Summary
Autophagy is an important catabolic process with roles in cell survival and cell death. It sequesters cytosol and organelles within double-membrane autophagosomes that deliver their contents to lysosomes for degradation. Autophagosome biogenesis is coordinated by the autophagy-related protein 4 (Atg4) family of C54 endopeptidases (Atg4A-Atg4D). These enzymes prime and then later delipidate the autophagosome marker, Atg8. Here, we show that one family member, Atg4D, is cleaved by caspase-3 in vitro and in apoptotic cells. Atg4D is a poor priming and delipidation enzyme in vitro, but truncated ΔN63 Atg4D displays increased activity against the Atg8 paralogue, γ-aminobutyric acid receptor-associated protein-like 1 (GABARAP-L1). In living cells, ΔN63 Atg4D stimulates the delipidation of GABARAP-L1, whereas siRNA silencing of the gene expressing Atg4D abrogates GABARAP-L1 autophagosome formation and sensitises cells to starvation and staurosporine-induced cell death. Interestingly, Atg4D overexpression induces apoptosis, which is preceded by the caspase-independent recruitment of Atg4D to mitochondria and is facilitated by a putative C-terminal Bcl-2 homology 3 (BH3) domain. Atg4D also acquires affinity for damaged mitochondria in cells treated with hydrogen peroxide. These data suggest that Atg4D is an autophagy regulator that links mitochondrial dysfunction with apoptosis.
Keywords: Autophagy, Autophagin, Atg4, Atg8, LC3, GABARAP-L1
Introduction
Macroautophagy (autophagy, hereafter) is an important catabolic process that promotes cell survival during episodes of nutrient or growth-factor stress (Lum et al., 2005). Autophagy has important roles in development and homeostasis (Levine and Klionsky, 2004; Lockshin and Zakeri, 2004; Qu et al., 2007). It recycles toxic or redundant components by sequestering cytoplasm within double-membrane autophagosomes that deliver their contents to lysosomes (Yorimitsu and Klionsky, 2005). Autophagy prevents disease by removing damaged organelles, misfolded protein aggregates and some pathogens (Shintani and Klionsky, 2004). Under certain conditions, autophagy also contributes to cell death [type II or autophagic cell death (Leist and Jaattela, 2001)], by causing cellular vacuolisation and cellular dysfunction (Lum et al., 2005; Yu et al., 2004). Notably, the regulation of autophagy is directly coupled to apoptosis (Lum et al., 2005; Pattingre and Levine, 2006), and its capacity both to protect and to kill cells suggests that autophagy will prove to be an important therapeutic target for neurodegenerative diseases and cancer (Botti et al., 2006; Jin and White, 2007; Kondo and Kondo, 2006; Shintani and Klionsky, 2004).
Autophagosome biogenesis is coordinated by the Beclin-1/class III phosphatidylinositol 3 kinase (PI3K) complex [autophagy-related protein 6 (Atg6) or vacuolar protein sorting 34 (VPS34) in yeast] (Lee et al., 2007), and requires parallel, ubiquitin-like conjugation pathways. The first pathway forms an isopeptide bond between Atg5 and Atg12 [Atg7 is the E1-type enzyme (Kim et al., 1999); Atg10 is the E2-type conjugation enzyme (Shintani et al., 1999)]. In the second pathway, Atg8 is a modifier of the lipid phosphatidylethanolamine (PE) [here, Atg7 is referred to as E1, and Atg3 as E2 (Ichimura et al., 2000)]. Atg5-Atg12 binds to Atg16 (Suzuki et al., 2001), and associates transiently with membranes to target lipidated Atg8 to the nascent autophagosome. Atg8 remains attached throughout autophagosome biogenesis, but is removed from the outer autophagosomal membrane before fusion with the lysosome (Mizushima et al., 2001). The C54 endopeptidase, Atg4, is a crucial regulatory component of the autophagosome biogenesis pathway (Scherz-Shouval et al., 2007): it cleaves the C-terminus of newly synthesised Atg8 to reveal a glycine residue (priming) required for covalent attachment to PE (lipidation), and later removes PE from outer-membrane-bound Atg8 (delipidation) to facilitate autophagosome-lysosome fusion and Atg8 recycling.
Yeast express single genes encoding Atg4 and Atg8; however, in mammals there are four assumed Atg4 paralogues [autophagins Atg4A-Atg4D (Marino et al., 2003)] and several Atg8-related proteins [e.g. MAP1A/B light chain 3 (LC3), γ-aminobutyric acid receptor-associated protein (GABARAP), GABARAP-like 1 (GABARAP-L1, also known as Atg8L) and GABARAP-like 2 (GABARAP-L2, also known as Golgi-associated ATPase enhancer GATE-16)]. Studies suggest that Atg4B is likely to be the principle human Atg4 paralogue: it rescues the inability of yeast ΔATG4 strains to process aminopeptidase I (Marino et al., 2003), it primes each of the human Atg8-related proteins tested so far in vitro [LC3; GABARAP; GABARAP-L1; GATE-16 (Hemelaar et al., 2003; Tanida et al., 2004)], and is an efficient delipidation enzyme for LC3 and GABARAP in living cells (Tanida et al., 2004). Human ATG4C can rescue the yeast ΔATG4 aminopeptidase I processing deficiency, suggesting that it might also be a bone fide mammalian Atg4 paralogue (Marino et al., 2003). However, Tanida and colleagues have demonstrated that Atg4C lacks LC3 and GABARAP-L1 priming activity in vitro (Tanida et al., 2004). Accordingly, the mouse knockout of Atg4C has only a minor autophagy deficiency (Marino et al., 2007). Interestingly, although both Atg4A and Atg4D were unable to rescue the yeast ΔATG4 mutant [data not shown in Marino et al. (Marino et al., 2003)], human Atg4A has been shown independently to be an efficient priming enzyme for GATE-16 in vitro (Scherz-Shouval et al., 2003; Scherz-Shouval et al., 2007), but to be ineffective towards LC3 [data not shown in Scherz-Shouval et al. (Scherz-Shouval et al., 2003)]. Together these data suggest that Atg4B may be a universal regulator of Atg8 in mammalian cells, but that other Atg4 family members may be specific for individual Atg8 paralogues. Clearly there is a need for a more complete understanding of mammalian Atg4 family regulation and Atg8 substrate specificity and for information regarding the relative roles of Atg4 and Atg8 during development, homeostasis and disease prevention.
Evidence suggests that there is significant regulatory interplay between autophagy and apoptosis (e.g. Baehrecke, 2005; Holler et al., 2000; Martin et al., 2007; Scott et al., 2007; Shimizu et al., 2004). Intriguingly, caspases have been found to be required for autophagy in Drosophila (Hou et al., 2008; Martin et al., 2007), suggesting a direct link between protein cleavage and autophagic stimulation. Evidence also suggests that some autophagy regulators can signal to the apoptotic machinery. Atg5 overexpression is toxic to human cells (Pyo et al., 2005), and toxicity is enhanced following calpain-cleavage, which releases an N-terminal fragment that induces apoptosis by triggering mitochondrial cytochrome c release (Yousefi et al., 2006). Initially identified as a binding partner for anti-apoptotic Bcl-2 (Liang et al., 1998), Beclin-1 contains a Bcl-2 homology-3 (BH3) domain, facilitating binding to the crucial hydrophobic cleft of anti-apoptotic Bcl-2 or Bcl-XL (Maiuri et al., 2007; Oberstein et al., 2007). Binding inhibits autophagy and potentially lowers the apoptotic threshold (Maiuri et al., 2007; Pattingre et al., 2005). Hence, de-repressor BH3-only proteins and BH3 mimetics are predicted to co-stimulate apoptosis and autophagy by competing with Beclin-1 for binding to Bcl-2 or Bcl-XL (Maiuri et al., 2007), further emphasising the close regulatory links between these distinct pathways.
