Summary
The Hippo pathway, identified in Drosophila and conserved in vertebrates, regulates tissue growth by promoting cell cycle exit and apoptosis. In addition to their well-characterised overproliferation phenotype, adult Drosophila epithelial cells mutant for the kinases Hippo and Warts have hypertrophic apical domains. Here we examine the molecular basis of this apical hypertrophy and its impact on cell proliferation. In the wing imaginal disc epithelium, we observe increased staining for members of the apical polarity complexes aPKC and Crumbs as well as adherens junction components when Hippo activity is compromised, while basolateral markers are not affected. This increase in apical proteins is correlated with a hypertrophy of the apical domain and adherens junctions. The cell surface localisation of the Notch receptor is also increased in mutant clones, opening the possibility that aberrant receptor signalling may participate in overgrowth of hpo-deficient tissue. Interestingly, however, although the polarity determinant Crumbs is required for the accumulation of apical proteins, this does not appear to significantly contribute to the overproliferation defect elicited by loss of Hippo signalling. Therefore, Hippo signalling controls growth and apical domain size by distinct mechanisms.
Keywords: Hippo signalling, Polarity, Proliferation
Introduction
In an epithelium, the optimisation of contact between neighbouring cells arises through the polarised architecture of each cell, which have several membrane domains separated by distinct types of cellular junctions (Tepass et al., 2001). In Drosophila epithelia, the membrane of a cell is divided in apical and basolateral domains. Those domains are separated from each other by the zonula adherens (ZA), an actin-rich region that forms a belt around the cell and where adherens junctions (AJs) are located (see supplementary material Fig. S1G). The main components of the ZA are the transmembrane adhesion protein Drosophila E-cadherin [DE-cad; also known as Shotgun (Shg)] and its scaffold protein Armadillo (Arm), the homolog of β-catenin.
The apical domain of the cell is subdivided into a free apical domain and the subapical region (SAR). The free apical domain is on the cell surface that is facing the external milieu. It is composed of a brush of microvilli whose formation depends on the apical localisation of the cadherin Cad99C (D'Alterio et al., 2005; Schlichting et al., 2006). Just below this free apical domain, the SAR is characterised by the presence of two apical polarity complexes (Tepass et al., 2001). The Bazooka (Baz) complex contains proteins such as aPKC (atypical protein kinase C), Par6 (Partitioning defect 6), Baz (the homolog of Par3), and the small GTPase Cdc42. The Crumbs (Crb) complex is composed of the transmembrane protein Crb, Stardust (Sdt) and Drosophila Patj. Another type of cell-cell junction, the septate junction (SJ), is located basally to the ZA. The Scribble (Scrib) polarity complex, which consists of Scrib and Disc large (Dlg), is localised to the SJs, whereas the Lethal (2) giant larvae (Lgl) protein is present on all the lateral membranes (Tepass et al., 2001). These polarity determinants are highly conserved in mammals (Knust and Bossinger, 2002; Nelson, 2003).
How the polarity complexes localise at the appropriate plasma membrane compartment or how they regulate polarity remains unclear, but a hierarchy between them has started to emerge (Bilder et al., 2003; Tanentzapf and Tepass, 2003). The Baz complex appears to be the primary apical determinant. The Scrib complex prevents the assembly of the Baz complex in the basolateral part of the cell. In turn, the Baz complex antagonises the function of the Scrib complex by recruiting the Crb complex. These reciprocal inhibitions ensure a strict separation of the apical and basolateral domain. The Lgl protein has a unique role in the establishment and maintenance of this apicobasal polarity. On the one hand, localisation of Lgl to the basolateral membrane requires the Scrib complex (Bilder et al., 2000). On the other hand, Lgl is excluded from the apical membrane through its phosphorylation by aPKC (Betschinger et al., 2003; Plant et al., 2003). Finally, Lgl antagonises Crb function (Bilder et al., 2003; Tanentzapf and Tepass, 2003). Thus, the Lgl protein seems to act as an integrator of both signals from the apical and basolateral domains. Lgl, scrb and dlg are also known to function as tumour suppressor genes (Bilder et al., 2000), supporting the notion of a link between loss of polarity and overproliferation.
The Hippo pathway regulates tissue growth and proliferation by promoting cell cycle exit and apoptosis. The core of this pathway is composed of two kinases, Hippo (Hpo) and Warts (Wts), and of the scaffold proteins Salvador (Sav) and Mob as Tumour Suppressor (Mats) (Harvey and Tapon, 2007). Activation of the pathway inhibits the activity of the Yorkie (Yki) transcriptional co-activator, triggering the downregulation of its target genes (Cyclin E, the Drosophila Inhibitor of Apoptosis 1 and the bantam miRNA) (Dong et al., 2007; Huang et al., 2005). The atypical cadherin Fat (Ft) as well as the FERM proteins Merlin (Mer) and Expanded (Ex) have been linked with the activation of this pathway, although the mechanistic details of this process remain unclear (Bennett and Harvey, 2006; Cho et al., 2006; Hamaratoglu et al., 2006; Silva et al., 2006; Willecke et al., 2006). Recently, the transcription factor Scalloped has been shown to bind to Yki and mediate DIAP1 regulation (Goulev et al., 2008; Wu et al., 2008; Zhang et al., 2008; Zhao et al., 2008a). Like wts, hpo and sav, mutations in ft, in Mer and ex, as well as yki gain-of-function, induce hyperplastic growth. The mutant cells have a proliferative advantage over wild-type cells but are able to differentiate. As a result, imaginal discs with mutant clones show outgrowths with extra folding of tissue but maintain their epithelial nature. In addition, adult epithelial cells lacking the activity of the Hpo pathway have hypertrophic apical domains (Justice et al., 1995; Wu et al., 2003).
The consequences of this apical hypertrophy have not been characterised, leading us to investigate in more detail the cell polarity defect elicited by Hpo pathway inactivation and its impact on cell proliferation. In the wing imaginal disc epithelium, we observed an increase of the apical polarity proteins aPKC, Crb and DE-cad which belong respectively to the Baz and Crb polarity complexes, and to the AJs, as well as an increase of the cell surface receptor Notch (N), when Hpo pathway activity is compromised. Basolateral markers (Dlg and Dystroglycan) are not affected. The phenotype is characteristic of the Hpo pathway since apical hypertrophy is not observed when other pathways controlling cell growth or inducing cell sorting are compromised. Furthermore, this increase in apical polarity protein localisation is correlated with a hypertrophy of the apical domain and AJs. Interestingly, although accumulation of apical proteins is dependent on the apical determinant Crb, it is not required for overgrowth in response to loss of wts. Thus, the apical hypertrophy and overproliferative phenotypes of hpo-deficient cells are two distinct processes, suggesting that there is no straightforward link between an increase of surface receptors and an increase in signalling output.
Results
Generation of a hpo null allele
Different groups have generated a variety of hpo mutant alleles using chemical mutagenesis (Harvey et al., 2003; Jia et al., 2003; Udan et al., 2003; Wu et al., 2003). We had noted in previous studies that the various alleles differed considerably in strength (Polesello and Tapon, 2007). We therefore decided to clarify the relative properties of the strongest known hpo alleles. First, we generated a null allele of hpo, hpo5.1, by imprecise excision of the P-element G3315 (inserted in the 5′UTR of the hpo gene; supplementary material Fig. S1A). This excision allele, which removes most of the hpo coding sequence, represents a bona fide protein null for hpo. To test the strength of hpo5.1, we compared it with three other hpo alleles. The presumed null allele hpoBF33 is a point mutation introducing a premature stop codon in position 174, resulting in the production of a truncated protein (Jia et al., 2003). hpo42-47 is an in-frame deletion of six amino acids (166-171) in the Hpo kinase domain, N166 being required for ATP binding (Wu et al., 2003). hpoJM1 is a single amino acid substitution of a critical amino acid (G181E) in the kinase domain (Jia et al., 2003). Thus, the alleles hpo42-47 and hpoJM1 produce kinase-dead versions of Hpo.
Using an antibody directed against the C-terminal part of the Hpo kinase domain (Polesello et al., 2006), we examined Hpo staining in clones of cells mutant for each of the four hpo alleles described above. Mutant cells carrying the hpo5.1 mutation, or the hpoBF33 mutation, show no Hpo staining above background (supplementary material Fig. S1B,B′,D,D′). The antibody allows us to distinguish cells carrying one or two copies of wild-type (WT) hpo (one or two copies of GFP, respectively; supplementary material Fig. S1B,B′). By contrast, Hpo staining is not reduced in hpoJM1 or hpo42-47 clones (supplementary material Fig. S1C,C′,E,E′). The staining even appears more intense in some clones. This could be due either to increased Hpo stability in those mutants, or to altered localisation of the kinase-dead proteins. We also generated clones of hpo mutant cells in the eye (eyFLP driver), and assessed the viability of flies carrying those clones as a measure of the relative strength of the alleles. The expected fraction of flies containing mutant clones should be 50% of the total population. hpo42-47 or hpoJM1 clone-containing flies represented only 2.1% (4/190) or 13.1% (28/214) of the population, respectively, whereas hpo5.1 or hpoBF33 clone-containing flies were 34.3% (34/99) or 36.4% (52/143) of the population, respectively (supplementary material Fig. S1F). This result indicates that kinase-dead alleles (hpoJM1 and hpo42-47) have a stronger effect than null alleles (hpoBF33 and hpo5.1). This suggests that the kinase-dead alleles may have a dominant-negative effect, possibly interfering with activation of the downstream kinase Wts. Nevertheless, hpoJM1 and hpo5.1 behaved similarly in all subsequent experiments.
