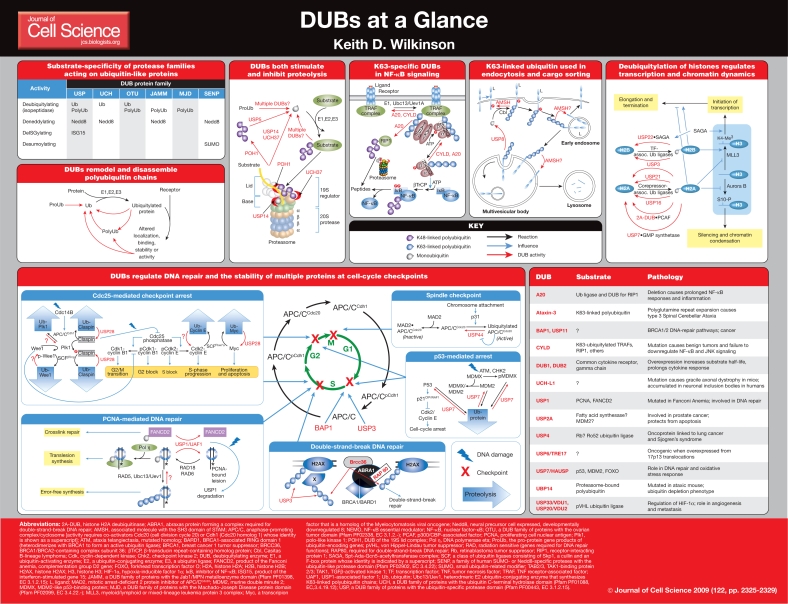
Introduction
The discovery of protein ubiquitylation three decades ago was the beginning of our understanding of a new mechanism by which proteins are marked for assembly into macromolecular complexes or movement between cellular compartments. The finding that ubiquitylation (the covalent attachment of the small protein ubiquitin to other proteins) targeted proteins for degradation earned Avram Hershko, Aaron Ciechanover and Irwin Rose the Nobel Prize in Chemistry in 2004 (Wilkinson, 2004). This early work spawned a large number of studies that investigated the role of ubiquitin and several other ubiquitin-like proteins (UBLs) as targeting signals in virtually all aspects of cellular protein metabolism (Chen, 2005; Cohn and D'Andrea, 2008; Saksena et al., 2007; Weake and Workman, 2008). The best understood example of ubiquitylation is the marking of proteins for delivery to the 26S proteasome, resulting in their degradation (Chiba and Tanaka, 2004; Guo et al., 2007; Hershko and Ciechanover, 1998; Schwartz and Hochstrasser, 2003; Varshavsky et al., 1989).
Similar to all regulated targeting pathways, the process of ubiquitylation is reversible. The enzymes that reverse the modification of proteins by ubiquitin are collectively known as deubiquitylating enzymes (DUBs) (Amerik and Hochstrasser, 2004; D'Andrea and Pellman, 1998; Wilkinson, 1997). Enzymes that reverse the modification by UBLs are similarly named: desumoylating enzymes remove SUMO (small ubiquitin-related modifier), deneddylating enzymes remove NEDD8 (neural precursor cell expressed, developmentally downregulated 8) and de-ISGylating enzymes remove ISG15 (interferon-stimulated gene product 15).
Figure 1.
There are many fine reviews describing the complex enzymatic mechanisms that regulate the conjugation of ubiquitin and UBLs to target proteins; the reader is referred to these for details of the ubiquitylation machinery (Belgareh-Touze et al., 2008; Dye and Schulman, 2007; Hochstrasser, 2007; Starita and Parvin, 2006). Briefly, ubiquitin is first thiol-esterified at its C-terminus by the action of the E1 ubiquitin-activating enzyme and then is subsequently transferred to one of several E2 ubiquitin-conjugating enzymes that act as carrier proteins. Finally, ubiquitin is transferred to a lysine residue of the target protein through the combined action of the E2-ubiquitin thiol ester and one of hundreds of E3 ubiquitin ligases. The ubiquitylation signal that is attached can consist of a single ubiquitin, multiple ubiquitins or a polyubiquitin chain in which successive ubiquitin molecules are assembled by the ubiquitylation of ubiquitin itself (Baboshina and Haas, 1996; Chau et al., 1989; Hofmann and Pickart, 1999; Koegl et al., 1999; Tokunaga et al., 2009; Wu-Baer et al., 2003). Because any of the seven lysine residues of ubiquitin, or its amino terminus, can be modified by a subsequent ubiquitin to form a polyubiquitin chain, there is a huge variation in the structure of polyubiquitin signals that can be attached. A polyubiquitin chain can involve linkages to the same lysine residue on each ubiquitin moiety to yield a homogeneous chain, or it can involve linkages to different lysine residues on different ubiquitin moeities, which results in a heterogeneous linear or branched chain.
