Summary
With every cell division, peroxisomes duplicate and are segregated between progeny cells. Here, we discuss the different modes of peroxisome multiplication and the machinery that is involved in each case. Peroxisomes have been considered by many to be peripheral to mainstream cell biology. However, this is changing in response to the recent finding that peroxisomes obtain membrane constituents from the endoplasmic reticulum, making them the latest branch of the endomembrane system to be identified. Furthermore, the observations that peroxisome and mitochondrial biogenesis can occur in a coordinated manner, and that these organelles share factors for their multiplication, demonstrate previously unanticipated aspects of cellular organisation.
Keywords: ER, Dynamin, Peroxin, Peroxisome
Introduction
Eukaryotic cells are extensively subdivided into functionally distinct membrane-bound compartments called organelles. Each type of organelle houses a unique set of proteins that creates a specific environment, and the presence of all these specialised environments within a single cell allows for a great variety of functions to occur simultaneously. The processes that occur within organelles are integral to cellular metabolism. Most anabolic and catabolic processes are not confined to a single organelle, and an extensive exchange of metabolites occurs between distinct organelles and the cytoplasm. This interdependence of cellular compartments requires a coordinated formation and turnover of organelles.
Peroxisomes are organelles that are surrounded by a single membrane and are found in most eukaryotic cells. The metabolic pathways contained within peroxisomes vary between cell types and organisms; in humans, they are involved in a variety of catabolic and anabolic processes, including β-oxidation of a wide range of fatty acids that are not oxidised by mitochondria, and synthesis of plasmalogens and bile acids (for a review, see Wanders and Waterham, 2006). Characteristic of peroxisomes is the presence of hydrogen-peroxide-producing oxidases (hence the name peroxisome) and catalase, which breaks down the hydrogen peroxide.
The importance of the peroxisome for human health is emphasised by the existence of inherited disorders that are caused by peroxisomal dysfunction (Wanders and Waterham, 2005). In the most severe cases, the complete organelle is absent, and these individuals usually die in their first year. Genetic approaches in yeast species and Chinese hamster ovary (CHO) cells have led to the identification of many factors involved in peroxisomal biogenesis. These factors, called peroxins, are evolutionarily conserved and are defective in the most devastating of the human peroxisomal disorders (Weller et al., 2003), including Zellweger syndrome and neonatal adrenoleukodystrophy (NALD).
All peroxisomal proteins are encoded in the nucleus. Proteins that are destined for the peroxisomal lumen (or matrix) are synthesised on free (non-bound) ribosomes in the cytosol, and fold and assemble prior to peroxisomal import. Lumenal proteins contain a short peroxisome targeting signal (PTS) that is recognised by PTS-specific receptors in the cytosol. These receptors deliver their cargo by docking on the peroxisomal membrane. A large number of peroxins are required for the posttranslational import of lumenal proteins, and these are currently under intensive study (reviewed by Brown and Baker, 2008). Peroxisomal membrane proteins (PMPs) were also thought to be imported directly from the cytosol, but it is now clear that this is not true for all PMPs: at least some have been shown to traverse the endoplasmic reticulum (ER) en route to peroxisomes (see below).
Peroxisomes double in number during cell growth and, in yeast, are segregated between mother and daughter cells before cell division. This process has been termed `peroxisome replication' and is the subject of this Commentary. Peroxisome replication is distinct from peroxisome proliferation, which is defined as the increase in peroxisome number that is seen in response to changes in environmental or intracellular conditions (Fagarasanu et al., 2007). The mechanism of peroxisome multiplication has long been a subject of debate, and two opposing models have dominated the field. In the first, peroxisomes multiply by growth and division (reviewed by Lazarow, 2003). In the second, peroxisomes form de novo (Fig. 1) (reviewed by Tabak et al., 2003; Tabak et al., 2006). There is evidence to support both models and this seeming contradiction between them has been resolved only recently by the introduction of new live-cell imaging approaches (Kim et al., 2006; Motley and Hettema, 2007) (and see below).
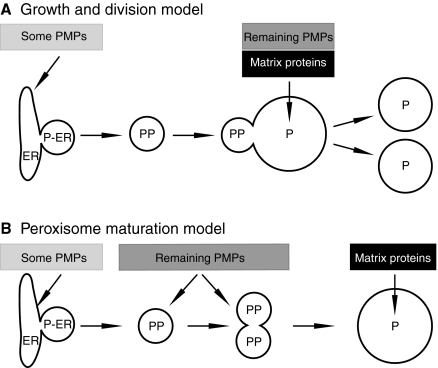
Fig. 1.
Models for peroxisome multiplication and biogenesis of peroxisomal membrane proteins (PMPs). (A) Growth and division model. A specialised region of the ER, the peroxisomal ER (P-ER) (Mullen and Trelease, 2006), which contains a subset of PMPs buds off to form a pre-peroxisome (PP) that fuses with peroxisomes (P), resulting in membrane expansion. Lumenal proteins and additional PMPs are imported from the cytosol directly into peroxisomes. Subsequently, peroxisomes divide. (B) Peroxisome maturation model. The peroxisomal ER buds from the ER and fuses homotypically to produce larger membrane structures. Additional PMPs are imported, and only once the import machinery has been inserted will lumenal protein import occur and the organelle mature into a metabolically active peroxisome. Shaded boxes indicate the stage at which proteins are proposed to be imported.