We have carried out the first functional characterisation of human Atg4D. This study was prompted by the identification of a canonical DEVD (aspartic acid, glutamic acid, valine, aspartic acid) caspase motif within the divergent N-terminus of Atg4D, suggesting that this enzyme might be regulated by caspases in healthy cells and/or in cells undergoing apoptosis. We have found that Atg4D is indeed a substrate for caspase-3 during apoptosis, and that truncated Atg4D displays increased priming and delipidation activities towards GABARAP-L1 in vitro. Our data also show that Atg4D is an important cell-survival factor because silencing Atg4D expression sensitises cells to starvation and staurosporine-induced cell death. Our studies also suggest that caspase cleavage renders Atg4D toxic to mammalian cells. Atg4D-mediated apoptosis is facilitated by a putative C-terminal BH3 domain and is preceded by recruitment of Atg4D to mitochondria, suggesting that Atg4D plays an important role at the interface between autophagy and apoptosis.
Results
Atg4D is cleaved by caspase-3 at DEVD63K
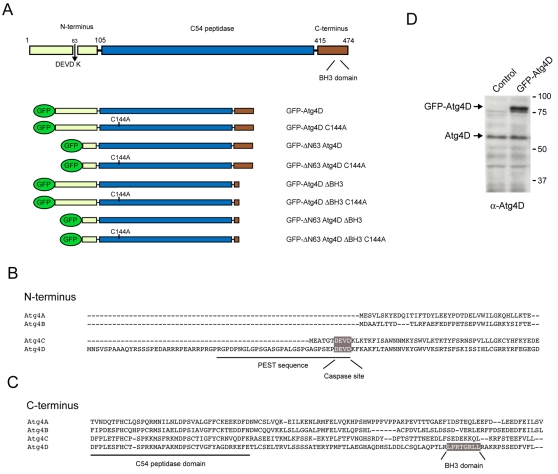
The mammalian autophagin family comprises four presumed paralogues, Atg4A-Atg4D. These display sequence identity within the central, C54 peptidase domain (Marino et al., 2003), and have divergent N- and C-termini that may confer substrate specificity and/or encode for specific regulatory properties (Fig. 1A-C). Overall, closest pair-wise sequence identity is shared by Atg4A and Atg4B and by Atg4C and Atg4D (Fig. 1B,C) (Marino et al., 2003). The extended N-terminus of Atg4D contains a canonical caspase cleavage sequence (DEVD63K) (Fig. 1B). This sequence is conserved in Atg4C, but is missing from Atg4A and Atg4B. The N-terminus of Atg4D is also notable for the presence of a putative PEST (proline-, glutamic-acid-, serine- and threonine-rich) sequence that terminates at the DEVD63K motif (Fig. 1B), suggesting that the protein has a short half-life. These interesting features prompted us to investigate the roles of Atg4D in autophagy, and its regulation during apoptosis. First, we used the reverse transcription polymerase chain reaction (RT-PCR) to assess the relative expression levels of ATG4 and ATG8 paralogues in HeLa, A431 and MCF-7 cell-lines: expression of all ATG4 and ATG8 paralogues is similar, with the exception of ATG4B and GABARAP-L1, which are underrepresented in HeLa and MCF-7 cells, respectively (supplementary material Fig. S1). We then raised a polyclonal antibody against the divergent C-terminus of Atg4D to monitor protein expression and to determine its localisation in human cell-lines. This antibody recognised a weak band on SDS-PAGE gels at the expected Mr (∼54 kDa) (Fig. 1D), and stained the cytoplasm uniformly, with a notable enrichment at the centrosome (supplementary material Fig. S2). This staining pattern was identical in cells overexpressing GFP-Atg4D (supplementary material Fig. S2).
Fig. 1.
Domain structure and alignment of human autophagins, and expression of Atg4D. (A) Domain structure of human Atg4D showing the location of the features described in parts (B) and (C), and cartoons depicting the various Atg4D constructs used in this study. C144 is an essential cysteine in the active site triad of Atg4D. (B) Alignment of the N-termini of the four presumed, human Atg4 paralogues (Atg4A-Atg4D). Atg4C and Atg4D have extended N-termini, containing putative caspase cleavage motifs (DEVD). (C) Alignment of the C-termini of human Atg4A-Atg4D. Atg4D contains a putative BH3 domain towards the end of the molecule (see text for details). (D) Immunoblot of HeLa cell extracts using polyclonal anti-Atg4D antiserum. Control and GFP-Atg4D transfected cell lysates are shown. A band at the expected molecular mass of the endogenous protein (∼54 kDa) is indicated, as is a ∼78 kDa band representing GFP-Atg4D.
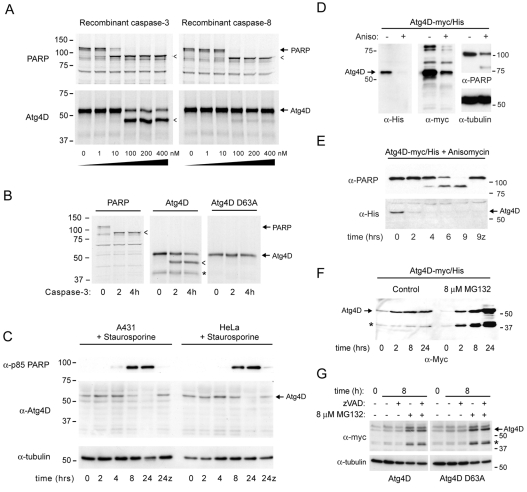
To determine whether the DEVD63K sequence of Atg4D is a valid caspase site, 35S-labelled Atg4D was translated in vitro, and subjected to cleavage using increasing concentrations of recombinant caspase-3 or caspase-8 (Fig. 2A). As a positive control we used human poly-(ADP-ribose) polymerase (PARP), which contains an equivalent caspase-cleavage motif (DEVD213G). Caspase-3 and caspase-8 efficiently cleaved PARP in vitro (Fig. 2A). Caspase-3 also cleaved Atg4D very efficiently in vitro, generating a ∼47 kDa product, but very minimal Atg4D cleavage was detected using caspase-8 (Fig. 2A). To confirm that cleavage occurred at DEVD63K, we mutated the critical aspartic acid to alanine and tested this mutant against recombinant caspase-3 (Fig. 2B). As anticipated, cleavage was completely blocked in the Atg4D DEVA63K mutant (Fig. 2B). To test whether Atg4D was subject to caspase cleavage in living cells, we induced apoptosis by staurosporine treatment of A431 and HeLa cells and immunoblotted cell lysates for Atg4D and for caspase-cleaved PARP (p85) (Fig. 2C). Significant PARP cleavage was detected at around 8 hours, at which point the intensity of Atg4D had diminished. Atg4D could no longer be detected 24 hours following addition of staurosporine (Fig. 2C). Importantly, loss of Atg4D was prevented by the caspase inhibitor zVAD.fmk (Fig. 2C).
Fig. 2.
