Summary
The double lipid bilayer of the nuclear envelope (NE) remains intact during closed mitosis. In the fission yeast Schizosaccharomyces pombe, the intranuclear mitotic spindle has envelope-embedded spindle pole bodies (SPB) at its ends. As the spindle elongates and the nucleus divides symmetrically, nuclear volume remains constant but nuclear area rapidly increases by 26%. When Ran-GTPase function is compromised in S. pombe, nuclear division is strikingly asymmetrical and the newly synthesized SPB is preferentially associated with the smaller nucleus, indicative of a Ran-dependent SPB defect that interferes with symmetrical nuclear division. A second defect, which specifically influences the NE, results in breakage of the NE upon spindle elongation. This defect, but not asymmetric nuclear division, is partially rescued by slowing spindle elongation, stimulating endoplasmic reticulum (ER) proliferation or changing conformation of the ER membrane. We propose that redistribution of lipid within the ER-NE network is crucial for mitosis-specific NE changes in both open and closed mitosis.
Keywords: Ran GTPase, Nuclear division, Fission yeast, Endoplasmic reticulum, Spindle pole body
Introduction
During interphase in all eukaryotic cells the double lipid bilayer of the nuclear envelope (NE) physically separates the chromosomes, and chromosome-related processes, from the cytoplasm and increases in area by 59% (Lim et al., 2007) as the nuclear volume doubles in preparation for mitosis (reviewed by Hetzer et al., 2005; Lim et al., 2007; Winey et al., 1997). In the open mitosis of animal cells, NE breakdown allows the spindle microtubules that are nucleated by the cytoplasmic centrosomes to attach to and then separate the chromosomes. In the closed mitosis of yeast, the centrosome equivalents, called spindle pole bodies (SPBs), are embedded in the NE and nucleate the formation of an intranuclear spindle (Ding et al., 1997). As the spindle elongates in anaphase B, nuclear volume remains constant but division of the roughly spherical nucleus into two smaller spheres, which occurs in less than 5 minutes, requires a rapid increase of 26% in NE area (Lim et al., 2007).
The nucleus, often thought of as a freestanding organelle, is actually a specialized region of the endoplasmic reticulum (ER) (Voeltz et al., 2002): the outer NE is continuous with both the ER and the inner NE (Hetzer et al., 2005), providing a means by which lipids and membrane proteins can move between the sheet form of the ER at the nuclear periphery and the tubular form of the ER, which, in animal cells, extends throughout the cytoplasm and, in yeast, is primarily at the cell periphery (Pidoux and Armstrong, 1993; Voeltz et al., 2002).
The Ran GTPase influences many cellular functions (Quimby and Dasso, 2003), including nucleocytoplasmic transport, NE reformation after mitosis in animal cells (Hetzer et al., 2005), and NE structure in fission (Demeter et al., 1995) and budding (Ryan et al., 2003) yeast. However, the mechanism by which the Ran GTPase influences mitosis-specific NE changes during both open and closed mitosis remains unknown. We have previously shown that, when the Ran system is mis-regulated in the fission yeast Schizosaccharomyces pombe, cells arrest with NEs that have lost their integrity, and that this defect is concomitant with and depends on the passage through mitosis (Demeter et al., 1995).
The predictions of a biophysical model of the fission-yeast nucleus (Lim et al., 2007) and experimental observations of abnormal nuclear shapes seen when microtubule bundles lacking SPBs at their ends cause thin tethers to protrude from the spherical nucleus (Khodjakov et al., 2004; Tange et al., 2002; Toya et al., 2007; West et al., 1998; Zheng et al., 2007) raise the possibility that the SPB influences the mechanical properties of the NE to prevent tether formation during normal mitosis and ensure symmetric nuclear division. Because of limitations on the mechanical strength of lipid bilayers and their ability to stretch in response to pressure exerted by the elongating spindle, our computational model incorporates a lipid reservoir (Lim et al., 2007). Because most cellular lipids are synthesized in the ER, and because the ER is continuous with the NE, it might serve as this lipid reservoir (Lim et al., 2007) that is necessary to accommodate the slow doubling in nuclear volume during interphase in all eukaryotes and the rapid increase in NE area as the spherical nucleus divides into two smaller spheres during closed mitosis.
Because of its multiple functions, conventional genetic screens have identified only a small number of direct Ran targets (Demeter and Sazer, 1998). However, our recent computational model of the fission-yeast nucleus (Lim et al., 2007) made experimentally testable predictions about the possible roles for the ER in NE growth and the SPB in nuclear-shape changes during division.
Results
The Ran-GTPase system is required for symmetric nuclear division
To precisely map with respect to cell-cycle events the previously described loss of NE integrity in the temperature-sensitive RanGEF mutant pim1-d1 (Demeter et al., 1995; Sazer and Nurse, 1994), we monitored nuclear size, shape and integrity in cells producing GFP-Nsp1p and GFP–NLS–β-gal (NLS represents nuclear localization signal) (Yoshida and Sazer, 2004) to visualize the nuclear periphery and the nuclear volume, respectively (see Materials and Methods).
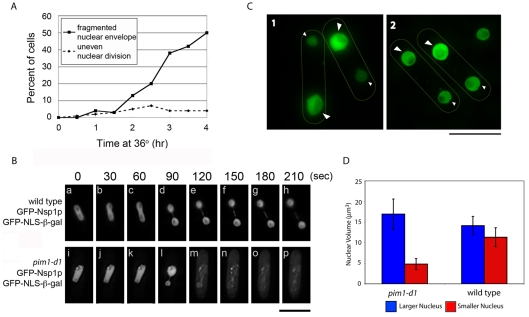
Consistent with previous results using fixed cells (Demeter et al., 1995), a live-cell timecourse analysis showed that, after 4 hours at the restrictive temperature, approximately 50% of pim1-d1 mutant cells had fragmented NEs (Fig. 1A). Strikingly, approximately 7% of the population had one nucleus that was larger than normal and one smaller than normal at incubation times longer than 2 hours (Fig. 1A).