Peroxisome function and matrix protein import have been covered extensively in recent reviews (Brown and Baker, 2008; Platta and Erdmann, 2007b; Rayapuram and Subramani, 2006) and are not discussed here. Instead, we focus on peroxisome replication and the factors involved; peroxisome proliferation will be referred to only in the context of organelle crosstalk. Our discussion centres on the budding yeast Saccharomyces cerevisiae, and findings in other model systems are also outlined if relevant.
Two modes of peroxisome multiplication
Organelles can multiply by growth and division or by de novo synthesis (Lowe and Barr, 2007). As described above, time-lapse microscopy of dividing S. cerevisiae cells has revealed that the number of peroxisomes doubles prior to every cell division. After doubling in number, peroxisomes are distributed between mother and daughter cells (Hoepfner et al., 2001). In dividing S. cerevisiae and Hansenula polymorpha cells, peroxisomes multiply by fission (Motley and Hettema, 2007; Nagotu et al., 2008a; Nagotu et al., 2008b).
Segregation of peroxisomes between mother and daughter cells is achieved by balanced but opposing mechanisms: some peroxisomes are retained in the mother cell, and those that are not retained are transported along actin cables to the bud. Transport depends on the class-V myosin Myo2p (Hoepfner et al., 2001) and the integral membrane protein Inp2, which functions as the peroxisomal receptor for Myo2 (Fagarasanu et al., 2006). Retention of peroxisomes in the mother cell requires the peroxisomal peripheral membrane protein Inp1, which is thought to anchor peroxisomes to the cell periphery (Fagarasanu et al., 2005).
Peroxisomes can also form de novo. Mammalian cells with a defect in the peroxins PEX3, PEX16 or PEX19 (Table 1) lack any peroxisomal structures, but peroxisomes reappear after genetic complementation (Kim et al., 2006; Matsuzono et al., 1999; South et al., 2000; Voorn-Brouwer et al., 2001). The same phenomenon has been observed in pex3 and pex19 mutants in various yeast species (Hoepfner et al., 2005; Hohfeld et al., 1991; Kragt et al., 2005; Tam et al., 2005). In contrast to the situation in yeasts, de novo peroxisome formation and multiplication of peroxisomes by fission can occur simultaneously in mammalian cells (Kim et al., 2006). Whether yeasts are able to do this under certain conditions is not known.
Table 1.
Factors involved in various aspects of peroxisome formation and maintenance in S. cerevisiae
| Functional class | S. cerevisiae gene |
|---|---|
| Import of peroxisomal matrix proteins | PEX1, PEX2, PEX4, PEX5, PEX6, PEX7, PEX8, PEX10, PEX12, PEX13, PEX14, PEX15, PEX17, PEX18, PEX21, PEX22, DJP1 |
| PMP biogenesis and de novo formation | PEX3, PEX19a |
| Peroxisome morphology, replication and proliferation | VPS1, DNM1, CAF4, MDV1, FIS1, PEX11, PEX25, PEX27, PEX28, PEX29, PEX30, PEX31, PEX32 |
| Peroxisome inheritance | INP1, INP2, MYO2, polarised actin cytoskeleton |
PEX16 is absent from S. cerevisiae. In Yarrowia lipolytica it is involved in peroxisome proliferation. In mammals, it is involved in peroxisomal membrane protein biogenesis and de novo formation. For other species-specific differences in factors involved in peroxisome formation and maintenance, we refer readers to Kiel et al. (Kiel et al., 2006)
In S. cerevisiae mutants with an inheritance defect, many cells within a culture initially lack peroxisomes (Fig. 2) (Motley and Hettema, 2007), although these cells eventually form peroxisomes de novo. The presence of this delay is in contrast to other yeast compartments; for instance, when inheritance of vacuoles fails, vacuoles reform de novo before cytokinesis occurs (Gomes de Mesquita et al., 1996; Wang et al., 1996). This delay suggests that the time that is needed for de novo peroxisome formation exceeds the duration of the cell cycle when cells are growing rapidly. When the growth rate decreases, the proportion of cells containing peroxisomes increases (Fig. 2). Therefore, in order to maintain a peroxisome population, S. cerevisiae must multiply peroxisomes by fission and segregate them efficiently.
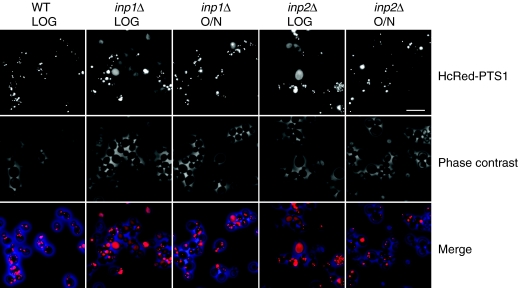
Fig. 2.
Segregation defects lead to a temporary absence of peroxisomes. S. cerevisiae cells constitutively expressing HcRed peroxisome targeting signal type 1 (HcReD-PTS1) were inoculated into glucose medium and grown until their doubling time was 2 hours (after 5 hours of growth at low cell density; LOG) or increased to more than 4 hours (after growing cells overnight). Cells were imaged using fluorescence microscopy (top panel) and phase contrast (middle panels) and the images are merged in the bottom panels. Up to 50% of cells in inp1 or inp2 cultures lack peroxisomes under conditions of rapid growth. When the growth rate decreases, however, the proportion of cells containing peroxisomes increases. The fluorescent images are coloured red and the phase contrast images blue; Adobe Photoshop was used to produce the merged image. Scale bar: 10 μm.