Ubiquitylation is a versatile and dynamic targeting signal. The use of a protein, rather than a small molecule, to modify a target protein confers a large interaction surface that can be recognized by specific receptors. In addition, the many different polymeric forms of ubiquitin allow for structural variation of the signal. Structurally different forms of polyubiquitin are thought to target proteins for different cellular fates. For example, early work showed that K48-, K29-, and K11-linked polyubiquitin chains can target proteins for degradation by the proteasome (Chau et al., 1989; Jin et al., 2008; Koegl et al., 1999); K63-linked polyubiquitin chains participate in DNA repair and signaling kinase complexes (Deng et al., 2000; Spence et al., 1995); monoubiquitin and K63-linked chains are involved in targeting cell-surface proteins for internalization and endosomal sorting (Hicke and Riezman, 1996; Springael et al., 1999); and monoubiquitylation of histones can influence chromatin structure and transcription (Levinger and Varshavsky, 1980). Recent mass spectrometry analysis of ubiquitylated proteins shows that chains with multiple linkages can be attached to a single protein (Bish et al., 2008; Crosas et al., 2006; Kim et al., 2007; Kirkpatrick et al., 2005; Mayor et al., 2005; Xu and Peng, 2008), although the specific pathways in which these more complex polyubiquitin chains are involved remain poorly understood.
In this article and its accompanying poster, I summarize our understanding of the metabolic function of DUBs and discuss their roles in regulating several ubiquitin-dependent processes. Here, I use the term DUBs to refer only to those enzymes that act on ubiquitin. Much less is known about the enzymes that act on UBLs (Hay, 2007; Love et al., 2007; Mikolajczyk et al., 2007; Reverter et al., 2005; Sulea et al., 2006) and they will not be discussed here. Although much of what is known about DUBs was first observed in yeast, the yeast pathways or enzyme names are not emphasized. The poster illustrates the role of over 20 of nearly 100 mammalian DUBs that act on ubiquitin (Nijman et al., 2005), and concentrates on the DUBs about which something is known regarding their physiology or pathology. For pathways where the substrate or the process regulated is known in some detail, specific examples are provided. It should be noted that a role for DUBs has been implied in many other contexts, such as apoptosis, Parkinson's disease and neuronal-inclusion-body diseases, although in many cases the precise DUB involved has not been identified. Space limitations restrict the inclusion of these aspects in the poster.
DUBs are numerous and specific
The nearly 100 putative mammalian DUBs are grouped into five different families (Amerik and Hochstrasser, 2004; D'Andrea and Pellman, 1998; Wilkinson, 1997). Four of these families are thiol proteases: the ubiquitin-C-terminal hydrolases (UCHs), ubiquitin-specific proteases (USPs), ovarian-tumor (OTU) domain DUBs and Machado-Joseph domain (MJD) DUBs. The DUBs of a fifth family contain a Jab1/MPN metalloenzyme (JAMM) domain and act as zinc-dependent metalloproteases.
The large number of gene families, each with multiple members, suggests that selective pressure to evolve such catalysts has occurred numerous times. In addition, this diversity implies that considerable substrate specificity exists. This assumption is supported by the finding that the mutation, deletion or downregulation of specific DUBs induces very limited and specific cellular phenotypes and pathologies (Shanmugham and Ovaa, 2008; Singhal et al., 2008). For example, the mutation or deletion of the major neuronal DUB in mammals, UCH-L1 (ubiquitin C-terminal hydrolase L1), causes a localized axonal dystrophy but few other overt effects (Setsuie and Wada, 2007). A benign tumor syndrome of hair follicles known as cylindromatosis is caused by the mutation of CYLD, a USP-family DUB named after the disease it causes. Although the major defects caused by mutation of CYLD are limited to the nuclear factor-κB (NF-κB) pathway, (Courtois, 2008) this DUB has also been shown to have important roles in cell-cycle regulation (Stegmeier et al., 2007). Interfering with the function of USP1 mainly causes DNA-repair defects (Cohn and D'Andrea, 2008), whereas deleting USP14 in mice results in ataxia, a movement disorder characterized by uncoordinated motions (Crimmins et al., 2006).