Disruption of the Hpo pathway elicits accumulation of apical and AJ proteins in imaginal disc cells
We first tested whether Hpo is required for normal distribution of the polarity complexes by generating FLP/FRT mitotic clones of mutant cells in wing imaginal discs. In hpo mutant cells, Cad99C, which localises to the apical microvilli (D'Alterio et al., 2005; Schlichting et al., 2006), shows a brighter and broader staining (Fig. 1A,A′). This indicates that the microvillus domain of the hpo mutant cells is larger than that of wild-type (WT) cells, since Cad99C overexpression is sufficient to induce growth of the apical microvilli (D'Alterio et al., 2005; Schlichting et al., 2006). As for Cad99C, stainings for aPKC and Crb revealed that, when hpo is mutated, there is an increased staining for members of the two apical polarity complexes (Fig. 1B-C′). The membrane domains where those proteins are localised also appear to be broader (see supplementary material Fig. S2A,A′,C,C′,G,G′ for XY sections and higher magnification). Phospho-Moesin (P-Moe), which co-immunoprecipitates with Crb in embryos (Medina et al., 2002), showed a similar pattern (Fig. 1D,D′; see supplementary material Fig. S2D,D′ for XY section), as well as the apically localised Notch receptor (supplementary material Fig. S3B,B′).
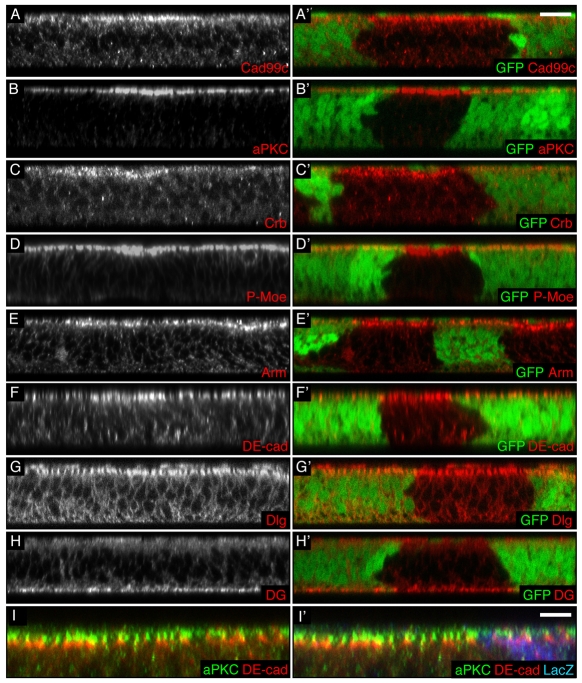
Fig. 1.
hpo loss of function induces an increase in apical polarity proteins in wing imaginal cells. (A-H′) Transverse sections of third instar wing discs containing clones of hpo mutant cells (marked by absence of GFP). Apical is to the top in this and all subsequent XZ sections. Scale bar: 10 μm. (A′-H′) Merged images of A-H with GFP images (green). (I,I′) Transverse section of a third instar wing disc containing clones of hpo5.1 mutant cells (marked by absence of β-gal) stained for aPKC (green) and DE-cad (red). (I′) Merged image of I with β-gal (blue). Scale bar: 5 μm. hpo mutant cells have an increased apical staining of the following apical determinants: Cad99c (A,A′), aPKC (B,B′), Crb (C,C′), P-Moe (D,D′), Arm (E,E′) and DE-cad (F,F′). By contrast, they do not show an increase of the septate junction protein Dlg (G,G′) or the basal marker Dystroglycan (DG; H,H′). Despite their respective increase, the proteins aPKC and DE-cad remain in non-overlapping membrane domains (I,I′).
The AJs in hpo mutant cells are also characterised by increased protein localisation at the membrane, visualised by staining for Arm and DE-cad (Fig. 1E-F′; supplementary material Fig. S2E-F′ for XY sections). By contrast, the basolateral complexes are not affected by loss of Hpo signalling, as visualised by Dlg (lateral marker) and Dystroglycan (basal marker) stainings (Fig. 1G-H′; see supplementary material Fig. S2B,B′ for XY section). Although appearing broader, the aPKC and DE-cad domains remain mostly non-overlapping in hpo mutant cells, as in WT cells (Fig. 1I,I′). This suggests that the broader and more intense apical stainings observed in cells lacking hpo do not result from a mixing of the different apical polarity subdomains but might be caused by an enlargement of each subdomain of the apical region.
The accumulation of DE-cad and some apical signalling receptors had been noted for ft and mer;ex clones, respectively (Feng and Irvine, 2007; Jaiswal et al., 2006; Maitra et al., 2006). However, this was not believed to be a general property of Hpo pathway components. We therefore proceeded to test whether every level of the Hpo pathway showed the same disruption in the distribution of the polarity complexes. Clones of cells mutant for mer;ex or ft (Fig. 2A-A″; data not shown), which have been suggested to be upstream elements of the network, as well as clones of cells mutant for wts (Fig. 2B-B″), the Hpo downstream kinase, were generated. As in hpo mutant clones, cells mutant for those genes show a brighter and broader staining for apical complex members, as well as a broadening of the Notch (N) receptor staining (supplementary material Fig. S3A-C′), but a normal distribution of basolateral markers. Overexpression clones for Yki, the downstream effector of the pathway whose gain of function mimics Hpo pathway loss of function, yielded the same phenotype (Fig. 2C,C′). Furthermore, activating the Hpo pathway induces the reverse phenotype as cells overexpressing ex or hpo show a reduction in apical localisation of aPKC (Fig. 2D,D′,F,F′), DE-cad (Fig. 2E,E′) and N (supplementary material Fig. S3F,F′).
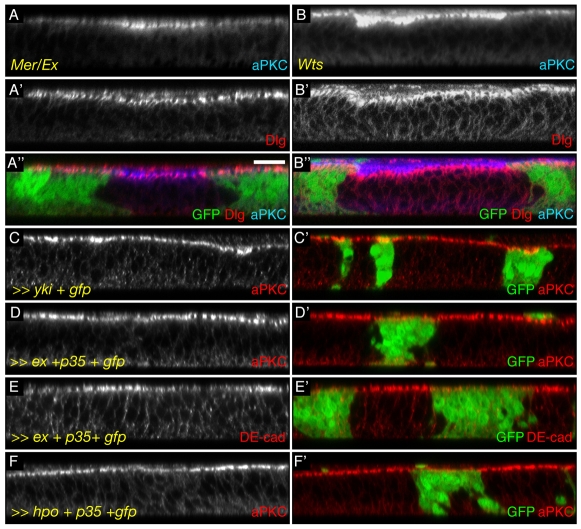
Fig. 2.
Inactivation of Hpo pathway members induces apical protein accumulation. (A-F′) Transverse sections of wing discs. Scale bar: 10 μm. (A″,B″) Merged images of A,B, A′,B′, respectively and of GFP. C′-F′ Merged images of C-F with GFP (green). Grey or red: stainings for aPKC (C-D′,F,F′) or for DE-cad (E,E′). (A-A″) Cells mutant for mer;ex show an increase in the apical markers aPKC (grey in A, blue in A″) but not of the lateral marker Dlg (grey in A′, red in A″). (B-B″) Similarly, wts mutant cells show an increase in aPKC (grey in B, blue in B″) but not in Dlg (grey in B′, red in B″). (C,C′) Mimicking Hpo pathway inactivation by overexpressing yki also leads to an increase in apical aPKC (cells marked by GFP). (D-F′) Activating the Hpo pathway, by overexpressing the caspase inhibitor p35 and either ex (D-E′) or hpo (F,F′), leads to a reduction in apically localised aPKC (D,D′,F,F′) or DE-cad (E,E′) (cells marked by GFP).
Hpo pathway mutant cells have apical membrane and AJ hypertrophy
The observed increase of apically localised polarity proteins in Hpo pathway-deficient cells could be the cause of the reported apical bulging observed by transmission electron microscopy (Justice et al., 1995) and scanning electron microscopy (Wu et al., 2003) in adult epidermal cells mutant for wts and hpo. We decided to investigate this further in wing imaginal discs by correlative transmission electron microscopy (TEM). To examine whether the apical domain of cells lacking a functioning Hpo pathway was expanded, we dissected third instar imaginal discs overexpressing yki and gfp under the control of the engrailed (en) promoter. The overgrown yki-overexpressing compartment can be identified using GFP (Fig. 3A). The discs retain their gross morphology after embedding in resin (Fig. 3B; compare with 3A). Thus, by correlating the transmitted light and GFP image with the TEM micrographs, the yki-overexpressing cells can be identified in transverse sections of the resin-embedded sample (Fig. 3C, transverse section cut across the region indicated by the dotted red line drawn on Fig. 3A,B) and it is possible to compare characteristics of control and yki-overexpressing cells in the same TEM section. The thickness of the wing pouch region appeared slightly reduced in the yki-expressing area, which might indicate a mild loss of stratification as a result of the apical polarity defect.
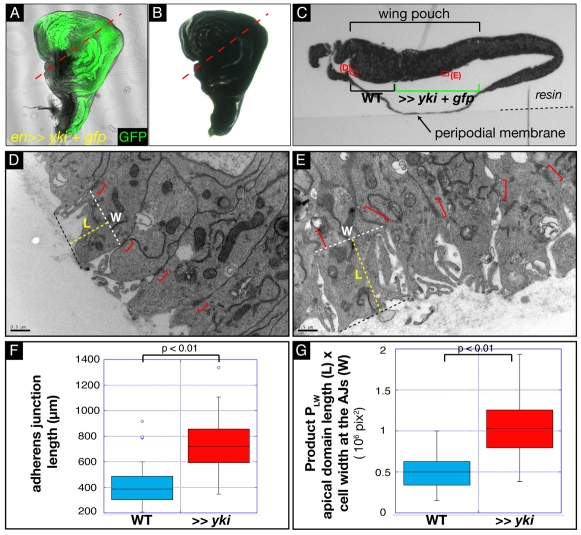
Fig. 3.