Caspase cleavage and cellular turnover of Atg4D. (A) 35S-labelled human Atg4D translated in vitro and subjected to caspase cleavage using increasing concentration of recombinant caspase-3 and caspase-8 (90 minutes at 30°C). Autoradiographs of SDS-PAGE gels are shown. Human PARP is shown as a positive control. Cleavage products are indicated by arrowheads. (B) Site-directed mutagenesis of the aspartate at position 63 (D63A) prevented caspase cleavage of Atg4D. 35S-labelled, recombinant wild type and caspase-resistant (D63A) Atg4D incubated for increasing times with 100 nM caspase-3. Rapid cleavage of wild type but not D63A was seen (arrowhead). An additional band was observed at all time-points in the wild-type Atg4D samples (*). (C) Cleavage of Atg4D in living cells undergoing staurosporine-induced apoptosis. Human A431cells (to the left) and HeLa cells (to the right) were incubated with 1 μM staurosporine for increasing lengths of time, and extracts were immunoblotted for caspase-cleaved p85 PARP and Atg4D. Tubulin is shown as a loading control. By 24 hours, no Atg4D was detected, and this was prevented by the caspase inhibitor zVAD.fmk (z). (D-G) Proteasome-mediated turnover of human Atg4D. (D) HeLa cells transiently expressing myc/His-tagged Atg4D (Atg4D-myc/His) were incubated with anisomycin (6 hours), and extracts were immunoblotted with antibodies recognising myc, His, PARP and α-tubulin. The signal for Atg4D was diminished in anisomycin-treated samples. (E) HeLa cells transiently expressing Atg4D-myc/His were treated with anisomycin for up to 9 hours in the absence or presence of zVAD.fmk (z), and immunoblotted for PARP and Atg4D. Loss of Atg4D-myc/His preceded PARP cleavage and was not prevented by zVAD.fmk. (F-G) HeLa cells transiently expressing wild-type or caspase-resistant (G) Atg4D-myc/His were incubated in the absence or presence of the proteasome inhibitor MG132. MG132 caused the accumulation of full-length and ∼42 kDa Atg4D (*) both in wild-type cells and in cells expressing caspase-resistant Atg4D, in the absence or presence of zVAD.fmk.
The putative N-terminal PEST sequence (amino acids 32-63) suggested that Atg4D might be subject to proteasomal degradation. Accordingly, treatment of HeLa cells transiently expressing myc/His-tagged Atg4D with the protein synthesis inhibitor anisomycin, caused rapid loss of expressed protein (Fig. 2D). Degradation preceded anisomycin-induced apoptosis (determined by PARP cleavage; Fig. 2D,E), and was not prevented by zVAD.fmk (Fig. 2E). To test for proteasomal involvement in Atg4D turnover, we incubated cells transiently expressing Atg4D-myc/His in the absence or presence of the proteasome inhibitor MG132, and compared protein levels over 24 hours (Fig. 2F). MG132 caused accumulation of Atg4D (Fig. 2F), and also of a 42 kDa product (Fig. 2F) that was also sometimes seen when wild-type Atg4D was translated in vitro (see Fig. 2B). We reasoned that this product might be generated by caspase cleavage at an alternative site. To test this, we transiently expressed wild-type and D63A Atg4D-myc/His in HeLa cells, and incubated them in the absence or presence of zVAD.fmk alone or in combination with MG132 (Fig. 2G). The 42 kDa Atg4D was detected at similar levels in wild-type and in cells expressing D63A Atg4D (Fig. 2G), and was also observed in the presence of zVAD.fmk (Fig. 2G), suggesting that caspases are not needed for Atg4D processing to its 42 kDa form.
Caspase cleavage of Atg4D stimulates GABARAP-L1 priming and delipidation
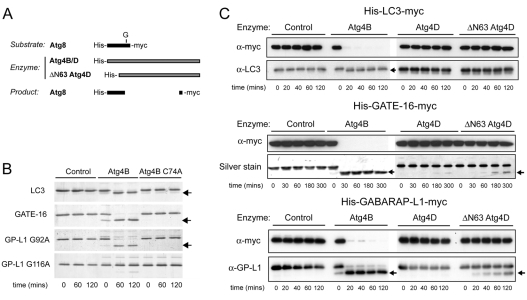
To investigate the role of Atg4D as a regulator of Atg8, we performed in vitro priming assays using recombinant Atg4D, and dual-tagged Atg8 paralogues (see Fig. 3A). For these assays, recombinant Atg4B was used as a positive control (Fig. 3B). As previously reported (Hemelaar et al., 2003; Tanida et al., 2004), LC3, GABARAP-L1 and GATE-16 were each rapidly cleaved by Atg4B, as judged by gel shift and loss of the C-terminal myc tag (Fig. 3B,C). Site-directed mutagenesis confirmed that GABARAP-L1 cleavage occurs at glycine 116, and cleavage of each Atg8 paralogue was prevented using active-site mutant Atg4B C74A (Fig. 3B). By contrast, full-length Atg4D was inactive against all substrates tested (Fig. 3C). Surprisingly, when these assays were repeated using the caspase-cleavage mimic ΔN63 Atg4D, we detected priming activity towards GABARAP-L1 (Fig. 3C). Very weak priming activity was also detected towards GATE-16, although no LC3 priming was observed (Fig. 3C). Note that ΔN63 Atg4D priming of GABARAP-L1 did not run to completion (Fig. 3C).
Fig. 3.
Action of Atg4D and ΔN63 Atg4D in Atg8 priming assays. (A) Schematic of the reagents used for in vitro assays. Atg8 paralogues were double-tagged (6xHis-Atg8-myc). (B) Priming efficiency of Atg4B and its C74A active-site mutant against LC3, GATE-16 and GABARAP-L1 (G92A and G116A mutants). Arrows mark the positions of primed products. (C) In vitro Atg8 priming assays. Recombinant Atg8 paralogues were incubated with Atg4B, Atg4D and ΔN63 Atg4D and analysed by immunoblotting and/or by silver staining. Priming products are indicated by arrows.
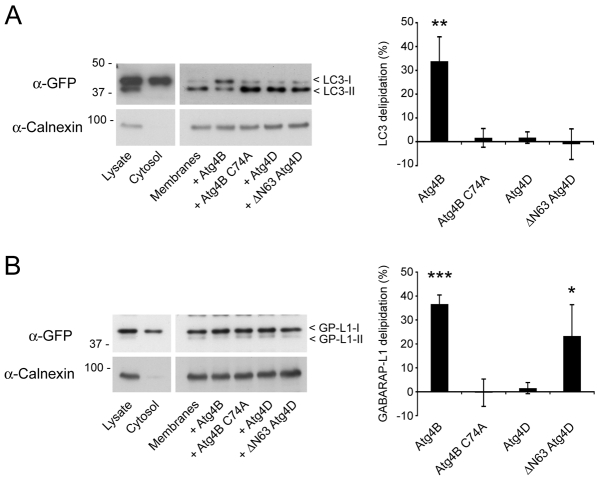
We next investigated the capacity of Atg4B, Atg4D and ΔN63 Atg4D to delipidate Atg8 paralogues by incubating autophagosome-enriched membrane fractions from cell-lines stably expressing YFP-LC3 or GFP-GABARAP-L1 with recombinant Atg4 (Fig. 4). Addition of Atg4B resulted in efficient delipidation of LC3 and GABARAP-L1, as judged by gel shift from more mobile, lipidated forms (Atg8-II) to slower running unmodified protein (Atg8-I) (Fig. 4A,B). As expected, this activity was blocked using the Atg4B C74A mutant (Fig. 4A,B). Full-length Atg4D was not able to delipidate LC3 or GABARAP-L1 on autophagosomes (Fig. 4A,B); however, significant GABARAP-L1 delipidation (Fig. 4B), but not LC3 delipidation (Fig. 4A), was observed when membranes were incubated with ΔN63 Atg4D. These observations suggest that upon caspase cleavage, Atg4D acquires GABARAP-L1 priming and delipidation activity, indicating that caspases stimulate Atg4D-coordinated autophagy.