Fig. 1.
pim1-d1 (RanGEF) mutant cells undergo asymmetric nuclear division followed by NE breakage. (A,B) pim1-d1 and wild-type cells with GFP-Nsp1p and GFP–SV40-NLS–β-gal were grown to log phase at 25°C and shifted to 36°C for the indicated times. (A) The number of pim1-d1 mutant cells with fragmented NEs (squares, solid lines) or uneven-sized daughter nuclei (diamonds, dashed lines) were counted using fluorescence microscopy of living cells every 30 minutes after a temperature shift to 36°C for 4 hours. (B) Nuclear division was monitored by time-lapse deconvolution microscopy of living cells that were pre-incubated at 36°C for approximately 2 hours and then maintained at 36°C during observation using a heated stage. Wild-type cells (a-h) undergo symmetric division of the nucleus (see supplementary material Movie 1) but pim1-d1 cells (i-p) (see supplementary material Movie 2) undergo uneven nuclear division (l) and then break, after which the previously nuclear GFP–NLS–β-gal is distributed throughout the cell (m-p). (C) pim1-d1 (1) or wild-type (2) cells expressing the nuclear reporter GFP–SV40-NLS–β-gal were grown to log phase at the permissive temperature of 25°C and then shifted to the restrictive temperature of 36°C for 4 hours. Large arrowheads indicate the larger nucleus; small arrowheads indicate the smaller nucleus. (D) The nuclear volumes of the larger and smaller nuclei in binucleated cells as shown in C (n≥22). Error bars represent s.e.m. Scale bars: 10 μm.
Time-lapse microscopy revealed that this uneven nuclear division was not a rare terminal phenotype but a transient state that precedes NE breakage: wild-type cells underwent roughly equal nuclear division at mitosis (Fig. 1Ba-h; supplementary material Movie 1) but pim1-d1 mutant cells (Fig. 1Bi-p; supplementary material Movie 2) consistently underwent an asymmetric nuclear division (Fig. 1Bl) immediately followed by loss of NE integrity (Fig. 1Bm-p). The Ran-GTPase system is therefore necessary for the symmetrical division of the nucleus during mitosis and for the integrity of the NE early in anaphase B.
After asymmetric nuclear division in the pim1-d1 mutant, the larger and smaller nuclei are dramatically different in volume
We quantified nuclear volume (see Materials and Methods) in the sister nuclei of binucleated wild-type cells and found a small difference of 2.7±1.5 μm3 between the average volumes of the larger (14.1 μm3) and smaller (11.3 μm3) nuclei (Fig. 1C2,D). Because the largest difference in nuclear volume in wild-type cells was 1.8-fold, we defined asymmetric division as a ≥twofold difference in size between the larger and smaller nuclei. Using this criterion to examine asymmetric nuclear division in binucleated pim1-d1 mutant cells at 36°C, the average volume difference was 12.1±3.4 μm3 (Fig. 1C1,D), indicating a striking asymmetry that is uniform within the population.
In the pim1-d1 (Ran GEF) temperature-sensitive mutant, the NE breaks immediately after the nucleus divides asymmetrically, the point in the cell cycle at which there is a rapid 26% increase in NE surface area (Lim et al., 2007).
The SPB synthesized in pim1-d1 mutant cells at the restrictive temperature preferentially localizes in the smaller nucleus after asymmetric division
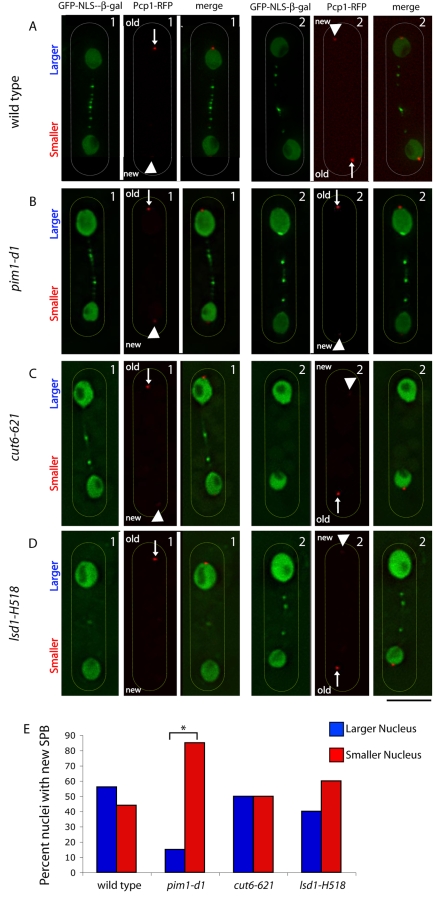
To test our prediction that the SPB influences nuclear size and shape during division (Lim et al., 2007), we monitored the localization of the new SPB, assembled in pim1-d1 cells at the restrictive temperature, with respect to nuclear size after asymmetric division, using a previously described experimental strategy to distinguish between the new and the old SPB (Grallert et al., 2004). Wild-type (Fig. 2A) and pim1-d1 mutant (Fig. 2B) cells expressing GFP–NLS–β-Gal (Yoshida and Sazer, 2004) and Pcp1p-RFP (Grallert et al., 2004), which encodes the SPB component Pcp1p fused to a slow-folding version of red fluorescent protein (RFP), were starved and then re-fed under conditions that enrich for binucleated cells (see Materials and Methods). We found no correlation between SPB age and nuclear size in wild-type cells (Fig. 2A,E); however, in pim1-d1 cells in which the new and old SPB could be clearly distinguished (Fig. 2B), the new SPB preferentially localized in the small nucleus (Fig. 2B,E). This contrasts with the random distribution of the new SPB between the large and small nucleus in cut6-621 (Fig. 2C,E) or lsd1-H518 (Fig. 2D,E) mutants (Saitoh et al., 1996), which maintain NE integrity. These mutants have defects in fatty-acid metabolism, but the relationship between this biochemical defect and the morphological and other defects in the cells is not known.
Fig. 2.
After asymmetric division in pim1-d1 mutant cells, the new SPB is preferentially associated with the smaller nucleus. Wild-type (A), pim1-d1 (B), cut6-621 (C) or lsd1-H518 (D) cells with the nuclear reporter GFP–SV40-NLS–β-gal and the SPB reporter Pcp1p-RFP were grown to log phase, starved of nitrogen for 16 hours at the permissive temperature of 25°C, resuspended in complete medium and shifted to the restrictive temperature of 34°C for 4 hours (A,B) or resuspended in complete medium at 25°C for 30 minutes and shifted at 36°C for 4 hours (C,D). The larger and smaller nuclei were distinguished in binucleated cells in which the difference in RFP fluorescence intensity allowed identification of the new and old SPB. Arrowheads indicate new SPB; arrows indicate old SPB; duplicate cells for each sample are indicated as 1 and 2. (E) The proportion of cells in which the new SPB localized to the larger or smaller nucleus was determined (n≥30). *P=0.0005. Scale bar: 5 μm.