Correlative transmission electron microscopy of yki-overexpressing wing imaginal cells reveals apical membrane hypertrophy. (A) Transmitted light and GFP merged image of a third instar wing disc overexpressing Yki under the engrailed promoter. The disc was attached to an etched grid coverslip to stabilise its shape. (B) Same disc as in A now resin-embedded and prepared for TEM. The disc retains its gross morphology, allowing us, by comparing A and B, to distinguish the yki-overexpressing cells (green in A) and the WT cells. The dotted red line indicates the transverse section shown in C. (C) Semi-thin transverse section of the en>>yki disc, viewed in transmitted light. The different features of the discs, such as the peripodial membrane and the pouch, are distinguishable. By comparing A,B and C, it is possible to locate the position of WT and yki-overexpressing cells. The red rectangles indicate the position of the higher magnification images shown in D and E. (D,E) High magnification micrographs of control cells (D) and of yki-overexpressing cells (E). Scale bar: 0.5 μm. The AJs are indicated by red brackets. The white dashed lines indicate the width at the AJs (W). The yellow dashed lines give a measure of apical domain length (L). The product PLW of L×W gives an indication of relative apical domain size. (F) Box and whisker plot of AJ measurements. The P-value from a Mann-Whitney test is indicated. (G) Box and whisker plot of relative apical domain size, assessed by calculating the PLW product. The P-value from a Mann-Whitney test is indicated.
AJs are clearly identifiable in high magnification images of the apical region in control WT cells and yki-overexpressing cells (Fig. 3D,E), and are similar in morphology to embryonic AJs (Bachmann et al., 2008). To monitor apical hypertrophy, we measured AJs (see supplementary material Table S1). The mean size of AJs in WT cells is 411.70 nm, compared with 735.03 nm for those in yki-overexpressing cells (Fig. 3F). Thus, the AJs of yki-overexpressing cells are 1.78 times wider than those of WT cells.
The total surface of the apical domain is much more difficult to accurately determine than AJ length because of the presence of the microvilli. However, because of the cylindrical nature of imaginal disc cells, we can consider that the apical surface is proportional to the product of the width by the length of the apical domain. We therefore measured cell width (W) at the AJs and the distance (L) between the AJ and the top-most apical membrane level (Fig. 3D,E). We then calculated the product PLW of those two values, which gives us a relative measure of subapical region size. yki-overexpressing cells have a significantly larger apical domain [PLW(yki)=10.6×106] compared with control cells [PLW(control)=5.1×106] (Fig. 3G; for details, see supplementary material Table S1). Together with our immunofluorescence data, this suggests that deregulation of the Hpo pathway elicits an accumulation of apical and AJ proteins correlated with an increase in apical domain size and AJ width.
The accumulation of apical determinants resulting from loss of Hpo signalling may involve new Yki target genes
Yki is a transcriptional co-activator. It is therefore probable that the apical domain hypertrophy we observe in Hpo pathway loss-of-function cells is elicited by misregulation of one or more Yki target genes. Combined overexpression of the known Yki targets DIAP1, cycE and bantam (ban) increases the proliferation rate (Nolo et al., 2006) but overexpressing those three targets under the control of the patched promoter does not lead to an increase in aPKC staining at the apical domain of the cells (supplementary material Fig. S4B,B′). However, overexpressing Yki with the same promoter does induce the accumulation of apical proteins (supplementary material Fig. S4A,A′). This suggests that there could be another transcriptional target of Yki responsible for the apical hypertrophy.
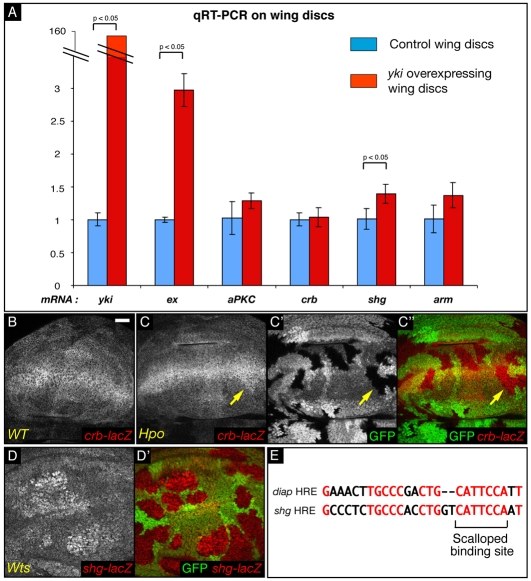
Consequently, we determined whether any of the main polarity genes are targets of the Hpo pathway by conducting quantitative RT-PCR experiments on yki-overexpressing imaginal wing discs and on control discs (Fig. 4A). The yki-expressing discs had a robust increase in yki mRNA level compared with control discs. mRNA levels of ex, a known transcriptional target of the Hpo pathway (Hamaratoglu et al., 2006), were approximately threefold higher, as expected. We proceeded to measure mRNA levels for aPKC, crb, shg and arm. Whereas the crb mRNA level did not change in yki-overexpressing discs (1.04±0.14 fold), shg mRNA increased substantially [1.39±0.14 fold; P(yki-control)=0.013]. aPKC and arm mRNA levels were slightly, but not significantly, elevated (respectively 1.29±0.12 fold and 1.37±0.19 fold). Since the increase in ex mRNA, a well-documented target of the Hpo pathway, is relatively modest, it is possible that the effect on aPKC and arm mRNA levels might be biologically relevant but too subtle to be detected as significant in this experiment.
Fig. 4.
Transcriptional regulation of polarity genes in loss-of-function clones for the Hpo pathway. (A) Graph of mRNA levels of different polarity genes measured by qRT-PCR in yki-overexpressing and WT discs. (B-D′) XY sections of third instar wing discs. Scale bar: 20 μm. C″ is a merged image of C (red) and C′ (green). D′ is a merged image of D with that of GFP (green). Grey or red: anti-β-galactosidase staining. Compared with WT cells (A), hpo mutant cells (B,B′), do not show an overall change in β-gal staining (monitoring crb-lacZ activity). A weak increase could occasionally be observed close to the dorsoventral boundary (arrow). By contrast, wts cells (C,C′) show a marked increase in shg-lacZ staining. (E) Alignment between the Hippo response element (HRE) identified in the DIAP1 locus (Wu et al., 2008) and a putative HRE found in the first intron of the shg gene (positions 369-395). The Scalloped binding site is indicated. Conserved nucleotides are shown in red.
crb and shg transcription can also be visualised using a crb-lacZ and a shg-lacZ enhancer trap (Herranz et al., 2006; Tepass et al., 1996). crb is mainly expressed in a broad stripe at the dorsoventral boundary and in a fainter band at the anteroposterior boundary of the disc (Fig. 4B) (Herranz et al., 2006). In discs containing hpo clones (Fig. 4C-C″), a slight increase in staining could occasionally be observed (arrow) but the overall pattern of crb-lacZ was not significantly changed. By contrast, the shg enhancer trap, which has a more uniform expression in the disc, showed a marked increase in wts mutant cells (Fig. 4D,D′). Thus, for the two genes tested, the enhancer traps recapitulate the qRT-PCR results. ft clones were also shown to have elevated shg-lacZ (Jaiswal et al., 2006). Furthermore, in the first intron of the shg gene (position 369 to 395) we found a sequence with a high homology to the Hpo response element (HRE) (Wu et al., 2008) (Fig. 4E). The Scalloped-binding site in this putative shg HRE is conserved in other Drosophila species (data not shown). This suggests that shg may be directly regulated by the activity of Yki and Scalloped, prompting us to investigate, in more detail, the role of DE-cad in apical hypertrophy.
Our TEM analysis suggested that the AJs are wider than normal but are properly formed in yki-overexpressing cells (Fig. 3F). Furthermore, transverse sections of wing imaginal discs stained for DE-cad and Arm show a proper junctional co-localisation of both proteins (supplementary material Fig. S5A-A″). Finally, the level of DE-cad in Rab5/7/11-positive vesicles appears normal in hpo clones (supplementary material Fig. S5B,B′; and data not shown). These data suggest that in Hpo-pathway-deficient cells excessive DE-cad is correctly targeted to the membrane. To study whether the increase in DE-cad in hpo mutant cells is sufficient to induce apical hypertrophy, we overexpressed shg under the control of the patched (ptc) promoter. This elicits a duplication of the notum (data not shown) through titration of Arm by excess DE-cad, resulting in a wingless loss-of-function phenotype (Sanson et al., 1996). To circumvent this problem, we expressed both shg and arm under the control of the patched promoter. Cells overexpressing those apical polarity determinants do not show an increase in aPKC staining (supplementary material Fig. S5C,C′). Occasionally, shg- and arm-overexpressing cells delaminate and lose polarity, as seen by membrane staining of aPKC around the whole cell surface (supplementary material Fig. S5D,D′). These results show that shg and arm overexpression does not phenocopy Hpo pathway loss of function and suggest that increases of DE-cad and Arm in Hpo-pathway-deficient cells is not sufficient to induce apical hypertrophy. Thus, whereas shg might be a direct target of the Hpo pathway, the apical expansion phenotype may involve additional Yki targets.
