Summary
The β4 integrin is expressed in epithelial cells, a few other cell types and in some carcinomas. Despite this restricted expression pattern and the functional importance of β4 integrin in epithelial and carcinoma biology, little is known about how its expression is regulated. Here, we assessed the epigenetic regulation of β4 integrin based on the presence of a large CpG island in the β4-integrin gene promoter. We separated basal (β4+) and luminal (β4–) epithelial cells from the mammary glands of K14-eGFP mice and demonstrated that the β4-integrin promoter is unmethylated in basal cells and methylated in luminal cells. We also observed that expression of β4 integrin and E-cadherin is lost during the epithelial-to-mesenchymal transition (EMT) of mammary gland cells induced by transforming growth factor beta (TGFβ), which is coincident with de novo DNA methylation, a decrease in active histone modifications (H3K9Ac and H3K4me3) and an increase in the repressive histone modification H3K27me3. Furthermore, TGFβ withdrawal promotes a mesenchymal-to-epithelial transition (MET) and triggers the re-expression of β4 integrin and E-cadherin. Intriguingly, demethylation at either promoter is not obligatory for transcriptional reactivation after TGFβ withdrawal. However, both H3K9Ac and H3K4me3 modifications are restored during the MET, and H3K27me3 is reduced, strongly suggesting that reversible histone modifications rather than DNA demethylation are the predominant factors in reactivating expression of these genes. Our data indicate that complex epigenetic modifications contribute to the regulation of the β4 integrin and E-cadherin.
Keywords: Epigenetic, Epithelial mesenchymal transition, Integrin
Introduction
The α6β4 integrin (hereafter referred to as `β4' because there is only one β4 integrin) was identified originally as an epithelial-specific integrin that is expressed primarily on basal epithelial cells where it functions as a receptor for laminins of the basement membrane (Mercurio, 1995; Wilhelmsen et al., 2006). It is not expressed in most luminal epithelial cells. Expression of this integrin is maintained in some epithelial-derived cancers (carcinomas), and numerous studies have highlighted its impact on tumor formation, migration, invasion, survival and angiogenesis (Bachelder et al., 1999; Falcioni et al., 1986; Jauliac et al., 2002; Lipscomb et al., 2005; Nikolopoulos et al., 2004; O'Connor et al., 1998; Rabinovitz and Mercurio, 1997; Raymond et al., 2007; Santoro et al., 2003; Sehgal et al., 2006; Shaw et al., 1997; Trusolino et al., 2001; Weaver et al., 2002). Interestingly, however, its expression in carcinomas is heterogeneous. For example, we reported recently that ∼30% of invasive human breast carcinomas express β4 and that its expression is associated with basal-like tumors (Lu et al., 2008). Thus, β4 expression is restricted to a limited number of cell types, including basal epithelial cells, some carcinoma cells and a limited number of other cell types. Despite these restricted patterns of expression, little is known about the mechanisms that regulate β4 expression. Given the functional importance of β4 in epithelial cell biology and the impact it can have on the behavior of carcinoma cells, elucidating such mechanisms is an issue of paramount importance.
Epigenetic modification, including both DNA methylation and histone modifications, is emerging as a dynamic and efficacious mechanism that regulates gene expression during development, differentiation and carcinogenesis (Neely and Workman, 2002; Jaenisch and Bird, 2003; Jones and Baylin, 2007). The highly regulated expression of β4 in normal epithelia and its restricted expression in carcinomas make it an ideal candidate for being regulated epigenetically. Surprisingly, however, there has been little effort to investigate the potential epigenetic regulation of β4. Our analysis of the gene expressing β4 revealed that both the human and murine β4 proximal promoters contain CpG islands, which are targets for methylation. This observation prompted us to assess the epigenetic regulation of β4 using the mouse mammary gland as a model epithelium. We demonstrate here that the methylation status of the β4 promoter correlates with the spatial localization of β4 in the mammary gland. In addition, our data reveal that the expression of β4 can be regulated dynamically as a function of the epithelial-to-mesenchymal transition (EMT) induced by transforming growth factor beta (TGFβ) and that this dynamic regulation is associated with interplay between DNA methylation and histone modifications of the β4 promoter. Although β4 expression is restored during a mesenchymal-to-epithelial transition (MET), its promoter remains hypermethylated, but histone modifications are reversed. Interestingly, a similar pattern of regulation was observed for E-cadherin.
Results
Methylation status of the β4 promoter correlates with the spatial localization of the β4 integrin in the mouse mammary gland
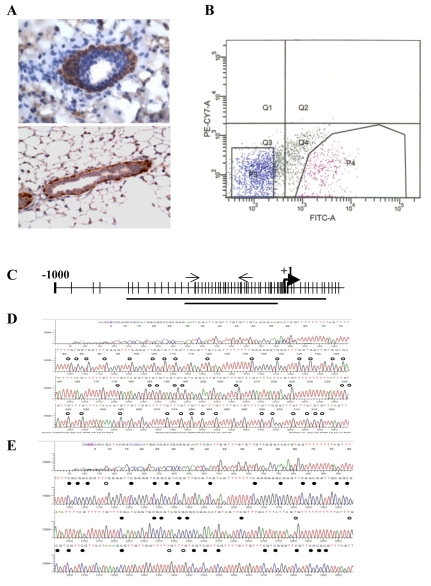
The mouse mammary epithelium comprises an outer layer of basal cells in contact with the basement membrane and an inner layer of luminal cells. Interestingly, β4 is expressed only in basal cells and not in luminal cells (top panel, Fig. 1A). In pursuit of a mechanism to account for this differential localization, we observed that the proximal promoter region of the mouse β4 gene (Takaoka et al., 1998) contains a CpG island that extends into the first exon (Fig. 1C). To assess the methylation status of the β4 promoter in the mammary gland experimentally, we obtained transgenic mice that express enhanced green fluorescent protein (eGFP) under the control of the cytokeratin 14 promoter (K14-eGFP) (Hirakawa et al., 2007) because K14 is expressed only in basal epithelial cells (bottom panel, Fig. 1A). Mammary glands were isolated from these mice, and single-cell suspensions were separated by fluorescence-activated cell sorting (FACS) into GFP+ and GFP– populations (Fig. 1B). GFP– cells include luminal cells and stromal cells that also lack β4 expression. Genomic DNA isolated from these populations was used to analyze the methylation status of a 400-bp region within the CpG island of the β4 gene promoter by bisulfite conversion, followed by nested PCR, TA cloning and sequencing. Consistent with the β4 expression pattern in the mammary gland (top panel, Fig. 1A), the β4 CpG island is unmethylated in GFP+ cells (Fig. 1D), but it is hypermethylated in GFP– cells (Fig. 1E). This inverse correlation between expression of β4 and methylation of the β4 promoter indicates that DNA methylation contributes to its restricted expression pattern in the mammary gland.
Fig. 1.
