Hydrogenotrophic methanogenesis by moderately acid-tolerant methanogens of a methane-emitting acidic peat. Appl. Environ. Microbiol. 69:74-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inagaki, F., U. Tsunogai, M. Suzuki, A. Kosaka, H. Machiyama, K. Takai, T. Nunoura, K. H. Nealson, and K. Horikoshi. 2004. Characterization of c-1-metabolizing prokaryotic communities in methane seep habitats at the Kuroshima Knoll, Southern Ryukyu Arc, by analyzing pmoA, mmoX, mxaF, mcrA, and 16s rRNA genes. Appl. Environ. Microbiol. 70:7445-7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Juottonen, H., P. E. Galand, and K. Yrjala. 2006. Detection of methanogenic archaea in peat: comparison of PCR primers targeting the mcrA gene. Res. Microbiol. 157:914-921. [DOI] [PubMed] [Google Scholar]
- 23.Karr, E. A., J. M. Ng, S. M. Belchik, W. M. Sattley, M. T. Madigan, and L. A. Achenbach. 2006. Biodiversity of methanogenic and other archaea in the permanently frozen Lake Fryxell, Antarctica. Appl. Environ. Microbiol. 72:1663-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kotsyurbenko, O. R., K. J. Chin, M. V. Glagolev, S. Stubner, M. V. Simankova, A. N. Nozhevnikova, and R. Conrad. 2004. Acetoclastic and hydrogenotrophic methane production and methanogenic populations in an acidic west-Siberian peat bog. Environ. Microbiol. 6:1159-1173. [DOI] [PubMed] [Google Scholar]
- 25.Leclerc, M., J. P. Delgenes, and J. J. Godon. 2004. Diversity of the archaeal community in 44 anaerobic digesters as determined by single strand conformation polymorphism analysis and 16s rDNA sequencing. Environ. Microbiol. 6:809-819. [DOI] [PubMed] [Google Scholar]
- 26.Lee, C., J. Kim, K. Hwang, V. O'Flaherty, and S. Hwang. 2009. Quantitative analysis of methanogenic community dynamics in three anaerobic batch digesters treating different wastewaters. Water Res. 43:157-165. [DOI] [PubMed] [Google Scholar]
- 27.Liu, W. T., O. C. Chan, and H. H. P. Fang. 2002. Characterization of microbial community in granular sludge treating brewery wastewater. Water Res. 36:1767-1775. [DOI] [PubMed] [Google Scholar]
- 28.Lueders, T., K. J. Chin, R. Conrad, and M. Friedrich. 2001. Molecular analyses of methyl-coenzyme m reductase alpha-subunit (mcrA) genes in rice field soil and enrichment cultures reveal the methanogenic phenotype of a novel archaeal lineage. Environ. Microbiol. 3:194-204. [DOI] [PubMed] [Google Scholar]
- 29.Lueders, T., and M. W. Friedrich. 2003. Evaluation of PCR amplification bias by terminal restriction fragment length polymorphism analysis of small-subunit rRNA and mcrA genes by using defined template mixtures of methanogenic pure cultures and soil DNA extracts. Appl. Environ. Microbiol. 69:320-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luton, P. E., J. M. Wayne, R. J. Sharp, and P. W. Riley. 2002. The mcrA gene as an alternative to 16s rRNA in the phylogenetic analysis of methanogen populations in landfill. Microbiology 148:3521-3530. [DOI] [PubMed] [Google Scholar]
- 31.Marchesi, J. R., A. J. Weightman, B. A. Cragg, R. J. Parkes, and J. C. Fry. 2001. Methanogen and bacterial diversity and distribution in deep gas hydrate sediments from the Cascadia Margin as revealed by 16s rRNA molecular analysis. FEMS Microbiol. Ecol. 34:221-228. [DOI] [PubMed] [Google Scholar]
- 32.Metje, M., and P. Frenzel. 2007. Methanogenesis and methanogenic pathways in a peat from subarctic permafrost. Environ. Microbiol. 9:954-964. [DOI] [PubMed] [Google Scholar]
- 33.Nozhevnikova, A. N., C. Holliger, A. Ammann, and A. J. B. Zehnder. 1997. Methanogenesis in sediments from deep lakes at different temperatures (12-70 degrees C). Water Sci. Technol. 36:57-64. [Google Scholar]
- 34.Nunoura, T., H. Oida, T. Toki, J. Ashi, K. Takai, and K. Horikoshi. 2006. Quantification of mcrA by quantitative fluorescent PCR in sediments from methane seep of the Nankai Trough. FEMS Microbiol. Ecol. 58:659-665. [DOI] [PubMed] [Google Scholar]
- 35.Peters, V., and R. Conrad. 1995. Methanogenic and other strictly anaerobic bacteria in desert soil and other oxic soils. Appl. Environ. Microbiol. 61:1673-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Purdy, K. J., D. B. Nedwell, and T. M. Embley. 2003. Analysis of the sulfate-reducing bacterial and methanogenic archaeal populations in contrasting Antarctic sediments. Appl. Environ. Microbiol. 69:3181-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raskin, L., L. K. Poulsen, D. R. Noguera, B. E. Rittmann, and D. A. Stahl. 1994. Quantification of methanogenic groups in anaerobic biological reactors by oligonucleotide probe hybridization. Appl. Environ. Microbiol. 60:1241-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raskin, L., J. M. Stromley, B. E. Rittmann, and D. A. Stahl. 1994. Group-specific 16s ribosomal RNA hybridization probes to describe natural communities of methanogens. Appl. Environ. Microbiol. 60:1232-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rastogi, G., D. R. Ranade, T. Y. Yeole, M. S. Patole, and Y. S. Shouche. 2008. Investigation of methanogen population structure in biogas reactor by molecular characterization of methyl-coenzyme M reductase A (mcrA) genes. Bioresource Technol. 99:5317-5326. [DOI] [PubMed] [Google Scholar]