The increase in apically localised proteins is not a general consequence of cell sorting defects or overgrowth
Several growth-promoting manipulations in Drosophila epithelial cells have been shown to induce `cell-sorting' defects. In these situations, the mutant cells appear to adhere preferentially to neighbours of the same genotype and tend to minimise contacts with WT neighbours, leading to formation of round clones. This property, which is shared by Hpo pathway mutants (Harvey and Tapon, 2007), cells expressing oncogenic Ras (Prober and Edgar, 2000) and an activated form of the Dpp receptor Thickveins (Tkv) (Nellen et al., 1996), remains little understood but is believed to result from altered cell-cell adhesion properties. We therefore wished to determine whether the apical protein accumulation in Hpo pathway mutants is a consequence of this cell-sorting defect. We generated rasv12-overexpressing cells in the wing disc (supplementary material Fig. S4C,C′). aPKC staining in those cells was not detectably changed compared with WT cells. We conclude that increased levels of apical determinants is a specific consequence of loss of Hpo signalling, and is not common to all cells with a sorting defect.
We tested whether accumulation of apical proteins is merely a consequence of the overgrowth elicited by loss of Hpo signalling or a specific consequence of loss of this pathway. Overexpressing cyclin D (cycD) and cyclin-dependent kinase 4 (cdk4) in the wing imaginal disc leads to an increase in cell proliferation by accelerating cell growth (Datar et al., 2000). However, those cells do not show a brighter or broader staining of aPKC (supplementary material Fig. S4D,D′). Furthermore, as previously discussed, overexpressing the known Hpo pathway targets cycE, DIAP1 and ban strongly promotes proliferation, but does not lead to an increase in aPKC staining at the apical domain of the cells (supplementary material Fig. S4B,B′). Thus, the apical hypertrophy seen in cells mutant for the Hpo pathway is not a general property of all overproliferating tissues.
Signalling consequences of apical determinant accumulation
Maitra et al. reported that several receptors, such as Smo, Egfr and N accumulate on the surface of mer;ex double clones, probably because of an imbalance between endo- and exocytosis in these cells (Maitra et al., 2006). The same study also contends that N signalling is elevated in mer;ex clones, on the basis of dominant genetic interactions. This finding is at odds with observations from several groups indicating that N signalling is disrupted in hpo clones in the ovarian follicular epithelium (Meignin et al., 2007; Polesello and Tapon, 2007; Yu et al., 2008). We therefore decided to further explore N signalling in wing discs.
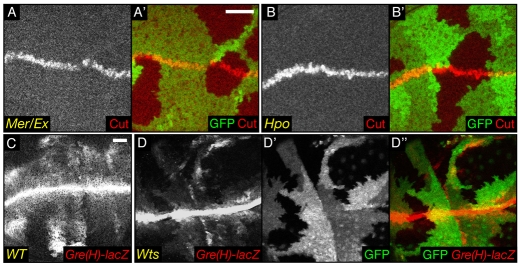
We confirmed that N is apically upregulated in all Hpo pathway members that we tested, and that this phenotype is not specific to mer;ex clones (supplementary material Fig. S3). We looked at the expression of Cut, a direct target of N that is expressed in a thin stripe at the dorsoventral (DV) boundary of the wing disc (de Celis and Bray, 1997; de Celis et al., 1996). Cut staining can be used to monitor an increase in N activity (Childress et al., 2006). We thus generated clones for mer;ex or hpo. When the Cut expression domain crossed mutant cells, we did not observe a broader Cut stripe (Fig. 5A-B′). On the contrary, there appeared to be a slight reduction in its width. To confirm that result, we also used the Gre(H)-lacZ reporter to monitor N target gene expression (Furriols and Bray, 2001). In WT wing discs, this reporter is highly expressed in a stripe at the DV boundary, which is the region of highest N activity. It is also expressed more faintly in patches in the pouch of the wing disc, a region of lower N activity (Fig. 5C). Clones of wts mutant cells expressing the Gre(H)-lacZ reporter and located in the pouch clearly showed a decrease in β-galactosidase (β-gal) staining. In rare clones that straddle the DV boundary, there was a decrease in the high N activity region (Fig. 5D-D″). The E(spl)m8-lacZ reporter gives similar results to Gre(H)-lacZ (not shown). This suggests that, as in ovaries, N signalling is impaired in Hpo pathway loss-of-function wing disc clones. Thus, whereas apical proteins, including signalling receptors, accumulate on the surface of mutant cells, depending on the receptor, this may result in increased activation (if the receptor signals at the surface) or decreased activation (if the receptor needs to be internalised to signal).
Fig. 5.
Downregulation of Notch activity in Hpo signalling-deficient cells. (A-D′) XY sections of wing imaginal discs. (A′,B′) Merged images of A and B with GFP (green). (D″) The merged images of D (red) and D′ (green). Scale bar: 20 μm. mer;ex (A,A′) and hpo (B,B′) mutant cells do not show an increase in Cut staining at the dorsoventral boundary. Compared with WT cells (C), wts mutant cells (D,D′) show a decrease in β-galactosidase staining monitoring the activity of the Notch reporter Gre(H)-lacZ in the pouch, and also more weakly at the dorsoventral boundary.
Is apical determinant accumulation required for cell proliferation?
The fact that receptors aberrantly accumulate on the surface of Hpo-pathway-deficient cells opens the possibility that the overproliferation in Hpo pathway mutants is due, at least in part, to apical hypertrophy leading to abnormal signalling (Maitra et al., 2006). For example, excess Egfr activation would be predicted to promote both proliferation and cell survival (Baker and Yu, 2001; Dominguez et al., 1998). Finally, Crb overexpression can induce hyperplastic growth in wing discs (Lu and Bilder, 2005). To determine the relationship between apical expansion and overproliferation, we took advantage of the fact that crb mutant cells in the wing disc do not lose apicobasal polarity and delaminate. Furthermore, Tepass and colleagues reported that Crb is required for apical membrane growth in the retina (Pellikka et al., 2002). We therefore reasoned that removing Crb might prevent Hpo pathway-induced apical membrane hypertrophy.
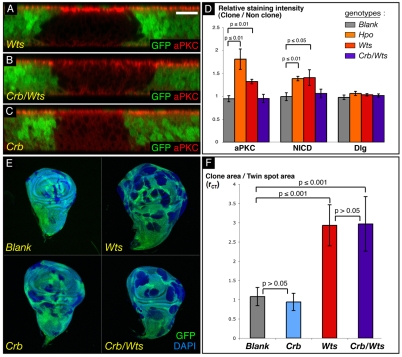
As anticipated, apical proteins failed to accumulate in crb/wts cells. NICD and aPKC stainings are not fully homogeneous in crb/wts clones, with occasional clones still showing slightly increased staining in a row of cells at the periphery (supplementary material Fig. S3D,D′), whereas in most of the clones, stainings are weaker than in WT cells, as can be seen in transverse sections (Fig. 6B; supplementary material Fig. S3D,D′). As expected, aPKC and NICD apical stainings are slightly reduced in crb clones (Fig. 6C; supplementary material Fig. S3E,E′).
Fig. 6.
The Hpo pathway controls proliferation independently of cell polarity. (A-C) Transverse sections of wing imaginal discs containing mutant clones of different genotypes (marked by absence of GFP) and stained for aPKC. Scale bar: 10 μm. Cells mutated for wts show an increase in aPKC staining (A), whereas cells mutant for crb show less apical aPKC (C). As for crb mutant cells, crb/wts cells have less aPKC on their apical surface (B). (D) Quantification of the apical determinant accumulation in hpo or wts mutant cells, as well as its rescue in crb/wts mutant cells, compared with WT cells. Histogram of the ratio of staining intensities for aPKC, NICD and Dlg between clones of WT, hpo, wts or crb/wts cells, and non-mutant cells of the same wing disc. P-values from Mann-Whitney tests are shown on the graph. (E,F) Quantification of the proliferative advantage of wts, crb and crb/wts mutant cells compared with WT cells. (E) Sample of wing discs used to measure and compare the clonal (absence of GFP) and twin spot areas (two copies of GFP); DAPI (blue) is used to visualise disc outlines. (F) Graph of rCT ratio between clonal and twin spot areas, for each genotype. Blank clones are clones of WT cells. P-values from Mann-Whitney tests are shown on the graph.
To quantify the extent of the apical determinant increase in wts, hpo and crb/wts mutant cells, we generated hpo, wts and crb/wts clones in wing discs and measured the staining intensity ratio between mutant cells (no GFP) and non-mutant cells (one or two copies of GFP). We calculated this ratio (r) for aPKC, NICD and Dlg stainings (Fig. 6D; supplementary material Table S2). As expected, for discs containing control (WT) clones, the ratio approached 1 for the three stainings. Dlg staining intensity in all conditions tested was not significantly altered, suggesting that the septate junctions are not affected. aPKC and N levels were increased in hpo and wts mutant cells compared with WT cells confirming our previous observations in transverse sections (Fig. 1, Fig. 2B-B″, Fig. 6A; supplementary material Fig. S3B-C′). Both stainings were slightly less in crb mutant cells (Fig. 6C; supplementary material Fig. S3E,E′). In crb/wts clones, the levels of both aPKC and NICD were at WT levels, indicating that loss of crb can indeed rescue the apical determinant and receptor accumulation observed in wts mutant tissue.
To determine if apical protein accumulation has an impact on the overgrowth phenotype of Hpo pathway loss of function, we compared the growth rates of clones mutant for crb, wts or crb/wts, in density-controlled crosses. When mutant clones and WT twin spots are generated using the FLP/FRT system and a heat-shock Flipase transgene, both mutant (GFP-negative) and WT (two copies of GFP) cells are born at the same time from the same recombination event. Thus, comparing the size of the mutant and WT areas after a fixed time interval (72 hours) allows us to compare the growth rate of mutant cells with that of normal cells. We therefore dissected wing discs containing clones of the relevant genotypes, acquired images in the GFP channel and calculated the ratio rCT between the total clonal area and the total twin spot area for the different genotypes (Fig. 6E,F; supplementary material Table S3).