Slowing down spindle elongation influences nuclear division in pim1-d1 mutant cells
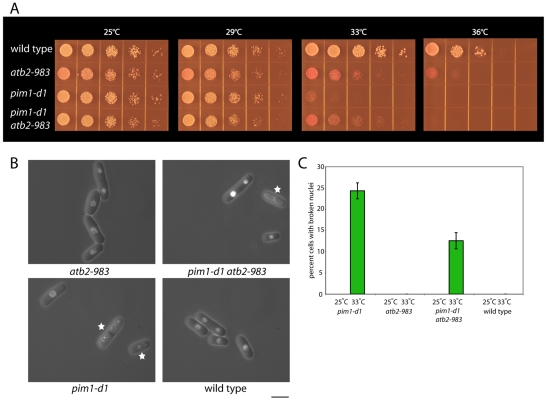
To test the hypothesis that the pim1-d1 mutant undergoes NE breakage because it is unable to efficiently increase NE area at mitosis, we slowed its rate of spindle elongation by introducing the temperature-sensitive atb2-983 mutation in α2-tubulin, which doubles the length of anaphase B (Asakawa et al., 2006).
When wild-type, pim1-d1 or atb2-983 single-mutant, or pim1-d1 atb2-983 double-mutant strains were incubated at a range of temperatures from 25°C to 36°C, we observed partial suppression of the pim1-d1 growth defect in the double mutant at 33°C (Fig. 3A). To test whether this rescue correlated with a reduction in NE breakage, we quantified NE integrity in cells incubated at 25°C or 33°C (Fig. 3B). At 25°C, none of the cells in these four strains had broken NEs (Fig. 3C). At 33°C, no wild-type or atb2-983 cells had broken NEs. For pim1-d1 cells, 24.2% had broken NEs, but this proportion was decreased by half to 12.6% in the pim1-d1 atb2-983 mutant (Fig. 3B,C). This rescue was not due to preventing entry into or completion of mitosis, because, at 33°C, the percentage of binucleated cells in atb2-983 cells was 13.7%, compared with 10.0% in wild-type cells. We also found that introduction of the atb2-983 mutation did not reduce the frequency of asymmetric nuclear division in pim1-d1 cells (4.3±0.6% in pim1-d1, 4.7±0.4 in pim1-d1 atb2-983).
Fig. 3.
Slowing spindle elongation partially rescues the defects in growth and NE integrity in pim1-d1 cells. (A) Wild-type, atb2-983, pim1-d1 and pim1-d1 atb2-983 double-mutant strains were grown in EMM, spotted onto EMM plates with the vital dye phloxine B, which accumulates in dead cells turning them dark pink, and incubated at the indicated temperatures for 3-5 days. (B) Wild type, pim1-d1, atb2-983 and pim1-d1 atb2-983 double-mutant strains containing the nuclear reporter GFP–SV40-NLS–β-gal and the NE reporter GFP-Nsp1p were incubated at 33°C for 15 hours on EMM plates and NE integrity monitored by fluorescence microscopy. Star indicates cell with broken nucleus. (C) Quantification of the proportion of cells in which the NE is broken (n=200). The error bars represent s.e.m. Scale bar: 5 μm.
NE breakage in pim1-d1 cells is partially rescued by proliferation of the ER membrane
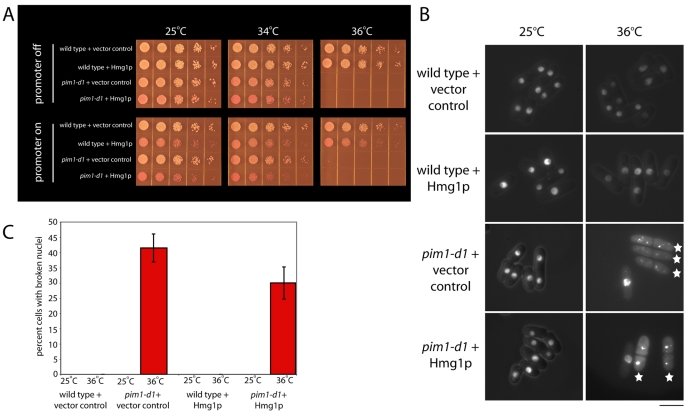
Because the ER and the NE comprise a single membrane system, the ER is the most likely source of the rapid 26% increase in NE area needed to accommodate the division of the nucleus into two smaller nuclei at anaphase B (Lim et al., 2007). To test the possibility that the ER represents the proposed lipid reservoir necessary for the rapid increase in NE area at mitosis (Lim et al., 2007), we investigated whether artificially increasing the area of ER membrane by overproduction of the ER-membrane protein HMG-CoA reductase (encoded by the Saccharomyces cerevisiae HMG1 gene) (Lum and Wright, 1995) would protect pim1-d1 mutant cells from NE breakage at mitosis.
HMG1 expression decreased viability (Fig. 4A) but did not impact the NE in wild-type or pim1-d1 cells at 25°C (Fig. 4B,C). It did reduce the proportion of pim1-d1 cells with fragmented NEs at 36°C from 41.5±4.5% in the vector control to 30.0±5.3% (Fig. 4B,C) but did not increase cell viability when compared to the vector control, as would be expected owing to its toxicity. HMG1 did not protect the NE by preventing entry into or completion of mitosis; after 27 hours of expression at 25°C, the frequency of binucleated cells was 15.3% compared to 15.8% in the vector control. We also found that HMG1 overexpression for 30 hours (26 hours at 25°C and 4 hours at 34°C) did not rescue the defect of asymmetric nuclear division in pim1-d1 cells (7.8±0.2% assymetric in pim1-d1, 7.0±1.0% assymetric in pim1-d1 + HMG1).
Fig. 4.
Increasing ER membrane by overexpression of HMG1, which encodes the S. cerevisiae ER-membrane protein Hmg1p (HMG CoA reductase), partially rescues the NE-integrity defects of pim1-d1 cells. (A) Wild-type or pim1-d1 cells carrying the plasmid pPL283 (Lum and Wright, 1995) expressing S. cerevisiae HMG1 from the thiamine-repressible nmt1 gene promoter or a vector control were grown in EMM with thiamine to log phase. HMG1 expression was induced by removal of thiamine for a total of 26 hours, the first 22 of which were at the permissive temperature of 25°C and the last 4 of which were at the restrictive temperature of 36°C. Cells were spotted onto EMM plates with the vital dye phloxine B, which accumulates in dead cells turning them dark pink, with or without thiamine and incubated at the indicated temperatures for 3-5 days. (B) The wild-type and pim1-d1 strains described in A with the nuclear reporter GFP–SV40-NLS–β-gal and the NE reporter GFP-Nsp1p were incubated without thiamine at 25°C for 22 hours and then for 4 additional hours at 25°C or 36°C, and NE integrity monitored by fluorescence microscopy. Stars indicate cells with a broken nucleus. (C) Quantification of the proportion of cells in B in which the NE is broken (n=200). The error bars represent s.e.m. Scale bar: 5 μm.