A significant aspect of specificity is the ability of DUBs to recognize and act on different types of polyubiquitin. The catalytic domain of all DUBs contains a binding site for ubiquitin, and several DUBs bind ubiquitin at submicromolar concentrations. Many other DUBs, however, bind ubiquitin only very weakly (Reyes-Turcu and Wilkinson, 2009). Some DUBs have additional binding sites with affinity for the target protein that is ubiquitylated (Ventii and Wilkinson, 2008); for example, USP7 binds to a peptide sequence present in its substrates p53, MDM2 (murine double minute 2, an oncoprotein) and the Epstein Barr nuclear antigen-1 (Hu et al., 2006). It is clear that differently linked polyubiquitin chains have different structures, and it is thought that some DUBs can distinguish between them. For example, the DUBs CYLD and A20, which are involved in downregulating the NF-κB response, only disassemble K63-linked polyubiquitin chains, the type that is assembled on the signaling components of the NF-κB pathway (Courtois, 2008; Heyninck and Beyaert, 2005). Recent structures of CYLD and A20 suggest that these proteins achieve specific cleavage of K63-linked polyubiquitin chains by recognizing the unique surfaces of ubiquitin that are juxtaposed in this type of polyubiquitin. Similar conclusions are supported by a co-crystal structure containing the JAMM domain of the DUB AMSH (associated molecule with the SH3 domain of STAM) and K63-linked diubiquitin (Sato et al., 2008).
DUBs associate with ubiquitin ligases, scaffold proteins and substrate adaptors
In contrast to the specificity of DUBs that is apparent in vivo, assays carried out using artificial substrates in vitro often indicate that DUBs show little specificity. This can, in part, be attributed to the qualitative nature of many assays that do not measure the rates of substrate cleavage, although this factor alone cannot fully explain the apparent lack of specificity. A more likely explanation is that most DUBs contain additional protein interaction domains (which are utilized in vivo but not in in vitro) that direct the binding of DUBs to specific scaffolds or substrate adaptors and thereby confer substrate specificity. Thus, it is thought that in vivo specificity is determined mostly by the colocalization of the DUB and its substrates, and that adaptors are necessary for many DUBs to bind to their substrates (Marfany and Denuc, 2008; Ventii and Wilkinson, 2008). For example, USP1 is known to form a complex with a non-proteolytic subunit, UAF1, and the degradation of UAF1 leads to proteolysis of USP1 and consequent defects in the DNA repair functions that USP1 is involved in regulating (Cohn and D'Andrea, 2008). Similarly, the proteasome-associated DUBs USP14, UCH37 and POH must all be associated with the proteasome for significant DUB activity (Schmidt et al., 2005; Ventii and Wilkinson, 2008).
Another surprising observation is that several DUBs have been found to associate with ubiquitin ligases, which suggests that DUBs have a role in regulating ubiquitylation. The proteasome has both ubiquitin ligases and DUBs that associate with it (Crosas et al., 2006), and several DUB-ligase pairs interact directly, including BRCC36-BRCA1, BAP1-BRCA1, USP4-Ro52, USP7-MDM2, USP8-GRAIL, USP20-pVHL, USP33-pVHL and USP44-APC (Kee and Huibregtse, 2007; Marfany and Denuc, 2008; Ventii and Wilkinson, 2008). One explanation for these associations may be that the associated DUBs counteract the tendency of ubiquitin ligases to autoubiquitylate in the absence of other substrates. Another purpose that the interaction might serve is to target the DUB for degradation via the ligase-catalyzed ubiquitylation of the associated DUB. In at least some cases, the two interaction partners are indeed transregulated by each other. For example, in the absence of their substrates, the ubiquitin ligases MDM2 and Ro52 (Sjogren's syndrome associated autoantigen) become autoubiquitylated, and this is reversed by the activity of their associated DUBs, USP7 and USP4, respectively (Clegg et al., 2008; Meulmeester et al., 2005; Wada and Kamitani, 2006). Conversely, USP4 can be ubiquitylated by Ro52 and subsequently degraded. However, another function of these interactions might be to enforce the substrate specificity of ubiquitylation: the action of the DUB might `proofread' ubiquitylation and prevent the assembly of inappropriate ubiquitin linkages. The DUB A20, which contains both a ligase and a DUB domain on the same polypeptide, is the most extreme example of this. Its apparent role is to remodel the polyubiquitin chains that are generated on RIP1 (receptor-interacting protein 1) during tumor necrosis factor (TNF)-mediated stimulation of the NF-κB pathway. Removing the K63-linked polyubiquitin downregulates signaling, and assembling a K48-linked chain on RIP1 drives its degradation, further damping signaling (Heyninck and Beyaert, 2005).