Methylation of the CpG island in the β4 promoter correlates inversely with β4 expression in the mouse mammary gland. (A) Immunohistochemistry images of β4 (top panel) and green fluorescent protein (GFP; bottom panel) both show intense staining of the basal cells of mammary gland ducts from a mouse harboring an eGFP reporter driven by the keratin K14 promoter. (B) Mammary gland single-cell suspension was prepared from the fourth mammary glands of K14-eGFP reporter mice. Basal mammary epithelial cells (P4, high eGFP signal) and luminal cells (P3, low eGFP signal) were collected individually by FACS. (C) Schematic illustration of the CpG island of the mouse β4 promoter as predicted by ABI Methyl Primer Express v1.0. Lines represent regions amplified by PCR for TA cloning. Transcription start site is indicated as +1 and arrows represent the primer set used for ChIP PCR. (D,E) Methylation status of β4 promoter CpG island in GFP+ (β4+) cells (D) and GFP– (β4–) cells (E) isolated from K14-eGFP mice. Genomic DNA from GFP+ (β4+) and GFP– (β4–) cells was treated with sodium bisulfite, and nested PCR was performed to amplify a 400-bp fragment from the CpG island of the β4 promoter, then cloned into a pGEM-T vector for sequencing. D and E represent one of the six clones randomly picked from GFP+ (β4+) and GFP– (β4–) cells, respectively. White oval, unmethylated CpG; black oval, methylated CpG.
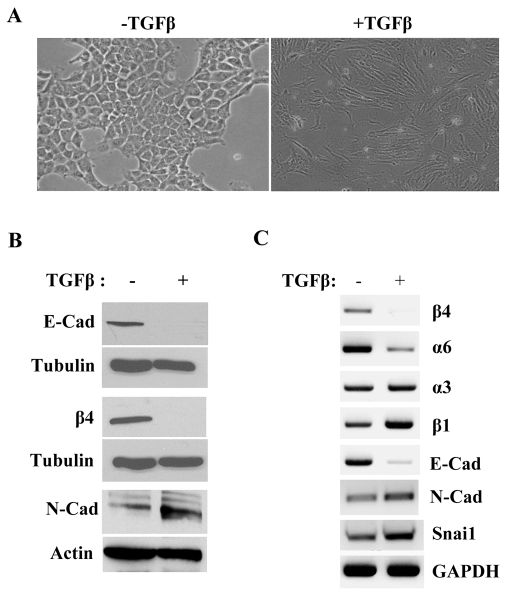
Given the relationship between β4 expression and promoter methylation that we observed in vivo and in vitro, we sought to establish an experimental system in which to study the epigenetic regulation of β4 more rigorously. NMuMG cells were derived from a mouse mammary gland (David et al., 1981) and express β4 (Fig. 2B,C). Treatment of these cells with TGFβ (5 ng/ml) results in an EMT, as reported previously (Gal et al., 2008). Indeed, we observed that TGFβ treatment for 48 hours resulted in the acquisition of a mesenchymal morphology (Fig. 2A), loss of E-cadherin expression (Fig. 2B,C) and increased expression of mesenchymal genes, including those encoding N-cadherin and Snai1 (Fig. 2C). Although Snail1 expression is evident in untreated cells, TGFβ stimulation results in a significant increase in expression that correlates with diminished E-cadherin expression (Fig. 2C). Interestingly, there is also loss of both β4 mRNA and protein expression during this EMT (Fig. 2B,C). Expression of the α6 integrin (hereafter referred to as α6) subunit, which is the heterodimeric partner of β4 (Hemler et al., 1989), is also repressed by TGFβ treatment, but expression of the α3 and β1 integrin subunits is slightly increased (Fig. 2C). These data led us to investigate further the molecular mechanisms regulating β4 expression during this EMT and to assess their relationship to the regulation of E-cadherin.
Fig. 2.
TGFβ-induced EMT in NMuMG cells alters integrin expression. (A) NMuMG cells were incubated for 2 days in either the absence (–TGFβ) or presence of 5 ng/ml TGFβ (+TGFβ), and phase-contrast photographic images were obtained. (B) Extracts from the cells described in A were immunoblotted for E-cadherin (E-Cad), β4 integrin (β4) and N-cadherin (N-Cad). Tubulin or actin served as a loading control. (C) Total RNA isolated from the cells described in A was used for RT-PCR to assess expression of specific integrin subunits, E-cadherin (E-Cad), N-cadherin (N-Cad) and Snai1. GAPDH served as a loading control.
Promoter de novo DNA methylation is involved in repressing β4 expression during a TGFβ-induced EMT
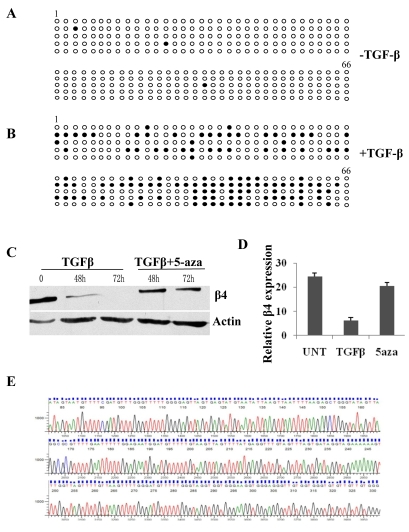
E-cadherin expression can be repressed by promoter methylation during the EMT (Dumont et al., 2008; Lombaerts et al., 2006). In the NMuMG EMT model, we also observed that de novo methylation occurred at the E-cadherin promoter (see later). Based on our in vivo data (Fig. 1), we hypothesized that TGFβ might also induce de novo methylation of the CpG island in the β4 promoter and, as a consequence, suppression of β4 expression. Genomic DNA from NMuMG cells with or without TGFβ stimulation was treated with bisulfite, followed by PCR amplification of the entire CpG island (which includes a total of 66 CpG dinucleotides), TA cloning and sequencing. We observed that the β4 promoter is completely unmethylated in NMuMG cells in the absence of TGFβ treatment (Fig. 3A). However, the β4 promoter becomes partially methylated after TGFβ treatment (Fig. 3B), which is consistent with the role of de novo methylation in transcriptional repression. To test the functional importance of this methylation, we treated NMuMG cells simultaneously with TGFβ and 5-aza-2′-deoxycytidine (5-aza), an inhibitor of DNA methylation (Jones and Taylor, 1980), for up to 72 hours. Interestingly, 5-aza treatment prevented the loss of β4 protein (Fig. 3C) and mRNA (Fig. 3D) caused by TGFβ. These data indicate that de novo DNA methylation of the β4 promoter during TGFβ treatment is essential for the repression of β4 expression.
Fig. 3.