To investigate whether the rescue of NE breakage by ER proliferation or by slowing spindle elongation was additive, we overexpressed HMG1 in pim1-d1 atb2-983 double-mutant cells using experimental conditions in which we could clearly monitor the influence of HMG1 overexpression and of the two different temperature-sensitive mutations on NE integrity (HMG1 was overexpressed for 30 hours, 26 hours at 25°C and 4 hours at 36°C). Consistent with our previous results, we found that, under these conditions, HMG1 overexpression reduced NE breakage in pim1-d1 cells by approximately 10 percentage points, from 46.0±2.1% to 36.6±1.4%. Introduction of the atb2-983 mutation into this strain further reduced the frequency of NE breakage to 20.8±1.0%.
NE breakage in pim1-d1 cells is partially rescued by altering ER-membrane conformation
To more specifically test the possibility that the nuclear-division defects in pim1-d1 mutants are caused by an inability to add sufficient lipid to the NE at mitosis, we sought a way to specifically increase the area of NE membrane. Because reticulon-family proteins stabilize the tubular form of the ER at the expense of the sheet form in vivo and in vitro (Voeltz et al., 2006), we reasoned that they were good candidates for this purpose. We deleted the rtn1 gene [also known as cwl1 (Godoy et al., 1996)], which encodes the only reticulon-like-domain-containing protein in the S. pombe genome (Oertle et al., 2003) and found that it was not essential for viability (Fig. 5B). Deletion of rtn1 did not impair the growth rate at any temperature (Fig. 5B) or alter cell-cycle progression, because the proportion of binucleated cells at 34°C was 16.5% in Δrtn1 and 13.2% in wild-type cells. However, deletion of rtn1 rescued growth of the pim1-d1 mutant at 34°C (Fig. 5B).
Fig. 5.
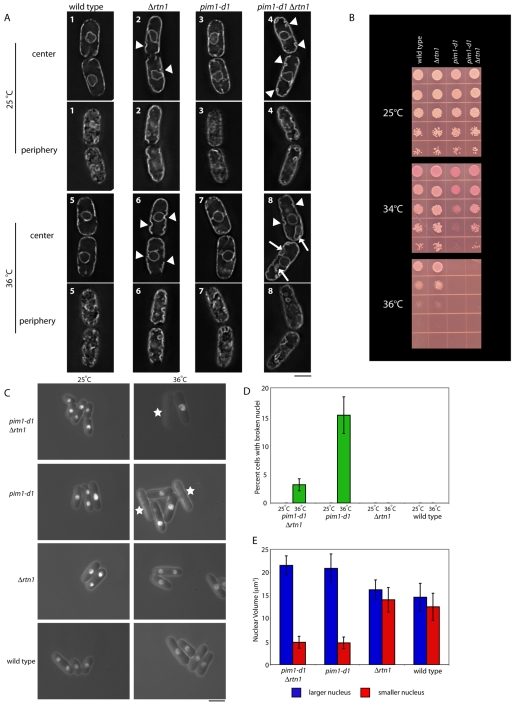
Changing ER-membrane conformation by deletion of rtn1, which encodes a reticulon ortholog, partially rescues the growth and NE breaking in pim1-d1 cells. (A) Wild-type, pim1-d1, Δrtn1 and pim1-d1 Δrtn1 double-mutant strains with the ER-membrane marker GFP-13g6 (Brazer et al., 2000) were incubated at 25°C in EMM for 22 hours (cells 1-4) or incubated at 25°C in EMM for 20 hours and then at 36°C for 2 hours (cells 5-8), and then observed using deconvolution microscopy to visualize ER conformation at the cell center or the cell periphery. Arrowhead indicates ER invagination; arrow indicates abnormal ER structure. (B) Wild-type, pim1-d1, Δrtn1 and pim1-d1 Δrtn1 double-mutant strains were spotted onto EMM plates with the vital dye phloxine B, which accumulates in dead cells turning them dark pink, and incubated at the indicated temperatures for 3-5 days. (C) Wild type, pim1-d1, Δrtn1 and pim1-d1 Δrtn1 double-mutant strains with the nuclear reporter GFP–SV40-NLS–β-gal and the NE reporter GFP-Nsp1p were incubated at the indicated temperatures, and NE integrity was monitored by fluorescence microscopy. Stars indicate cells with a broken nucleus. (D) Quantification of the proportion of cells in C in which the NE is broken (n=200). (E) Wild-type, pim1-d1, Δrtn1 and pim1-d1 Δrtn1 double-mutant strains with the nuclear reporter GFP–SV40-NLS–β-gal were incubated at 36°C for 2 hours, and nuclear volume of the larger and smaller nucleus in binucleated cells (n=20) determined using deconvolution microscopy. The error bars represent s.e.m. Scale bars: 5 μm.
Localization of the integral ER-membrane proteins GFP-13g6 (Brazer et al., 2000) in wild-type and pim1-d1 cells at the permissive temperature confirmed the previously described ER localization in wild-type cells (Pidoux and Armstrong, 1993) at the cell cortex, surrounding the nucleus and in a small number of tubules that connect these two ER forms (Fig. 5A1,A3). Deletion of rtn1 caused two major changes (Fig. 5A, compare cells 1 and 2, 3 and 4): (1) portions of the ER dissociated from the plasma membrane, forming cytoplasmic invaginations; and (2) the reticular organization at the cell periphery was significantly altered. This change is identical to that seen in budding-yeast reticulon mutants (Voeltz et al., 2006) and is consistent with a conversion of tubular ER to sheet ER at the cell periphery.
We then investigated whether deletion of rtn1 influenced the NE in the pim1-d1 background by monitoring the conformation of the ER (Fig. 5A), the integrity of the NE (Fig. 5C,D) and the volume of the sister nuclei (Fig. 5E) in binucleated pim1-d1 Δrtn1 double-mutant cells.
NE breakage in pim1-d1 cells that were incubated at the restrictive temperature was partially rescued by Δrtn1 (Fig. 5C,D). No wild-type, Δrtn1, pim1-d1 or pim1-d1 Δrtn1 cells at the permissive temperature of 25°C and no Δrtn1 or wild-type cells at the restrictive temperature of 36°C had broken NEs (Fig. 5C,D). After 3 hours at 34°C, the temperature at which Δrtn1 partially rescued the loss of viability (Fig. 5B), 20.7% of pim1-d1 mutants but only 4.5% of pim1-d1 Δrtn1 double-mutant cells lost NE integrity. After 2 hours at 36°C (Fig. 5C), Δrtn1 lowered NE breakage from 15.3% to 3.2% (Fig. 5D). At the restrictive temperature, both the absolute and relative sizes of the larger and the smaller nuclei in the pim1-d1 mutant were nearly identical to those of pim1-d1 Δrtn1 double-mutant cells (Fig. 5E).