Pathological conditions related to DUB dysfunction
Defects in DUB functions have been implicated in several pathological conditions, most notably cancer, neurological disease and microbial pathogenesis (de Pril et al., 2006; Rytkonen and Holden, 2007; Setsuie and Wada, 2007; Shackelford and Pagano, 2005; Singhal et al., 2008; Stuffers et al., 2008; Yang, 2007).
Based on the findings that DUBs have a role in regulating multiple cell-cycle and DNA repair checkpoints, in addition to cytokine-signaling and apoptosis pathways, it is likely that defects in DUB function could contribute to the development of cancer. Notably, mutations in CYLD cause cylindromatosis, and the translocation of the UBP6 coding region downstream of heterologous promoters is an oncogenic event that is found in many mesenchymal tumors. Furthermore, deletion of the gene encoding A20 in mice results in severe inflammation and cachexia (Singhal et al., 2008).
The potential role of DUBs in neurological disease is even less well understood. Mutation of USP14 in mice or ataxin-3 in humans causes ataxia (Crimmins et al., 2006; Duenas et al., 2006), whereas the S18Y allele of human UCH-L1 confers protection against sporadic Parkinson's disease. UCH-L1 is concentrated in a variety of neuronal inclusion bodies in humans, and loss-of-function mutations in this protein cause axonal degeneration in neurons that terminate at the Gracile nucleus, a region of the brainstem that receives dorsal-root fibers conveying sensory innervation of the leg and lower trunk (Setsuie and Wada, 2007). It is possible that interfering with DUB function leads to cellular stress that is not obvious in most tissues but has a major impact in the nervous system, as the death of a small number of neurons can have profound functional consequences.
Finally, it is notable that several bacteria (Rytkonen and Holden, 2007) and viruses (Lindner, 2007) have exploited the host-cell ubiquitin pathway by encoding DUBs that play a role in infection and pathogenesis. For example, the SARS coronavirus PLpro processing protease acts on a broad range of ubiquitylated and ISG15-modified host proteins and is required for viral replication (Ratia et al., 2008); the obligate intracellular bacterium, Burkholderia mallei, expresses and secretes a DUB inside infected macrophages (Shanks et al., 2009); and the ChlaDub1 expressed by Chlamydia trachomatis suppresses NF-κB activation (Le Negrate et al., 2008). Presumably these microbial DUBs confer a selective advantage on the pathogen by deubiquitylating host proteins and interfering with their normal cellular functions.
The above examples describe pathological conditions that are caused by expression of heterologous DUBs or by mutations of endogenous DUBs, although many other disease states or cellular functions have been shown to be modulated by DUBs. There are only a few DUB mutations that are currently known to cause disease, but it is very likely that more will be recognized in the future. It is also probable that other DUBs can modulate the effects of disease. Furthermore, as DUB-dependent processes are integral to many regulatory pathways, it is possible that DUBs will prove to be attractive drug targets in cases where the pathological lesions are caused by other mutations or damage events.
Perspectives
It is apparent that ubiquitin signals are pervasive, flexible and dynamic. Virtually every cellular process that requires temporally or spatially regulated protein-protein interactions is affected by ubiquitylation and deubiquitylation. In the past three years, numerous DUBs have been linked to some of the most vital of cellular functions and responses. These enzymes contribute greatly to the dynamic nature of the ubiquitin signal, and act by proofreading and disassembling ubiquitin chains with great specificity. This is achieved through the specificity of DUBs for their target proteins, the type of ubiquitin chain that they recognize and their cellular location. The associations of DUBs with ubiquitin ligases, scaffold proteins, substrates or substrate adaptors are also important factors in conferring this specificity.