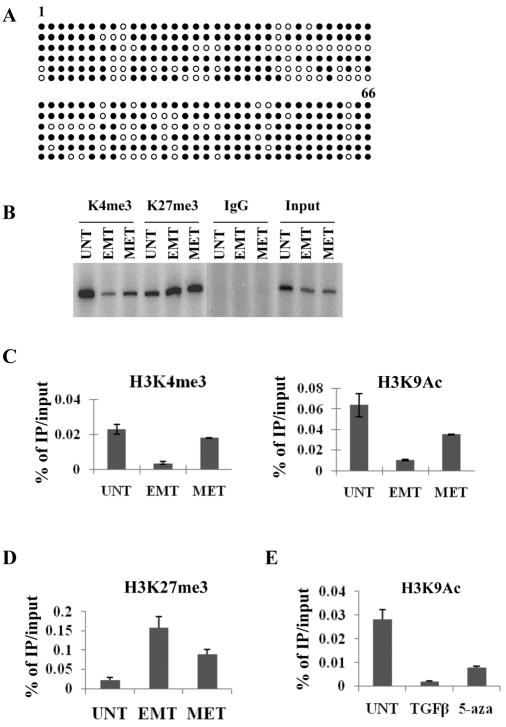
Promoter methylation is involved in β4 repression during TGFβ-induced EMT. (A,B) Methylation status of the entire CpG island in the β4 promoter in the absence (A) or presence (B) of TGFβ for 11 days. Each circle represents a CpG dinucleotide from the first (1) to the last (66) CpG of randomly selected clones that were sequenced. White circles, unmethylated CpG; black circles, methylated CpG. (C) Immunoblot analysis of β4 expression in the presence of TGFβ alone or TGFβ plus 5-aza for the times indicated. (D) Analysis of β4 mRNA expression by SYBR Green qRT-PCR in the absence of TGFβ (UNT), with TGFβ alone (TGFβ) or with TGFβ and 5-aza (5aza) for 48 hours. Results are reported as the relative expression normalized to that of GAPDH. (E) Bisulfite conversion and methylation analysis of integrin α6 promoter after TGFβ treatment for 11 days as described in A.
Given that α6 and β4 form a functional heterodimer (Hemler et al., 1989), and α6 expression also decreases during the EMT (Fig. 2C), we asked whether DNA methylation is also involved in regulating the α6 subunit. The α6 proximal promoter also contains a CpG island, but we observed that it remains unmethylated after TGFβ treatment using bisulfite conversion and TA cloning (Fig. 3E). These data suggest that distinct mechanism(s) are involved in regulating α6 and β4 promoter activity.
TGFβ withdrawal is sufficient to induce MET and restore β4 expression
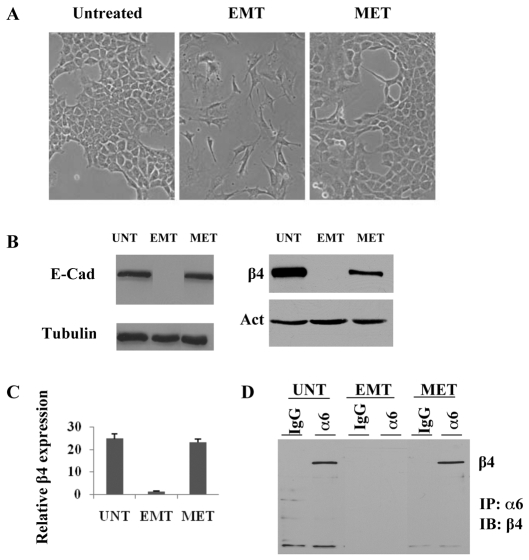
Upon withdrawal of TGFβ, NMuMG cells undergo dramatic morphological changes indicative of a MET (Fig. 4A) and consequently restore expression of E-cadherin (Fig. 4B), consistent with a previous report (Gal et al., 2008). Intriguingly, β4 expression is also restored during this MET (Fig. 4B,C), and β4 can be co-immunoprecipitated with an antibody against α6 (Fig. 4D). This observation enabled us to dissect the molecular mechanisms by which the β4 promoter is dynamically regulated by physiological stimuli. The recent identification of DNA demethylase activity in Arabidopsis (Penterman et al., 2007; Agius et al., 2006) led us to hypothesize that a putative DNA demethylase might be recruited to the methylated promoters and that it causes promoter demethylation and transcriptional reactivation after TGFβ withdrawal. Surprisingly, however, TGFβ withdrawal (Fig. 4A) did not result in the demethylation of the CpG island in either the β4 (Fig. 5A) or E-cadherin promoters (Fig. 6A), although expression of both genes is reactivated, as shown in Fig. 4. This paradoxical methylation phenomenon is not caused by cell aging-related hypermethylation because the β4 promoter remains completely unmethylated in untreated cells of the same passage (data not shown).
Fig. 4.
TGFβ withdrawal reverses EMT and restores both E-cadherin and β4 integrin expression. (A) Phase-contrast photomicrographs of NMuMG cells either untreated (UNT), treated with TGFβ for 11 days (EMT) or treated with TGFβ for 11 days and subsequently in the absence of TGFβ for 13 days (MET). (B) Immunoblot analysis of E-cadherin (E-cad) and β4 integrin (β4) expression under conditions described in A. Tubulin or actin served as a loading control. (C) Analysis of β4 mRNA expression normalized to that of GAPDH in the cells described in A by SYBR Green qRT-PCR. (D) Extracts from the cells described in A were immunoprecipitated with either a nonspecific IgG or a monoclonal antibody against α6 (GOH3), and the immunoprecipitates were blotted with a polyclonal antibody against β4.
Fig. 5.
Complexity of epigenetic remodeling of the β4 promoter during the EMT and MET. (A) Methylation status of the entire CpG island in the β4 promoter in cells that underwent a MET (see Fig. 4A). Each circle represents a CpG dinucleotide from the first (1) to the last (66) CpG of six clones that were sequenced. White circles, unmethylated CpG; black circles, methylated CpG. (B-D) ChIP analyses of histone modifications of the β4 promoter using antibodies specific for H3K4me3, H3K9Ac or H3K27me3. NMuMG cells were treated as described in the legend to Fig. 4A and then processed for ChIP as described in Materials and Methods. ChIP results were quantified by `hot' PCR for H3K4me3 or H3K27me3 (B) or by using SYBR Green real-time PCR (qPCR) and reported as the percentage of immunoprecipitate (IP) to input (C,D). (E) ChIP qPCR analysis for the enrichment of H3K9Ac at the β4 promoter in NMuMG cells that were either untreated (UNT), treated with TGFβ for 3 days (TGFβ) or simultaneously treated with TGFβ and 5 μM 5-aza for 3 days (5-aza).
Fig. 6.
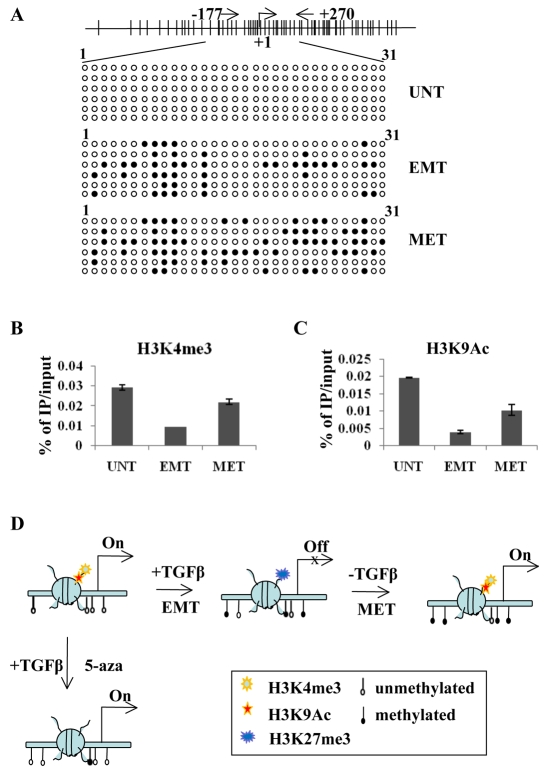
Epigenetic remodeling of the E-cadherin promoter after TGFβ withdrawal. (A) Methylation status of the CpG island in the E-cadherin promoter in cells, as described in Fig. 4A. (B,C) XhIP qPXP analyses of enrichment of H3K4me3 (B) or H3K9Ac (C) at the E-cadherin promoter under conditions described in Fig. 5C. (D) Proposed model of epigenetic regulation at the β4 promoter during the EMT and MET, as described in the text.
