Abstract
Identification and quantification of phylogenetically defined bacterial populations in the environment are often performed using molecular tools targeting 16S rRNA. Fluorescence in situ hybridization has been used to monitor the expression and processing of rRNA by targeting the 3′ tail of precursor 16S rRNA. To expand this approach, we employed reverse transcription of total RNA using primer S-D-Bact-0338-a-A-18. Length heterogeneity detected by slab gel analysis, denaturing high-performance liquid chromatography (DHPLC) was used to differentiate the 5′ tail of the precursor from mature 16S rRNA, and the relative abundance of the precursor compared to the abundance of mature 16S rRNA was shown to be a sensitive indicator of the physiologic state of Acinetobacter calcoaceticus ATCC 23055T. Our results demonstrate that this is a sensitive and reliable method with a detection limit of 10 ng of single-stranded DNA. The assay was also used to differentiate among precursor 16S rRNA levels with mixed pure cultures, as well as to examine the response of a mixed activated sludge culture exposed to fresh growth medium and the antibiotic chloramphenicol. The results of this study demonstrate that this assay is a novel reverse transcription assay that simultaneously measures the mature and precursor 16S rRNA pools for mixed bacterial populations in an engineered environment. Furthermore, collection of the reverse transcription products derived from activated sludge samples by the DHPLC approach enabled identification of the active bacterial genera. Comparison of 16S and precursor 16S rRNA clone library results indicated that the precursor 16S rRNA library is a more sensitive indicator for active bacteria in engineered environmental samples.
For nearly 20 years, the “full-cycle 16S rRNA approach” has been employed to identify, enumerate, and determine the spatial organization of bacterial populations in environmental samples without the need for cultivation (1). The results of these studies have profoundly impacted the view of microbial diversity as a regulator of the global biosphere (10). During the development, demonstration, and maturation of the use of 16S rRNA-targeted molecular biology tools for bacterial identification, a number of researchers expanded the value of these methods in an attempt to determine simultaneously the identity and physiological status of bacterial populations. For example, Poulsen and coworkers quantified the intensity of the fluorescent signal from whole-cell fluorescence in situ hybridizations (FISH), targeting 16S rRNA as a predictor of ribosome abundance in bacterial cells of young and mature biofilms (31).
The correlation between the cellular ribosome (rRNA) content and the growth rate was one of the earliest and most fundamental observations in microbial physiology (31). An approximately 10-fold increase in the ribosome level is observed when the doubling time of Escherichia coli decreases from 100 min to 24 min. During rapid growth (doubling time, <1 h), over 50% of the total RNA produced in E. coli is rRNA, which is remarkable given that there are only 14 promoters that are associated with the seven rrn operons, compared to the 2,000 promoters available (14). Recently, Cutter and Stroot demonstrated that there is good agreement between the specific ribosome synthesis rate and the specific growth rate for Acinetobacter calcoaceticus ATCC 23055T under different growth conditions using standard culture media (9).
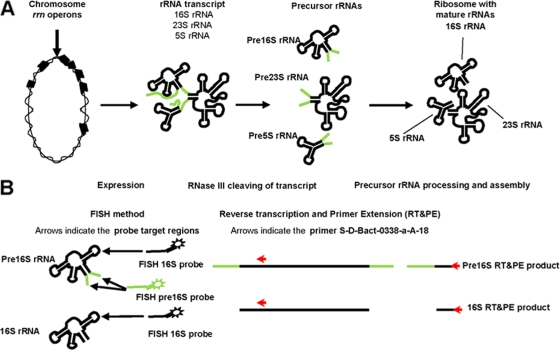
Ribosome genesis, as shown in Fig. 1A (8), is a central process in microbial growth. Currently, the ribosome genesis model of E. coli is the most complete, best-understood model and is assumed to describe ribosome genesis for most bacteria (11, 14, 18, 20, 23). In Borrelia burgdorferi and Helicobacter pylori, the 16S rRNA gene is located in a different operon than the 5S and 23S rRNA genes, which suggests that the rRNA processing in these bacteria may be different (28, 38). Ribosome genesis in E. coli begins with transcription from the rrn operon, producing a polycistronic rrn transcript that is subsequently processed to release precursor rRNA (pre-rRNA) molecules. Secondary processing and ribosome formation produce mature rRNA molecules. The secondary processing step is believed to be slower than the primary processing step, which results in an intracellular pool of pre-rRNAs.
FIG. 1.
(A) Schematic diagram of bacterial ribosome genesis. Chromosomal rrn operons are transcribed in a polycistronic transcript, which is processed to release precursor 16S rRNA fragments. The production of mature ribosomes involves final processing of pre-16S rRNA. (B) Schematic diagram of the FISH method and RT assay targeting the 16S rRNA pool. Extracts of total RNA contain pools of pre-16S rRNA and mature 16S rRNA which serve as targets for RT. Length heterogeneity is used to determine the relative abundance of pre-16S rRNA and mature 16S rRNA.
The response to favorable growth conditions is determined by measuring the pre-16S rRNA, an intermediate in ribosome genesis. Cangelosi and coworkers found that in E. coli, pre-16S rRNA (both pre-16S 3′ rRNA and pre-16S 5′ rRNA) was not detectable in stationary-phase cultures. However, a dramatic increase in the level of pre-16S rRNA (approximately 1/10 of the total 16S rRNA level) was noted when stationary-phase cultures were exposed to fresh medium (5). Licht et al. examined samples collected throughout an entire E. coli batch culture grown in Luria-Bertani (LB) medium. A 2-h transient peak in the pre-16S 5′ rRNA levels was observed immediately after inoculation (23). Oerther et al. observed that both A. calcoaceticus ATCC 23055T and E. coli ATCC 11755T displayed initial transient peak pre-16S 3′ rRNA levels when they were grown in fresh LB medium, and A. calcoaceticus ATCC 23055T had much higher pre-16S 3′ rRNA levels than E. coli. Unexpectedly, large variations in cellular levels of pre-16S 3′ rRNA were observed for individual A. calcoaceticus ATCC 23055T cells grown in filtered wastewater (27). Schmid and coworkers demonstrated that in situ measurements of pre-16S 3′ rRNA levels were a more sensitive indicator of the physiologic status of “Candidatus Brocadia anammoxidans” than measurements of mature rRNA obtained using conventional 16S and 23S rRNA-targeted fluorescently labeled oligonucleotide hybridization probes (33). Hawkins et al. suggested that pre-rRNA from Nitrobacter spp. could be used for in situ measurement of nitrite-oxidizing activity to improve implementation of traditional nitrification (17). Furthermore, pre-16S 3′ rRNA sequence information has been very important in taxonomic and ecological studies (16) and has been used to differentiate closely related strains in a broad diversity of bacterial genera and species, such as Sphingomonas (37), Frankia (15), and Pseudomonas avellanae (25).