In summary, studies in yeast and mammalian cells have shown that peroxisomes can multiply either by fission or by de novo formation. The predominant mode of multiplication varies among organisms and might be dictated by whether peroxisomes are present.
Factors that regulate peroxisome fission
Dynamin-related proteins (DRPs) are large GTPases that are implicated in membrane-remodelling events including organelle fission and fusion (Praefcke and McMahon, 2004). Two DRPs, Vps1 and Dnm1, are required for peroxisome fission in S. cerevisiae (Table 1) (Hoepfner et al., 2001; Kuravi et al., 2006; Motley and Hettema, 2007). Vps1 has a greater role in peroxisome replication (Fig. 3), whereas the contribution of Dnm1 is increased under conditions of peroxisome proliferation. Dnm1 is also involved in mitochondrial fission, in which its role has been well studied – it is recruited to the mitochondrial outer membrane by a complex comprising the tail-anchored protein Fis1 and the peripheral membrane proteins Mdv1 and Caf4. Once at the membrane, Dnm1 is thought to multimerise into a ring structure around a mitochondrial tubule, after which GTP hydrolysis might drive constriction and ultimately scission of the mitochondrial membranes (Westermann, 2008). Surprisingly, the same factors recruit Dnm1 to the peroxisomal membrane (Fig. 3). It is not known how Vps1 is recruited to peroxisomes, but its recruitment is independent of Fis1, Caf4 and Mdv1 (Motley et al., 2008). A requirement for Pex19 has been reported (Vizeacoumar et al., 2006). The fact that peroxisomes and mitochondria share common factors for their fission might facilitate the coordination of their biogenesis. For a more extensive overview of mitochondrial and peroxisomal crosstalk, see Camoes et al. (Camoes et al., 2008).
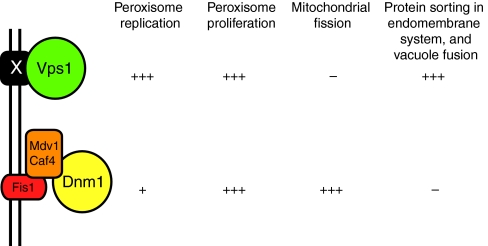
Fig. 3.
Schematic representation of the DRP-dependent fission machineries of S. cerevisiae peroxisomes. Under conditions of peroxisome replication, fission is mediated predominantly by the DRP Vps1. During peroxisome proliferation, both Vps1 and Dnm1 are involved in peroxisome fission. Whereas Dnm1 can act on both mitochondria and peroxisomes, Vps1 acts on various compartments of the endomembrane system as well as peroxisomes. X is an unknown factor through which Vps1 is proposed to bind to the peroxisomal membrane. –, + and +++ indicate level of involvement.
Other factors that are required for normal peroxisome abundance include the Pex11 family of proteins (Pex11, 25 and 27) (Rottensteiner et al., 2003) (Table 1). These peripheral membrane proteins associate tightly with the peroxisomal membrane and form homo- and hetero-oligomers. Peroxisome morphology is affected by overexpression or deletion of Pex11 family members, suggesting that they have a role in peroxisome fission. In S. cerevisiae, Pex11 is required only under conditions of peroxisome proliferation, whereas Pex25 and Pex27 contribute to peroxisome abundance under non-proliferative conditions (Rottensteiner et al., 2003; Tam et al., 2003).
EM analysis has shown that peroxisomes have a `beads-on-a-string' morphology in vps1 cells, suggesting that peroxisomes can recruit factors that induce constriction as well as tubulation (Hoepfner et al., 2001). A similar observation has been made in mammalian cells after DRP1 knockdown (Koch et al., 2004). A role for Pex11 in this process has been shown in H. polymorpha dnm1 cells, in which the emergence of a long tubular extension from the peroxisomal structure is dependent on the presence of Pex11 (Nagotu et al., 2008b). Whether Pex11 and its family members can deform the peroxisomal membrane to induce tubule formation and even constriction, or whether additional factors are required, awaits biochemical characterisation of the Pex11 family of proteins. Interestingly, a significant proportion of cells in a pex11/pex25/pex27 triple mutant culture lack peroxisomal membranes. The function of these proteins might therefore extend beyond a role in peroxisome fission (Rottensteiner et al., 2003).
Analysis of peroxisome replication in Arabidopsis thaliana has shown that peroxisome growth, elongation and fission are coordinated with the cell cycle (Lingard et al., 2008). A single DRP (DRP3a) was shown to be involved in both peroxisome and mitochondrial fission in a Fis1-dependent mechanism, which thereby resembles Dnm1-dependent peroxisome fission in yeasts. Furthermore, Pex11 isoforms were shown to have a role in peroxisome elongation and tubulation, and to interact with Fis1. On the basis of these data, a model has been proposed whereby Pex11 induces membrane tubulation and Fis1 recruitment; Fis1 then recruits DRP3a to the membrane and fission ensues (Lingard et al., 2008). A similar model has been proposed for the proliferation of peroxisomes in mammalian cells (Schrader and Fahimi, 2006). In line with this proposal, overexpression of Pex11 results in an increased association of the human Dnm1 homologue, DLP1, with peroxisomes in human fibroblasts (Li and Gould, 2003).