The metabolic functions of ubiquitylation and deubiquitylation parallel that of phosphorylation. There are estimated to be 500 or more each of kinases and ubiquitin ligases, whereas phosphatases and DUBs number around 100 each. Similar to the many kinases and phosphatases that have been studied for their therapeutic potential, DUBs have a role in numerous physiological and pathological processes. Thus it is obvious that opportunities abound for pharmacological intervention. As the picture is emerging that each DUB has a limited set of substrates, the selective interference of an individual DUB may have highly selective effects on the localization, stability and/or function of specific proteins. Therefore, drugs that inhibit the catalytic activity of specific DUBs, or that interfere with their interactions with other proteins, hold great promise for modulating the ubiquitin-dependent physiological processes that are involved in human disease.
K.D.W. is the recipient of grants GM030308 and GM066355 from the National Institutes of Health. Deposited in PMC for release after 12 months.
References
- Amerik, A. Y. and Hochstrasser, M. (2004). Mechanism and function of deubiquitinating enzymes. Biochim. Biophys. Acta 1695, 189-207. [DOI] [PubMed] [Google Scholar]
- Baboshina, O. V. and Haas, A. L. (1996). Novel multiubiquitin chain linkages catalyzed by the conjugating enzymes E2EPF and RAD6 are recognized by 26 S proteasome subunit 5. J. Biol. Chem. 271, 2823-2831. [DOI] [PubMed] [Google Scholar]
- Belgareh-Touze, N., Leon, S., Erpapazoglou, Z., Stawiecka-Mirota, M., Urban-Grimal, D. and Haguenauer-Tsapis, R. (2008). Versatile role of the yeast ubiquitin ligase Rsp5p in intracellular trafficking. Biochem. Soc. Trans. 36, 791-796. [DOI] [PubMed] [Google Scholar]
- Bish, R. A., Fregoso, O. I., Piccini, A. and Myers, M. P. (2008). Conjugation of complex polyubiquitin chains to WRNIP1. J. Proteome Res. 7, 3481-3489. [DOI] [PubMed] [Google Scholar]
- Chau, V., Tobias, J. W., Bachmair, A., Marriott, D., Ecker, D. J., Gonda, D. K. and Varshavsky, A. (1989). A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243, 1576-1583. [DOI] [PubMed] [Google Scholar]
- Chen, Z. J. (2005). Ubiquitin signalling in the NF-kappaB pathway. Nat. Cell Biol. 7, 758-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba, T. and Tanaka, K. (2004). Cullin-based ubiquitin ligase and its control by NEDD8-conjugating system. Curr. Protein Pept. Sci. 5, 177-184. [DOI] [PubMed] [Google Scholar]
- Clegg, H. V., Itahana, K. and Zhang, Y. (2008). Unlocking the Mdm2-p53 loop: ubiquitin is the key. Cell Cycle 7, 287-292. [DOI] [PubMed] [Google Scholar]
- Cohn, M. A. and D'Andrea, A. D. (2008). Chromatin recruitment of DNA repair proteins: lessons from the fanconi anemia and double-strand break repair pathways. Mol. Cell 32, 306-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtois, G. (2008). Tumor suppressor CYLD: negative regulation of NF-kappaB signaling and more. Cell Mol. Life Sci. 65, 1123-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crimmins, S., Jin, Y., Wheeler, C., Huffman, A. K., Chapman, C., Dobrunz, L. E., Levey, A., Roth, K. A., Wilson, J. A. and Wilson, S. M. (2006). Transgenic rescue of ataxia mice with neuronal-specific expression of ubiquitin-specific protease 14. J. Neurosci. 26, 11423-11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosas, B., Hanna, J., Kirkpatrick, D. S., Zhang, D. P., Tone, Y., Hathaway, N. A., Buecker, C., Leggett, D. S., Schmidt, M., King, R. W. et al. (2006). Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell 127, 1401-1413. [DOI] [PubMed] [Google Scholar]