We investigated the possible relationship between changes in ER conformation and NE integrity by monitoring ER-membrane morphology under conditions in which the Δrtn1 mutation had the greatest influence on NE integrity: 36°C for 2 hours (Fig. 5A, cells 5-8). In pim1-d1 cells, ER conformation resembled that of wild type at both the cell center and periphery (Fig. 5A). However, in pim1-d1 Δrtn1 cells, ER abnormalities, including invaginations from the cell periphery and abnormal sheet-like structures close to or in contact with the NE, were more frequent than in the Δrtn1 strain (Fig. 5A).
Discussion
The Ran-GTPase system is required for symmetric nuclear division and NE integrity at mitosis
We have previously shown that the Ran-GTPase system in fission yeast is essential for the integrity of the NE and report here that, in the pim1-d1 (RanGEF) temperature-sensitive mutant, the NE breaks immediately after the nucleus divides asymmetrically at early anaphase B (Fig. 1), the point in the cell cycle at which there is an increased demand for rapid NE growth (Lim et al., 2007).
The SPB influences nuclear size and shape during nuclear division
Consistent with the possibility that symmetric nuclear division depends on the SPB (Lim et al., 2007), we found that the SPB that is synthesized when Ran-GTPase function is compromised by the pim1-d1 (RanGEF) mutation is preferentially associated with the smaller daughter nucleus after asymmetric nuclear division (Fig. 2). The Ran GTPase is therefore directly or indirectly required for proper SPB structure and/or function, and the inability of pim1-d1 cells to maintain nuclear size and shape during division is likely to be caused by this as-yet-unidentified SPB defect. The structure and function of animal-cell centrosomes, which are functionally equivalent to yeast SPBs, are also influenced by the Ran-GTPase system, but in those cases reduction of RanGTP promotes loss of centrosome cohesion and thereby the production of multipolar spindles (Di Fiore et al., 2004).
Asymmetric division does not necessarily lead to NE breakdown
In the pim1-d1 (RanGEF) temperature-sensitive mutant, the NE breaks immediately after the nucleus divides asymmetrically, the point in the cell cycle at which there is a rapid 26% increase in NE surface area (Lim et al., 2007). However, under conditions in which NE fragmentation is reduced by slowing spindle elongation, increasing ER membrane or changing ER structure, we found no influence on the asymmetry of nuclear division. In contrast to the NE breakage after asymmetric division in pim1-d1 cells, the NE remains intact in cut6 and lsd1 single-mutant strains that have defects in fatty-acid metabolism and undergo unequal nuclear division by an unknown mechanism (Saitoh et al., 1996). Fission-yeast strains with mutations in the kinetochore proteins Mis6p or Mis12p undergo asymmetric nuclear division due to chromosome mis-segregation, but their NEs also remain intact (Goshima et al., 1999). Chromosome mis-segregation is unlikely to account for the asymmetric nuclear division of pim1-d1 cells because, at the restrictive temperature, these cells consistently underwent uneven nuclear division (Fig. 1B) but undergo equal chromosome segregation (Sazer and Nurse, 1994).
These data are consistent with the hypothesis that, in fission yeast, the Ran-GTPase system independently influences nuclear division via the SPB, and NE growth and integrity via the ER.
Regulation of NE growth and its coordination with spindle elongation is required for maintenance of NE integrity at mitosis
The observation that the pim1-d1 mutant undergoes NE breakage at the point in mitosis when there is a sharp increase in the demand for increased NE area (Lim et al., 2007) suggests that lipid might not be added in sufficient quantity and/or at sufficient speed to keep pace with spindle elongation. Consistent with this possibility, we previously showed that loss of NE integrity correlates temporally with and depends on the passage of cells through mitosis, and we show here that there are three conditions in which the loss of NE integrity and/or loss of viability in pim1-d1 mutant cells were partially rescued: slowing spindle elongation by the atb2-983 mutation in α2-tubulin (Fig. 3), increasing ER membrane by overproduction of the ER-membrane protein HMG CoA reductase (Fig. 4) and changing ER morphology from tubular to sheet by deletion of rtn1, which encodes a reticulon-like protein (Oertle et al., 2003) (Fig. 5). Because the Ran GTPase participates in many cellular processes (Quimby and Dasso, 2003) other than NE structure, it is not surprising that these nucleus-specific manipulations result in only partial rescue of pim1-d1 cell viability in the case of atb2-983 and rtn1 deletion, and no rescue in the case of HMG1 overexpression, which is somewhat toxic even at 25°C.
Together, these studies demonstrate the importance of proper regulation of increases in NE area and the coordination of these increases with spindle elongation in mitosis. Furthermore, they suggest that conformational changes in the ER-NE membrane system or lipid redistribution within it are crucial for these processes.
Fission yeast has a single reticulon ortholog, named rtn1, which is not essential for viability or nuclear division. In contrast to the situation in S. cerevisiae, deletion of the single rtn1 gene in fission yeast changes the conformation of the tubular ER at the cell periphery and causes it to detach from the cell periphery (Fig. 5). These conformational changes alter the distribution of lipid between the tubular and sheet forms, thereby promoting lipid incorporation into the NE portion of the ER network. They also facilitate the conversion of tubular to sheet ER at the time of nuclear division during closed mitosis, because deletion of rtn1 partially protects pim1-d1 nuclei from breakage upon spindle elongation and partially restores viability (Fig. 5). These data demonstrate that mitotic increases in NE area are normally coordinated with spindle elongation, and they are consistent with the hypothesis that lipid redistribution within the ER-NE network is crucial for these processes. It is notable that Δrtn1 cells do not have nuclei that are larger than normal, suggesting that an as-yet-unknown mechanism that regulates NE growth during interphase functions normally in the absence of Rtn1p.
Is there an evolutionarily conserved role for the Ran-GTPase system in NE structure?