The growth state of specific bacteria in mixed cultures can be determined by using FISH with probes that target two predominant molecules in ribosome genesis, namely pre-16S rRNA and 16S rRNA (Fig. 1B). Previously, we demonstrated that the level of pre-16S 3′ rRNA in individual cells of A. calcoaceticus ATCC 23055T is an indication of the growth state for three defined types of cells (36). Low levels of pre-16S rRNA (type I) corresponded to stationary-phase cultures. Intermediate levels of pre-16S rRNA (type II) corresponded to cultures diluted 20-fold into fresh medium and allowed to grow for 1 h. Elevated levels of pre-16S rRNA (type III) corresponded to cultures treated with the antibiotic chloramphenicol for a short period. Chloramphenicol has been reported to inhibit the secondary processing of pre-16S rRNA but does not inhibit expression of the rrn operon, which leads to buildup of the pre-16S rRNA levels relative to the 16S rRNA levels (39). Subsequently, type III cells were observed when A. calcoaceticus ATCC 23055T was exposed to primary effluents collected from conventional activated sludge treatment plants, which suggested that the growth of A. calcoaceticus ATCC 23055T was inhibited by an unidentified component of primary effluent present in the wastewater (36). However, this approach is laborious for mixed cultures, since it requires a unique pre-16S 3′ rRNA probe for each specific bacterium of interest. So far, only limited pre-16S 3′ rRNA sequence information is available to researchers from the Ribosomal Intergenic Spacer Sequence Collection (http://egg.umh.es/rissc), and the total number of sequences is significantly less than the available 16S rRNA sequence information (13). An alternative approach that would allow rapid determination of the growth state of abundant microbial populations in a mixed culture was desired. A new molecular biology method was developed that determines the growth state of an entire bacterial population by determining the level of pre-16S 5′ rRNA relative to the level of 16S rRNA.
A schematic diagram of the reverse transcription (RT)-primer extension (PE) assay is shown in Fig. 1B. The primer chosen, S-D-Bact-0338-a-A-18, targets a site that is found in precursor and mature 16S rRNA for all bacteria. Compared with agarose gel electrophoresis, slab gel analysis provided better resolution for detection of the pre-16S 5′ and 16S single-stranded DNA (ssDNA) derived from three types of pure cultures, as well as activated sludge cultures, based on size. Furthermore, a novel denaturing high-performance liquid chromatography (DHPLC) method was developed to identify specific bacteria of interest by collection of prominent pre-16S 5′ ssDNA for sequence analysis.
DHPLC is a promising technology that is primarily used to scan DNA mutations for clinical diagnostics in humans. Recently, it has been used for separation and identification of PCR-amplified fragments of genes coding for 16S rRNA for bacterial community analysis, monitoring, and identification (2, 4, 12, 24). In this study, we employed the DNASep cartridge, which uses alkylated nonporous polystyrene-polydivinylbenzene copolymer microspheres for high-performance nucleic acid separation. Under fully denaturing conditions and with a flow containing acetonitrile and triethylammonium acetate (TEAA), bacterial pre-16S 5′ and 16S ssDNA displayed different retention times due to size differences. This is because the positively charged ammonium ions of TEAA interact more favorably with the negatively charged phosphate ions of the larger DNA molecules than with the smaller DNA molecules, allowing more retention to the stationary phase in the cartridge. In addition, the FCW 200 fragment collector provided with the DHPLC system enabled fully automated collection of the prominent pre-16S DNA peaks of interest for reamplification, cloning, and sequencing.
MATERIALS AND METHODS
Bacterial cultures and defined cultivation conditions.
A. calcoaceticus ATCC 23055T (American Type Culture Collection) and E. coli ATCC 11755T were cultured in LB medium (which contained, per liter of water, 10 g tryptone, 5 g yeast extract, and 10 g NaCl), while Paracoccus denitrificans ATCC 13543 and Zoogloea ramigera ATCC 19623 were cultured in nutrient broth (which contained, per liter of water, 5 g peptone, 5 g NaCl, 2 g yeast extract, and 1 g beef extract). Stationary-phase cultures were prepared by cultivation on a shaker at 35°C overnight to obtain type I cells with low levels of pre-16S rRNA (36). To produce type II cells with intermediate levels of pre-16S rRNA, stationary-phase cultures were diluted 20-fold into fresh medium, and samples were removed after 1 h of growth. To produce type III cells with elevated levels of pre-16S rRNA, stationary-phase cultures were diluted 20-fold into fresh medium, allowed to grow for 1 h, and treated with the antibiotic chloramphenicol at a final concentration of 20 mg/liter, and samples were removed after 1 h of growth with the antibiotic. A stock concentration of chloramphenicol (2,000 mg/liter) was prepared with distilled and deionized water as the solvent.
Activated sludge and defined cultivation conditions.
Two sources of activated sludge were used. Fresh mixed liquor collected from the Mill Creek wastewater treatment plant was used first for agarose gel electrophoresis and slab gel analysis. Later, mixed liquor from a laboratory-scale nitrifying activated sludge bioreactor was used to examine the DHPLC method. Type I biomass with low levels of pre-16S rRNA was collected after 24 h of incubation with aeration at room temperature. To generate type II biomass with an intermediate level of pre-16S rRNA, a sample of activated sludge was diluted 20-fold into fresh LB medium and incubated for 2 h at 35°C on a rotary shaker. Type III biomass with elevated levels of pre-16S rRNA was generated by addition of chloramphenicol to a final concentration of 20 mg/liter and additional incubation for 2 h at 35°C on a rotary shaker.
Nucleic acid extraction.
DNA was extracted from activated sludge samples using a soil DNA extraction kit (MoBio, Carlsbad, CA) according to the manufacturer's instructions. RNA was extracted using the hot phenol-chloroform extraction method (34), with modifications (32). Extracted total RNA was further purified with an RNAqueous kit (Ambion, Austin, TX) used according to the manufacturer's instructions. Briefly, 30 μg of RNA (as determined using a spectrophotometer) was precipitated using LiCl and resuspended to obtain a final concentration between 2 and 2.5 μg/μl. Residual DNA was removed from the RNA samples by DNase I treatment with a DNA-free kit (Ambion, Austin, TX) used according to the manufacturer's instructions. Briefly, 2 μl of DNase I was used, and the incubation time was 1 h. Samples were collected and concentrated by centrifugation prior to storage at −80°C.
RT and PE.
The ArrayScript reverse transcriptase (Ambion, Austin, TX) was used according to the manufacturer's instructions. Briefly, ssDNA was synthesized by adding 20 μM primer S-D-Bact-0338-a-A-18 (5′ GCTGCCTCCCGTAGGAGT 3′) to 10 μg of total RNA, heating the preparation at 70°C for 5 min, cooling it on ice, adding 2 μl of 10× ArrayScript reverse transcriptase buffer, 4 μl of deoxynucleoside triphosphates (each deoxynucleoside triphosphate at 2.5 mM), 1 μl of Superase-in (20 U/μl), and 1 μl of ArrayScript reverse transcriptase (200 U/μl), and then incubating the mixture at 42°C for 2 h. For slab gel electrophoresis, primer S-D-Bact-0338-a-A-18 was labeled with the fluorochrome 6-carboxyfluorescein (FAM) on the 5′ end. The remaining RNA was removed by treating the samples with RNase A (Sigma, St. Louis, MO). Briefly, each reaction mixture was incubated at 85°C for 10 min to inactivate any RNase A inhibitor. Samples were quickly cooled by immersing them in an ice slurry for 5 min. Two volumes of RNase A cocktail was then added to 1 volume of the RT reaction mixture, and each mixture was incubated for 30 min at 37°C.