- D'Andrea, A. and Pellman, D. (1998). Deubiquitinating enzymes: a new class of biological regulators. Crit. Rev. Biochem. Mol. Biol. 33, 337-352. [DOI] [PubMed] [Google Scholar]
- de Pril, R., Fischer, D. F. and van Leeuwen, F. W. (2006). Conformational diseases: an umbrella for various neurological disorders with an impaired ubiquitin-proteasome system. Neurobiol. Aging 27, 515-523. [DOI] [PubMed] [Google Scholar]
- Deng, L., Wang, C., Spencer, E., Yang, L., Braun, A., You, J., Slaughter, C., Pickart, C. and Chen, Z. J. (2000). Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103, 351-361. [DOI] [PubMed] [Google Scholar]
- Duenas, A. M., Goold, R. and Giunti, P. (2006). Molecular pathogenesis of spinocerebellar ataxias. Brain 129, 1357-1370. [DOI] [PubMed] [Google Scholar]
- Dye, B. T. and Schulman, B. A. (2007). Structural mechanisms underlying posttranslational modification by ubiquitin-like proteins. Annu. Rev. Biophys. Biomol. Struct. 36, 131-150. [DOI] [PubMed] [Google Scholar]
- Guo, B., Yang, S. H., Witty, J. and Sharrocks, A. D. (2007). Signalling pathways and the regulation of SUMO modification. Biochem. Soc. Trans. 35, 1414-1418. [DOI] [PubMed] [Google Scholar]
- Hay, R. T. (2007). SUMO-specific proteases: a twist in the tail. Trends Cell Biol. 17, 370-376. [DOI] [PubMed] [Google Scholar]
- Hershko, A. and Ciechanover, A. (1998). The ubiquitin system. Annu. Rev. Biochem. 67, 425-479. [DOI] [PubMed] [Google Scholar]
- Heyninck, K. and Beyaert, R. (2005). A20 inhibits NF-kappaB activation by dual ubiquitin-editing functions. Trends Biochem. Sci. 30, 1-4. [DOI] [PubMed] [Google Scholar]
- Hicke, L. and Riezman, H. (1996). Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell 84, 277-287. [DOI] [PubMed] [Google Scholar]
- Hochstrasser, M. (2007). Ubiquitin ligation without a ligase. Dev. Cell 13, 4-6. [DOI] [PubMed] [Google Scholar]
- Hofmann, R. M. and Pickart, C. M. (1999). Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96, 645-653. [DOI] [PubMed] [Google Scholar]
- Hu, M., Gu, L., Li, M., Jeffrey, P. D., Gu, W. and Shi, Y. (2006). Structural basis of competitive recognition of p53 and MDM2 by HAUSP/USP7: implications for the regulation of the p53-MDM2 pathway. PLoS Biol. 4, e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, L., Williamson, A., Banerjee, S., Philipp, I. and Rape, M. (2008). Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell 133, 653-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee, Y. and Huibregtse, J. M. (2007). Regulation of catalytic activities of HECT ubiquitin ligases. Biochem. Biophys. Res. Commun. 354, 329-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H. T., Kim, K. P., Lledias, F., Kisselev, A. F., Scaglione, K. M., Skowyra, D., Gygi, S. P. and Goldberg, A. L. (2007). Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J. Biol. Chem. 282, 17375-17386. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick, D. S., Denison, C. and Gygi, S. P. (2005). Weighing in on ubiquitin: the expanding role of mass-spectrometry-based proteomics. Nat. Cell Biol. 7, 750-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koegl, M., Hoppe, T., Schlenker, S., Ulrich, H. D., Mayer, T. U. and Jentsch, S. (1999). A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 96, 635-644. [DOI] [PubMed] [Google Scholar]
- Le Negrate, G., Krieg, A., Faustin, B., Loeffler, M., Godzik, A., Krajewski, S. and Reed, J. C. (2008). ChlaDub1 of Chlamydia trachomatis suppresses NF-kappaB activation and inhibits IkappaBalpha ubiquitination and degradation. Cell Microbiol. 10, 1879-1892. [DOI] [PubMed] [Google Scholar]