Transcriptional reactivation of methylated genes is predominantly regulated by reversible histone modifications during a MET
The restoration of β4 and E-cadherin expression in the presence of sustained promoter methylation after TGFβ withdrawal led us to investigate other epigenetic mechanisms that could regulate gene expression. Histone H3 Lys4 trimethylation (H3K4me3) and H3 Lys9 acetylation (H3K9Ac) are two well-characterized epigenetic modifications for actively transcribed gene promoters (Santos-Rosa et al., 2002; Schneider et al., 2004). Using chromatin immunoprecipitation (ChIP) assays, we next investigated whether the transcriptional reactivation of the β4 gene is associated with the enrichment of H3K4me3 and H3K9Ac at the promoter in NMuMG cells without TGFβ treatment (UNT), with TGFβ treatment for 11 days (EMT) and after TGFβ withdrawal for 13 days (MET). Both H3K4me3 and H3K9Ac are enriched at the β4 promoter in untreated NMuMG cells that express β4 (UNT; Fig. 5B,C). Both H3K4me3 and H3K9Ac, however, are dramatically decreased at the β4 promoter after TGFβ treatment for 11 days (EMT; Fig. 5B,C). Interestingly, both modifications are restored at the β4 promoter after TGFβ withdrawal, but to a lesser intensity (MET; Fig. 5B,C), suggesting that the partially methylated promoter can be remodeled for activation through histone modifications, although other mechanisms cannot be excluded.
As we observed that active histone modifications play a dominant role in reactivating the β4 promoter during the MET regardless of its methylation status, we asked whether repressive histone modifications such as histone H3 Lys27 trimethylation (H3K27me3) also play a role in β4 transcriptional regulation in the NMuMG model system. ChIP studies demonstrated that enrichment of H3K27me3 is increased sevenfold during TGFβ-induced EMT (EMT; Fig. 5D). This repressive histone modification is reduced to ∼44% of its original level after TGFβ withdrawal (MET; Fig. 5D), indicating an interplay between distinct histone modifications and DNA methylation in regulating β4 promoter activity in response to distinct micro-environmental stimuli.
Next, ChIP assays were performed to investigate the effects of the DNA-demethylation agent 5-aza on H3K9Ac enrichment at the β4 promoter in cells either without treatment (UNT), with TGFβ treatment for 3 days (TGFβ) or simultaneously treated with TGFβ and 5 μM 5-aza for 3 days. Interestingly, TGFβ induced a dramatic reduction in the H3K9Ac level at the β4 promoter during this short timecourse of treatment, and DNA demethylation by 5-aza was able to partially prevent the reduction of histone acetylation (5-aza; Fig. 5E). All of the ChIP data indicate that repressive histone modifications such as H3K27me3 and histone deacetylation are coordinated with DNA methylation in repressing the β4 gene, and that loss of H3K27me3 and gain of H3K9Ac and H3K4me3 both contribute to reactivation of β4 expression. We also investigated the effect of 5-aza treatment on the EMT process itself. NMuMG cells were treated with TGFβ for 11 days to induce an EMT, then TGFβ was removed and the mesenchymal-like cells were either grown in fresh media without 5-aza, or treated with different doses of 5-aza. Although TGFβ withdrawal alone is sufficient to induce a MET, 5-aza did not facilitate a MET and, in fact, long-term treatment with 5-aza at 1 or 5 μM caused significant cell death (data not shown).
Finally, we asked whether the methylated E-cadherin promoter shares the same epigenetic regulation as that of β4 by using ChIP analyses. Interestingly, both H3K4me3 (Fig. 6B) and H3K9Ac (Fig. 6C) modifications of the E-cadherin promoter are recovered with a pattern similar to that of β4. Together, these data indicate that β4 and E-cadherin promoter activity is dynamically regulated by the interplay between CpG island methylation and histone modifications during TGFβ-mediated EMT and MET and that reversible histone modifications can override sustained DNA methylation after removal of TGFβ (MET).
Discussion
In this study, we establish that epigenetic mechanisms can regulate expression of β4 and that this regulation is dynamic and complex during the EMT and MET. Specifically, we demonstrate that the distinct expression of β4 in basal and luminal cells of the mouse mammary gland correlates strongly with methylation of the β4 promoter. Our data also reveal a dynamic regulation of β4 expression by TGFβ that is coincident with that of E-cadherin and involves both de novo DNA methylation and histone modifications. Together, our results provide an important advance in our understanding of how this unique integrin is regulated in normal tissue and, potentially, during cancer progression.
Given the multiple functions of β4 in normal epithelial homeostasis and its impact on the functions of carcinoma cells (Dowling et al., 1996; Lipscomb and Mercurio, 2005; Vanderneut et al., 1996; Nikolopoulos et al., 2004; Wilhelmsen et al., 2006), it is surprising that few, if any, studies, have addressed the mechanisms that regulate expression of the β4 gene. Interestingly, a high GC content in the β4 promoter was observed in its original cloning 10 years ago, but the possibility of epigenetic regulation was not pursued (Takaoka et al., 1998). We were struck by the presence of a large CpG island in our analysis of this promoter, an observation that provided the impetus for this study. However, analyzing the β4 promoter in vivo as a function of β4 expression is challenging because of the restricted and heterogeneous expression of this integrin. Indeed, our initial use of laser capture microscopy did not provide adequate resolution of β4+ and β4– cells. We were able to overcome this challenge by using K14-eGFP mice (Hirakawa et al., 2007), which express GFP in basal epithelial cells only. Our ability to separate basal from luminal cells of the mammary glands of these mice by FACS enabled us to demonstrate that the β4 promoter is hypermethylated in luminal cells, which lack β4 expression, and is unmethylated in basal cells, which express β4. These data provide strong correlative evidence that promoter methylation contributes to the regulation of β4 expression in vivo. However, as discussed below, our in vitro data indicate that the regulation of dynamic β4 expression during the EMT and MET also involves other epigenetic modifications.