Agarose gel electrophoresis.
RNA (1 μg) and ssDNA (6 μl of an RNase A-treated sample, which was equivalent to 1 μg of RNA) were electrophoresed for 2 h at 100 V using a 2% (wt/vol) agarose gel prepared in Tris-borate-EDTA buffer (90 mM Tris base, 90 mM boric acid, 2 mM EDTA; pH 8). After electrophoresis, staining was performed for 40 min at room temperature using the SYBR green I nucleic acid stain (Molecular Probes, Inc., Eugene, OR). Briefly, fresh stain was prepared by adding 20 μl of SYBR green I to 200 ml of Tris-borate-EDTA buffer. Digital images of gels were captured using a Kodak EDAS 290 imaging system (Eastman Kodak Co., Rochester, NY) with a SYBR green I filter.
Slab gel electrophoresis.
For slab gel electrophoresis, it was necessary to clean up all samples using the Wizard PCR Preps DNA purification system (Promega, Madison, WI) according to the manufacturer's instructions. For slab gel electrophoresis, 1 μl of an RNase A-treated sample was analyzed with an ABI Prism 377 DNA sequencer (Applied Biosystems, Foster City, CA) by the DNA Core Laboratory of the University of Cincinnati.
DHPLC analysis.
The pre-16S DNA and 16S ssDNA were analyzed using the WAVE system (Transgenomic, Omaha, NE). A gradient was formed by buffer A, which consisted of 0.1 M TEAA (pH 7.0), and buffer B, which consisted of 0.1 M TEAA and 25% acetonitrile (pH 7.0; analytical grade; Transgenomic, Omaha, NE). The following optimum conditions were used for separation of the RT-PE products: column temperature, 80.0°C; and 37.0% buffer B for 1 min, 42% buffer B for 16.7 min, and 58.7% buffer B for 0.1 min at a flow rate of 0.9 ml/min. An analysis was performed with Wave Navigator software, version 1.6.1.
Amplification and cloning of the 16S rRNA gene.
An aliquot of genomic DNA and RT-PE products collected from the WAVE system was used as a template for the PCR. PCR was performed with a 50-μl reaction mixture containing 1× PCR buffer, 200 μM of each deoxynucleoside triphosphate, 1.5 mM MgCl2, 0.025 U of Taq DNA polymerase/μl (Takara), and 0.2 μM of each primer. The primers used, S-D-Bact-0011-a-S-17 (5′ GTTTGATCCTGGCTCAG 3′) and S-D-Bact-0338-a-A-18, were specific for conserved bacterial 16S rRNA sequences. Amplification of DNA and pre-16S ssDNA was performed using an Applied Biosystems 2400 thermal cycler (Applied Biosystems, Foster City, CA) and the following program: an initial denaturation step of 94°C for 5 min, followed by 25 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s and then a final extension at 72°C for 7 min.
PCR products were ligated into the vector pCR 2.1 using a TOPO TA cloning kit, and the ligation products were used to transform E. coli DH5α-T1R cells according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). Transformants were detected by blue-white screening using kanamycin and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). Plasmids were sent to the DNA Sequencing Facility at Children's Hospital (Cincinnati, OH). Sequencing results were compared to the GenBank (NR) database online using the Basic Local Alignment Search Tool (BLAST) utility by the National Center for Biotechnology Information and The Ribosomal Database Project (43).
Nucleotide sequence accession numbers.
Partial 16S rRNA sequences have been deposited in the GenBank database under accession numbers FJ798586 to FJ798590.
RESULTS AND DISCUSSION
Development and testing of the RT-PE method. (i) RT-PE method detects type I, II, and III cells of A. calcoaceticus ATCC 23055T.
Samples were collected from three different cultures of A. calcoaceticus ATCC 23055T, which corresponded to three distinct types of cells (types I, II, and III) based on pre-16S 3′ rRNA levels (36). For each sample, RNA was extracted and pre-16S and 16S ssDNA products were generated using the method described above. With the RT-PE method, it was estimated that ssDNA would be generated from approximately 6% of the total RNA. This estimate is a product of the fraction of RNA that is rRNA (85%) (29) and the fraction of the rRNA that generates ssDNA (7%). The latter value was determined by comparing number of nucleotides at the 5′ end of the 16S rRNA that are available for our method (340 nucleotides [nt]) to the total number of nucleotides for all rRNA (4,570 nt). For example, 1 μg of RNA produces approximately 60 ng of 16S ssDNA from the 16S rRNA. The amount of pre-16S 5′ ssDNA derived from pre-16S 5′ rRNA is significantly lower for type I and II cells.
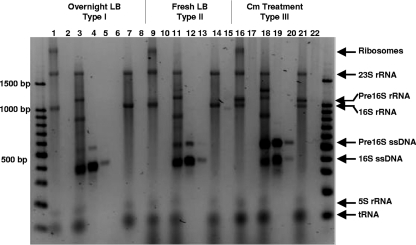
Figure 2 shows the results for RNA and ssDNA from three A. calcoaceticus ATCC 23055T cultures that contained type I (lanes 1 to 8), type II (lanes 9 to 15), or type III cells (lanes 16 to 22) and for various treatments of each culture that were analyzed by agarose gel electrophoresis. Although double-stranded DNA size markers are not appropriate for strict determination of sizes of RNA and ssDNA, they were used to describe the results and the sizes of individual bands, as indicated below.
FIG. 2.
Agarose gel electrophoresis of total RNA, products of RT obtained using primer S-D-Bact-0338-a-A-18, and products of RT reactions treated with RNase and DNase for A. calcoaceticus ATCC 23055T prepared as an overnight culture (type I), a culture after transfer to fresh medium (type II), and a culture treated with the antibiotic chloramphenicol after transfer to fresh medium (type III). The lanes contained total extracted RNA (lanes 1, 9, and 16), RNase A-treated RNA (lanes 2, 10, and 17), the products of RT reactions (lanes 3, 11, and 18), the products of RT reactions treated with RNase A (lanes 4, 12, and 19), a 10-fold dilution of the products of RT reactions treated with RNase A (lanes 5, 13, and 20), the products of RT reactions treated with RNase A and DNase I (lane 6), the products of RT reactions without primer S-D-Bact-0338-a-A-18 (lanes 7, 14, and 21), and the products of RT reactions without primer S-D-Bact-0338-a-A-18 treated with RNase A (lanes 8, 15, and 22). The lane on the left contained a 100-bp DNA size ladder, and the identities of bands are indicated on the right. Cm, chloramphenicol.