Fig. 4.
Action of Atg4D and ΔN63 Atg4D in Atg8 delipidation assays. Autophagosome-enriched membranes from cells stably expressing YFP-LC3 (A) and GFP-GABARAP-L1 (B) were incubated with recombinant Atg4B (wild type and C74A), Atg4D and ΔN63 Atg4D, and analysed by immunoblotting with anti-GFP antibodies. Lipidated LC3 (LC3-II) and GABARAP-L1 (GP-L1-II) migrated faster than the delipidated products (LC3-I and GP-L1-I, respectively). Graphs show means ± s.d. of three independent experiments. Student's t-test: *P<0.05, **P<0.01, *** P<0.001.
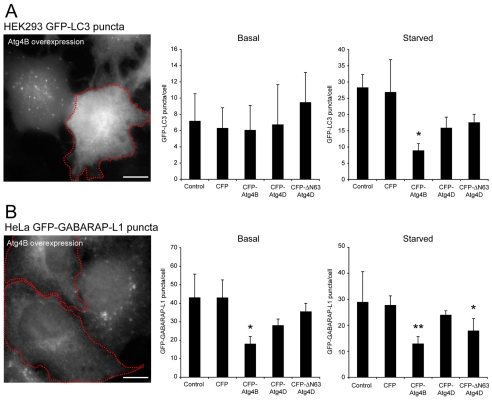
ΔN63 Atg4D overexpression restricts GABARAP-L1 autophagosome formation in living cells
To test whether Atg4D or ΔN63 Atg4D influence autophagosome biogenesis in living cells, we overexpressed CFP-tagged Atg4 constructs in cell-lines stably expressing GFP-LC3 or GFP-GABARAP-L1 and counted autophagosome numbers at basal levels and after autophagy stimulation (2 hours of amino acid and serum starvation) (Fig. 5). Overexpression of Atg4B depletes LC3-positive autophagosomes in HeLa cells (Tanida et al., 2004), either by over-stimulating Atg8 delipidation (Tanida et al., 2004), or by sequestering LC3 to prevent de novo autophagosome formation (Fujita et al., 2008). In starved HEK293 cells stably expressing GFP-LC3, we recorded a significant decrease in GFP-positive puncta upon overexpression of CFP-Atg4B (Fig. 5A). Similarly, CFP-Atg4B significantly reduced the relative numbers of puncta in starved and unstarved HeLa cells expressing GFP-GABARAP-L1 (Fig. 5B). Note that GFP-GABARAP-L1 puncta were always abundant in unstarved HeLa cells, and their relative numbers typically decreased during starvation (see Fig. 5B). Notably, puncta appearance changed from perinuclear and large to dispersed and small during starvation (see supplementary material Fig. S3), suggesting that in HeLa cells, GABARAP-L1 associates with distinct membrane compartments when converting between unmodified and lipidated forms. This is supported by immunoblots demonstrating that membranes from GFP-GABARAP-L1 stable HeLa cells are enriched for unmodified GABARAP-L1 (Fig. 4B). In addition, unlipidated GABARAP-L1 is resistant to salt washing, suggesting tight membrane association (supplementary material Fig. S4).
Fig. 5.
Overexpression supports a role for ΔN63 Atg4D in GABARAP-L1 processing in living cells. (A) HEK293 cells stably expressing GFP-LC3 and (B) HeLa cells stably expressing GFP-GABARAP-L1 were transiently transfected with CFP, CFP-Atg4B, CFP-Atg4D or CFP-ΔN63 Atg4D. Bar charts show autophagosome numbers (mean ± s.d. of three independent experiments; n>50 cells for each treatment) assessed under basal and starvation conditions (see Materials and Methods). Example images of GFP-LC3 and GFP-GABARAP-L1 puncta are shown to the left for Atg4B-expressing cells (red outline; scale bar: 10 μm). Additional representative images are shown in supplementary material Fig. S3. Student's t-test for values compared with CFP controls: *P<0.05, **P<0.01.
We repeated the above experiments in cells transiently overexpressing CFP-Atg4D or CFP-ΔN63 Atg4D (Fig. 5). In cells expressing either CFP-Atg4D or CFP-ΔN63 Atg4D changes in GFP-LC3 puncta numbers were not statistically significant (Fig. 5A). By contrast, in the GFP-GABARAP-L1 HeLa cell-line, CFP-ΔN63 Atg4D overexpression – but not Atg4D overexpression – significantly reduced puncta numbers during starvation (Fig. 5B). These observations support the proposed links between caspase cleavage of Atg4D and the stimulation of GABARAP-L1 processing in living cells.
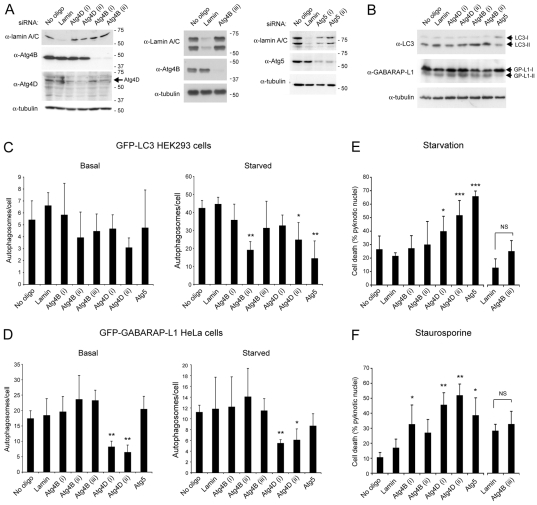
siRNA silencing of Atg4D expression constrains autophagy and sensitises cells to stress-induced death
In a further attempt to determine its roles in autophagy, we silenced expression of Atg4D by siRNA. We achieved a reasonable level of knockdown of Atg4D expression using two independent oligonucleotides (Fig. 6A), and near-complete depletion of Atg4B with three independent oligonucleotides (Fig. 6A). We also silenced Atg5 expression as an autophagy control (Fig. 6A). We assessed the impact of silencing these genes upon autophagy by comparing the ratios of unlipidated to lipidated LC3 and GABARAP-L1 in extracts from starved HeLa cells (Fig. 6B), and by counting puncta in cells expressing GFP-GABARAP-L1 and GFP-LC3 (Fig. 6C,D). In starved HeLa cells (1 hour starvation), Atg5 siRNA reduced the lipidation of LC3 and GABARAP-L1 (as judged using SDS-PAGE, by failure to shift to the more mobile “II” form) (Fig. 6B). However, silencing of the genes expressing Atg4D or Atg4B had no significant effect on LC3 or GABARAP-L1 lipidation (Fig. 6B). In starved GFP-LC3 HEK293 cells, Atg5 silencing significantly reduced autophagosome numbers (Fig. 6C). Atg4D silencing did not alter LC3 puncta numbers at basal level; however, one oligonucleotide significantly reduced LC3 puncta numbers in starved HEK293 cells (Fig. 6C). Importantly, significant reductions in GFP-GABARAP-L1 autophagosome numbers were recorded in HeLa cells silenced for Atg4D (both oligonucleotides) at basal levels and following starvation, although none of the other silencing oligonuceotides were effective (Fig. 6D). One possibility is that sufficient residual Atg5 remains following silencing to mediate GABARAP-L1 autophagosome formation. There were inconsistencies in the assessment of Atg8 lipidation and autophagosome formation by immunoblotting (Fig. 6B) and by microscopy (Fig. 6C,D), and we believe that counting autophagosome numbers by microscopy is the more sensitive assay.