- 40.Rocheleau, S., C. W. Greer, J. R. Lawrence, C. Cantin, L. Laramee, and S. R. Guiot. 1999. Differentiation of Methanosaeta concilii and Methanosarcina barkeri in anaerobic mesophilic granular sludge by fluorescent in situ hybridization and confocal scanning laser microscopy. Appl. Environ. Microbiol. 65:2222-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saengkerdsub, S., P. Herrera, L. Woodward, C. Anderson, D. J. Nisbet, and S. C. Ricke. 2007. Detection of methane and quantification of methanogenic archaea in faeces from young broiler chickens using real-time PCR. Lett. Appl. Microbiol. 45:629-634. [DOI] [PubMed] [Google Scholar]
- 42.Sawayama, S., K. Tsukahara, and T. Yagishita. 2006. Phylogenetic description of immobilized methanogenic community using real-time PCR in a fixed-bed anaerobic digester. Bioresource Technol. 97:69-76. [DOI] [PubMed] [Google Scholar]
- 43.Schulz, S., and R. Conrad. 1996. Influence of temperature on pathways to methane production in the permanently cold profundal sediment of Lake Constance. FEMS Microbiol. Ecol. 20:1-14. [Google Scholar]
- 44.Sekiguchi, Y., Y. Kamagata, K. Nakamura, A. Ohashi, and H. Harada. 1999. Fluorescence in situ hybridization using 16s rRNA-targeted oligonucleotides reveals localization of methanogens and selected uncultured bacteria in mesophilic and thermophilic sludge granules. Appl. Environ. Microbiol. 65:1280-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shigematsu, T., Y. Q. Tang, T. Kobayashi, H. Kawaguchi, S. Morimura, and K. Kida. 2004. Effect of dilution rate on metabolic pathway shift between aceticlastic and nonaceticlastic methanogenesis in chemostat cultivation. Appl. Environ. Microbiol. 70:4048-4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sizova, M. V., N. S. Panikov, T. P. Tourova, and P. W. Flanagan. 2003. Isolation and characterization of oligotrophic acido-tolerant methanogenic consortia from a sphagnum peat bog. FEMS Microbiol. Ecol. 45:301-315. [DOI] [PubMed] [Google Scholar]
- 47.Smith, K. S., and C. Ingram-Smith. 2007. Methanosaeta, the forgotten methanogen? Trends Microbiol. 15:150-155. [DOI] [PubMed] [Google Scholar]
- 48.Sorensen, A. H., V. L. Torsvik, T. Torsvik, L. K. Poulsen, and B. K. Ahring. 1997. Whole-cell hybridization of Methanosarcina cells with two new oligonucleotide probes. Appl. Environ. Microbiol. 63:3043-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Springer, E., M. S. Sachs, C. R. Woese, and D. R. Boone. 1995. Partial gene-sequences for the α-subunit of methyl coenzyme M reductase (mcrI) as a phylogenetic tool for the family Methanosarcinaceae. Int. J. Syst. Bacteriol. 45:554-559. [DOI] [PubMed] [Google Scholar]
- 50.Steinberg, L. M., and J. M. Regan. 2008. Phylogenetic comparison of the methanogenic communities from an acidic, oligotrohic fen and an anaerobic digester treating municipal wastewater sludge. Appl. Environ. Microbiol. 74:6663-6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ufnar, J. A., S. Y. Wang, D. F. Ufnar, and R. D. Ellender. 2007. Methanobrevibacter ruminantium as an indicator of domesticated ruminant fecal pollution in surface waters. Appl. Environ. Microbiol. 73:7118-7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whitman, W. B., T. L. Bowen, and D. R. Boone. 2006. The methanogenic bacteria, p. 165-207. In The prokaryotes, vol. 3.2. Springer-Verlag, New York, NY. [Google Scholar]
- 53.Wright, A. D. G., C. H. Auckland, and D. H. Lynn. 2007. Molecular diversity of methanogens in feedlot cattle from Ontario and Prince Edward Island, Canada. Appl. Environ. Microbiol. 73:4206-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu, Y., J. Kim, and S. Hwang. 2006. Use of real-time PCR for group-specific quantification of aceticlastic methanogens in anaerobic processes: population dynamics and community structures. Biotechnol. Bioeng. 93:424-433. [DOI] [PubMed] [Google Scholar]
- 55.Yu, Y., C. Lee, and S. Hwang. 2005. Analysis of community structures in anaerobic processes using a quantitative real-time PCR method. Water Sci. Technol. 52:85-91. [PubMed] [Google Scholar]
- 56.Yu, Y., C. Lee, J. Kim, and S. Hwang. 2005. Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol. Bioeng. 89:670-679. [DOI] [PubMed] [Google Scholar]
- 57.Zheng, D., and L. Raskin. 2000. Quantification of Methanosaeta species in anaerobic bioreactors using genus- and species-specific hybridization probes. Microb. Ecol. 39:246-262. [DOI] [PubMed] [Google Scholar]