The Ran-GTPase system influences NE structure and assembly in both open and closed mitosis, and is also required for nuclear pore complex (NPC) assembly (Demeter et al., 1995; Hetzer et al., 2005) (reviewed by Quimby and Dasso, 2003), but it is not know whether it does so by means of a common Ran target, because no nucleus-specific targets have been identified either in vivo or in vitro. In meiotic egg-extract systems, the Ran-GTPase system is required for NE vesicle targeting to the chromosomes after mitosis and subsequent fusion to reform the NE in vitro (reviewed by Hetzer et al., 2005; Mattaj, 2004). Ran-mediated vesicle fusion was also proposed to be essential for NPC assembly and NE growth in budding yeast (Ryan et al., 2003), but the kinetics and magnitude of NE growth at mitosis in yeast make it unlikely that it is mediated by vesicle fusion (Hetzer et al., 2005; Ryan et al., 2003). Alternatively, reports that some inner-NE-specific proteins relocalize to the tubular ER upon NE breakdown at mitosis in animal cells (Ellenberg et al., 1997; Yang et al., 1997) are consistent with previously proposed, and recently demonstrated, conversion of the sheet ER of the NE into tubular ER during open mitosis (Hetzer et al., 2005; Mattaj, 2004; Puhka et al., 2007). Conversely, we show that changing ER conformation from tubular to sheet in the pim1-d1 (RanGEF) mutant facilitates the rapid incorporation of large amounts of lipid into the fission-yeast NE during closed mitosis and propose that it might also be crucial for NE reformation after open mitosis in animal cells (Mattaj, 2004). In fact, while this study was in progress, Hetzer's group reported that tubular ER is required for NE reformation in vitro (Anderson and Hetzer, 2007) in a process mediated by chromatin. By contrast, during nuclear division in fission yeast, the mitotic chromosomes are highly condensed and, because at this stage of the cell cycle they are not in direct contact with the NE, are unlikely to directly mediate the increase in NE area. However, our data on the importance of reticulons in regulating NE growth are consistent with a recent report from Anderson and Hetzer showing that the removal of reticulons from tubular ER is rate limiting for animal-cell NE reformation in vitro (Anderson and Hetzer, 2008). Redistribution of lipid within the NE-ER network is therefore likely to represent a common step in the mitotic-specific changes to the NE during both open and closed mitosis.
Materials and Methods
Yeast strains, strain construction and cell culture
Standard methods and genetic techniques were used (Moreno et al., 1991), and strains are described in Table 1. Spotting experiments were performed by growing cells in Edinburgh minimal medium (EMM) to mid-log phase and spotting 106 cells and fivefold dilutions onto EMM plates with the pink vital dye phloxine B (Sigma) (that is excluded from living cells), supplements and thiamine as indicated. Gene expression from the nmt1 gene promoter (Forsburg and Sherman, 1997; Maundrell, 1990) was repressed by growth in EMM with 5 μg/ml thiamine. The Δrtn1 (rtn1-null) strain was generated by PCR-based targeted gene replacement using a natMX4 drug-resistance cassette (Bahler et al., 1998; Goldstein and McCusker, 1999). S. cerevisiae HMG1 expression was regulated by the nmt1 promoter on plasmid pPL283 (Lum and Wright, 1995).
Table 1.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| SS7 | h– leu1-32 | Our stock |
| SS446 | h– leu1-32 ura4-D18 ade6-M210 | Our stock |
| SS482 | h– leu1-32 ade6-M216 int::pREP3X-SV40NLS-GFP-lacZ | Our stock |
| SS483 | h– pim1-d1 leu1-32 ade6-M216 int::pREP3X-SV40NLS-GFP-lacZ | Our stock |
| SS749 | h– pim1-d1 leu1-32 ura4-D18 ade6-M210 | Our stock |
| SS777 | h+ pim1-d1 leu1-32 ura4-D18 int::pREP82X-GFP-nsp1 int::pREP3X-SV40NLS-GFP-lacZ | Our stock |
| SS817 | h– leu1-32 ura4-D18 int::pREP82X-GFP-nsp1 int::pREP3X-SV40NLS-GFP-lacZ | Our stock |
| SS1652 | h– pim1-d1 ura4-D18 ade6-M210 | Our stock |
| SS1758 | h– pim1-d1 leu1-32 | This study |
| SS1876 | h– pcp1.RFP::kanR | Iain Hagan, Paterson Institute for Cancer Research, Manchester, UK |
| SS1878 | h+ pim1-d1 leu1-32 ura4-D18 int::pREP82X-GFP-nsp1 int::pREP3X-SV40NLS-GFP-lacZ pREP3X | This study |
| SS1881 | h+ pim1-d1 leu1-32 ura4-D18 int::pREP82X-GFP-nsp1 int::pREP3X-SV40NLS-GFP-lacZ pPL283 (pREP3X-HMG1) | This study |
| SS1901 | h+ pcp1.RFP::kanR leu1-32 ura4-D18 int::pREP82X-GFP-nsp1 int::pREP3X-SV40NLS-GFP-NLS-lacZ | This study |
| SS1903 | h+ pim1-d1 pcp1.RFP::kanR leu1-32 ura4-D18 int::pREP82X-GFP-nsp1 int::pREP3X-SV40NLS-GFP-NLS-lacZ | This study |
| SS1933 | h– pim1-d1 atb2-983 leu1-32 ura4-D18 int::pREP82X-GFP-nsp1 int::pREP3X-SV40NLS-GFP-lacZ | This study |
| SS1938 | h+ atb2-983 leu1-32 ura4-D18 int::pREP82X-GFP-nsp1 int::pREP3X-SV40NLS-GFP-lacZ | This study |
| SS1952 | h+ rtn1::natMX4 leu1-32 ura4-D18 ade6-216 pREP41X-GFP-13g6 | This study |
| SS1962 | h+ pim1-d1 rtn1::natMX4 leu1-32 ura4-D18 ade6-M216 int::pREP3X-SV40NLS-GFP-lacZ | This study |
| SS1969 | h– rtn1::natMX4 leu1-32 ura4-D18 ade6-M216 int::pREP3X-SV40NLS-GFP-lacZ | This study |
| SS1756 | h+ cut6-621 leu1-32 ura4-D18 int::pREP82X-GFP-nsp1 int::pREP3X-SV40NLS-GFP-lacZ | Our stock |
| SS1757 | h– lsd1-H518 leu1-32 int::pREP82X-GFP-nsp1 int::pREP3X-SV40NLS-GFP-lacZ | Our stock |
| SS2058 | h+ pcp1.RFP::kanR leu1-32 ura4-D18 ade6-M210 | This study |
| SS2062 | h– leu1-32 ura4-D18 ade6-M210 pREP41X-GFP-13g6 | This study |
| SS2063 | h– pim1-d1 leu1-32 ura4-D18 ade6-M210 pREP41X-GFP-13g6 | This study |
| SS2064 | h– pim1-d1 rtn1::natMX4 leu1-32 ura4-D18 ade6- pREP41X-GFP-13g6 | This study |
| SS2065 | h– rtn1::natMX4 leu1-32 ura4-D18 ade6-M210 | This study |
| SS2066 | h– pim1-d1 rtn1::natMX4 leu1-32 ura4-D18 | This study |
| SS2079 | h– leu1-32 ura4-D18 int::pREP82X-GFP-nsp1 int::pREP3X-SV40NLS-GFP-lacZ pREP3X | This study |
| SS2080 | h– leu1-32 ura4-D18 int::pREP82X-GFP-nsp1 int::pREP3X-SV40NLS-GFP-lacZ pPL283 (pREP3X-HMG1) | This study |
| SS2131 | h+ cut6-621 pcp1.RFP::kanR leu1-32 ura4-D18 int::pREP82X-GFP-nsp1 int::pREP3X-SV40NLS-GFP-lacZ | This study |