Species-specific differences in the fission machinery seem to exist; for instance, peroxisome fission in H. polymorpha is mediated by Dnm1 rather than Vps1. Mammalian orthologues of Caf4 and Mdv1 have not been identified, but recently a tail-anchored protein, mitochondrial fission factor (MFF), was shown to be required for fission of both peroxisomes and mitochondria in Drosophila and mammalian cells (Gandre-Babbe and van der Bliek, 2008). Several additional fungal-specific PMPs have been identified (Pex23, 24, 28, 29, 30, 31 and 32) that, when absent or overexpressed, affect peroxisome morphology under conditions of peroxisome proliferation (reviewed by Thoms and Erdmann, 2005).
In summary, research in different model systems shows that some of the factors that are required for peroxisome fission are evolutionarily conserved. A model emerges whereby peroxisome membrane tubulation precedes fission (Platta and Erdmann, 2007a) and the first molecular connection between these two processes has been described (Lingard et al., 2008). Future research will shed light on the extent of overlap between the fission events of peroxisome proliferation and replication.
Role of the ER in peroxisome formation
The origin of the peroxisomal membrane is a long-standing debate in the field. Most peroxisome biogenesis mutants cannot import matrix proteins, but PMPs still assemble in membranous structures. There are three yeast mutants, pex3, pex16 and pex19, that appear to lack any residual peroxisomal membrane structures. In these mutants, PMPs are unstable and several PMPs have been shown to be mislocalised to the cytoplasm. Genetic complementation of these mutants results in the reappearance of peroxisomes.
The existence of a `seed compartment' from which peroxisomes can form de novo was postulated long before this compartment was definitively identified. Several papers showed that a small amount of some endogenous PMPs, as well as tagged or truncated PMPs, were targeted to membrane structures in the absence of pex3, pex19 or pex16. Were these PMPs accumulating in a peroxisomal precursor? As some of these proteins were found in mitochondria, others in the ER and yet others in unidentified membrane structures, the significance of these findings was difficult to judge (reviewed by Lazarow, 2003).
It was only relatively recently that the ER was shown to be the precursor for peroxisomes that are formed de novo. Below we discuss the key experiments that led to this conclusion. This topic has been reviewed extensively (Tabak et al., 2008; van der Zand et al., 2006; Titorenko and Mullen, 2006; Mullen and Trelease, 2006).
One of the strongest data sets is provided by Hoepfner et al. (Hoepfner et al., 2005), who show that a short burst of Pex3-YFP expression in pex3 cells initiates peroxisome formation from the ER via a multistep maturation process. Pex3-YFP was found to associate first with the ER, and then to concentrate into ER-associated punctae. These punctae dissociate from the ER and eventually become peroxisomes that can import matrix proteins (Fig. 1B). Hoepfner and coworkers proposed that, during de novo formation, pre-peroxisomal vesicles bud from the ER membrane and fuse homotypically, thereby forming larger pre-peroxisomal structures that acquire additional PMPs. Only once the PMPs, including the import machinery for lumenal proteins, have been incorporated into the newly forming peroxisomal structure can the lumenal proteins be imported, thereby completing the maturation process (Kunau, 2005; Tabak et al., 2006; Tabak et al., 2003). The whole process takes 4-5 hours (see Fig. 1). This ER-to-peroxisome maturation pathway has been observed by others in both S. cerevisiae and H. polymorpha (Haan et al., 2006; Tam et al., 2005). Biochemical studies by Rachubinski and coworkers in the yeast Yarrowia lipolytica also support the finding that peroxisomes form via a maturation pathway (Titorenko et al., 1996; Titorenko and Mullen, 2006).
Of equal significance are the studies of the integral membrane protein PEX16 in mammalian cells by Kim et al. (Kim et al., 2006). When an ER signal anchor sequence was appended to PEX16, forcing it into the ER, it still trafficked to peroxisomes and supported de novo peroxisome formation in human fibroblasts with a PEX16 mutation (Kim et al., 2006). These experiments implicate the ER in de novo peroxisome formation.
The requirement for ER-to-peroxisome transport is not restricted to de novo formation (Fig. 1). Pex16-GFP accumulates in the ER and peroxisomes upon overexpression in COS cells and, although the ER accumulation is a result of overexpression, the authors show that ER-localised Pex16-GFP transfers to peroxisomes over time. Tagged Pex3 (either full-length or truncated) can transfer from ER to existing peroxisomes in S. cerevisiae (Hoepfner et al., 2005; Tam et al., 2005; Motley and Hettema, 2007). In addition, in Arabidopsis and the yeast Pichia pastoris there is evidence to support a role of the ER in PMP sorting (Motley and Hettema, 2007).
The process by which material is transported from ER to peroxisomes is not understood. Most studies have focussed on de novo peroxisome formation and transport has been proposed to be vesicular (see above). De novo peroxisome formation in mammalian cells is not inhibited by a block in either coatomer protein complex I (COPI)- or COPII-mediated transport (Kim et al., 2006; Matsuzono et al., 1999; South et al., 2000; Voorn-Brouwer et al., 2001). Neither have other factors that are important for membrane trafficking events in the endomembrane system (SNAREs, Rab GTPases, coat proteins and others) been implicated in de novo peroxisome biogenesis. Therefore, it seems likely that a specific, yet-to-be-identified machinery is required for transport during de novo peroxisome formation.