As expected, the ratio approached 1 for discs containing control (WT) clones [rCT(control)=1.08]. Discs containing crb clones have a ratio very similar to that of control clones, showing that crb mutant cells have neither a growth advantage nor a growth defect in the wing. As previously shown, wts clones have a growth advantage over WT tissue [rCT(wts)=2.93; P(wts-control)=1.29E-08]. Finally, clones of crb/wts mutant cells also have a growth advantage over their twin spot [rCT(crb/wts)=2.96; P(crb/wts-control)=6.65E-09]. Importantly, the growth advantages of wts and crb/wts clones are almost identical [P(wts-crb/wts)=0.95]. In addition, the rate of apoptosis is the same in wts clones and crb/wts double clones, as assayed by anti-active caspase-3 staining (not shown). These results suggest that the apical hypertrophy is not necessary for the overgrowth phenotype elicited by disruption of Hpo signalling.
Apical constriction occurs in hpo or wts mutant cells but cannot account for apical determinant accumulation
The hypertrophy of the apical domain we observe in the absence of wts or hpo could either be due to a net expansion of the apical membrane or a change in cell shape. For instance, constriction at the ZA could increase the apparent levels of aPKC staining by inducing the bulging out of the apical membrane. Our TEM measurements (Fig. 3H; supplementary material Table S1) took this possibility into account since we measured the width as well as the length of the apical domain. Nevertheless, to confirm this in an independent manner, we quantified apical constriction in wts, hpo and crb/wts mutant clones. We generated positively labelled clones of cells mutant for hpo, wts or crb/wts stained for DE-cad and filamentous actin (supplementary material Fig. S6A-C″). We calculated the ratio (rACS) between apical cell surface at the level of the ZA in mutant areas and in WT areas (supplementary material Fig. S6D, Table S4). hpo and wts mutant cells were mildly constricted compared with WT cells since the apical cell surface ratio was below one [rACS(hpo)=0.84; rACS(wts)=0.88]. Thus, apical constriction could participate in the apical hypertrophy phenotype. However, in crb/wts cells, which do not show apical protein accumulation, we observed a similar constriction to that in wts cells [rACS(crb/wts)=0.92; P(wts-crb/wts)=0.45]. We noted that some areas in crb/wts clones had discontinuous DE-cad staining, preventing us from estimating cell number in these areas, even though cell densities appeared even higher than in other parts of the clones. Thus, the apical constriction in crb/wts clones is probably slightly underestimated, suggesting that, despite the fact that aPKC staining is normal in crb/wts clones, the cells are in fact more constricted than in wts tissue. As suggested by the TEM measurements, these results show that, whereas ZA constriction may contribute to the apical membrane phenotype, this is primarily due to an increase in apical determinants.
Discussion
Polarity and the Hpo pathway
We have examined, at the molecular level, apicobasal polarity in cells mutant for the Hpo tumour-suppressor pathway. We show that cells in which the Hpo pathway is inactivated stain more intensely for apical polarity, ZA components and the N receptor, which is correlated with an increase in apical domain size and adherens junction width. By contrast, basal polarity proteins are unaffected (Figs 1, 2 and 3). This altered polarity is manifest in disruptions at every level of the Hpo pathway, from upstream elements (e.g. mer;ex clones) to cells overexpressing Yki. Though wts and hpo mutant cells are mildly constricted, our results suggest that the apical hypertrophy phenotype is a consequence of increased levels of apical determinants (Fig. 3G; supplementary material Fig. S6). Increase in the apical domain staining of several cell surface receptors had been reported for mer;ex mutant clones (Maitra et al., 2006), leading to the suggestion that Ex and Mer control Hpo activity indirectly via other signalling pathways. Our data indicate that apical determinant accumulation is a general consequence of loss of Hpo signalling. This suggests that Ex and Mer are indeed bona fide members of the Hpo pathway.
How does apical hypertrophy arise?
We can suggest several possible causes for the apical protein expansion phenotype. First, loss of basal determinants such as Scrib or Dlg is known to increase the apical domain (Bilder and Perrimon, 2000). However, the basal polarity machinery appears undisturbed in hpo pathway clones, which rules out this possibility (Figs 1 and 2).
Second, one or several polarity determinants may be transcriptional targets of Yki. At the very least, the fact that Yki overexpression (but not that of known pathway targets cycE, DIAP1 and ban) is sufficient for apical hypertorphy suggests that the effect of Hpo pathway loss of function on polarity is transcriptional (supplementary material Fig. S4). The shg gene, which encodes the ZA-associated protein DE-cad, could be a target, since shg transcriptional activity is increased in wts and ft clones as well as in yki-overexpressing discs (Fig. 4) (Jaiswal et al., 2006). In fact, a putative Hpo response element [HRE (Wu et al., 2008)] containing a conserved Scalloped (Sd) binding site is present in the shg first intron. Yki, with its partner Sd, could therefore be directly responsible for the increased transcriptional activity of shg. However, whereas increased DE-cad could account for broader junctions, it would not necessarily be predicted to lead to an expanded apical domain. Indeed, combined DE-cad and Arm overexpression is not sufficient to drive apical expansion (supplementary material Fig. S5). The apical determinant Crb has been reported to promote apical membrane growth by antagonising the FERM domain protein Yurt, although the mechanism remains to be defined (Laprise et al., 2006). Interestingly, our results indicate that Crb activity is necessary for apical protein accumulation upon loss of wts (Fig. 6). The mild increase in crb transcription we observe in some hpo clones may not be sufficient to explain this phenotype (Fig. 4). It will therefore be interesting to see whether Hpo signalling can modulate Crb or Yurt activity.
Finally, apical hypertrophy might occur as a result of an imbalance between exo- and endocytosis. This interesting possibility is supported by two lines of evidence. Firstly, mer;ex clones have decreased rates of N endocytosis (Maitra et al., 2006). Secondly, hpo mutant clones in ovarian follicle cells have ectopic accumulation of the endosomal sorting protein Hrs (HGF-regulated tyrosine kinase substrate) (Yu et al., 2008). Altered vesicle trafficking would be predicted to lead to inappropriate delivery of polarity proteins, which might account for apical determinant accumulation. So far, no known Yki targets could account for such a phenotype, but future research should resolve this important issue. It is interesting to note that many regulators of vesicle trafficking also appear to function as tumour suppressor genes in Drosophila (Hariharan and Bilder, 2006).
Apical protein accumulation and cell surface signalling
mer;ex cells have been reported to increase apical levels of different signalling receptors such as Egfr, the Hedgehog receptor Smoothened and N (Maitra et al., 2006). We found that hpo or wts mutant cells had more N receptor at their apical membrane (supplementary material Fig. S3). This led us and others (Maitra et al., 2006) to speculate that the presence of more signalling receptors at the apical surface, where many signalling events occur, could participate in the hpo overgrowth phenotype by triggering ectopic signalling. However, the fact that crb/wts mutant cells overproliferate to the same extent as wts mutant cells while not having increased apical N or aPKC suggests that apical hypertrophy and proliferation are separate processes (Fig. 6).
Does the presence of higher levels of receptor at the cell surface necessarily equate to increased signalling? Despite this accumulation of N receptors, N signalling is downregulated in follicle cells mutant for the Hpo pathway (Meignin et al., 2007; Polesello and Tapon, 2007; Yu et al., 2008). Our results show that in epithelial imaginal wing cells, N signalling activity is decreased in Hpo pathway loss-of-function cells, despite increased N at the apical surface (Fig. 5; supplementary material Fig. S3). We can hypothesise that reduced N endocytosis blocks its cleavage by γ-secretase, which normally occurs in late endosomes and is a crucial step for N receptor activation (Le Borgne, 2006). Thus, depending on whether a receptor requires internalisation for activation or signalling, we can expect distinct outcomes (activation or blockage) in Hpo pathway mutant tissue.
Epithelial architecture and cancer
In human cancer, loss of polarity is a feature of invasive tumours (Wodarz and Nathke, 2007). Indeed, imaginal cells mutant for scrib or dlg are highly invasive, suggesting that loss of polarity can promote invasion of surrounding tissue. By contrast, wts mutations failed to collaborate with oncogenic ras or scrib mutations in an eye disc invasion model (Igaki et al., 2006; Pagliarini and Xu, 2003). However, in ovarian follicle cells, wts mutations have been reported to induce invasion and to potentiate the invasive phenotype of dlg mutant cells (Zhao et al., 2008b). Thus, perturbation of epithelial architecture induced by loss of Hpo pathway signalling may well participate in the invasive behaviour of some tumour types.