There are several bands for the extracted RNA of each sample (Fig. 2, lanes 1, 9, and 16). There are no bands for extracted RNA treated with RNase A (lanes 2, 10, and 17). Therefore, all bands for the extracted RNA samples are RNA bands. The largest band is an intact ribosome band, and the next four most abundant bands are bands for 23S rRNA (1,700 bp), 16S rRNA (1,000 bp), 5S rRNA (150 bp), and tRNA (≤100 bp). The extra band present in the type II and III samples is a pre-16S rRNA band (1,100 bp).
The results for the untreated RT-PE products are shown in Fig. 2, lanes 3, 11, and 18. Compared to the results for the extracted RNA samples, several additional bands are present; these bands are bands for ssDNA (400 and 550 bp) and ssDNA-rRNA duplexes (1,200 and 1,300 bp). Two additional bands (700 and 850 bp) probably correspond to ssDNA-rRNA duplexes with degraded pre-16S rRNA or 16S rRNA. However, degraded pre-16S or 16S rRNA was not detected in samples obtained with the RNA extraction procedure or the negative controls for the RT-PE procedure (no primer added in the RT-PE procedure) (lanes 7, 14, and 21).
The RNase A-treated RT-PE products are shown in Fig. 2, lanes 4, 12, and 19, and two prominent bands (400 and 550 bp) are present. To better estimate the ratio of the two bands, the results for a 10-fold dilution of the RNase A-treated RT-PE products are shown in lanes 5, 13, and 20. The larger band is absent for the type I cells due to dilution (lane 5), while both bands are present for the type II and type III cells (lanes 13 and 20). In addition, the level of the larger RT-PE band is increased higher in the chloramphenicol-treated samples (lanes 19 and 20), which is consistent with the level of pre-16S rRNA (1,100 bp) present (lanes 1, 9, and 16) and our previous FISH results (36).
Lane 6 in Fig. 2 shows that DNase I treatment of the RNase A-treated RT-PE products derived from overnight cultures removes both bands (lane 4) and confirms that the RT-PE method produces ssDNA from rRNA. The results for the negative control samples (no primer added in the RT-PE procedure) are shown in lanes 7, 14, and 21. These samples are similar to the extracted RNA samples (lanes 1, 9, and 16); however, the largest band (intact ribosomes) is absent. These negative control samples were treated with RNase A (lanes 8, 15, and 22), which removed all RNA, and produced no DNA bands.
The gel shown in Fig. 2 provides evidence that our RT-PE method produced ssDNA from the rRNA present. In addition, our results suggest that pre-16S 5′ and 16S ssDNA can be derived from three distinct types of cells and that they can be differentiated based on size. Although two distinct bands for the RT-PE product could be distinguished by agarose gel electrophoresis, slab gel analysis was used to determine their exact sizes and abundance.
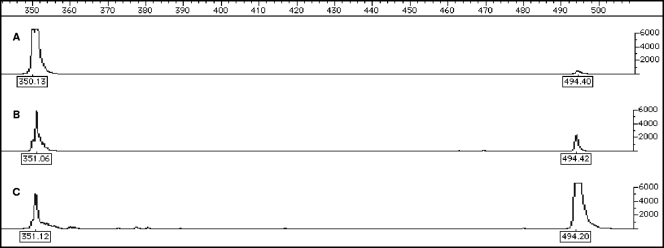
Electropherograms were generated using the FAM signal for A. calcoaceticus ATCC 23055T samples and are shown in Fig. 3, which shows the results for cultures that contained either type I cells (Fig. 3A), type II cells (Fig. 3B), or type III cells (Fig. 3C). The sizes of the 16S and pre-16S ssDNA for A. calcoaceticus ATCC 23055T were 351 and 494 nt, which are consistent with the sizes predicted from sequence information available for a closely related strain, Acinetobacter sp. strain ADP1 (www.genoscope.cns.fr). These results are consistent with the agarose gel electrophoresis results (Fig. 2).
FIG. 3.
Electropherograms of FAM-labeled RT products for type I (A), type II (B), and type III (C) cultivation conditions for A. calcoaceticus ATCC 23055T. For each electropherogram, the scale for size (in nucleotides) is at the top, and the FAM signal intensity scale is on the right.
(ii) RT-PE method detects type I, II, and III cells in activated sludge from a treatment plant.
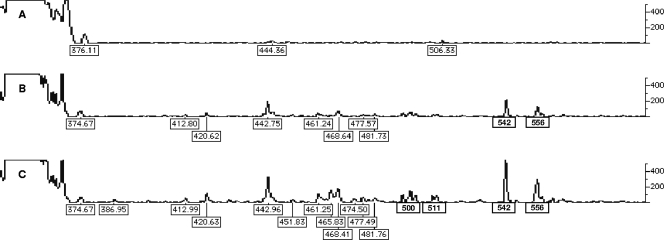
After promising results were obtained by applying the RT-PE method to A. calcoaceticus ATCC 23055T, the RT-PE method was applied to activated sludge samples that corresponded to type I, II, and III cells (Fig. 4A, B, and C, respectively). Like the electropherograms for A. calcoaceticus ATCC 23055T, the electropherograms for the activated sludge samples showed a trend for increased amounts of pre-16S 5′ ssDNA RT-PE products for type II and type III cells. As shown in Fig. 4C, 14 pre-16S 5′ ssDNA peaks were detected, whose sizes ranged from 412 to 556 nt, while the 16S ssDNA peaks consisted of the large, irregular peak between 316 and 373 nt. This suggests that the pre-16S and 16S ssDNA were derived from specific binding of the primer to the targeted site in the pre-16S and 16S rRNA. For slab gel analysis, the effective range of the FAM signal is between 100 and 10,000 for peak height or 2 orders of magnitude. The ratio of the heights of the pre-16S 5′ ssDNA peaks to the heights of 16S ssDNA peaks for the preparations derived from activated sludge is extremely low and comparable only to the Mycobacterium mucogenicm results (0.01) (data not shown). M. mucogenicum grows very slowly, with an optimal doubling time of 4 h, compared to a doubling time of 20 min for E. coli. This suggests that the bacteria in the activated sludge sample were growing slowly, which presents a problem for slab gel analysis because there is limited length heterogeneity for the pre-16S and 16S ssDNA. This limited length heterogeneity prevents identification and measurement of specific RT-PE products and subsequently makes determination of the pre-16S 5′ ssDNA/16S ssDNA ratio impossible. The pre-16S-5′ ssDNA/16S ssDNA ratio is an indicator of how a microbial population is growing. In order to address this problem, the DHPLC method was developed to separate, quantify, and sequence the RT-PE products. The sequence of an RT-PE product, which is a fraction of the 16S rRNA gene, can then be used to identify the bacterial genus associated with the pre-16S ssDNA of interest.
FIG. 4.
Electropherograms of FAM-labeled RT products for type I (A), type II (B), and type III (C) cultivation conditions for samples of activated sludge from a treatment plant. For each electropherogram, the size of each peak (in nucleotides) is indicated below the peak, and the FAM signal intensity scale is on the right.