| SS2132 | h+ lsd1-H518 pcp1.RFP::kanR leu1-32 ura4-D18 ade6-M210 int::pREP82X-GFP-nsp1 int::pREP3X-SV40NLS-GFP-lacZ | This study |
Fluorescence microscopy
Nuclear-division and -integrity analysis
Cells expressing the fluorescently labeled NPC protein GFP-Nsp1p, to visualize the NE, and/or a fluorescently labeled soluble nuclear protein GFP–NLS–β-gal, to visualize the nuclear compartment (Yoshida and Sazer, 2004), were examined using either a Zeiss Axioskop fluorescence microscope, from which images were captured by a DVD 1300 black and white CCD camera using QED software (Media Cybernetics, Silver Springs, MD), or a DeltaVision deconvolution microscope system (Applied Precision, Issaquah, WA), to collect images of living cells in a heated cell chamber maintained at 36°C, using a Photometrics CoolSnap HQ camera (Roper Scientific, Tucson, AZ). For time-lapse studies, early mitotic cells were identified on the basis of their oblong-shaped nuclei, and images were collected every 5 seconds until nuclear division was completed. For analysis of nuclear volume and SPB localization, stacks of 0.2 μm z sections were projected two dimensionally using the maximum-intensity protocol and analyzed using SoftWoRx3.3.
Nuclear-volume analysis
To calculate nuclear volume in live cells independent of absolute fluorescence intensity, the distribution of GFP–NLS–β-gal was used to define the periphery of the nucleus by creating two-dimensional (2D) polygons in each z section that were visually verified and then assembled into a 3D model of the nucleus upon which nuclear volume was calculated.
Distinguishing the old and the new SPBs
Using a previously described protocol (Grallert et al., 2004) with minor modifications [including substitution of MSL (Egel et al., 1994) for EMM2 medium], strains with GFP–NLS–β-gal (Yoshida and Sazer, 2004) and Pcp1-RFP (the SPB protein Pcp1p fused to a slow-folding version of RFP) (Grallert et al., 2004) were grown at the permissive temperature of 25°C to log phase, starved of nitrogen at the permissive temperature of 25°C for 16 hours, then re-fed at the indicated restrictive temperature for 4 hours. Cells in which the old (brightly fluorescent) and new (dimly fluorescent) SPB could be clearly distinguished by differences in RFP signal intensity were used to determine the relationship between SPB inheritance and nuclear size.
Statistical analysis
A chi-square test (χ2) was performed to determine whether the distribution of the new SPB between the large and small nucleus was significantly different from random. Because only two phenotypic classes (small or large nucleus) were studied, Yates' correction for continuity was applied. Using 0.05 as the alpha (α) level of significance and one degree of freedom, the probability (P) was calculated for each sample.
Supplementary Material
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/122/14/2464/DC1
We thank Makoto Umeda for contributions and advice, and Richard Atkinson, Ursula Fleig and Xiangwei He for manuscript comments and technical advice. This material is based in part on work supported by the National Science Foundation under Grants No. 0344471 and 0744945 (both to S.S.). Any opinions, findings and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation. Y.G. was supported by National Institutes of Health grants IMSD R25 GM56929 and 5F31GM 076862, and N.N.P. was supported by National Institutes of Health training grant T32 DK07328. Deposited in PMC for release after 12 months.
References
- Anderson, D. and Hetzer, M. (2007). Nuclear envelope formation by chromatin-mediated reorganization of the endoplasmic reticulum. Nat. Cell Biol. 9, 1160-1166. [DOI] [PubMed] [Google Scholar]
- Anderson, D. J. and Hetzer, M. W. (2008). Reshaping of the endoplasmic reticulum limits the rate for nuclear envelope formation. J. Cell Biol. 182, 911-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakawa, K., Kume, K., Kanai, M., Goshima, T., Miyahara, K., Dhut, S., Tee, W. W., Hirata, D. and Toda, T. (2006). The V260I mutation in fission yeast alpha-tubulin Atb2 affects microtubule dynamics and EB1-Mal3 localization and activates the Bub1 branch of the spindle checkpoint. Mol. Biol. Cell 17, 1421-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler, J., Wu, J. Q., Longtine, M. S., Shah, N. G., McKenzie, A., 3rd, Steever, A. B., Wach, A., Philippsen, P. and Pringle, J. R. (1998). Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943-951. [DOI] [PubMed] [Google Scholar]
- Brazer, S. C., Williams, H. P., Chappell, T. G. and Cande, W. Z. (2000). A fission yeast kinesin affects Golgi membrane recycling. Yeast 16, 149-166. [DOI] [PubMed] [Google Scholar]
- Demeter, J. and Sazer, S. (1998). imp2, a new component of the actin ring in the fission yeast Schizosaccharomyces pombe. J. Cell Biol. 143, 415-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeter, J., Morphew, M. and Sazer, S. (1995). A mutation in the RCC1-related protein Pim1 results in nuclear envelope fragmentation in fission yeast. Proc. Natl. Acad. Sci. USA 92, 1436-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fiore, B., Ciciarello, M. and Lavia, P. (2004). Mitotic functions of the Ran GTPase network: the importance of being in the right place at the right time. Cell Cycle 3, 305-313. [PubMed] [Google Scholar]
- Ding, R., West, R. R., Morphew, M., Oakley, B. R. and McIntosh, J. R. (1997). The spindle pole body of Schizosaccharomyces pombe enters and leaves the nuclear envelope as the cell cycle proceeds. Mol. Biol. Cell 8, 1461-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egel, R., Willer, M., Kjaerulff, S., Davey, J. and Nielsen, O. (1994). Assessment of pheromone production and response in fission yeast by a halo test of induced sporulation. Yeast 10, 1347-1354. [DOI] [PubMed] [Google Scholar]
- Ellenberg, J., Siggia, E. D., Moreira, J. E., Smith, C. L., Presley, J. F., Worman, H. J. and Lippincott-Schwartz, J. (1997). Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J. Cell Biol. 138, 1193-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg, S. L. and Sherman, D. A. (1997). General purpose tagging vectors for fission yeast. Gene 191, 191-195. [DOI] [PubMed] [Google Scholar]