Materials and Methods
Fly stocks
w; FRT42D,hpoJM1/Cyo,P[act::GFP] and yw; FRT42D,hpoBF33/Cyo,P[act::GFP] were gifts from Jin Jiang (University of Texas Southwestern Medical Center, Dallas, TX) (Jia et al., 2003). w; FRT42D,hpo42-47/Cyo,P[w] and w; UAS::yki/TM3,Sb were gifts from Duojia Pan (Johns Hopkins University School of Medicine, Baltimore, MD) (Huang et al., 2005; Wu et al., 2003). yw; FRT82B,wtslatsX1/TM3,Sb was a gift from Tian Xu (Yale University School of Medicine, New Haven, CT) (Xu et al., 1995). yw,Mer4,FR19A; P[UbiMer],P[UbiGFP],FRT40A/Cyo,P[act::GFP] and w; exe1,FRT40A/Cyo,P[act::GFP]; hsFLP,MKRS/TM6b were gifts from Richard Fehon (Universityof Chicago, Chicago, IL) (Maitra et al., 2006). w; UAS::ex was obtained from Georg Halder (M. D. Anderson Cancer Center, Houston, TX) and is a construct by Peter Bryant (University of California, Irvine, CA) (Boedigheimer et al., 1997). yw,hsFLP; UAS::cycD,UAS::cdk4/SM6-6B was a gift from Bruce Edgar (Fred Hutchinson Cancer Research Center, Seattle, WA) (Datar et al., 2000). w; UAS::Cad,UAS::Arm/Cyo was a gift from Jean-Paul Vincent (National Institute for Medical Research, London, UK). w; E(spl)m8-lacZ and w; Gre(H)-lacZ were gifts from Sarah Bray (University of Cambridge, Cambridge, UK). yw; P[lacW]shgk03401/CyO (shg-lacZ in the text) was obtained from Emily Richardson (University College London, London, UK). w/hshid(Y); ptc>gal4,UAS::GFP/Cyo was a gift from Barry Thompson (CRUK London Research Institute, London, UK). w; crbM11.M2/TM3 (crb-lacZ in the text) and w; FRT,crb11a22/TM3Sb were gifts from Marco Milán (Institut de Recerca Biomedica, Barcelona, Spain) (Herranz et al., 2006). w; UAS::hpo was described by Pantalacci et al. (Pantalacci et al., 2003). The MARCM makers yw,tubGAL4,hsFLP,122,UAS::nucGFPmyc, FRT82B,CD21,y+,tubG80.LL3/TM6 and yw,tubGAL4,hsFLP,122,UAS::nucGFPmyc; FRT42D,CD21,tubG80.LL3/CyO were gifts from Gary Struhl (Columbia University, New York, NY). w; UAS::rasv12, yw,hsFLP; FRT42D,GFP, yw,hsFLP; FRT82B,GFP, yw,eyFLP; FRT42D,GFP, w; Act>cd2>Gal4,UAS::GFP were from Bloomington Stock Center.
FRT82B,crb11a22,wtslatsx1 double mutants were generated by meiotic recombination. w; FRT42D,hpo5.1/Cyo was generated by mobilisation of G3315 (Genexel). Sequencing of the PCR products revealed a deletion of 1983 bp that removes part of the 5′UTR, the coding start site, the whole first exon and part of the second exon of the hpo gene.
Immunostainings
Mosaic tissues were obtained with the FLP/FRT system with eyFLP drivers (see supplementary material Fig. S1), or hsFLP drivers in all other experiments. For loss-of-function experiments, 48-hour larvae were heat shocked for 50 minutes at 37°C and dissected 3 days after heat shock. For gain-of-function experiments (flip out), 72-hour larvae were heat shocked for 5-20 minutes at 37°C and dissected 2 days later. Some gain-of-function (supplementary material Fig. S4) experiments were carried out using ptc>gal4 drivers.
Tissues were processed as described by Polesello et al. (Polesello et al., 2006). β-galactosidase antibody (Promega) was used at 1/500 and NICD (C17.9C6) antibody, from Development Studies Hybridoma Bank (DSHB), at 1/500. Anti-DG antibody [a gift from Wu-Min Deng (Florida State University, Tallahassee, FL)] was used at 1/3000. Anti-Arm (N2 7A1 from DSHB) antibody was used at 1/10, anti-aPKC (Santa Cruz) at 1/500 and anti-E-cad at 1/20. Anti-Cad99C [a gift from Dorothea Godt (University of Toronto, Toronto, Canada)] was used at 1/3000, anti-P-Moe [a gift from Donald F. Ready (Purdue University, West Lafayette, IN)] was used at 1/500, and anti-Dlg (4F3 from DSHB) was used at 1/500. Anti-Cut (2B10 DSHB) was used at 1/20. Anti-Rab5 was used at 1/1000 [a gift from Akira Nakamura (RIKEN Center for Developmental Biology, Kobe, Japan)]. Rhodamine-phalloidin (Sigma) was used at 1/500. Secondary antibodies (Rhodamine red X-conjugated donkey anti-rabbit, anti-rat or anti-mouse, Cy5-conjugated donkey anti-mouse; from Jackson ImmunoResearch) were used at 1/500. Hoechst 33342 was added for 20 minutes. After washes, tissues were mounted in Vectashield (Vector). Fluorescence images were acquired on a Zeiss LSM510 confocal laser scanning microscope (25× and 40× lenses, zoom 1 or 2), either as XY acquisitions or transverse sections (XZ).
Correlative transmission electron microscopy (TEM) of wing imaginal discs
Third instar imaginal discs were dissected in fixing solution A (PBS + 4% PFA) and transferred on to an etched grid coverslip (Bellco, 1916-91012) coated with poly-L-lysine to aid correlation between light and EM imaging. Transmitted light and GFP fluorescence images of the discs, still dipped in fixing solution A, were acquired with a 10× lens on a Zeiss LSM510 confocal laser scanning microscope and a pinhole of 5 μm. After imaging, the discs were fixed for 2 hours in 4% PFA + 2.5% glutaraldehyde in 0.1 M phospate buffer, pH 7.4 at room temperature. They were then postfixed in 1.5% potassium ferrocyanide/1% osmium tetroxide for 1 hour and stained with 1% tannic acid in 0.05 M sodium cacodylate pH 7.4 for 45 minutes. Coverslips were dehydrated stepwise through an ethanol series, infiltrated with 50:50 propylene oxide:Epon followed by one change of pure resin every 24 hours for 7 days and then polymerised `en face' at 60°C overnight. Each wing disc was re-embedded perpendicularly and trimmed along the plane shown in Fig. 3A,B so as to incorporate both mutant and WT tissue. Ultrathin sections of ∼75 nm were collected using a UCT ultramicrotome (Leica Microsystems UK), post-stained with lead citrate and viewed using a Tecnai G2 Spirit 120 kV transmission electron microscope (FEI Company) with an SC1000 Orius CCD camera (Gatan UK). Approximately 110 high magnification images were taken along the entire disc for quantification of the apical domain.
Quantification of AJ width
Calculations of relative apical domain size of WT and yki-overexpressing cells were assessed by the product PLW (apical domain size multiplied by cell width) (graph in Fig. 3G). Cell width was measured at the AJs. Apical domain length was calculated by measuring the length of a perpendicular line drawn between the AJ level and the apical extremity of each cell. For WT and yki-overexpressing cells, the lengths of 40 to 50 AJs were estimated, using the segmented line tool of the ImageJ software (supplementary material Table S1). In order to measure the apical domain size of the cells and taking into account a possible difference in apical constriction of the cells, we measured the cell width (W) at the AJs and the distance (L) between the AJ and the top-most apical membrane level using the straight line tool of the ImageJ software (Fig. 3D,E). We then calculated the product PLW of those two values, which gives us a relative measure of subapical region size in the control and yki-expressing cells. The statistical significance of the difference between wild-type (WT) and mutant tissue was assessed by a Mann-Whitney non-parametric test. P<0.01 when the difference was significant. The AJs were of similar width in posterior vs anterior cells of an en-GAL4, UAS-GFP control disc (see supplementary material Table S1).
Quantitative RT-PCR
MS1096>> (control) and MS1096>>yki wing discs were dissected in PBS and snap-frozen in liquid nitrogen. Total RNA was isolated from them using the RNeasy kit (Qiagen) and treated with RQ1 DNase (Promega). First Strand cDNA Synthesis Kit for RT-PCR (AMV; Roche) and oligo-p(dT)15 primers were used to produce cDNAs from the extracted total RNA (1.5 μg). To measure mRNA levels, qPCRs were carried out on reverse-transcribed total mRNA using exon-specific primers. Real-time qPCR was performed with Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen) in 96-well plates and using the Chromo 4 Real-time qPCR Detection System (MJ Research). All reactions were performed four times. The relative amount of specific mRNAs under each condition was calculated after normalisation to the histone 3 transcript. The statistical significance of the difference between control and yki-overexpressing discs was assessed using a Student's t-test with significance at P<0.05.
Staining intensity ratio measurements
Z-stack images of the apical part of third instar imaginal wing discs containing WT, hpo, wts, or crb/wts clones and stained for aPKC, NICD or Dlg, were taken with a pinhole of 0.9 μm every 0.4 μm. Using the Bitplan Imaris imaging software, the intensity in all the Z-stack was assessed in different areas of mutant tissue (absence of GFP) or of non-mutant tissue (presence of GFP). For each disc, the staining intensity was measured in five to 15 areas of mutant tissue and in an equal number of non-mutant tissue areas. The chosen areas were representative of the heterogeneousity of the tissue sampled. The mean intensity of those areas was then calculated, as well as the intensity ratio between mutant and non-mutant tissue for each disc. Five discs were processed in the same way for each genotype and each staining. The statistical significance of the difference between WT clones and mutant clones was assessed by a Mann-Whitney non-parametric test. P<0.01 when the difference was significant. Supplementary Table S2 presents the ratios, called ri, of staining intensities for aPKC, NICD and Dlg between clones of WT (Blank), hpo, wts or crb/wts cells, and non-mutant cells of the same wing disc.
Standard growth conditions and size measurements
For size measurement experiments, 50 L1 larvae (0–4 hour collections) per vial were collected from agar plates 24 hours after egg deposition and were grown until larval stage L3 before dissection. Heat shocks were as described above. Images of the discs were acquired with a 10↔lens on a Zeiss LSM510 confocal laser scanning microscope and a pinhole of 5 μm. Measurements of the GFP negative area (clonal area) and twin spot area (two copies of GFP) were done by hand using Adobe Photoshop CS. Each area of interest was measured, with pixel as the unit, using the Polygonal Lasso Tool. All clonal areas of a disc were then added, and compared to the sum of the twin spot areas of the same disc. This process was repeated for several discs (5≤n≤16) and the results were averaged. The statistical significance of the difference between WT and mutant tissue was assessed by a Mann-Whitney non-parametric test. P<0.01 when the difference was significant. Supplementary Table S2 presents the ratios between the clonal area and the twin spot area, called rCT, for clones of WT (Blank), crb, wts or crb/wts cells.