A key finding in this study is the dynamic regulation of β4 expression that can occur in response to physiological stimuli can be mediated by epigenetic modifications. The use of NMuMG cells, which were derived from a mouse mammary gland (David et al., 1981), provided an ideal model system in which to study β4 regulation. TGFβ is known to induce an EMT in these cells (Gal et al., 2008), and we observed that this EMT is consistent with a loss of the epithelial marker E-cadherin and increased expression of mesenchymal markers, including N-cadherin and Snai1. Both β4 mRNA and protein expression were lost during TGFβ-induced EMT in our model. Although β4 has been implicated in epithelial cell migration by our group (Mercurio et al., 2001) and others (Santoro et al., 2003; Sehgal et al., 2006), we hypothesize that β4 expression is lost during this EMT because it is known that NMuMG cells produce a basement membrane in culture (David et al., 1981) and this basement membrane must be disrupted to facilitate migration. Indeed, recent studies have shown that suppression of basement membrane components can be a hallmark of the EMT of normal epithelia (Nakaya et al., 2008) and carcinoma cells (Takkunen et al., 2008). This consideration aside, the salient finding we made is that the β4 promoter is unmethylated in β4-expressing NMuMG cells and it is marked by active histone modifications (H3K4me3 and H3K9Ac) but only a basal level of repressive modification (H3K27me3). Similar to E-cadherin, β4 expression is also suppressed and its promoter becomes partially methylated during the TGFβ-induced EMT. This de novo DNA methylation is responsible for the initiation of silencing because co-treatment of 5-aza with TGFβ can prevent the loss of β4 transcriptionally and translationally probably through antagonizing the loss of histone acetylation, as suggested by our ChIP data. This de novo DNA methylation is correlated with a reduction in the enrichment of active histone modifications including H3K4me3 and H3K9Ac and a gain of repressive histone modification such as H3K27me3, as indicated by ChIP analyses. These data highlight how effectors of the EMT can regulate gene expression by epigenetic reprogramming. Aside from studies demonstrating that E-cadherin expression can be regulated by methylation and histone modifications (Lombaerts et al., 2006; Peinado et al., 2004), the hypothesis that epigenetic reprogramming plays a global role in executing the EMT is only just beginning to be pursued rigorously (Dumont et al., 2008).
Arguably, the most interesting finding we report is the relatively complex epigenetic regulation of β4 and E-cadherin that is manifested in response to TGFβ withdrawal. TGFβ withdrawal results in a MET (Gal et al., 2008) and a restoration of both β4 and E-cadherin expression. Surprisingly, however, both the β4 and E-cadherin promoters remain partially methylated during the MET even though both genes are transcriptionally activated, a finding that is consistent with a few earlier reports that gene reactivation can be independent of DNA demethylation (Abecassis et al., 2008; Cameron et al., 1999). The interesting question that arises from these data is why de novo methylation plays a role in the initial suppression of β4 expression by TGFβ (EMT) but not after TGFβ withdrawal (MET). It is conceivable that DNA methylation maintained after removal of TGFβ treatment recruits distinct factors and, therefore, functions differently from the de novo DNA methylation. Alternatively, sustained DNA methylation after TGFβ withdrawal might function as a cellular epigenetic memory for previous micro-environmental stimuli rather than permanently silencing the target gene. Remarkably, growth factors including TGFβ appear to induce epigenetic reprogramming, and some epigenetically reprogrammed genes including β4 can be reversed easily by changing the micro-environment, including removal of TGFβ in this model or inactivating a transcriptional repressor of β4 such as p63 (Carroll et al., 2006). These data raise the novel hypothesis that β4 expression might be suppressed during the early stages of tumorigenesis to facilitate basement membrane degradation and an EMT but reactivated in response to micro-environmental signals that promote aggressive behavior (Zahir et al., 2003).
Mechanistically, β4 expression during a MET is associated with the recovery of active histone modifications (H3K9Ac and H3K4me3) and a reduction in the repressive histone mark H3K27me3, although the enrichment is less than that in parental cells possibly because of the presence of partially methylated DNA. Based on these results, we propose a model that de novo DNA methylation can decrease active histone modifications including histone acetylation and H3K4 trimethylation and increase H3K27me3, then initiate transcriptional repression in response to micro-environmental stimuli such as TGFβ (Fig. 6D). This de novo DNA methylation can be blocked by treatment with 5-aza, preventing TGFβ-induced silencing of β4. We were surprised that no putative DNA demethylase is recruited to either the β4 or E-cadherin promoters for DNA demethylation when TGFβ is removed, indicating the complexity of transcriptional regulation during the MET. These data support the notion that promoter demethylation is not an obligatory event for transcriptional reactivation during the MET. It is plausible that additional transcription factors might be recruited to the methylated promoters maintained after TGFβ withdrawal and that these assist the reconfiguration of the chromatin by re-establishing reversible histone modifications such as H3K9 acetylation and H3K4me3. In fact, it is well recognized that histone acetylation catalyzed by histone acetyltransferases often coordinates with the ATP-dependent chromatin remodeling enzymes in both transcription initiation and elongation (Carey et al., 2006), and the complexity of epigenetic regulation during transcriptional initiation and elongation is obvious (Li et al., 2007). It would be interesting to investigate further whether this epigenetic coordination also occurs at the β4 and E-cadherin promoters during the MET. Based on this study, it appears that changes in histone modifications after TGFβ withdrawal are more important than DNA demethylation in determining the transcriptional activity at the β4 and E-cadherin promoters, although the DNA-demethylation agent 5-aza can antagonize the loss of histone acetylation in a short period of treatment. Our model is supported by previous observations showing that transcriptional reactivation can occur at methylated promoters and that DNA methylation in some cell lines does not always repress transcriptional activity (Abecassis et al., 2008; Cameron et al., 1999).
In summary, our data highlight the importance of epigenetic modifications in determining the pattern of β4 expression in tissues, and they indicate that such modifications are important for the dynamic regulation of β4 that can occur as a consequence of the EMT and probably in response to other micro-environmental stimuli.
Materials and Methods
Analysis of mouse mammary glands
FVB K14-eGFP reporter mice (Hirakawa et al., 2007) were obtained from Satoshi Hirakawa (Ehime University Graduate School of Medicine, Ehime, Japan). The fourth inguinal mammary glands of 10-week-old mice (six mice) were harvested. After removal of intra-gland lymph nodes, single epithelial cell suspensions were prepared as described previously (Sleeman et al., 2006). The eGFP+ and eGFP– epithelial cells were collected using a FACSVantage SE Cell Sorter (BD Biosciences).
Frozen sections from adult FVB mouse mammary glands were stained with a rat monoclonal antibody against β4 (346-11a; a gift from Rita Falcioni, Regina Elena Cancer Institute, Rome, Italy) using streptavidin-biotin (Vectastain Elite avidin-biotin complex kit, Vector Laboratories). Nonspecific background was diminished by 5% BSA (Sigma-Aldrich). Horseradish peroxidase was developed using 3,3-diaminobenzidine tetrahydrochloride (DakoCytomation), and the specimens were counterstained with hematoxylin. Immunostaining of eGFP protein was performed on formalin-fixed paraffin-embedded sections. The antibody binding sites were retrieved by steaming the slide in a citrate buffer (pH 6.0; Zymed/Invitrogen). The rabbit anti-eGFP antibody (sc. 8334; Santa Cruz Biotechnology) was used at a final concentration of 1 μg/ml.