Fig. 6.
siRNA-mediated Atg4D silencing sensitises cells to starvation and cell death. (A) Immunoblots of HeLa cells silenced for Atg4D, Atg4B and Atg5. Silencing was achieved over 72 hours and lamin was included as a negative control. An additional oligonucleotide for Atg4B is included (centre panel) because of inconsistencies in the responses of cells to Atg4B knockdown (see text). (B) Immunoblots of HeLa cells treated with the oligonucleotides indicated for 72 hours and subjected to starvation (1 hour). Lipidated Atg8 paralogues (LC3-II; GP-L1-II) migrate slightly further in SDS-PAGE. (C,D) Atg4 silencing abrogates GABARAP-L1 autophagosomes formation. HEK293 cells stably expressing GFP-LC3 (C) and HeLa cells stably expressing GFP-GABARAP-L1 (D) were depleted for Lamin, Atg4B, Atg4D or Atg5 (72 hours), then subjected or not to starvation (1 hour) before being fixed and analysed for autophagosome numbers. Silencing Atg4D but not Atg4B restricted GABARAP-L1 puncta formation. Silencing Atg5, Atg4B (one oligonucleotide from three) and Atg4D (one oligonucleotide from two) significantly reduced LC3 autophagosome formation in starved cells. Bar charts shows means ± s.d. of three independent experiments (150-450 cells for each treatment). (E) Depletion of Atg5 and Atg4D sensitises cells to starvation-induced cell death. HeLa cells were silenced as indicated (72 hours), then incubated for 5 hours in starvation media. (F) Depletion of Atg5 and Atg4D sensitised cells to staurosporine-induced cell death. HeLa cells were silenced as indicated (72 hours), then incubated for 5 hours with 1 μM staurosporine. For parts (E) and (F), cell death was assessed by DAPI fluorescence, scoring for pyknotic chromatin. Means ± s.d. are shown for three independent experiments. Student's t-test for values compared to the lamin controls: *P=0.5, **P=0.01, ***P=0.001.
We then tested the effect of silencing autophagy protein expression upon cell survival during prolonged starvation (5 hours). Silencing of the gene expressing Atg5 significantly sensitised HeLa cells to cell death during prolonged starvation (Fig. 6E), consistent with the essential role for this gene in autophagy (Mizushima et al., 2001). Importantly, silencing Atg4D expression – but not that of Atg4B – also significantly sensitised HeLa cells to starvation-induced cell death (Fig. 6E). We then assessed the effect of silencing genes expressing Atg4D, Atg4B and Atg5 upon staurosporine-induced apoptosis (Fig. 6F). Interestingly, cells silenced for Atg5, Atg4D (both oligonucleotides) and Atg4B [oligonucleotide (i) only] died at accelerated rates following staurosporine treatment (Fig. 6F). To rule out the possibility of off-targets effects of Atg4B oligonucleotide (i), we introduced a third Ag4B silencing oligonucleotide (see Fig. 6A). This reagent produced similar results to Atg4B oligonucleotide (ii) in that it failed to sensitise cells to starvation or staurosporine-induced cell death (Fig. 6E,F). In these experiments, cell death was assessed by monitoring the appearance of pyknotic chromatin, and was consistent with cells undergoing apoptosis (see example images in supplementary material Fig. S5). These data suggest that Atg4D – but not Atg4B – plays an important role in the survival response of cells depleted of nutrients and growth factors or following treatment with the apoptosis-inducing agent staurosporine.
Atg4D is recruited to mitochondria as a prelude to apoptosis
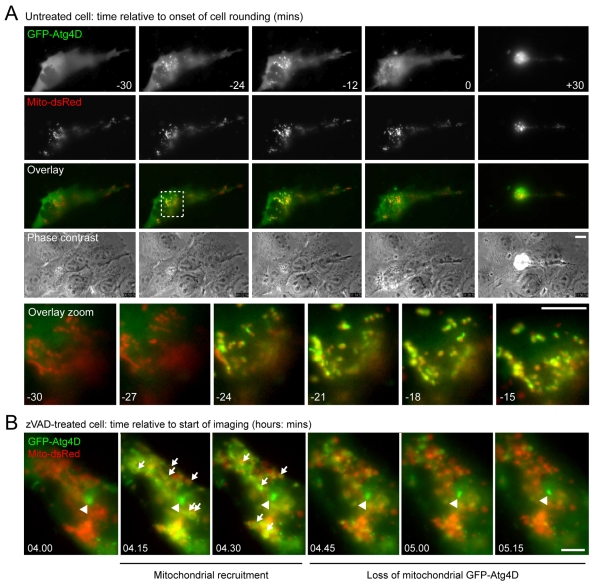
We noted that GFP-Atg4D transfected cells were often rounded, with condensed, fragmented chromatin – typical features of cells undergoing apoptosis. To investigate the mechanisms for Atg4D-induced cytotoxicity, we used time-lapse microscopy to image cells expressing GFP-Atg4D. In live-cell imaging experiments, apoptosis was often preceded by the sudden recruitment of GFP-Atg4D to mitochondria (Fig. 7A; supplementary material Movie 1). Recruitment was maximal between 12 and 24 minutes before cell rounding (Fig. 7A; supplementary material Movie 1), similar to the timing of SMAC release from mitochondria (data not shown) and concomitant with loss of mitochondrial membrane potential in apoptotic HeLa and A431 cells (data not shown). This suggests that recruitment of GFP-Atg4D correlated with mitochondrial dysfunction and release of pro-apoptotic factors. Interestingly, co-alignment of Atg4D with mitochondria was transient, lasting for ∼20 minutes, and at the point of cell rounding, most mitochondria had become depleted of GFP-Atg4D (Fig. 7A; supplementary material Movie 1). In late apoptotic cells, Atg4D formed aggregates that were distinct from the collapsed mitochondrial population (Fig. 7A; supplementary material Movie 1). We do not believe that mitochondrial recruitment is linked to caspase cleavage of Atg4D because: (i) mitochondrial recruitment was also observed in zVAD.fmk-treated, caspase-inhibited cells (Fig. 7B; supplementary material Movie 2), and (ii) recruitment was observed in cells expressing caspase-resistant GFP-Atg4D (DEVA63K) (data not shown). Of note, in zVAD.fmk-treated cells, GFP-Atg4D was recruited in rings around the outer surface of mitochondria, and remained associated with mitochondria for a similar length of time (∼20 minutes), before dissipating into the cytoplasm (Fig. 7B; supplementary material Movie 2). Importantly, mitochondrial recruitment was not necessary for the induction of apoptosis, because in cells treated with apoptosis-inducing agents such as staurosporine, only a subset of cells expressing GFP-Atg4D displayed Atg4D translocation prior to undergoing apoptosis (supplementary material Movie 3).