Quantification of apical constriction
hpo, wts, or crb/wts clones positively labelled with GFP were generated and stained for F-actin and Arm to visualise the apical side of the cells. For five different clones of each genotype, the clonal area (GFP) was measured as well as the number of cells constituting it, thus allowing us to calculate the mean apical cell surface in that clone. The same procedure was followed for an area of non-mutant tissue close to the clone. We then calculated the ratio between the apical cell surface in the clonal area and the apical cell surface in an adjacent non-mutant area, which provides us with an index of apical cell constriction. The results were then averaged and the statistical significance of the difference between different mutant tissues was assessed by a Mann-Whitney non-parametric test. P<0.01 when the difference was significant. Supplementary material Table S4 presents the ratios, called rACS, between apical cell surface in mutant tissue and apical cell surface in surrounding tissue, for clones of hpo, wts or crb/wts cells.
Genotypes
Fig. 1A,A′,C,C′, E-F′,I,I′: yw,hsFLP/w; FRT42D,GFP/FRT42D,hpo5.1.
Fig. 1B,B′,D,D',G-H′; Fig. 5B,B': yw,hsFLP/w; FRT42D,GFP/FRT42D,hpoJM1.
Fig. 2A-A″; Fig. 5A,A′: yw,Mer4; FRT40A,p[w+,Mer+],UbiGFP/FRT40A,exe1; MKRS,hsFLP/+.
Fig. 2B-B′; Fig. 6A: yw,hsFLP/w; FRT82B,GFP/FRT82B,wtslatsX1.
Fig. 2C,C′: yw,hsFLP/w; UAS::yki/Act>CD2>Gal4,UAS::GFP.
Fig. 2D-E′: yw,hsFLP/w; Act>CD2>Gal4,UAS::GFP,UAS::p35/+; UAS::ex.
Fig. 2F,F′: yw,hsFLP/w; UAS::hpo/Act>CD2>Gal4,UAS::GFP,UAS::p35.
Fig. 3A-E: w; en>Gal4,UAS::GFP/+; UAS::yki/+.
Fig. 3F,G; supplementary material Table S1: w; en>Gal4,UAS::GFP/+; UAS::yki/+, w; en>Gal4,UAS::GFP/+.
Fig. 4A: MS1096>Gal4/w, MS1096>Gal4/w;; UAS::yki/+.
Fig. 4B: yw,hsFLP/w; crb-LacZ/+; FRT42D,GFP/+.
Fig. 4C-C″: yw,hsFLP/w; crb-LacZ/+; FRT42D,GFP/FRT42D,hpo5.1.
Fig. 4D,D′: yw,hsFLP/w;shg-LacZ/+; FRT82B,GFP/FRT82B,wtslatsX1.
Fig. 5C: yw,hsFLP/w; Gre(H)-LacZ/+; FRT82B,GFP/+.
Fig. 5D-D″: yw,hsFLP/w; Gre(H)-LacZ/+; FRT82B,GFP/FRT82B,wtslatsX1.
Fig. 6B: yw,hsFLP/w; FRT82B,GFP/FRT82B,crb11a22,wtslatsX1.
Fig. 6C: yw,hsFLP/w; FRT82B,GFP/FRT82B,crb11a22.
Fig. 6D, supplementary material Table S2: yw,hsFLP/w; FRT82B,GFP/FRT82B, yw,hsFLP/w; FRT42D,GFP/FRT42D,hpo5.1, yw,hsFLP/w; FRT82B,GFP/FR-T82B,wtslatsX1, yw,hsFLP/w; FRT82B,GFP/FRT82B,crb11a22,wtslatsX1.
Fig. 6E,F, supplementary material Table S3: yw,hsFLP/w; FRT82B,GFP/FRT82B, yw,hsFLP/w; FRT82B,GFP/FRT82B,wtslatsX1, yw,hsFLP/w; FRT82B,GF-P/FRT82B,crb11a22,wtslatsX1, yw,hsFLP/w; FRT82B,GFP/FRT82B,crb11a22.
Supplementary Material
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/122/14/2360/DC1
We thank D. Pan, J. Jiang, R. Fehon, B. Edgar, S. Bray, H. McNeill, D. F. Ready, G. Halder, B. Thompson, E. Richardson, M. Milán, G. Struhl, A. Nakamura, J. P. Vincent, D. Godt, the Bloomington and Szeged stock centres for flies stocks and reagents. We are grateful to P. Jordan from the Light Microscopy Facility for technical help, D. Andersen for advice on qRT-PCR, and T. Gilbank, S. Murray and F. Earl for technical support. We thank B. Thompson and E. Chan for discussions and comments on the manuscript, and P. L. Bardet and J. P. Vincent for discussing unpublished data. The Tapon laboratory is supported by Cancer Research UK. The Pichaud laboratory is supported by the Medical Research Council. Deposited in PMC for release after 6 months.
References
- Bachmann, A., Draga, M., Grawe, F. and Knust, E. (2008). On the role of the MAGUK proteins encoded by Drosophila varicose during embryonic and postembryonic development. BMC Dev. Biol. 8, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, N. E. and Yu, S. Y. (2001). The EGF receptor defines domains of cell cycle progression and survival to regulate cell number in the developing Drosophila eye. Cell 104, 699-708. [DOI] [PubMed] [Google Scholar]
- Bennett, F. C. and Harvey, K. F. (2006). Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Curr. Biol. 16, 2101-2110. [DOI] [PubMed] [Google Scholar]
- Betschinger, J., Mechtler, K. and Knoblich, J. A. (2003). The Par complex directs asymmetric cell division by phosphorylating the cytoskeletal protein Lgl. Nature 422, 326-330. [DOI] [PubMed] [Google Scholar]
- Bilder, D. and Perrimon, N. (2000). Localization of apical epithelial determinants by the basolateral PDZ protein Scribble. Nature 403, 676-680. [DOI] [PubMed] [Google Scholar]
- Bilder, D., Li, M. and Perrimon, N. (2000). Cooperative regulation of cell polarity and growth by Drosophila tumor suppressors. Science 289, 113-116. [DOI] [PubMed] [Google Scholar]
- Bilder, D., Schober, M. and Perrimon, N. (2003). Integrated activity of PDZ protein complexes regulates epithelial polarity. Nat. Cell Biol. 5, 53-58. [DOI] [PubMed] [Google Scholar]
- Boedigheimer, M. J., Nguyen, K. P. and Bryant, P. J. (1997). Expanded functions in the apical cell domain to regulate the growth rate of imaginal discs. Dev. Genet. 20, 103-110. [DOI] [PubMed] [Google Scholar]
- Childress, J. L., Acar, M., Tao, C. and Halder, G. (2006). Lethal giant discs, a novel C2-domain protein, restricts notch activation during endocytosis. Curr. Biol. 16, 2228-2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, E., Feng, Y., Rauskolb, C., Maitra, S., Fehon, R. and Irvine, K. D. (2006). Delineation of a Fat tumor suppressor pathway. Nat. Genet. 38, 1142-1150. [DOI] [PubMed] [Google Scholar]
- D'Alterio, C., Tran, D. D., Yeung, M. W., Hwang, M. S., Li, M. A., Arana, C. J., Mulligan, V. K., Kubesh, M., Sharma, P., Chase, M. et al. (2005). Drosophila melanogaster Cad99C, the orthologue of human Usher cadherin PCDH15, regulates the length of microvilli. J. Cell Biol. 171, 549-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datar, S. A., Jacobs, H. W., de la Cruz, A. F., Lehner, C. F. and Edgar, B. A. (2000). The Drosophila cyclin D-Cdk4 complex promotes cellular growth. EMBO J. 19, 4543-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Celis, J. F. and Bray, S. (1997). Feed-back mechanisms affecting Notch activation at the dorsoventral boundary in the Drosophila wing. Development 124, 3241-3251. [DOI] [PubMed] [Google Scholar]
- de Celis, J. F., Garcia-Bellido, A. and Bray, S. J. (1996). Activation and function of Notch at the dorsal-ventral boundary of the wing imaginal disc. Development 122, 359-369. [DOI] [PubMed] [Google Scholar]
- Dominguez, M., Wasserman, J. D. and Freeman, M. (1998). Multiple functions of the EGF receptor in Drosophila eye development. Curr. Biol. 8, 1039-1048. [DOI] [PubMed] [Google Scholar]
- Dong, J., Feldmann, G., Huang, J., Wu, S., Zhang, N., Comerford, S. A., Gayyed, M. F., Anders, R. A., Maitra, A. and Pan, D. (2007). Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130, 1120-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, Y. and Irvine, K. D. (2007). Fat and expanded act in parallel to regulate growth through warts. Proc. Natl. Acad. Sci. USA 104, 20362-20367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furriols, M. and Bray, S. (2001). A model Notch response element detects Suppressor of Hairless-dependent molecular switch. Curr. Biol. 11, 60-64. [DOI] [PubMed] [Google Scholar]
- Goulev, Y., Fauny, J. D., Gonzalez-Marti, B., Flagiello, D., Silber, J. and Zider, A. (2008). SCALLOPED interacts with YORKIE, the nuclear effector of the hippo tumor-suppressor pathway in Drosophila. Curr. Biol. 18, 435-441. [DOI] [PubMed] [Google Scholar]
- Hamaratoglu, F., Willecke, M., Kango-Singh, M., Nolo, R., Hyun, E., Tao, C., Jafar-Nejad, H. and Halder, G. (2006). The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat. Cell Biol. 8, 27-36. [DOI] [PubMed] [Google Scholar]
- Hariharan, I. K. and Bilder, D. (2006). Regulation of imaginal disc growth by tumor-suppressor genes in Drosophila. Annu. Rev. Genet. 40, 335-361. [DOI] [PubMed] [Google Scholar]
- Harvey, K. and Tapon, N. (2007). The Salvador-Warts-Hippo pathway-an emerging tumour-suppressor network. Nat. Rev. Cancer 7, 182-191. [DOI] [PubMed] [Google Scholar]
- Harvey, K. F., Pfleger, C. M. and Hariharan, I. K. (2003). The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell 114, 457-467. [DOI] [PubMed] [Google Scholar]