EMT studies using NMuMG cells
Immortalized mouse mammary gland epithelial cells (NMuMG) were purchased from the ATCC and maintained in DMEM (high glucose) medium containing 10% fetal bovine serum, 5 mg/l insulin, and 1% streptomycin and penicillin at 37°C in an incubator supplied with 5% CO2. TGFβ (5 ng/ml) (R&D) was added directly into the culture medium when cells reached ∼70% confluence for the time periods indicated in the figure legends. In some experiments, 5-aza (5 μM; Sigma-Aldrich) was added together with TGFβ. For long-term treatment with TGFβ (EMT), cells were passed into fresh DMEM medium containing 5 ng/ml TGFβ. For the TGFβ-withdrawal experiments, cells that had been incubated in the presence of TGFβ were passed into fresh DMEM medium without TGFβ for an additional 13 days (TGFβ withdrawal or MET).
To assess the expression of specific integrin subunits, E-cadherin, N-cadherin and Snai1 during the EMT by RT-PCR, total RNA (2 μg) was extracted by RNeasy (Qiagen) and reverse transcribed with SuperScriptIII Reverse Transcription system (Invitrogen), and then PCR amplified using either the HotStar Taq Master Mix system (Qiagen) or Power SYBR Green Master Mix (Applied Biosystems), as indicated in the figure legends. The following primer sets were used for RT-PCR: integrin β4 forward 5′-TGTGTTCCAGGTGTTTGAGC-3′ and reverse 5′-TTTCTCATCATTGCGGTTCA-3′; integrin α6 forward 5′-CGGGATATGCCTCAAGGTTA-3′ and reverse 5′-TGCCTTTTTGAATTGGAAGG-3′; E-cadherin forward 5′-GAGGAACCCACAGCCTCATA-3′ and reverse 5′-GCTGGCTCAAATCAAAGTCC-3′; integrin α3 forward 5′-GCTTCTTCTCCATGCCAGAG-3′ and reverse 5′-GGGGTTTCCTAGCTCACACA-3′; integrin β1 forward 5′-TCACATGCAGGTTTGGAAAA-3′ and reverse 5′-TCACAATGGCACACAGGTTT-3′; N-cadherin forward 5′-CGGTTTCACTTGAGAGCACA-3′ and reverse 5′-TGATGATGTCCCCAGTCTCA-3′; Snai1 forward 5′-TAGAGCTGACCTCGCTGTCC-3′ and reverse 5′-ACAGGTCGTGCAGACACAAG-3′.
For quantitative RT-PCR (qRT-PCR), reverse-transcribed cDNA was used for real-time PCR using Power SYBR Green Master Mix (Applied Biosystems) with the HT7900 Fast Real Time PCR machine (Applied Biosystems). Standard curves were generated using serial dilutions of cDNA with arbitrary copy numbers. The relative expression of each mRNA was normalized to that of glyceraldehyde-3-phosphate dehydrogenase (GAPDH). All experiments were performed in triplicate. Primer set for β4: forward 5′-CGATGACAACCGACCTATTG-3′ and reverse 5′-TTCTCAATAAGCAGCATCCG-3′. Primer set for GAPDH: forward 5′-ATCATCCCTGCCTCTACTGG-3′ and reverse 5′-GTCAGGTCCACCACTGACAC-3′.
To assess expression of β4 and E-cadherin by immunoblotting, whole-cell extracts (30 μg protein) were resolved by 8% SDS-PAGE and blotted with a polyclonal antibody against β4 (Rabinovitz et al., 1999) or the polyclonal antibody against E-cadherin (clone DECMA-1; Sigma). To assess expression of the α6β4 heterodimer, cell extracts (100 μg protein) were precipitated with a monoclonal antibody against α6 (GOH3; Chemicon), and the precipitates were blotted using the polyclonal antibody against β4.
DNA methylation analysis
Genomic DNA (∼1 μg) was treated with sodium bisulfite using the EpiTect system (Qiagen) following the manufacturer's instructions. Bisulfite-converted DNAs (∼50 ng) were used as templates for PCR amplification of the entire CpG island in the β4 proximal promoter, which consists of 696 base pairs (bp) that contain 66 CpG dinucleotides (Fig. 1C). The PCR primers were: forward primer: 5′-GAATTTAGTGGTTAGTAGTTTAAGAG-3′ and reverse primer: 5′-AAAAACCAAAACCTACCAATCTTC-3′. For E-cadherin promoter methylation analysis, a fragment of 447 bp that contains 31 CpG dinucleotides was amplified within the CpG island spanning from –177 to +270 relative to the transcriptional initiation site (+1) (shown at the top of Fig. 6A), using primer set: 5′-AGGAAGTTGGGAAGTTTTTTAA-3′ and 5′-TCCCAACTTTCTTAAAAAAAAAA-3′. For analysis of DNA methylation from mouse mammary glands, nested PCR was performed using a second primer set that amplifies a 400-bp fragment from the CpG island with the following primers: forward primer: 5′-GGTTTGTGTTGTAGGGAAGAG-3′ and reverse primer: 5′-CTCTCCAACCCTATTATCCC-3′. All PCR products were purified from 1.5% agarose gels using Gel Extraction Kit (Qiagen) and cloned into the pGEM-T Easy vector (Promega). Six to ten randomly selected clones were selected for sequencing (Genewiz).
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed according to a published mammalian protocol (Forsberg et al., 2000) with some modifications. Briefly, cells (∼1×107) were crosslinked with formaldehyde at a final concentration of 1% for 15 minutes at room temperature, and glycine (125 mM) was then added to quench the reaction for 5 minutes with gentle agitation. Cells were washed twice with cold PBS, harvested and extracted in 1 ml of a buffer containing 150 mM NaCl, 50 mM Tris-HCl, pH 7.5, 5 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.5% Nonidet P-40 and protease inhibitors. Cell extracts were centrifuged at 6,700 × g for 1 minute to pellet nuclei. Nuclei were washed once with extraction buffer and resuspended in 600 μl of ChIP buffer (150 mM NaCl, 50 mM Tris-HCl, pH 7.5, 1 mM EDTA, 1% Triton X-100, 0.1% Nonidet P-40). Chromatin was sonicated using a microtip sonicator (Sonicator 3000; Misonix) with five 10-second bursts to generate chromatin fragments of 300-1000 bp.
Soluble chromatin was isolated by centrifugation and pre-cleared with nonspecific IgGs, followed by Protein A-Sepharose (BioVision). The chromatin (200 μl) was incubated with either 2% anti-H3K4me3 (Abcam), anti-H3K9Ac (Abcam), anti-H3K27me3 (Upstate) or nonspecific IgG at a final volume of 600 μl in ChIP buffer for 2 hours at 4°C. An aliquot of 20 μl (10%) of pre-cleared chromatin served as input. Protein A-Sepharose beads (20 μl; blocked by BSA and sonicated salmon sperm DNA) were used to precipitate the chromatin. The beads were washed once with ChIP buffer, twice with wash buffer I (50 mM Tris, 1 M NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, pH 7.5), twice with wash buffer II (10 mM Tris, 0.25 M NaCl, 1 mM EDTA, 0.5% Nonidet P-40, 0.5% sodium deoxycholate, pH 7.5), and once with Tris-EDTA (TE). Next, beads were eluted in 200 μl of buffer C (50 mM Tris-HCl, pH7.5, 10 mM EDTA, 1% SDS) by shaking at 65°C in a Thermomixer (Eppendorf) for 10 minutes. Immunoprecipitated chromatin or input DNA was deproteinated by proteinase K digestion at 42°C for 5 hours, and de-crosslinked in NaCl (final concentration 0.5 M) at 65°C overnight. DNA was extracted with phenol-chloroform–isoamyl-alcohol. Finally, glycogen (10 μg) was added and DNA was precipitated with 1 ml ethanol. Immunoprecipitated DNA was dissolved in 20 μl of TE (80 μl for input DNA).