Development and testing of the DHPLC method. (i) Detection of type I, II, and III cells of A. calcoaceticus ATCC 23055T and E. coli ATCC 11755T by the DHPLC method.
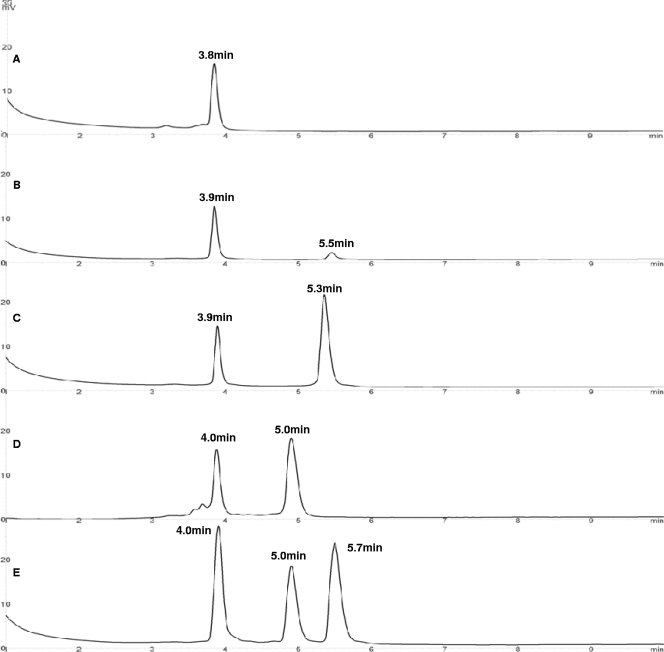
Chromatography results for RT-PE products derived from type I, II and III cells of A. calcoaceticus ATCC 23055T, type III cells of E. coli ATCC 11755T, and a mixture of the two preparations are shown in Fig. 5. The height of each peak was expressed as absorbance (in mV) and was dependent on the amount of DNA injected. The 16S and pre-16S 5′ ssDNA derived from each culture were present, as shown by two prominent peaks. Similar to previous results (36), chromatography results show that pre-16S 5′ ssDNA derived from type I A. calcoaceticus ATCC 23055T cells exhibited the lowest absorbance, while type II A. calcoaceticus ATCC 23055T cells exhibited slightly greater absorbance than type I cells. Type III A. calcoaceticus ATCC 23055T cells exhibited absorbance that was substantially greater than the absorbance exhibited by type I or type II cells.
FIG. 5.
Chromatograms of RT-PE products for type I (A), type II (B), type III (C) A. calcoaceticus ATCC 23055T cells, type III E. coli ATCC 11755T cells (D), and a combination of A. calcoaceticus ATCC 23055T and E. coli ATCC 11755T cells (E). For each chromatogram, the retention time of each peak is indicated above the peak, and the UV absorbance scale (in mV) is on the left.
In Fig. 5E, the individual chromatogram peaks in order of increasing retention time represent 16S ssDNA derived from A. calcoaceticus ATCC 23055T and E. coli ATCC 11755T (4 min), pre-16S 5′ ssDNA derived from E. coli ATCC 11755T (5 min), and pre-16S 5′ ssDNA derived from A. calcoaceticus ATCC 23055T (5.7 min). The retention times are consistent with pre-16S 5′ ssDNA sizes. For E. coli ATCC 11755T, the size is 444 nt (44), and for A. calcoaceticus ATCC 23055T, it is 494 nt. These results demonstrate that there is clear separation of two species based on distinct differences in retention times of pre-16S 5′ ssDNA, thus providing relative qualitative and quantitative characterization for species in a pre-16S ssDNA complex. The identities of the species correlated with their characteristic retention times and were confirmed by identification by sequence analysis. No cross contamination was observed.
The sensitivity and reproducibility of the DHPLC method were studied by injecting different concentrations of RT-PE products derived from A. calcoaceticus ATCC 23055T type III cells (Table 1). Peaks were eluted from the column with highly reproducible retention times, and the detection limit was 10 ng of ssDNA.
TABLE 1.
Reproducibility and detection limit of the DHPLC methoda
| cDNA mass (ng) | Retention time (min)b
|
Peak height (mV)c
|
||
|---|---|---|---|---|
| 16S cDNA | Pre-16S cDNA | 16S cDNA | Pre-16S cDNA | |
| 360 | 3.8 | 5.4 | 32.1 | 38.1 |
| 180 | 3.9 | 5.3 | 14.7 | 21.2 |
| 60 | 3.9 | 5.4 | 5.8 | 8.0 |
| 40 | 3.9 | 5.4 | 3.4 | 4.9 |
| 20 | 3.9 | 5.4 | 1.6 | 2.5 |
| 10 | 3.9 | 5.4 | 0.5 | 1.0 |
| 2 | NDd | ND | ND | ND |
ssDNA was derived from different amounts of type III A. calcoaceticus ATCC 23055T cultures.
The means ± standard deviations for 16S cDNA and pre-16S cDNA were 3.88 ± 0.04 and 5.38 ± 0.04 min, respectively.
The R2 values for 16S cDNA and pre-16S cDNA, calculated from a calibration curve for cDNA mass versus 16S and pre-16S cDNA peak height, were 0.998 and 0.996, respectively.
ND, not detectable.
(ii) Detection of type I and III cells in activated sludge from a bioreactor by the DHPLC method.
After promising results were obtained with the DHPLC method as described above, the 16S and pre-16S ssDNA derived from the lab-scale activated sludge samples that corresponded to type I and type III cells were detected by agarose gel electrophoresis and by DHPLC (Fig. 6).
FIG. 6.
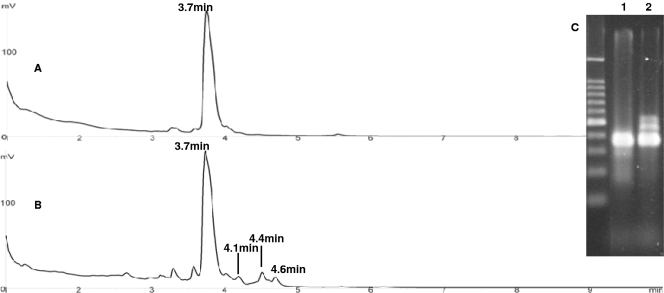
Chromatograms of RT-PE products for type I (A) and type III (B) cultivation conditions for activated sludge samples from a bioreactor. (C) Agarose gel electrophoresis analysis of RT-PE products from the same samples for comparison. The lanes contained samples for type I (lane 1) and type III (lane 2) cultivation conditions. The lane on the left contained a 100-bp DNA size ladder. For each chromatogram, the retention time of each peak is indicated above the peak, and the UV absorbance scale (in mV) is on the left.