It remains to be established whether transport from ER to existing peroxisomes depends on the same mechanism as de novo formation. Although a vesicular mode of transport between ER and peroxisomes would be in line with membrane trafficking in the endomembrane system, these putative vesicular structures have not been identified. We in the field should therefore be open to the possibility that alternative modes of transport are involved. For instance, peroxisomes have been observed in close apposition to the ER (Novikoff and Novikoff, 1972). Contact sites have been observed between the ER and many organelles, and these sites have been suggested to mediate non-vesicular transfer of lipids (Levine, 2004). Recently, transfer of phospholipids has been found to occur between ER and peroxisomes (Raychaudhuri and Prinz, 2008). This transfer is independent of Pex3 or any of the Sec proteins (which are required for membrane trafficking from the ER) and is considered to be non-vesicular. The contribution of this non-vesicular transfer to peroxisomal membrane growth is unknown.
Another possibility is that PMPs are transported via membrane continuities between the ER and peroxisomes. These continuities have been observed in mouse dendritic cells using immunogold electron microscopy. Although the original interpretation of the images was that peroxisomes might form from ER lamellae (Geuze et al., 2003; Tabak et al., 2003; Tabak et al., 2008), an alternative interpretation is that these lamellae fuse with existing peroxisomes, forming a bridge along which PMPs and lipids exchange.
In summary, peroxisomes can multiply by de novo formation and by growth and division. There is evidence that strongly supports a role for the ER in these processes. The mechanism of transport between ER and peroxisomes has not been characterised and needs further investigation.
Targeting of PMPs
The route that PMPs take to reach peroxisomes is another hotly debated topic. Most studies in mammalian cells support a model whereby PMPs are inserted directly into peroxisomes (Fujiki et al., 2006). Radioactive pulse-chase experiments show that newly synthesised PMPs are recovered in the cytosolic fraction before they associate with peroxisomes (Imanaka et al., 1996). Both in vitro and in vivo studies support the view that Pex19 acts as a soluble PMP receptor and chaperone that delivers newly synthesised PMPs from the cytosol to Pex3 on the peroxisomal membrane (Fang et al., 2004; Jones et al., 2004; Matsuzono and Fujiki, 2006; Matsuzono et al., 2006; Sacksteder et al., 2000). Pex16 is the only other factor known to be involved in PMP biogenesis.
How is the import machinery for PMPs inserted? There seems to be an interdependence of factors. Pex16 and Pex19 are required for Pex3 insertion and Pex19 and Pex3 are required for Pex16 insertion (Matsuzaki and Fujiki, 2008). The steady-state localisation of Pex3 and Pex16 is in the peroxisomal membrane. In vitro studies and studies using semi-permeabilised CHO cells suggest that Pex3 and Pex16 are inserted directly into the peroxisomal membrane. However, Kim et al. (Kim et al., 2006) show that a large proportion of peroxisomes form de novo in mammalian fibroblasts, and that PEX16 can traffic from ER to peroxisomes and can stimulate insertion of PMPs, including PEX3, into the ER. Therefore, we consider it very likely that PEX3 traffics from the ER to peroxisomes in mammalian cells during de novo peroxisome formation. The fraction of PEX3 that is inserted into the ER would then depend on the relative activities of the two peroxisome multiplication pathways.
Although S. cerevisiae peroxisomes multiply by fission, there is no evidence for direct import of PMPs into peroxisomes in yeast. It has even been suggested (Tabak et al., 2008) that all PMPs traffic via the ER. It is clear that some proteins, including Pex3 and Pex22 can traffic via the ER to peroxisomes (Kragt et al., 2005; Tam et al., 2005; Hoepfner et al., 2005; Motley and Hettema, 2007; Halbach et al., 2009). However, it should be borne in mind that many of these studies, including our own, were done using mutant cells and overexpression of tagged PMPs. These experiments might overemphasise the role of the ER. The lack of a Pex16 orthologue in S. cerevisiae means that we cannot necessarily extrapolate data from mammalian cells to S. cerevisiae and raises the question of the identity of the Pex3 insertion machinery in this yeast. Careful quantitative experimentation is required to distinguish whether all yeast PMPs travel via the ER, or whether yeast peroxisomes resemble mammalian peroxisomes in that they can import PMPs directly.
Conclusions and perspectives
In this Commentary, we have discussed our views on how peroxisomes multiply, and the pathways taken by PMPs. It is clear that peroxisomes can multiply in two ways, by growth and division and by de novo formation from the ER, depending on the organism and cellular conditions. Both modes of multiplication rely on the ER as the donor compartment for membrane constituents. With the ER established as being central to peroxisome biogenesis, many questions remain. Which routes do PMPs follow? Are Pex3, Pex16 and Pex19 the only components of the PMP targeting and insertion machinery? How does S. cerevisiae insert PMPs without Pex16? What is the nature of the peroxisomal ER? How are membrane constituents transported from the ER to peroxisomes? Currently, we can only hypothesise about the nature of the transport intermediates between ER and peroxisomes.
Another question relates to how peroxisome number is regulated under different conditions. We know the identity of a few of the key players, allowing us to tackle an analysis of this process at the molecular level. What is clear is that there is currently very little mechanistic or molecular insight into this question, but with new assays available and a basic framework to test we expect that progress will be rapid in the coming years.
E.H.H. is supported by a Wellcome Trust Senior Research Fellowship in Basic Biomedical Sciences. Deposited in PMC for release after 6 months.