All immunoprecipitated DNAs were initially amplified by PCR and resolved in agarose gels. `Hot' PCRs were performed by using 0.25 μCi 32P-dCTP (PerkinElmer) per PCR reaction and resolved into 10% polyacrylamide gel that was dried and imaged by a Phosphoimager scanner (Storm 860, Molecular Dynamics). For quantitative ChIP assays, real time-PCR (qPCR) was performed using Power SYBR Green Master Mix in a HT7900 Fast Real Time PCR machine as described above (Applied Biosystems). The primer set from the β4 proximal promoter is: forward 5′-CCTGCCGCAAGAGTAAGATT-3′ and reverse 5′-GACTGGGGCCTCTAGGTTTC-3′. The primer set for the E-cadherin proximal promoter is: forward 5′-AGACAGGGGTGGAGGAAGTT-3′ and reverse primer 5′-ACCAGTGAGCAGCGCAGAG-3′. All real-time PCR reactions were performed in triplicate, and the relative enrichment of each histone modification was reported as the percentage of immunoprecipitate to input. All ChIP experiments were performed at least twice, and the variation was less than 20%.
This work was supported by NIH Grants CA1107548 and CA80789 (A.M.M.) and T32130807 (X.Y.), an ACS Postdoctoral Fellowship Grant (X.Y.) and a Susan G. Komen Breast Cancer Foundation Grant PDF0600265 (S.L.). Deposited in PMC for release after 12 months.
References
- Abecassis, I., Maes, J., Carrier, J. L., Hillion, J., Goodhardt, M., Medjber, K., Wany, L., Lanotte, M. and Karniguian, A. (2008). Re-expression of DNA methylation-silenced CD44 gene in a resistant NB4 cell line: rescue of CD44-dependent cell death by cAMP. Leukemia 22, 511-520. [DOI] [PubMed] [Google Scholar]
- Agius, F., Kapoor, A. and Zhu, J. K. (2006). Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proc. Natl. Acad. Sci. USA 103, 11796-11801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachelder, R. E., Ribick, M. J., Marchetti, A., Falcioni, R., Soddu, S., Davis, K. R. and Mercurio, A. M. (1999). p53 inhibits alpha 6 beta 4 integrin survival signaling by promoting the caspase 3-dependent cleavage of AKT/PKB. J. Cell Biol. 147, 1063-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron, E. E., Bachman, K. E., Myohanen, S., Herman, J. G. and Baylin, S. B. (1999). Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat. Genet. 21, 103-107. [DOI] [PubMed] [Google Scholar]
- Carey, M., Li, B. and Workman, J. L. (2006). RSC exploits histone acetylation to abrogate the nucleosomal block to RNA polymerase II elongation. Mol. Cell 24, 481-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll, D. K., Carroll, J. S., Leong, C. O., Cheng, F., Brown, M., Mills, A. A., Brugge, J. S. and Ellisen, L. W. (2006). p63 regulates an adhesion programme and cell survival in epithelial cells. Nat. Cell Biol. 8, 551-561. [DOI] [PubMed] [Google Scholar]
- David, G., Van der Schueren, B. and Bernfield, M. (1981). Basal lamina formation by normal and transformed mouse mammary epithelial cells duplicated in vitro. J. Natl. Cancer Inst. 67, 719-728. [PubMed] [Google Scholar]
- Dowling, J., Yu, Q. C. and Fuchs, E. (1996). Beta-4 integrin is required for hemidesmosome formation, cell adhesion and cell survival. J. Cell Biol. 134, 559-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont, N., Wilson, M. B., Crawford, Y. G., Reynolds, P. A., Sigaroudinia, M. and Tlsty, T. D. (2008). Sustained induction of epithelial to mesenchymal transition activates DNA methylation of genes silenced in basal-like breast cancers. Proc. Natl. Acad. Sci. USA 105, 14867-14872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcioni, R., Kennel, S. J., Giacomini, P., Zupi, G. and Sacchi, A. (1986). Expression of tumor antigen correlated with metastatic potential of Lewis lung carcinoma and B16 melanoma clones in mice. Cancer Res. 46, 5772-5778. [PubMed] [Google Scholar]
- Forsberg, E. C., Downs, K. M. and Bresnick, E. H. (2000). Direct interaction of NF-E2 with hypersensitive site 2 of the beta-globin locus control region in living cells. Blood 96, 334-339. [PubMed] [Google Scholar]
- Gal, A., Sjoblom, T., Fedorova, L., Imreh, S., Beug, H. and Moustakas, A. (2008). Sustained TGF beta exposure suppresses Smad and non-Smad signalling in mammary epithelial cells, leading to EMT and inhibition of growth arrest and apoptosis. Oncogene 27, 1218-1230. [DOI] [PubMed] [Google Scholar]
- Hemler, M. E., Crouse, C. and Sonnenberg, A. (1989). Association of the VLA alpha 6 subunit with a novel protein: a possible alternative to the common VLA beta 1 subunit on certain cell lines. J. Biol. Chem. 264, 6529-6535. [PubMed] [Google Scholar]
- Hirakawa, S., Brown, L. F., Kodama, S., Paavonen, K., Alitalo, K. and Detmar, M. (2007). VEGF-C-induced lymphangiogenesis in sentinel lymph nodes promotes tumor metastasis to distant sites. Blood 109, 1010-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch, R. and Bird, A. (2003). Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 33 Suppl, 245-254. [DOI] [PubMed] [Google Scholar]
- Jauliac, S., Lopez-Rodriguez, C., Shaw, L. M., Brown, L. F., Rao, A. and Toker, A. (2002). The role of NFAT transcription factors in integrin-mediated carcinoma invasion. Nat. Cell Biol. 4, 540-544. [DOI] [PubMed] [Google Scholar]
- Jones, P. A. and Taylor, S. M. (1980). Cellular differentiation, cytidine analogs and DNA methylation. Cell 20, 85-93. [DOI] [PubMed] [Google Scholar]
- Jones, P. A. and Baylin, S. B. (2007). The epigenomics of cancer. Cell 128, 683-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B., Carey, M. and Workman, J. L. (2007). The role of chromatin during transcription. Cell 128, 707-719. [DOI] [PubMed] [Google Scholar]
- Lipscomb, E. A. and Mercurio, A. M. (2005). Mobilization and activation of a signaling competent alpha6beta4integrin underlies its contribution to carcinoma progression. Cancer Metastasis Rev. 24, 413-423. [DOI] [PubMed] [Google Scholar]
- Lipscomb, E. A., Simpson, K. J., Ring, J. E., Dugan, A. S. and Mercurio, A. M. (2005). The α6β4 integrin maintains the survival of human breast carcinoma cells in vivo. Cancer Res. 65, 10970-10976. [DOI] [PubMed] [Google Scholar]