Fig. 7.
GFP-Atg4D is recruited to mitochondria as a prelude to apoptotic cell death. (A) Time-lapse analysis of an A431 cell transiently co-expressing GFP-Atg4D (green) and Mito-dsRed (red). GFP-Atg4D was rapidly recruited to mitochondria ∼24 minutes before cell rounding and apoptosis (note, yellow signal in the zoomed panels). These sequences were taken from supplementary material Movie 1. Scale bars: 10 μm. (B) Caspase activity is not required for mitochondrial recruitment of GFP-Atg4D. Time-lapse analysis of mitochondrial GFP-Atg4D recruitment in a zVAD.fmk-treated A431 cell. Note the transient appearance of rings of GFP-Atg4D (green) in proximity to mitochondria (red) (arrows). Centrosomal GFP-Atg4D is indicated by an arrowhead. These sequences were taken from supplementary material Movie 2. Scale bar: 10 μm.
The C-terminus of Atg4D contains a proposed BH3 domain that participates in cell death
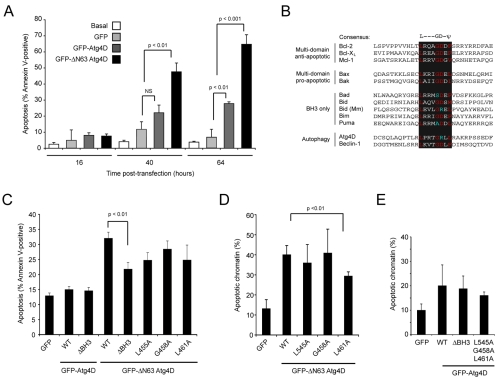
We next analysed the effects of long-term overexpression of Atg4D constructs on cell viability. HeLa cells were transfected with GFP, GFP-Atg4D or GFP-ΔN63 Atg4D and scored for the numbers of GFP-expressing apoptotic cells over a 64-hour period by staining with fluorescent annexin V (Fig. 8A). A higher proportion of cells expressing GFP-Atg4D were apoptotic than cells expressing GFP alone (27.81±1.1% compared with 6.86±5.0% at 64 hours). However, cells expressing GFP-ΔN63 Atg4D were much more likely to be annexin V positive (64.74±5.9%), suggesting that ΔN63 Atg4D is significantly more toxic (Fig. 8A). Importantly, we also observed mitochondrial recruitment of GFP-ΔN63 Atg4D as a prelude to cell death in live-cell imaging experiments (supplementary material Fig. S6), suggesting that the toxicities mediated by GFP-Atg4D and GFP-ΔN63 Atg4D are equivalent.
Fig. 8.
Assessment of Atg4D-induced cytotoxicity. (A) Overexpression of Atg4D and ΔN63 Atg4D triggers apoptosis. HeLa cells expressing GFP, GFP-Atg4D or GFP-ΔN63 Atg4D were labelled with fluorescent annexin V at 16, 40 and 64 hours post-transfection. Means ± s.d. are shown for three independent experiments with the P-values for Student's t-test. (B-E) Atg4D contains a putative C-terminal BH3 domain that facilitates overexpression-induced apoptosis. (B) Alignment of BH3 domains from Bcl-2 family members with pro- or anti-apoptotic roles, Beclin-1 and Atg4D (Mm, Mus musculus). (C) Analysis of apoptosis in cells overexpressing GFP-Atg4D, GFP-ΔN63 Atg4D and BH3 mutants by annexin-V-binding (40 hours). (D,E) Analysis of apoptosis in cells overexpressing GFP-Atg4D, GFP-ΔN63 Atg4D and BH3 mutants by scoring pyknotic chromatin (40-hour expression). Data show means ± s.d. of three independent experiments, scored blind. P-values for Student's t-test are shown.
Analysis of the divergent C-terminus of Atg4D suggested the presence of a BH3 domain (Fig. 8B). BH3 domains function as pro-apoptotic triggers upon insertion into the hydrophobic cleft of multi-domain Bcl-2 family members. BH3 domain sequences are shown in Fig. 8B. Note that, in common with mouse Bid, the aspartic acid in the core consensus sequence of Atg4D is substituted for arginine (Fig. 8B). To investigate the potential role of this sequence in Atg4D-mediated cytotoxicity, we overexpressed wild-type GFP-Atg4D, GFP-ΔN63 Atg4D and similar constructs with the BH3 domain deleted (ΔBH3) or with alanine substituted for key residues, and then scored apoptosis in the transfected population after 40 hours using annexin V binding (Fig. 8C) and by assessing chromatin morphology (Fig. 8D,E). We recorded a statistically significant reduction in cell death in GFP-ΔN63 Atg4D lacking the C-terminal BH3 domain (ΔBH3; Fig. 8C), and a significant reduction in cell death in cells expressing L461A GFP-ΔN63 Atg4D (pyknotic chromatin assay) (Fig. 8D). However, we did not observe significant differences in toxicity of GFP-Atg4D constructs lacking the BH3 domain (Fig. 8C,E). Together, these data suggest that C-terminal determinants that may or may not constitute a BH3 domain contribute to Atg4D-mediated cytotoxicity.
Autophagy is not required for Atg4D-mediated cytotoxicity
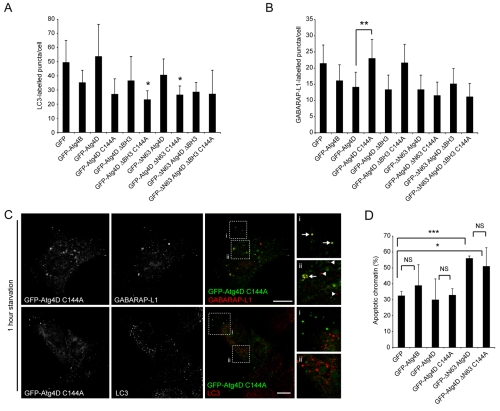
One alternative explanation for the cytotoxic effect of Atg4D overexpression is a change in the basal rate of autophagy, causing reduced cell viability. So, to examine the relationships between Atg4-mediated autophagy and apoptosis, we first tested the autophagy response in HeLa cells transiently expressing GFP, GFP-Atg4B or GFP-Atg4D, or expressing GFP-tagged Atg4D, ΔN63 Atg4D and ΔBH3 Atg4D constructs without or with C144A point mutations (an active site mutation equivalent to Atg4B C74A) (Fig. 1A; Fig. 9A,B). As described in Fig. 5, overexpression of Atg4B reduced LC3 autophagosome numbers in starved HEK293 cells, but wild-type Atg4D had no effect (Fig. 9A). Interestingly, we did observe reductions in LC3-labelled autophagosomes in HeLa cells overexpressing Atg4D C144A mutants, suggesting that this mutation causes dominant-negative regulation of LC3 processing (Fig. 9A). We then repeated this approach by staining for GABARAP-L1-labelled puncta (Fig. 9B). In contrast to the LC3 data, accumulation of GABARAP-L1 puncta during starvation was not prevented by overexpression of C144A mutant constructs (Fig. 9B). Instead, in cells expressing Atg4D C144A, GABARAP-L1 labelled puncta were significantly increased (Fig. 9B). On close inspection of cells expressing GFP-Atg4D C144A, we observed peripheral GFP-labelled cytoplasmic puncta that were reminiscent of autophagosomes in both their size and distribution (Fig. 9C). Importantly, these structures co-aligned with a subpopulation of GABARAP-L1-labelled puncta (top panels, Fig. 9C), but did not co-localise with LC3-labelled autophagosomes (bottom panels, Fig. 9C). These observations support the specificity of Atg4D for GABARAP-L1, and suggest that active-site cysteine mutants of Atg4D become locked in complex with GABARAP-L1, consistent with a recent study of LC3 processing by Atg4B C74A (Fujita et al., 2008). We believe that these Atg4D-C144A- and GABARAP-L1-positive structures accounted for the increase in relative puncta numbers in the automated scoring system that we have used.