References
- Brown, L. A. and Baker, A. (2008). Shuttles and cycles: transport of proteins into the peroxisome matrix (review). Mol. Membr. Biol. 25, 363-375. [DOI] [PubMed] [Google Scholar]
- Camoes, F., Bonekamp, N. A., Delille, H. K. and Schrader, M. (2008). Organelle dynamics and dysfunction: a closer link between peroxisomes and mitochondria. J. Inherit. Metab. Dis. 32, 163-180. [DOI] [PubMed] [Google Scholar]
- Fagarasanu, A., Fagarasanu, M., Eitzen, G. A., Aitchison, J. D. and Rachubinski, R. A. (2006). The peroxisomal membrane protein Inp2p is the peroxisome-specific receptor for the myosin V motor Myo2p of Saccharomyces cerevisiae. Dev. Cell 10, 587-600. [DOI] [PubMed] [Google Scholar]
- Fagarasanu, A., Fagarasanu, M. and Rachubinski, R. A. (2007). Maintaining peroxisome populations: a story of division and inheritance. Annu. Rev. Cell Dev. Biol. 23, 321-344. [DOI] [PubMed] [Google Scholar]
- Fagarasanu, M., Fagarasanu, A., Tam, Y. Y., Aitchison, J. D. and Rachubinski, R. A. (2005). Inp1p is a peroxisomal membrane protein required for peroxisome inheritance in Saccharomyces cerevisiae. J. Cell Biol. 169, 765-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, Y., Morrell, J. C., Jones, J. M. and Gould, S. J. (2004). PEX3 functions as a PEX19 docking factor in the import of class I peroxisomal membrane proteins. J. Cell Biol. 164, 863-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki, Y., Matsuzono, Y., Matsuzaki, T. and Fransen, M. (2006). Import of peroxisomal membrane proteins: the interplay of Pex3p- and Pex19p-mediated interactions. Biochim. Biophys. Acta 1763, 1639-1646. [DOI] [PubMed] [Google Scholar]
- Gandre-Babbe, S. and van der Bliek, A. M. (2008). The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol. Biol. Cell 19, 2402-2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuze, H. J., Murk, J. L., Stroobants, A. K., Griffith, J. M., Kleijmeer, M. J., Koster, A. J., Verkleij, A. J., Distel, B. and Tabak, H. F. (2003). Involvement of the endoplasmic reticulum in peroxisome formation. Mol. Biol. Cell 14, 2900-2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes de Mesquita, D. S., van den Hazel, H. B., Bouwman, J. and Woldringh, C. L. (1996). Characterization of new vacuolar segregation mutants, isolated by screening for loss of proteinase B self-activation. Eur. J. Cell Biol. 71, 237-247. [PubMed] [Google Scholar]
- Haan, G. J., Baerends, R. J., Krikken, A. M., Otzen, M., Veenhuis, M. and van der Klei, I. J. (2006). Reassembly of peroxisomes in Hansenula polymorpha pex3 cells on reintroduction of Pex3p involves the nuclear envelope. FEMS Yeast Res. 6, 186-194. [DOI] [PubMed] [Google Scholar]
- Halbach, A., Rucktaschel, R., Rottensteiner, H. and Erdmann, R. (2009). The N-domain of Pex22p can functionally replace the Pex3p N-domain in targeting and peroxisome formation. J. Biol. Chem. 284, 3906-3916. [DOI] [PubMed] [Google Scholar]
- Hoepfner, D., van den Berg, M., Philippsen, P., Tabak, H. F. and Hettema, E. H. (2001). A role for Vps1p, actin, and the Myo2p motor in peroxisome abundance and inheritance in Saccharomyces cerevisiae. J. Cell Biol. 155, 979-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoepfner, D., Schildknegt, D., Braakman, I., Philippsen, P. and Tabak, H. F. (2005). Contribution of the endoplasmic reticulum to peroxisome formation. Cell 122, 85-95. [DOI] [PubMed] [Google Scholar]
- Hohfeld, J., Veenhuis, M. and Kunau, W. H. (1991). PAS3, a Saccharomyces cerevisiae gene encoding a peroxisomal integral membrane protein essential for peroxisome biogenesis. J. Cell Biol. 114, 1167-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanaka, T., Shiina, Y., Takano, T., Hashimoto, T. and Osumi, T. (1996). Insertion of the 70-kDa peroxisomal membrane protein into peroxisomal membranes in vivo and in vitro. J. Biol. Chem. 271, 3706-3713. [DOI] [PubMed] [Google Scholar]
- Jones, J. M., Morrell, J. C. and Gould, S. J. (2004). PEX19 is a predominantly cytosolic chaperone and import receptor for class 1 peroxisomal membrane proteins. J. Cell Biol. 164, 57-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel, J. A., Veenhuis, M. and van der Klei, I. J. (2006). PEX genes in fungal genomes: common, rare or redundant. Traffic 7, 1291-1303. [DOI] [PubMed] [Google Scholar]
- Kim, P. K., Mullen, R. T., Schumann, U. and Lippincott-Schwartz, J. (2006). The origin and maintenance of mammalian peroxisomes involves a de novo PEX16-dependent pathway from the ER. J. Cell Biol. 173, 521-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch, A., Schneider, G., Luers, G. H. and Schrader, M. (2004). Peroxisome elongation and constriction but not fission can occur independently of dynamin-like protein 1. J. Cell Sci. 117, 3995-4006. [DOI] [PubMed] [Google Scholar]