- Lombaerts, M., van Wezel, T., Philippo, K., Dierssen, J. W., Zimmerman, R. M., Oosting, J., van Eijk, R., Eilers, P. H., van de Water, B., Cornelisse, C. J. et al. (2006). E-cadherin transcriptional downregulation by promoter methylation but not mutation is related to epithelial-to-mesenchymal transition in breast cancer cell lines. Br. J. Cancer 94, 661-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, S., Simin, K., Khan, A. and Mercurio, A. M. (2008). Analysis of integrin beta4 expression in human breast cancer: association with basal-like tumors and prognostic significance. Clin. Cancer Res. 14, 1050-1058. [DOI] [PubMed] [Google Scholar]
- Mercurio, A. (1995). Receptors for the laminins: achieving specificity through cooperation. Trends Cell Biol. 5, 419-423. [DOI] [PubMed] [Google Scholar]
- Mercurio, A. M., Rabinovitz, I. and Shaw, L. M. (2001). The alpha 6 beta 4 integrin and epithelial cell migration. Curr. Opin. Cell Biol. 13, 541-545. [DOI] [PubMed] [Google Scholar]
- Nakaya, Y., Sukowati, E. W., Wu, Y. and Sheng, G. (2008). RhoA and microtubule dynamics control cell-basement membrane interaction in EMT during gastrulation. Nat. Cell Biol. 10, 765-775. [DOI] [PubMed] [Google Scholar]
- Neely, K. E. and Workman, J. L. (2002). The complexity of chromatin remodeling and its links to cancer. Biochim. Biophys. Acta 1603, 19-29. [DOI] [PubMed] [Google Scholar]
- Nikolopoulos, S. N., Blaikie, P., Yoshioka, T., Guo, W. and Giancotti, F. G. (2004). Integrin beta4 signaling promotes tumor angiogenesis. Cancer Cell 6, 471-483. [DOI] [PubMed] [Google Scholar]
- O'Connor, K. L., Shaw, L. M. and Mercurio, A. M. (1998). Release of cAMP gating by the alpha6beta4 integrin stimulates lamellae formation and the chemotactic migration of invasive carcinoma cells. J. Cell Biol. 143, 1749-1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado, H., Ballestar, E., Esteller, M. and Cano, A. (2004). Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol. Cell. Biol. 24, 306-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penterman, J., Zilberman, D., Huh, J. H., Ballinger, T., Henikoff, S. and Fischer, R. L. (2007). DNA demethylation in the Arabidopsis genome. Proc. Natl. Acad. Sci. USA 104, 6752-6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovitz, I. and Mercurio, A. M. (1997). The integrin alpha6beta4 functions in carcinoma cell migration on laminin-1 by mediating the formation and stabilization of actin-containing motility structures. J. Cell Biol. 139, 1873-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovitz, I., Toker, A. and Mercurio, A. M. (1999). Protein kinase C-dependent mobilization of the alpha6beta4 integrin from hemidesmosomes and its association with actin-rich cell protrusions drive the chemotactic migration of carcinoma cells. J. Cell Biol. 146, 1147-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond, K., Kreft, M., Song, J. Y., Janssen, H. and Sonnenberg, A. (2007). Dual Role of alpha6beta4 integrin in epidermal tumor growth: tumor-suppressive versus tumor-promoting function. Mol. Biol. Cell 18, 4210-4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro, M. M., Gaudino, G. and Marchisio, P. C. (2003). The MSP receptor regulates alpha6beta4 and alpha3beta1 integrins via 14-3-3 proteins in keratinocyte migration. Dev. Cell 5, 257-271. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa, H., Schneider, R., Bannister, A. J., Sherriff, J., Bernstein, B. E., Emre, N. C., Schreiber, S. L., Mellor, J. and Kouzarides, T. (2002). Active genes are tri-methylated at K4 of histone H3. Nature 419, 407-411. [DOI] [PubMed] [Google Scholar]
- Schneider, R., Bannister, A. J., Myers, F. A., Thorne, A. W., Crane-Robinson, C. and Kouzarides, T. (2004). Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat. Cell Biol. 6, 73-77. [DOI] [PubMed] [Google Scholar]
- Sehgal, B. U., DeBiase, P. J., Matzno, S., Chew, T. L., Claiborne, J. N., Hopkinson, S. B., Russell, A., Marinkovich, M. P. and Jones, J. C. (2006). Integrin beta4 regulates migratory behavior of keratinocytes by determining laminin-332 organization. J. Biol. Chem. 281, 35487-35498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, L. M., Rabinovitz, I., Wang, H. H., Toker, A. and Mercurio, A. M. (1997). Activation of phosphoinositide 3-OH kinase by the alpha6beta4 integrin promotes carcinoma invasion. Cell 91, 949-960. [DOI] [PubMed] [Google Scholar]
- Sleeman, K. E., Kendrick, H., Ashworth, A., Isacke, C. M. and Smalley, M. J. (2006). CD24 staining of mouse mammary gland cells defines luminal epithelial, myoepithelial/basal and non-epithelial cells. Breast Cancer Res. 8, R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaoka, A. S., Yamada, T., Gotoh, M., Kanai, Y., Imai, K. and Hirohashi, S. (1998). Cloning and characterization of the human beta4-integrin gene promoter and enhancers. J. Biol. Chem. 273, 33848-33855. [DOI] [PubMed] [Google Scholar]
- Takkunen, M., Ainola, M., Vainionpaa, N., Grenman, R., Patarroyo, M., Garcia de Herreros, A., Konttinen, Y. T. and Virtanen, I. (2008). Epithelial-mesenchymal transition downregulates laminin alpha5 chain and upregulates laminin alpha4 chain in oral squamous carcinoma cells. Histochem. Cell Biol. 130, 509-525. [DOI] [PubMed] [Google Scholar]
- Trusolino, L., Bertotti, A. and Comoglio, P. M. (2001). A signaling adapter function for alpha6beta4 integrin in the control of HGF-dependent invasive growth. Cell 107, 643-654. [DOI] [PubMed] [Google Scholar]
- Vanderneut, R., Krimpenfort, P., Calafat, J., Niessen, C. M. and Sonnenberg, A. (1996). Epithelial detachment due to absence of hemidesmosomes in integrin beta-4 null mice. Nat. Genet. 13, 366-369. [DOI] [PubMed] [Google Scholar]
- Weaver, V. M., Lelievre, S., Lakins, J. N., Chrenek, M. A., Jones, J. C., Giancotti, F., Werb, Z. and Bissell, M. J. (2002). beta4 integrin-dependent formation of polarized three-dimensional architecture confers resistance to apoptosis in normal and malignant mammary epithelium. Cancer Cell 2, 205-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelmsen, K., Litjens, S. H. and Sonnenberg, A. (2006). Multiple functions of the integrin alpha6beta4 in epidermal homeostasis and tumorigenesis. Mol. Cell. Biol. 26, 2877-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahir, N., Lakins, J. N., Russell, A., Ming, W., Chatterjee, C., Rozenberg, G. I., Marinkovich, M. P. and Weaver, V. M. (2003). Autocrine laminin-5 ligates alpha6beta4 integrin and activates RAC and NFkappaB to mediate anchorage-independent survival of mammary tumors. J. Cell Biol. 163, 1397-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]