- Levinger, L. and Varshavsky, A. (1980). High-resolution fractionation of nucleosomes: minor particles, “whiskers,” and separation of mononucleosomes containing and lacking A24 semihistone. Proc. Natl. Acad. Sci. USA 77, 3244-3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner, H. A. (2007). Deubiquitination in virus infection. Virology 362, 245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, K. R., Catic, A., Schlieker, C. and Ploegh, H. L. (2007). Mechanisms, biology and inhibitors of deubiquitinating enzymes. Nat. Chem. Biol. 3, 697-705. [DOI] [PubMed] [Google Scholar]
- Marfany, G. and Denuc, A. (2008). To ubiquitinate or to deubiquitinate: it all depends on the partners. Biochem. Soc. Trans. 36, 833-838. [DOI] [PubMed] [Google Scholar]
- Mayor, T., Lipford, J. R., Graumann, J., Smith, G. T. and Deshaies, R. J. (2005). Analysis of polyubiquitin conjugates reveals that the Rpn10 substrate receptor contributes to the turnover of multiple proteasome targets. Mol. Cell Proteomics 4, 741-751. [DOI] [PubMed] [Google Scholar]
- Meulmeester, E., Pereg, Y., Shiloh, Y. and Jochemsen, A. G. (2005). ATM-mediated phosphorylations inhibit Mdmx/Mdm2 stabilization by HAUSP in favor of p53 activation. Cell Cycle 4, 1166-1170. [DOI] [PubMed] [Google Scholar]
- Mikolajczyk, J., Drag, M., Bekes, M., Cao, J. T., Ronai, Z. and Salvesen, G. S. (2007). Small ubiquitin-related modifier (SUMO)-specific proteases: profiling the specificities and activities of human SENPs. J. Biol. Chem. 282, 26217-26224. [DOI] [PubMed] [Google Scholar]
- Nijman, S. M., Luna-Vargas, M. P., Velds, A., Brummelkamp, T. R., Dirac, A. M., Sixma, T. K. and Bernards, R. (2005). A genomic and functional inventory of deubiquitinating enzymes. Cell 123, 773-786. [DOI] [PubMed] [Google Scholar]
- Ratia, K., Pegan, S., Takayama, J., Sleeman, K., Coughlin, M., Baliji, S., Chaudhuri, R., Fu, W., Prabhakar, B. S., Johnson, M. E. et al. (2008). A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc. Natl. Acad. Sci. USA 105, 16119-16124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverter, D., Wu, K., Erdene, T. G., Pan, Z. Q., Wilkinson, K. D. and Lima, C. D. (2005). Structure of a complex between Nedd8 and the Ulp/Senp protease family member Den1. J. Mol. Biol. 345, 141-151. [DOI] [PubMed] [Google Scholar]
- Reyes-Turcu, F. E. and Wilkinson, K. D. (2009). Polyubiquitin binding and disassembly by deubiquitinating enzymes. Chem. Rev. 109, 1495-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rytkonen, A. and Holden, D. W. (2007). Bacterial interference of ubiquitination and deubiquitination. Cell Host Microbe 1, 13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksena, S., Sun, J., Chu, T. and Emr, S. D. (2007). ESCRTing proteins in the endocytic pathway. Trends Biochem. Sci. 32, 561-573. [DOI] [PubMed] [Google Scholar]
- Sato, Y., Yoshikawa, A., Yamagata, A., Mimura, H., Yamashita, M., Ookata, K., Nureki, O., Iwai, K., Komada, M. and Fukai, S. (2008). Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature 455, 358-362. [DOI] [PubMed] [Google Scholar]
- Schmidt, M., Hanna, J., Elsasser, S. and Finley, D. (2005). Proteasome-associated proteins: regulation of a proteolytic machine. Biol. Chem. 386, 725-737. [DOI] [PubMed] [Google Scholar]
- Schwartz, D. C. and Hochstrasser, M. (2003). A superfamily of protein tags: ubiquitin, SUMO and related modifiers. Trends Biochem. Sci. 28, 321-328. [DOI] [PubMed] [Google Scholar]
- Setsuie, R. and Wada, K. (2007). The functions of UCH-L1 and its relation to neurodegenerative diseases. Neurochem. Int. 51, 105-111. [DOI] [PubMed] [Google Scholar]
- Shackelford, J. and Pagano, J. S. (2005). Targeting of host-cell ubiquitin pathways by viruses. Essays Biochem. 41, 139-156. [DOI] [PubMed] [Google Scholar]