- Kragt, A., Voorn-Brouwer, T., van den Berg, M. and Distel, B. (2005). Endoplasmic reticulum-directed Pex3p routes to peroxisomes and restores peroxisome formation in a Saccharomyces cerevisiae pex3Delta strain. J. Biol. Chem. 280, 34350-34357. [DOI] [PubMed] [Google Scholar]
- Kunau, W. H. (2005). Peroxisome biogenesis: end of the debate. Curr. Biol. 15, R774-R776. [DOI] [PubMed] [Google Scholar]
- Kuravi, K., Nagotu, S., Krikken, A. M., Sjollema, K., Deckers, M., Erdmann, R., Veenhuis, M. and van der Klei, I. J. (2006). Dynamin-related proteins Vps1p and Dnm1p control peroxisome abundance in Saccharomyces cerevisiae. J. Cell Sci. 119, 3994-4001. [DOI] [PubMed] [Google Scholar]
- Lazarow, P. B. (2003). Peroxisome biogenesis: advances and conundrums. Curr. Opin. Cell Biol. 15, 489-497. [DOI] [PubMed] [Google Scholar]
- Levine, T. (2004). Short-range intracellular trafficking of small molecules across endoplasmic reticulum junctions. Trends Cell Biol. 14, 483-490. [DOI] [PubMed] [Google Scholar]
- Li, X. and Gould, S. J. (2003). The dynamin-like GTPase DLP1 is essential for peroxisome division and is recruited to peroxisomes in part by PEX11. J. Biol. Chem. 278, 17012-17020. [DOI] [PubMed] [Google Scholar]
- Lingard, M. J., Gidda, S. K., Bingham, S., Rothstein, S. J., Mullen, R. T. and Trelease, R. N. (2008). Arabidopsis PEROXIN11c-e, FISSION1b, and DYNAMIN-RELATED PROTEIN3A cooperate in cell cycle-associated replication of peroxisomes. Plant Cell 20, 1567-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe, M. and Barr, F. A. (2007). Inheritance and biogenesis of organelles in the secretory pathway. Nat. Rev. Mol. Cell Biol. 8, 429-439. [DOI] [PubMed] [Google Scholar]
- Matsuzaki, T. and Fujiki, Y. (2008). The peroxisomal membrane protein import receptor Pex3p is directly transported to peroxisomes by a novel Pex19p- and Pex16p-dependent pathway. J. Cell Biol. 183, 1275-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzono, Y. and Fujiki, Y. (2006). In vitro transport of membrane proteins to peroxisomes by shuttling receptor Pex19p. J. Biol. Chem. 281, 36-42. [DOI] [PubMed] [Google Scholar]
- Matsuzono, Y., Kinoshita, N., Tamura, S., Shimozawa, N., Hamasaki, M., Ghaedi, K., Wanders, R. J., Suzuki, Y., Kondo, N. and Fujiki, Y. (1999). Human PEX19: cDNA cloning by functional complementation, mutation analysis in a patient with Zellweger syndrome, and potential role in peroxisomal membrane assembly. Proc. Natl. Acad. Sci. USA 96, 2116-2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzono, Y., Matsuzaki, T. and Fujiki, Y. (2006). Functional domain mapping of peroxin Pex19p: interaction with Pex3p is essential for function and translocation. J. Cell Sci. 119, 3539-3550. [DOI] [PubMed] [Google Scholar]
- Motley, A. M. and Hettema, E. H. (2007). Yeast peroxisomes multiply by growth and division. J. Cell Biol. 178, 399-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley, A. M., Ward, G. P. and Hettema, E. H. (2008). Dnm1p-dependent peroxisome fission requires Caf4p, Mdv1p and Fis1p. J. Cell Sci. 121, 1633-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen, R. T. and Trelease, R. N. (2006). The ER-peroxisome connection in plants: development of the “ER semi-autonomous peroxisome maturation and replication” model for plant peroxisome biogenesis. Biochim. Biophys. Acta 1763, 1655-1668. [DOI] [PubMed] [Google Scholar]
- Nagotu, S., Krikken, A. M., Otzen, M., Kiel, J. A., Veenhuis, M. and van der Klei, I. J. (2008a). Peroxisome fission in Hansenula polymorpha requires Mdv1 and Fis1, two proteins also involved in mitochondrial fission. Traffic 9, 1471-1484. [DOI] [PubMed] [Google Scholar]
- Nagotu, S., Saraya, R., Otzen, M., Veenhuis, M. and van der Klei, I. J. (2008b). Peroxisome proliferation in Hansenula polymorpha requires Dnm1p which mediates fission but not de novo formation. Biochim Biophys Acta 1783, 760-769. [DOI] [PubMed] [Google Scholar]
- Novikoff, P. M. and Novikoff, A. B. (1972). Peroxisomes in absorptive cells of mammalian small intestine. J. Cell Biol. 53, 532-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platta, H. W. and Erdmann, R. (2007a). Peroxisomal dynamics. Trends Cell Biol. 17, 474-484. [DOI] [PubMed] [Google Scholar]
- Platta, H. W. and Erdmann, R. (2007b). The peroxisomal protein import machinery. FEBS Lett. 581, 2811-2819. [DOI] [PubMed] [Google Scholar]
- Praefcke, G. J. and McMahon, H. T. (2004). The dynamin superfamily: universal membrane tubulation and fission molecules? Nat. Rev. Mol. Cell Biol. 5, 133-147. [DOI] [PubMed] [Google Scholar]
- Rayapuram, N. and Subramani, S. (2006). The importomer-a peroxisomal membrane complex involved in protein translocation into the peroxisome matrix. Biochim. Biophys. Acta 1763, 1613-1619. [DOI] [PubMed] [Google Scholar]