- Godoy, C., Arellano, M., Diaz, M., Duran, A. and Perez, P. (1996). Characterization of cwl1+, a gene from Schizosaccharomyces pombe whose overexpression causes cell lysis. Yeast 12, 983-990. [DOI] [PubMed] [Google Scholar]
- Goldstein, A. L. and McCusker, J. H. (1999). Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15, 1541-1553. [DOI] [PubMed] [Google Scholar]
- Goshima, G., Saitoh, S. and Yanagida, M. (1999). Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes Dev. 13, 1664-1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grallert, A., Krapp, A., Bagley, S., Simanis, V. and Hagan, I. M. (2004). Recruitment of NIMA kinase shows that maturation of the S. pombe spindle-pole body occurs over consecutive cell cycles and reveals a role for NIMA in modulating SIN activity. Genes Dev. 18, 1007-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer, M. W., Walther, T. C. and Mattaj, I. W. (2005). Pushing the envelope: structure, function, and dynamics of the nuclear periphery. Ann. Rev. Cell Dev. Biol. 21, 347-380. [DOI] [PubMed] [Google Scholar]
- Khodjakov, A., La Terra, S. and Chang, F. (2004). Laser microsurgery in fission yeast: role of the mitotic spindle midzone in anaphase B. Curr. Biol. 14, 1330-1340. [DOI] [PubMed] [Google Scholar]
- Lim, H. W. G., Huber, G., Torii, Y., Hirata, A., Miller, J. and Sazer, S. (2007). Vesicle-like biomechanics governs important aspects of nuclear geometry in fission yeast. PLoS ONE 2(9), e948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum, P. Y. and Wright, R. (1995). Degradation of HMG-CoA reductase-induced membranes in the fission yeast, Schizosaccharomyces pombe. J. Cell Biol. 131, 81-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj, I. W. (2004). Sorting out the nuclear envelope from the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 5, 65-69. [DOI] [PubMed] [Google Scholar]
- Maundrell, K. (1990). nmt1 of fission yeast. J. Biol. Chem. 265, 10857-10864. [PubMed] [Google Scholar]
- Moreno, S., Klar, A. and Nurse, P. (1991). Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795-823. [DOI] [PubMed] [Google Scholar]
- Oertle, T., Klinger, M., Stuermer, C. A. and Schwab, M. E. (2003). A reticular rhapsody: phylogenic evolution and nomenclature of the RTN/Nogo gene family. FASEB J. 17, 1238-1247. [DOI] [PubMed] [Google Scholar]
- Pidoux, A. L. and Armstrong, J. (1993). The BiP protein and the endoplasmic reticulum of Schizosaccharomyces pombe: fate of the nuclear envelope during cell division. J. Cell Sci. 105, 1115-1120. [DOI] [PubMed] [Google Scholar]
- Puhka, M., Vihinen, H., Joensuu, M. and Jokitalo, E. (2007). Endoplasmic reticulum remains continuous and undergoes sheet-to-tubule transformation during cell division in mammalian cells. J. Cell Biol. 179, 895-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quimby, B. B. and Dasso, M. (2003). The small GTPase Ran: interpreting the signs. Curr. Opin. Cell. Biol. 15, 338-344. [DOI] [PubMed] [Google Scholar]
- Ryan, K. J., McCaffery, J. M. and Wente, S. R. (2003). The Ran GTPase cycle is required for yeast nuclear pore complex assembly. J. Cell Biol. 160, 1041-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh, S., Takahashi, K., Nabeshima, K., Yamashita, Y., Nakaseko, Y., Hirata, A. and Yanagida, M. (1996). Aberrant mitosis in fission yeast mutants defective in fatty acid synthetase and acetyl CoA carboxylase. J. Cell Biol. 134, 949-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazer, S. and Nurse, P. (1994). A fission yeast RCC1-related protein is required for the mitosis to interphase transition. EMBO J. 13, 606-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tange, Y., Hirata, A. and Niwa, O. (2002). An evolutionarily conserved fission yeast protein, Ned1, implicated in normal nuclear morphology and chromosome stability, interacts with Dis3, Pim1/RCC1 and an essential nucleoporin. J. Cell Sci. 115, 4375-4385. [DOI] [PubMed] [Google Scholar]
- Toya, M., Sato, M., Haselmann, U., Asakawa, K., Brunner, D., Antony, C. and Toda, T. (2007). Gamma-tubulin complex-mediated anchoring of spindle microtubules to spindle-pole bodies requires Msd1 in fission yeast. Nat. Cell Biol. 9, 646-653. [DOI] [PubMed] [Google Scholar]
- Voeltz, G. K., Rolls, M. M. and Rapoport, T. A. (2002). Structural organization of the endoplasmic reticulum. EMBO Rep. 3, 944-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz, G. K., Prinz, W. A., Shibata, Y., Rist, J. M. and Rapoport, T. A. (2006). A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124, 573-586. [DOI] [PubMed] [Google Scholar]
- West, R. R., Vaisberg, E. V., Ding, R., Nurse, P. and McIntosh, J. R. (1998). cut11+: a gene required for cell cycle-dependent spindle pole body anchoring in the nuclear envelope and bipolar spindle formation in Schizosaccharomyces pombe. Mol. Biol. Cell 9, 2839-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey, M., Yarar, D., Giddings, T. H., Jr and Mastronarde, D. N. (1997). Nuclear pore complex number and distribution throughout the Saccharomyces cerevisiae cell cycle by three-dimensional reconstruction from electron micrographs of nuclear envelopes. Mol. Biol. Cell 8, 2119-2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, L., Guan, T. and Gerace, L. (1997). Integral membrane proteins of the nuclear envelope are dispersed throughout the endoplasmic reticulum during mitosis. J. Cell Biol. 137, 1199-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, M. and Sazer, S. (2004). Nucleocytoplasmic transport and nuclear envelope integrity in the fission yeast Schizosaccharomyces pombe. Methods Cell Sci. 33, 226-238. [DOI] [PubMed] [Google Scholar]
- Zheng, L., Schwartz, C., Magidson, V., Khodjakov, A. and Oliferenko, S. (2007). The spindle pole bodies facilitate nuclear envelope division during closed mitosis in fission yeast. PLoS Biol. 5, e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.