Figure 6C shows agarose gel electrophoresis results for RT-PE products derived from type I (lane 1) and type III (lane 2) activated sludge cultures. Double-stranded size markers were used to describe the results. Several DNA bands ranging in size from 300 bp to 500 bp were detected by agarose gel electrophoresis. For type I cells, only 16S ssDNA (400 nt) was present. When transferred into fresh medium and treated with chloramphenicol, type III cells exhibited high levels of pre-16S 5′ ssDNA (between 400 and 500 bp), which is consistent with our pure-culture results (Fig. 2). The smaller bands below 300 bp are probably due to incorrect processing of mature 16S rRNA during ribosome genesis. For examining pre-16S 5′ ssDNA, incorrect processing of mature 16S rRNA is not a problem because the DHPLC method can separate the pre-16S rRNA from mature 16S rRNA based on retention times.
For the chromatography results, 16S ssDNA derived from activated sludge cells with a retention time of 3.7 min was designated P1 (343 nt), while pre-16S 5′ ssDNA derived from activated sludge cells with retention times of 4.1, 4.4, and 4.6 min were designated PRE1 (375 nt), PRE2 (400 nt), and PRE3 (416 nt), respectively. These DHPLC results suggest that chloramphenicol treatment “reveals” bacterial species in activated sludge samples that are expressing rrn genes.
The peaks with retention times less than 3.7 min are probably peaks for degraded 16S ssDNA and may be indicative of a large fraction of decaying biomass in activated sludge samples. Our analysis showed that the peaks observed for triplicate samples were eluted from the column with particular retention times with reliable and reproducible mean values (3.7, 4.2, 4.5, and 4.7 min for P1, PRE1, PRE2, and PRE3, respectively). The standard deviation for all peaks is 0.06, which indicates a high degree of reproducibility for this method. Compared with agarose gel electrophoresis, the DHPLC method provides better resolution for sensitive quantification of RT-PE products under different growth conditions.
Microbial community characterization for type I and type III activated sludge from a bioreactor.
Until now, it has not been clear whether the pre-16S rRNA and 16S rRNA libraries are significantly different (26). In the current study, the 16S rRNA library was generated from type I activated sludge cells and represented the total community structure. Three pre-16S rRNA libraries were generated by collecting, reamplifying, cloning, and sequencing PRE1, PRE2, and PRE3 peaks (Table 2). Clones sharing at least 95% sequence identity with one another were placed in the same taxonomic unit (i.e., genus).
TABLE 2.
Community structure of the activated sludge sample from a bioreactor as determined by using 16S rRNA genesa
| Clone library | No. of clones sequenced | No. (%) of clones in the following bacterial genera:
|
|||||
|---|---|---|---|---|---|---|---|
| Paracoccus | Zoogloea | Hydrogenophaga | Flavobacterium | Pedobacter | Unidentified bacterium | ||
| 16S rRNA gene | 197 | 2 (1.0) | 170 (86.3) | 8 (4.1) | 8 (4.1) | 1 (0.5) | 8 (4.1) |
| PRE1 | 100 | 46 (46) | 36 (36) | 0 (0) | 0 (0) | 14 (14) | 4 (4) |
| PRE2 | 100 | 10 (10) | 60 (60) | 0 (0) | 22 (22) | 0 (0) | 8 (8) |
| PRE3 | 97 | 19 (19.6) | 75 (77.3) | 0 (0) | 1 (1.0) | 0 (0) | 2 (2.1) |
pre-16S 5′ ssDNA from PRE1, pre-16S 5′ ssDNA from PRE2, and pre-16S 5′ ssDNA from PRE3 were used for analysis. The accession numbers for Paracoccus, Zoogloea, Hydrogenophaga, Flavobacterium, and Pedobacter are AB025188 (>98% identity), DQ413157 (>97% identity), DQ413146 (>97% identity), AF087062 (>97% identity), and AF270943 (>95% identity), respectively.
BLAST results revealed that a bacterial species related to Zoogloea spp. was the predominant species in the 16S rRNA libraries (86.3%). This is not surprising since Zoogloea spp. have commonly been found in wastewater sludge samples (19). Van Loosdrecht and coworkers stated that Zoogloea could accumulate storage polymers (polyhydroxyalkanoates) for later consumption (42), which may explain why Zoogloea spp. are the predominant active members of the PRE2 and PRE3 clone libraries (60% and 77.3%, respectively).
The second abundant genus present in each of the three precursor clone libraries is Paracoccus. The Paracoccus population represents only 1% of the 16S rRNA clones, but it represents a much higher percentage of the pre-16S rRNA clones. The difference reflects the finding that the size of the Paracoccus population increased compared with the size of the total community in the activated sludge samples. Therefore, cells of Paracoccus spp. are the active cells that positively respond to chloramphenicol in this enrichment medium, and their growth rate increased significantly when substrate and excess electron acceptors were available. Additional support comes from high potential of Paracoccus to adapt to various carbon and energy sources and accumulate storage polymers (3).
Previous studies have shown that both Paracoccus spp. and Zoogloea spp. exhibit versatile metabolism and are able to perform denitrification. These active bacterial functions correlated well with the performance of our lab-scale bioreactor, which was operated for nitrification and denitrification (21, 40).
Species of several other genera were also detected in the activated sludge clone libraries. For instance, increasing numbers of active Pedobacter cells were detected in PRE1 peaks. Active Flavobacterium cells were observed in PRE2 peaks. Members of the genera Pedobacter and Flavobacterium are commonly isolated from activated sludge samples (30, 35). A comparison of 16S rRNA and pre-16S rRNA libraries supports the hypothesis that the pre-16S rRNA libraries are more sensitive indicators of active bacteria.
Investigation of ribosome genesis in P. denitrificans and Z. ramigera.
Since Paracoccus and Zoogloea are the predominant genera that are detected in the activated sludge clone libraries and since members of both genera respond positively to chloramphenicol treatment by increasing their pre-16S rRNA levels, ribosome genesis in P. denitrificans and Z. ramigera was studied by using the RT-PE approach and the DHPLC method.