- Shanks, J., Burtnick, M. N., Brett, P. J., Waag, D. M., Spurgers, K. B., Ribot, W. J., Schell, M. A., Panchal, R. G., Gherardini, F. C., Wilkinson, K. D. et al. (2009). Burkholderia mallei tssM encodes a putative deubiquitinase that is secreted and expressed inside infected RAW 264.7 murine macrophages. Infect. Immun. 77, 1636-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugham, A. and Ovaa, H. (2008). DUBs and disease: activity assays for inhibitor development. Curr. Opin. Drug Discov. Devel. 11, 688-696. [PubMed] [Google Scholar]
- Singhal, S., Taylor, M. C. and Baker, R. T. (2008). Deubiquitylating enzymes and disease. BMC Biochem. 9 Suppl. 1, S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence, J., Sadis, S., Haas, A. L. and Finley, D. (1995). A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol. Cell. Biol. 15, 1265-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springael, J. Y., Galan, J. M., Haguenauer-Tsapis, R. and Andre, B. (1999). NH4+-induced down-regulation of the Saccharomyces cerevisiae Gap1p permease involves its ubiquitination with lysine-63-linked chains. J. Cell Sci. 112, 1375-1383. [DOI] [PubMed] [Google Scholar]
- Starita, L. M. and Parvin, J. D. (2006). Substrates of the BRCA1-dependent ubiquitin ligase. Cancer Biol. Ther. 5, 137-141. [DOI] [PubMed] [Google Scholar]
- Stegmeier, F., Sowa, M. E., Nalepa, G., Gygi, S. P., Harper, J. W. and Elledge, S. J. (2007). The tumor suppressor CYLD regulates entry into mitosis. Proc. Natl. Acad. Sci. USA 104, 8869-8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuffers, S., Brech, A. and Stenmark, H. (2008). ESCRT proteins in physiology and disease. Exp. Cell Res. 315, 1619-1626. [DOI] [PubMed] [Google Scholar]
- Sulea, T., Lindner, H. A. and Menard, R. (2006). Structural aspects of recently discovered viral deubiquitinating activities. Biol. Chem. 387, 853-862. [DOI] [PubMed] [Google Scholar]
- Tokunaga, F., Sakata, S., Saeki, Y., Satomi, Y., Kirisako, T., Kamei, K., Nakagawa, T., Kato, M., Murata, S., Yamaoka, S. et al. (2009). Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat. Cell Biol. 11, 123-132. [DOI] [PubMed] [Google Scholar]
- Varshavsky, A., Bachmair, A., Finley, D., Gonda, D. K. and Wunning, I. (1989). Targeting of proteins for degradation. Biotechnology 13, 109-143. [PubMed] [Google Scholar]
- Ventii, K. H. and Wilkinson, K. D. (2008). Protein partners of deubiquitinating enzymes. Biochem. J. 414, 161-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada, K. and Kamitani, T. (2006). UnpEL/Usp4 is ubiquitinated by Ro52 and deubiquitinated by itself. Biochem. Biophys. Res. Commun. 342, 253-258. [DOI] [PubMed] [Google Scholar]
- Weake, V. M. and Workman, J. L. (2008). Histone ubiquitination: triggering gene activity. Mol. Cell 29, 653-663. [DOI] [PubMed] [Google Scholar]
- Wilkinson, K. D. (1997). Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J. 11, 1245-1256. [DOI] [PubMed] [Google Scholar]
- Wilkinson, K. D. (2004). Ubiquitin: a Nobel protein. Cell 119, 741-745. [DOI] [PubMed] [Google Scholar]
- Wu-Baer, F., Lagrazon, K., Yuan, W. and Baer, R. (2003). The BRCA1/BARD1 heterodimer assembles polyubiquitin chains through an unconventional linkage involving lysine residue K6 of ubiquitin. J. Biol. Chem. 278, 34743-34746. [DOI] [PubMed] [Google Scholar]
- Xu, P. and Peng, J. (2008). Characterization of polyubiquitin chain structure by middle-down mass spectrometry. Anal. Chem. 80, 3438-3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J. M. (2007). Emerging roles of deubiquitinating enzymes in human cancer. Acta Pharmacol. Sin. 28, 1325-1330. [DOI] [PubMed] [Google Scholar]