Fig. 9.
Dissociation of Atg4D-mediated autophagy from overexpression-induced apoptosis. (A,B) HeLa cells transiently expressing the constructs shown (refer to Fig. 1A) were fixed and labelled with anti-LC3 antibodies (A) or GABARAP-L1 antibodies (B). Autophagosome numbers were counted in three independent experiments (>100 cells; bars represent means ± s.d.) In (A), means are compared with GFP control; *P<0.5 using the Student's t-test. (C) Inactive Atg4D colocalises with a population of GABARAP-L1-positive puncta. Confocal images are shown of starved (1 hour) HeLa cells transiently expressing GFP-Atg4D C144A, fixed and stained with antibodies against LC3 and GABARAP-L1. GFP-positive puncta were observed and these colocalised with a population of GABARAP-L1-positive puncta (arrows in zoom panels) but not with LC3 puncta (GABARAP-L1 puncta not co-labelled with GFP-Atg4D C144A are indicated by arrowheads). Scale bar: 10 μm. (D) Atg4D endopeptidase activity is not required for overexpression-induced apoptosis. HeLa cells transiently overexpressing (64 hours) GFP-Atg4B, GFP-Atg4D, GFP-ΔN63 Atg4D and Atg4D C144A constructs (see Fig. 1A) were scored for pyknotic chromatin. Data show means ± s.d. of three independent experiments. Student's t-test: *P<0.5, **P<0.01, ***P<0.001. NS, not significant.
If the cytotoxic effect of Atg4D overexpression is linked to changes in autophagic capacity, then we would predict that: (i) overexpression of Atg4B – which has a marked impact on both LC3 and GABARAP-L1 puncta formation (see Fig. 5) – would be significantly more toxic than overexpression of Atg4D and ΔN63 Atg4D; and (ii) overexpression of C144A mutants of Atg4D would be significantly more toxic than overexpression of unmodified equivalent constructs. To test these predictions, we overexpressed GFP-tagged Atg4B, Atg4D, ΔN63 Atg4D and C144A mutants in HeLa cells for 64 hours, and counted cell death in fixed cells by scoring for pyknotic chromatin (Fig. 9D). Overexpression of GFP-Atg4B and GFP-Atg4D did not elevate cell death significantly above background, suggesting that a general reduction in autophagy efficiency is not sufficient to induce cell death in this context (Fig. 9D). Moreover, although overexpression of GFP-ΔN63 Atg4D did cause a significant increase in cell death, neither GFP-Atg4D C144A nor GFP-ΔN63 Atg4D C144A was more toxic than its unmodified counterpart (Fig. 9D). These data suggest that the cytotoxic effect of GFP-ΔN63 Atg4D is unlikely to occur as a consequence of altered autophagy.
Atg4D is recruited to mitochondria in response to oxidative damage
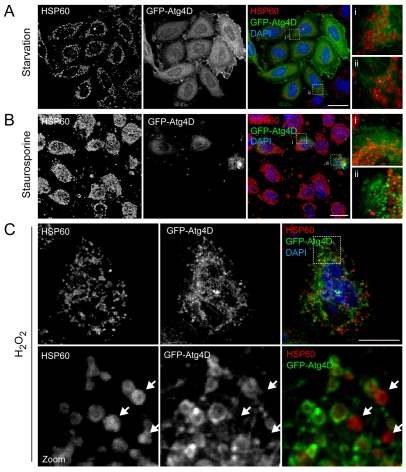
The above observations suggest that Atg4D-mediated cytotoxicity does not result from altered autophagy, but may instead be caused by mitochondrial targeting of Atg4D via its proposed C-terminal BH3 domain. The factors driving mitochondrial recruitment of Atg4D in pre-apoptotic cells remain undetermined. One possible explanation is that Atg4D acquires affinity for damaged mitochondria, perhaps as a component of a targeted pathway for mitochondrial autophagy (mitophagy). To test this, we treated cells transiently expressing GFP-Atg4D with reagents known to induce mitochondrial stress, autophagy and ultimately apoptosis (serum- and nutrient-free media, staurosporine and low concentrations of H2O2), and analysed Atg4D and mitochondrial distribution by confocal microscopy. In starved cells, we did not observe mitochondrial Atg4D recruitment, although a subpopulation of cells contained bright GFP-Atg4D puncta that lay adjacent to mitochondria (Fig. 10A). Similarly, in viable, staurosporine-treated cells, GFP-Atg4D labelling remained cytoplasmic with no evidence of mitochondrial co-alignment (Fig. 10B). Late apoptotic cells in the staurosporine-treated population did accumulate aggregates of Atg4D; however, these did not co-align with mitochondria (Fig. 10B). Strikingly, in a sub-fraction of H2O2-treated cells, GFP-Atg4D was localised to rings around the outside of mitochondria (Fig. 10C). Cells with clear evidence of mitochondria-associated GFP-Atg4D typically had distorted nuclei and distended mitochondria, suggesting that these would soon have succumbed to oxidative damage (Fig. 10C). These observations suggest that oxidative stress is one plausible trigger for mitochondrial Atg4D recruitment, and we are currently exploring the molecular basis and physiological consequences of this phenomenon.
Fig. 10.
Analysis of Atg4D localisation in response to cellular stress. HeLa cells transiently expressing GFP-Atg4D were starved (A), treated with staurosporine (1 μM) (B) or treated with hydrogen peroxide (10 nM) (C) for 5 hours, then fixed and labelled with antibodies against HSP60. In a sub-population of starved cells, small GFP-Atg4D puncta were observed in the vicinity of mitochondria (Aii); however, no evidence of mitochondrial Atg4D localisation was observed in starved or staurosporine-treated cells (A,B). Note that in late apoptotic cells Atg4D formed aggregates, but these did not colocalise with mitochondria (Bii; see also supplementary material Movie 1). (C) GFP-Atg4D relocates to mitochondria in a sub-population of H2O2-treated HeLa cells. Mitochondria not labelled with GFP-Atg4D are indicated with arrows. In parts (A) and (B) zoomed panels are shown to the right. Scale bars: 10 μm (A,B), 5 μm (C).
Discussion
Autophagy protects cells from environmental stress (Cuervo, 2004; Levine and Klionsky, 2004), and is regulated by the Atg gene family. Here, we have studied the roles of the endopeptidase Atg4D as a regulator of autophagy and apoptosis. Our data reveal that Atg4D contributes to starvation-induced autophagy, with activity demonstrated towards the Atg8 paralogue GABARAP-L1 (Atg8L). Importantly, Atg4D contains a caspase site within its N-terminus, meaning that Atg4D acts at the regulatory interface between autophagy and apoptosis.