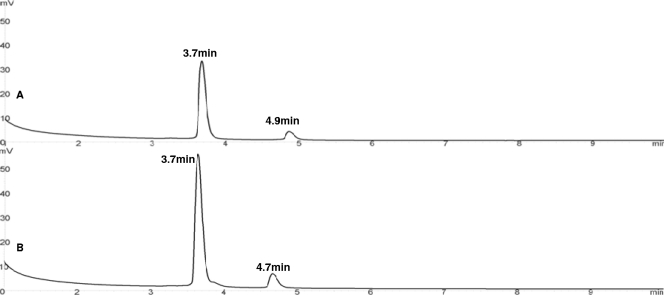
Pre-16S 5′ ssDNA derived from P. denitrificans was detected with a retention time of 4.9 min, while the pre-16S 5′ ssDNA RT-PE product derived from Z. ramigera was detected with a retention time of 4.7 min (Fig. 7A and B, respectively). However, in activated sludge samples, three pre-16S 5′ ssDNA RT-PE products for each genus were detected (PRE1, PRE2, and PRE3). A number of possible reasons for this are discussed below. First, P. denitrificans and Z. ramigera are different than the species that we detected in the activated sludge sample; as a result, the sizes of pre-16S 5′ rRNAs are different. This suggests that the DHPLC method is able to differentiate bacteria at the species level based on the pre-16S rRNA sequences, which is an important complement for 16S rRNA gene sequencing for the study of bacterial diversity and phylogeny. Studies have shown that pre-16S rRNA genes exhibit higher degrees of sequence and length variation that can be exploited to compare closely related bacterial strains than 16S rRNA genes (13, 25). Second, most bacteria can harbor multiple rrn operons which have sequence heterogeneities. For instance, E. coli possesses 7 rrn operons (7), while Bacillus subtilis possesses 10 rrn operons (22). Battermann and coworkers found that there are at least three chromosomal rrn operons in Paracoccus sp. strain OL18 (3). Regulation of multiple rrn operons depends largely on distinct growth conditions (6). In our studies, the precursor 16S rRNA length heterogeneity might have occurred because P. denitrificans and Z. ramigera were grown under different conditions, mixed-culture conditions (Fig. 6) and pure-culture conditions (Fig. 7). A mixed-culture environment, such as the activated sludge system, is dynamic (41), while pure cultures cultivated in nutrient-rich media under ideal environmental conditions exhibit a consistent growth response. Subsequently, multiple rrn operons were regulated and processed differently under these conditions, which led to the pre-16S rRNA heterogeneous products. This finding supports the conclusion that our assay could be used to study bacterial phylogenetic responses to different growth conditions. It also suggests that the RT-PE approach may work well for other bacteria cultured under ideal laboratory conditions, but the utility of this approach for evaluation of the growth response of bacteria present in environmental samples is considerably more difficult to predict. Another limitation of the method is that we assume that chloramphenicol impacts all bacterial species equally; however, differences in susceptibility to chloramphenicol among bacterial species also have the potential to bias the results.
FIG. 7.
Chromatograms of RT-PE products from type III P. denitrificans ATCC 13543 (A) and Z. ramigera ATCC 19623 (B). For each chromatogram, the retention time of each peak is indicated above the peak, and the UV absorbance scale (in mV) is on the left.
Conclusions.
A new molecular biology method coupled with a DHPLC approach was developed to identify the active microbial populations present in an environmental sample, such as activated sludge. The ssDNA derived from A. calcoaceticus ATCC 23055T cells, E. coli ATCC 11755T cells, and a mixture of the two types of cells were successfully tested with the DHPLC method. Our results demonstrate that this method is a sensitive and reliable method with a detection limit of 10 ng of ssDNA. Subsequently, this novel and robust method was successfully applied to activated sludge samples that represented two defined growth states. Compared with agarose and slab gel electrophoresis, the DHPLC method provides better resolution for sensitive identification and quantification of 16S rRNA RT-PE products under different growth conditions. Comparison of the 16S rRNA and pre-16S rRNA community structures suggested that pre-16S rRNA is a more sensitive indicator for active bacteria. These results suggest that the RT-PE method coupled with the DHPLC method has the potential to determine active bacterial populations in activated sludge and other environmental samples.
Footnotes
Published ahead of print on 24 April 2009.
REFERENCES
- 1.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barlaan, E. A., M. Sugimori, S. Furukawa, and K. Takeuchi. 2005. Profiling and monitoring of microbial populations by denaturing high-performance liquid chromatography. J. Microbiol. Methods 61:399-412. [DOI] [PubMed] [Google Scholar]
- 3.Battermann, A., C. Disse-Kromker, and B. Dreiseikelmann. 2003. A functional plasmid-borne rrn operon in soil isolates belonging to the genus Paracoccus. Microbiology 149:3587-3593. [DOI] [PubMed] [Google Scholar]
- 4.Belda, E., V. Sentandreu, and F. J. Silva. 2004. Identification and separation of PCR products based on their GC content by denaturing high-performance liquid chromatography. J. Chromatogr. B 811:263-268. [DOI] [PubMed] [Google Scholar]
- 5.Cangelosi, G. A., and W. H. Brabant. 1997. Depletion of pre-16S rRNA in starved Escherichia coli cells. J. Bacteriol. 179:4457-4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Condon, C., S. French, C. Squires, and C. L. Squires. 1993. Depletion of functional ribosomal RNA operons in Escherichia coli causes increased expression of the remaining intact copies. EMBO J. 12:4305-4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Condon, C., D. Liveris, C. Squires, I. Schwartz, and C. Squires. 1995. rRNA operon multiplicity in Escherichia coli and the physiological implications of rrn inactivation. J. Bacteriol. 177:4152-4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Court, D. 1993. RNA processing and degradation by RNase III, p. 71-116. In J. Belasco and G. Brawerman (ed.), Control of messenger RNA stability. Academic Press, San Diego, CA.
- 9.Cutter, M. R., and P. G. Stroot. 2008. Determination of specific growth rate by measurement of specific rate of ribosome synthesis in growing and nongrowing cultures of Acinetobacter calcoaceticus. Appl. Environ. Microbiol. 74:901-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeLong, E. E., and N. R. Pace. 2001. Environmental diversity of Bacteria and Archaea. Syst. Biol. 50:470-478. [PubMed] [Google Scholar]
- 11.Dennis, P. P., M. Ehrenberg, and H. Bremer. 2004. Control of rRNA synthesis in Escherichia coli: a systems biology approach. Microbiol. Mol. Biol. Rev. 68:639-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Domann, E., G. Hong, C. Imirzalioglu, S. Turschner, J. Kuhle, C. Watzel, T. Hain, H. Hossain, and T. Chakraborty. 2003. Culture-independent identification of pathogenic bacteria and polymicrobial infections in the genitourinary tract of renal transplant recipients. J. Clin. Microbiol. 41:5500-5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia-Martinez, J., I. Bescos, J. J. Rodriguez-Sala, and F. Rodriguez-Valera. 2001. RISSC: a novel database for ribosomal 16S-23S RNA genes spacer regions. Nucleic Acids Res. 29:178-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gourse, R. L., and M. Nomura. 1996. Prokaryotic rRNA gene expression, p. 373-394. In R. A. Zimmermann and A. E. Dahlberg (ed.), Ribosomal RNA: structure, evolution, processing, and function in protein biosynthesis. CRC Press, Boca Raton, FL.
- 15.Gtari, M., L. Brusetti, A. Cherif, A. Boudabous, and D. Daffonchio. 2007. Heteroduplex structures in 16S-23S rRNA intergenic transcribed spacer PCR products reveal ribosomal interoperonic polymorphisms within single Frankia strains. J. Appl. Microbiol. 103:1031-1040. [DOI] [PubMed] [Google Scholar]
- 16.Gurtler, V., and V. A. Stanisich. 1996. New approaches to typing and identification of bacteria using the 16S-23S rDNA spacer region. Microbiology 142:3-16. [DOI] [PubMed] [Google Scholar]
- 17.Hawkins, S. A., K. G. Robinson, A. C. Layton, and G. S. Sayler. 2006. A comparison of ribosomal gene and transcript abundance during high and low nitrite oxidizing activity using a newly designed real-time PCR detection system targeting the Nitrobacter spp. 16S-23S intergenic spacer region. Environ. Eng. Sci. 23:521-532. [Google Scholar]
- 18.Jemiolo, D. K. 1996. Processing of prokaryotic ribosomal RNA, p. 453-468. In R. A. Zimmermann and A. E. Dahlberg (ed.), Ribosomal RNA: structure, evolution, processing, and function in protein biosynthesis. CRC Press, Boca Raton, FL.