- Herranz, H., Stamataki, E., Feiguin, F. and Milan, M. (2006). Self-refinement of Notch activity through the transmembrane protein Crumbs: modulation of gamma-secretase activity. EMBO Rep. 7, 297-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J., Wu, S., Barrera, J., Matthews, K. and Pan, D. (2005). The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 122, 421-434. [DOI] [PubMed] [Google Scholar]
- Igaki, T., Pagliarini, R. A. and Xu, T. (2006). Loss of cell polarity drives tumor growth and invasion through JNK activation in Drosophila. Curr. Biol. 16, 1139-1146. [DOI] [PubMed] [Google Scholar]
- Jaiswal, M., Agrawal, N. and Sinha, P. (2006). Fat and Wingless signaling oppositely regulate epithelial cell-cell adhesion and distal wing development in Drosophila. Development 133, 925-935. [DOI] [PubMed] [Google Scholar]
- Jia, J., Zhang, W., Wang, B., Trinko, R. and Jiang, J. (2003). The Drosophila Ste20 family kinase dMST functions as a tumor suppressor by restricting cell proliferation and promoting apoptosis. Genes Dev. 17, 2514-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice, R. W., Zilian, O., Woods, D. F., Noll, M. and Bryant, P. J. (1995). The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 9, 534-546. [DOI] [PubMed] [Google Scholar]
- Knust, E. and Bossinger, O. (2002). Composition and formation of intercellular junctions in epithelial cells. Science 298, 1955-1959. [DOI] [PubMed] [Google Scholar]
- Laprise, P., Beronja, S., Silva-Gagliardi, N. F., Pellikka, M., Jensen, A. M., McGlade, C. J. and Tepass, U. (2006). The FERM protein Yurt is a negative regulatory component of the Crumbs complex that controls epithelial polarity and apical membrane size. Dev. Cell 11, 363-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Borgne, R. (2006). Regulation of Notch signalling by endocytosis and endosomal sorting. Curr. Opin. Cell Biol. 18, 213-222. [DOI] [PubMed] [Google Scholar]
- Lu, H. and Bilder, D. (2005). Endocytic control of epithelial polarity and proliferation in Drosophila. Nat. Cell Biol. 7, 1232-1239. [DOI] [PubMed] [Google Scholar]
- Maitra, S., Kulikauskas, R. M., Gavilan, H. and Fehon, R. G. (2006). The tumor suppressors Merlin and Expanded function cooperatively to modulate receptor endocytosis and signaling. Curr. Biol. 16, 702-709. [DOI] [PubMed] [Google Scholar]
- Medina, E., Williams, J., Klipfell, E., Zarnescu, D., Thomas, G. and Le Bivic, A. (2002). Crumbs interacts with moesin and beta(Heavy)-spectrin in the apical membrane skeleton of Drosophila. J. Cell Biol. 158, 941-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meignin, C., Alvarez-Garcia, I., Davis, I. and Palacios, I. M. (2007). The salvador-warts-hippo pathway is required for epithelial proliferation and axis specification in Drosophila. Curr. Biol. 17, 1871-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nellen, D., Burke, R., Struhl, G. and Basler, K. (1996). Direct and long-range action of a DPP morphogen gradient. Cell 85, 357-368. [DOI] [PubMed] [Google Scholar]
- Nelson, W. J. (2003). Adaptation of core mechanisms to generate cell polarity. Nature 422, 766-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolo, R., Morrison, C. M., Tao, C., Zhang, X. and Halder, G. (2006). The bantam microRNA is a target of the hippo tumor-suppressor pathway. Curr. Biol. 16, 1895-1904. [DOI] [PubMed] [Google Scholar]
- Pagliarini, R. A. and Xu, T. (2003). A genetic screen in Drosophila for metastatic behavior. Science 302, 1227-1231. [DOI] [PubMed] [Google Scholar]
- Pantalacci, S., Tapon, N. and Leopold, P. (2003). The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat. Cell Biol. 5, 921-927. [DOI] [PubMed] [Google Scholar]
- Pellikka, M., Tanentzapf, G., Pinto, M., Smith, C., McGlade, C. J., Ready, D. F. and Tepass, U. (2002). Crumbs, the Drosophila homologue of human CRB1/RP12, is essential for photoreceptor morphogenesis. Nature 416, 143-149. [DOI] [PubMed] [Google Scholar]
- Plant, P. J., Fawcett, J. P., Lin, D. C., Holdorf, A. D., Binns, K., Kulkarni, S. and Pawson, T. (2003). A polarity complex of mPar-6 and atypical PKC binds, phosphorylates and regulates mammalian Lgl. Nat. Cell Biol. 5, 301-308. [DOI] [PubMed] [Google Scholar]
- Polesello, C. and Tapon, N. (2007). Salvador-warts-hippo signaling promotes Drosophila posterior follicle cell maturation downstream of notch. Curr. Biol. 17, 1864-1870. [DOI] [PubMed] [Google Scholar]
- Polesello, C., Huelsmann, S., Brown, N. H. and Tapon, N. (2006). The Drosophila RASSF homolog antagonizes the hippo pathway. Curr. Biol. 16, 2459-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prober, D. A. and Edgar, B. A. (2000). Ras1 promotes cellular growth in the Drosophila wing. Cell 100, 435-446. [DOI] [PubMed] [Google Scholar]
- Sanson, B., White, P. and Vincent, J. P. (1996). Uncoupling cadherin-based adhesion from wingless signalling in Drosophila. Nature 383, 627-630. [DOI] [PubMed] [Google Scholar]
- Schlichting, K., Wilsch-Brauninger, M., Demontis, F. and Dahmann, C. (2006). Cadherin Cad99C is required for normal microvilli morphology in Drosophila follicle cells. J. Cell Sci. 119, 1184-1195. [DOI] [PubMed] [Google Scholar]
- Silva, E., Tsatskis, Y., Gardano, L., Tapon, N. and McNeill, H. (2006). The tumor-suppressor gene fat controls tissue growth upstream of expanded in the hippo signaling pathway. Curr. Biol. 16, 2081-2089. [DOI] [PubMed] [Google Scholar]
- Tanentzapf, G. and Tepass, U. (2003). Interactions between the crumbs, lethal giant larvae and bazooka pathways in epithelial polarization. Nat. Cell Biol. 5, 46-52. [DOI] [PubMed] [Google Scholar]
- Tepass, U., Gruszynski-DeFeo, E., Haag, T. A., Omatyar, L., Torok, T. and Hartenstein, V. (1996). shotgun encodes Drosophila E-cadherin and is preferentially required during cell rearrangement in the neurectoderm and other morphogenetically active epithelia. Genes Dev. 10, 672-685. [DOI] [PubMed] [Google Scholar]
- Tepass, U., Tanentzapf, G., Ward, R. and Fehon, R. (2001). Epithelial cell polarity and cell junctions in Drosophila. Annu. Rev. Genet. 35, 747-784. [DOI] [PubMed] [Google Scholar]
- Udan, R. S., Kango-Singh, M., Nolo, R., Tao, C. and Halder, G. (2003). Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat. Cell Biol. 5, 914-920. [DOI] [PubMed] [Google Scholar]
- Willecke, M., Hamaratoglu, F., Kango-Singh, M., Udan, R., Chen, C. L., Tao, C., Zhang, X. and Halder, G. (2006). The fat cadherin acts through the hippo tumor-suppressor pathway to regulate tissue size. Curr. Biol. 16, 2090-2100. [DOI] [PubMed] [Google Scholar]
- Wodarz, A. and Nathke, I. (2007). Cell polarity in development and cancer. Nat. Cell Biol. 9, 1016-1024. [DOI] [PubMed] [Google Scholar]
- Wu, S., Huang, J., Dong, J. and Pan, D. (2003). hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell 114, 445-456. [DOI] [PubMed] [Google Scholar]
- Wu, S., Liu, Y., Zheng, Y., Dong, J. and Pan, D. (2008). The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev. Cell 14, 388-398. [DOI] [PubMed] [Google Scholar]
- Xu, T., Wang, W., Zhang, S., Stewart, R. A. and Yu, W. (1995). Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development 121, 1053-1063. [DOI] [PubMed] [Google Scholar]
- Yu, J., Poulton, J., Huang, Y. C. and Deng, W. M. (2008). The hippo pathway promotes Notch signaling in regulation of cell differentiation, proliferation, and oocyte polarity. PLoS ONE 3, e1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Ren, F., Zhang, Q., Chen, Y., Wang, B. and Jiang, J. (2008). The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev. Cell 14, 377-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, B., Ye, X., Yu, J., Li, L., Li, W., Li, S., Yu, J., Lin, J. D., Wang, C. Y., Chinnaiyan, A. M. et al. (2008a). TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 22, 1962-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, M., Szafranski, P., Hall, C. A. and Goode, S. (2008b). Basolateral junctions utilize warts signaling to control epithelial-mesenchymal transition and proliferation crucial for migration and invasion of Drosophila ovarian epithelial cells. Genetics 178, 1947-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.