- Raychaudhuri, S. and Prinz, W. A. (2008). Nonvesicular phospholipid transfer between peroxisomes and the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 105, 15785-15790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottensteiner, H., Stein, K., Sonnenhol, E. and Erdmann, R. (2003). Conserved function of pex11p and the novel pex25p and pex27p in peroxisome biogenesis. Mol. Biol. Cell 14, 4316-4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacksteder, K. A., Jones, J. M., South, S. T., Li, X., Liu, Y. and Gould, S. J. (2000). PEX19 binds multiple peroxisomal membrane proteins, is predominantly cytoplasmic, and is required for peroxisome membrane synthesis. J. Cell Biol. 148, 931-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader, M. and Fahimi, H. D. (2006). Growth and division of peroxisomes. Int. Rev. Cytol. 255, 237-290. [DOI] [PubMed] [Google Scholar]
- South, S. T., Sacksteder, K. A., Li, X., Liu, Y. and Gould, S. J. (2000). Inhibitors of COPI and COPII do not block PEX3-mediated peroxisome synthesis. J. Cell Biol. 149, 1345-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabak, H. F., Murk, J. L., Braakman, I. and Geuze, H. J. (2003). Peroxisomes start their life in the endoplasmic reticulum. Traffic 4, 512-518. [DOI] [PubMed] [Google Scholar]
- Tabak, H. F., Hoepfner, D., Zand, A., Geuze, H. J., Braakman, I. and Huynen, M. A. (2006). Formation of peroxisomes: present and past. Biochim. Biophys. Acta 1763, 1647-1654. [DOI] [PubMed] [Google Scholar]
- Tabak, H. F., van der Zand, A. and Braakman, I. (2008). Peroxisomes: minted by the ER. Curr. Opin. Cell Biol. 20, 393-400. [DOI] [PubMed] [Google Scholar]
- Tam, Y. Y., Torres-Guzman, J. C., Vizeacoumar, F. J., Smith, J. J., Marelli, M., Aitchison, J. D. and Rachubinski, R. A. (2003). Pex11-related proteins in peroxisome dynamics: a role for the novel peroxin Pex27p in controlling peroxisome size and number in Saccharomyces cerevisiae. Mol. Biol. Cell 14, 4089-4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam, Y. Y., Fagarasanu, A., Fagarasanu, M. and Rachubinski, R. A. (2005). Pex3p initiates the formation of a preperoxisomal compartment from a subdomain of the endoplasmic reticulum in Saccharomyces cerevisiae. J. Biol. Chem. 280, 34933-34939. [DOI] [PubMed] [Google Scholar]
- Thoms, S. and Erdmann, R. (2005). Dynamin-related proteins and Pex11 proteins in peroxisome division and proliferation. FEBS J. 272, 5169-5181. [DOI] [PubMed] [Google Scholar]
- Titorenko, V. I. and Mullen, R. T. (2006). Peroxisome biogenesis: the peroxisomal endomembrane system and the role of the ER. J. Cell Biol. 174, 11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titorenko, V. I., Eitzen, G. A. and Rachubinski, R. A. (1996). Mutations in the PAY5 gene of the yeast Yarrowia lipolytica cause the accumulation of multiple subpopulations of peroxisomes. J. Biol. Chem. 271, 20307-20314. [DOI] [PubMed] [Google Scholar]
- van der Zand, A., Braakman, I., Geuze, H. J. and Tabak, H. F. (2006). The return of the peroxisome. J. Cell Sci. 119, 989-994. [DOI] [PubMed] [Google Scholar]
- Vizeacoumar, F. J., Vreden, W. N., Fagarasanu, M., Eitzen, G. A., Aitchison, J. D. and Rachubinski, R. A. (2006). The dynamin-like protein Vps1p of the yeast Saccharomyces cerevisiae associates with peroxisomes in a Pex19p-dependent manner. J. Biol. Chem. 281, 12817-12823. [DOI] [PubMed] [Google Scholar]
- Voorn-Brouwer, T., Kragt, A., Tabak, H. F. and Distel, B. (2001). Peroxisomal membrane proteins are properly targeted to peroxisomes in the absence of COPI- and COPII-mediated vesicular transport. J. Cell Sci. 114, 2199-2204. [DOI] [PubMed] [Google Scholar]
- Wanders, R. J. and Waterham, H. R. (2005). Peroxisomal disorders I: biochemistry and genetics of peroxisome biogenesis disorders. Clin. Genet. 67, 107-133. [DOI] [PubMed] [Google Scholar]
- Wanders, R. J. and Waterham, H. R. (2006). Biochemistry of mammalian peroxisomes revisited. Annu. Rev. Biochem. 75, 295-332. [DOI] [PubMed] [Google Scholar]
- Wang, Y. X., Zhao, H., Harding, T. M., Gomes de Mesquita, D. S., Woldringh, C. L., Klionsky, D. J., Munn, A. L. and Weisman, L. S. (1996). Multiple classes of yeast mutants are defective in vacuole partitioning yet target vacuole proteins correctly. Mol. Biol. Cell 7, 1375-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller, S., Gould, S. J. and Valle, D. (2003). Peroxisome biogenesis disorders. Annu. Rev. Genomics Hum. Genet. 4, 165-211. [DOI] [PubMed] [Google Scholar]
- Westermann, B. (2008). Molecular machinery of mitochondrial fusion and fission. J. Biol. Chem. 283, 13501-13505. [DOI] [PubMed] [Google Scholar]