- 19.Juretschko, S., A. Loy, A. Lehner, and M. Wagner. 2002. The microbial community composition of a nitrifying-denitrifying activated sludge from an industrial sewage treatment plant analyzed by the full-cycle rRNA approach. Syst. Appl. Microbiol. 25:84-99. [DOI] [PubMed] [Google Scholar]
- 20.Kaczanowska, M., and M. Ryden-Aulin. 2007. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol. Mol. Biol. Rev. 71:477-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kocur, M. 1984. Genus Paracoccus Davis 1969, 384, p. 399-402. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 1. Williams & Wilkins, Baltimore, MD. [Google Scholar]
- 22.LaFauci, G., R. L. Widom, R. L. Eisner, E. D. Jarvis, and R. Rudner. 1986. Mapping of rRNA genes with integrable plasmids in Bacillus subtilis. J. Bacteriol. 165:204-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Licht, T. R., T. Tolker-Nielsen, K. Holmstrom, K. A. Krogfelt, and S. Molin. 1999. Inhibition of Escherichia coli precursor-16S rRNA processing by mouse intestinal contents. Environ. Microbiol. 1:23-32. [DOI] [PubMed] [Google Scholar]
- 24.Muyzer, G., E. C. Dewaal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S ribosomal RNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Natalini, E., and M. Scortichini. 2007. Variability of the 16S-23S rRNA gene internal transcribed spacer in Pseudomonas avellanae strains. FEMS Microbiol. Lett. 271:274-280. [DOI] [PubMed] [Google Scholar]
- 26.Nogales, B., E. R. B. Moore, E. Llobet-Brossa, R. Rossello-Mora, R. Amann, and K. N. Timmis. 2001. Combined use of 16S ribosomal DNA and 16S rRNA to study the bacterial community of polychlorinated biphenyl-polluted soil. Appl. Environ. Microbiol. 67:1874-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oerther, D. B., J. Pernthaler, A. Schramm, R. Amann, and L. Raskin. 2000. Monitoring precursor 16S rRNAs of Acinetobacter spp. in activated sludge wastewater treatment systems. Appl. Environ. Microbiol. 66:2154-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ojaimi, C., B. E. Davidson, I. Saintgirons, and I. G. Old. 1994. Conservation of gene arrangement and an unusual organization of rRNA genes in the linear chromosomes of the Lyme disease spirochaetes Borrelia burgdorferi, B. garinii, and B. afzellii. Microbiology 140:2931-2940. [DOI] [PubMed] [Google Scholar]
- 29.Osawa, S. 1968. Ribosome formation and structure. Annu. Rev. Biochem. 37:109-130. [DOI] [PubMed] [Google Scholar]
- 30.Park, M., S. Lu, S. H. Ryu, B. S. Chung, W. Park, C. Kim, and C. O. Jeon. 2006. Flavobacterium croceum sp. nov., isolated from activated sludge. Int. J. Syst. Evol. Microbiol. 56:2443-2447. [DOI] [PubMed] [Google Scholar]
- 31.Poulsen, L. K., G. Ballard, and D. A. Stahl. 1993. Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl. Environ. Microbiol. 59:1354-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raskin, L., L. K. Poulsen, D. R. Noguera, B. E. Rittmann, and D. A. Stahl. 1994. Quantification of methanogenic groups in anaerobic biological reactors by oligonucleotide probe hybridization. Appl. Environ. Microbiol. 60:1241-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmid, M. C., B. Maas, A. Dapena, K. V. de Pas-Schoonen, J. V. de Vossenberg, B. Kartal, L. van Niftrik, I. Schmidt, I. Cirpus, J. G. Kuenen, M. Wagner, J. S. S. Damste, M. Kuypers, N. P. Revsbech, R. Mendez, M. S. M. Jetten, and M. Strous. 2005. Biomarkers for in situ detection of anaerobic ammonium-oxidizing (anammox) bacteria. Appl. Environ. Microbiol. 71:1677-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stahl, D. A., B. Flesher, H. R. Mansfield, and L. Montgomery. 1988. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl. Environ. Microbiol. 54:1079-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steyn, P. L., P. Segers, M. Vancanneyt, P. Sandra, K. Kersters, and J. J. Joubert. 1998. Classification of heparinolytic bacteria into a new genus, Pedobacter, comprising four species: Pedobacter heparinus comb. nov., Pedobacter piscium comb. nov., Pedobacter africanus sp. nov. and Pedobacter saltans sp. nov. Proposal of the family Sphingobacteriaceae fam. nov. Int. J. Syst. Bacteriol. 48:165-177. [DOI] [PubMed] [Google Scholar]
- 36.Stroot, P. G., and D. B. Oerther. 2003. Elevated precursor 16S rRNA levels suggest the presence of growth inhibitors in wastewater. Water Sci. Technol. 47:241-250. [PubMed] [Google Scholar]
- 37.Tokajian, S., S. Al-Medawar, and F. Hashwa. 2008. Use of the 16S-23S ribosomal genes spacer region for the molecular typing of sphingomonads. Can. J. Microbiol. 54:668-676. [DOI] [PubMed] [Google Scholar]
- 38.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. X. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weldman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, J. M. Weidman, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 39.Tomlins, R., and J. Ordal. 1971. Precursor ribosomal ribonucleic acid and ribosome accumulation in vivo during the recovery of Salmonella typhimurium from thermal injury. J. Bacteriol. 107:134-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Unz, R. F. 1984. Genus IV. Zoogloea Itzigsohn 1868,30, p. 214-219. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 1. Williams & Wilkins, Baltimore, MD. [Google Scholar]
- 41.van Aalast-van Leeuwen, M. A., M. A. Pot, M. C. M. Van Loosdrecht, and J. J. Heijnen. 1997. Kinetic modeling of poly(beta-hydroxybutyrate) production and consumption by Paracoccus pantotrophus under dynamic substrate supply. Biotechnol. Bioeng 55:773-782. [DOI] [PubMed] [Google Scholar]
- 42.Van Loosdrecht, M. C. M., M. A. Pot, and J. J. Heijnen. 1997. Importance of bacterial storage polymers in bioprocess. Water Sci. Technol. 35:41-47. [Google Scholar]
- 43.Wang, Q., G. M. Garrity, J. M. Tiedje, and J. R. Cole. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Young, A. R., and A. J. Steitz. 1978. Complementary sequences 1700 nucleotides apart form a ribonuclease III cleavage site in Escherichia coli ribosomal precursor RNA. Proc. Natl. Acad. Sci. USA 75:3593-3597. [DOI] [PMC free article] [PubMed] [Google Scholar]