We have used a combination of in vitro biochemical assays with recombinant proteins, and experiments in living cells to investigate the specificity of Atg4D for Atg8 substrates, and to evaluate the effects of Atg4D cleavage upon autophagy and cell viability. Our data suggest that recombinant full-length Atg4D has little or no priming or delipidation activity against LC3, GATE-16 and GABARAP-L1 in vitro, and has minimal impact on LC3 or GABARAP-L1 processing in living cells. However, unlike its native counterpart, caspase-cleaved Atg4D (ΔN63 Atg4D) can prime and delipidate GABARAP-L1 effectively. One possibility is that full-length Atg4D is auto-inhibited. In this regard, the publication of the crystal structure of Atg4B in complex with LC3 while this manuscript was in final revision is particularly significant (Satoo et al., 2009): most notably, the N-terminus of free Atg4B was shown to occlude the catalytic site of the enzyme (Satoo et al., 2009). Accordingly, and in common with Atg4D, an N-terminal truncation of Atg4B displays enhanced Atg8-priming activity in vitro (Satoo et al., 2009). This would provide a plausible mechanistic explanation for the gain of priming and delipidation activity that we have observed following caspase-cleavage of Atg4D. Our data therefore imply that caspases have the potential to stimulate autophagy via Atg4D cleavage, an observation in keeping with evidence linking caspases to the induction of autophagy programmes in Drosophila (Hou et al., 2008; Martin et al., 2007).
We have also assessed the impact of siRNA silencing of autophagy regulators upon autophagy and cell survival during starvation and staurosporine-induced apoptosis. Consistent with the proposed roles for Atg4D in GABARAP-L1 processing, Atg4D silencing had a major impact upon GABARAP-L1 puncta formation (basal and starvation conditions). If the primary role of autophagy is protection from cellular stress, then one would predict that its inhibition would advance cell death during withdrawal of nutrients or growth factors. Importantly, Atg4D (but not Atg4B) silencing markedly sensitised HeLa cells to starvation-induced cell death, suggesting that Atg4D-mediated autophagy contributes to the survival response in starved cells. We have also shown that silencing genes expressing Atg5 and Atg4D markedly sensitises cells to staurosporine-induced apoptosis, suggesting that autophagy acts as a protective mechanism during staurosporine treatment. Interestingly, HeLa cells silenced for Atg5 have been reported elsewhere to be resistant to staurosporine (Yousefi et al., 2006), and while we cannot currently explain why our data differ from those of Yousefi et al. (Yousefi et al., 2006), we note that Atg5 silencing has been shown in an independent study to sensitise HeLa cells to staurosporine (Pyo et al., 2005).
One unexpected finding here was that overexpression of caspase-cleaved ΔN63 Atg4D was potently cytotoxic. Live-cell imaging suggested that Atg4D-mediated toxicity is associated with recruitment to mitochondria before induction of apoptosis. Importantly, recruitment was not blocked by zVAD.fmk, and was still observed in cells expressing caspase-resistant Atg4D (data not shown), suggesting that mitochondrial targeting is independent of caspase cleavage of Atg4D. Two obvious questions arise from these observations: (i) Does Atg4D cause release of mitochondrial factors, perhaps by targeting the Bax or Bak pathways to trigger outer mitochondrial membrane permeabilisation? (ii) Is Atg4D responding to mitochondrial damage, perhaps in a futile attempt to target dysfunctional mitochondria for autophagy? Close inspection of the C-terminus of Atg4D revealed the presence of a putative BH3 domain, and removal or mutagenesis of this domain reduced, but did not eliminate, ΔN63-Atg4D-mediated cytotoxicity. BH3-related domains have been identified in several other proteins that are not otherwise related to the Bcl-2 family, suggesting the potential for interactions between apoptotic Bcl-2 proteins and a wide range of regulatory factors [examples include BRCC2 (Broustas et al., 2004), BNIP3 and BNIP3L (also known as Nix) (Chen et al., 1999), and HER-2 (Strohecker et al., 2008)]. Importantly, cell death mediated by Atg4D overexpression did not result from a change in levels of autophagy, because dominant-negative active site (C144A) mutants did not alter Atg4D-mediated cytotocixity. Importantly, treatment of cells with low concentrations of H2O2 stimulated mitochondrial recruitment of Atg4D, suggesting that Atg4D may have affinity for factor(s) associated with or released from damaged mitochondria. Interestingly, the ubiquitin ligase Parkin has been shown to demarcate damaged mitochondria for autophagic clearance (Narendra et al., 2008). We are currently investigating the molecular basis for Atg4D mitochondrial recruitment, and whether Atg4D contributes to the elimination of dysfunctional mitochondria.
Here, we have shown that human Atg4D contributes to autophagy by targeting the Atg8 paralogue, GABARAP-L1, and in doing so protects cells from starvation and staurosporine-induced cell death. Atg4D is a caspase substrate whose cleavage generates a truncated product that has enhanced GABARAP-L1 priming activity in vitro, and induces cell death when overexpressed in human cells. Intriguingly, Atg4D-mediated cell death is preceded by the rapid recruitment of Atg4D to the surface of mitochondria, and is facilitated by a putative C-terminal BH3 domain. Together, these data suggest that Atg4D is an important factor at the regulatory interface between autophagy and apoptosis.
Materials and Methods
Reagents
Unless otherwise stated, reagents were obtained from Sigma (Poole, UK). Stock solutions of anisomycin (5 mg/ml), staurosporine (1 mM), DAPI (4′,6-diamidino-2-phenylindole; 1 mg/ml), MG132 (10 mM) were stored at –20°C. Recombinant caspases were obtained from Alexis Biochemicals, and were stored at –80°C. Alexa Fluor 594-labelled annexin V was from Molecular Probes.
cDNA constructs
Human ESTs for the following proteins (accession numbers in parentheses) were obtained from the IMAGE consortium cDNA bank (Geneservice): Atg4D (AW024282); Atg4B (BQ276441); GATE-16 (AA085122); LC3 (BQ962882); GABARAP-L1 (CA455333). Atg4D and ΔN63 Atg4D were PCR amplified and cloned in frame into pEG/Y/CFP-C2 (Clontech), pcDNA3.1myc/His and ptrcHisB (Invitrogen). Site-directed mutagenesis was carried out by the QuickChange method (Stratagene) according to the manufacturer's instructions. For in vitro Atg8 priming experiments, Atg8 paralogues were cloned into ptrcHisB and a myc tag was subsequently inserted at the 3′ end to generate His-Atg8-myc. Mito-dsRed was from Clontech. All constructs were verified by sequencing. Transient transfections were carried out using Fugene 6 (Roche) or Genejuice (EMD Biosciences) according to the manufacturers' instructions.
Antibodies
The sheep anti-human Atg4D antibody was raised against a recombinant GST-fusion of the C-terminus of human Atg4D (amino acids 399-474), and affinity-purified against His-tagged, recombinant full-length Atg4D. The sheep anti-human GABARAP-L1 was raised against recombinant, GST-tagged full-length GABARAP-L1. The following antibodies were also used: polyclonal anti-Atg4B [from Yasuo Uchiyama, Tokyo (Yoshimura et al., 2006)]; polyclonal anti-Atg5 (Cell Signaling); monoclonal anti-myc (9E10: Sigma); polyclonal anti-His (Biorad); polyclonal anti-pericentrin (Abcam); monoclonal anti-PARP (Calbiochem); polyclonal anti-p85 PARP (Promega); monoclonal anti-α-tubulin clone B512 (Sigma); polyclonal anti-lamin A/C (Cell Signaling); polyclonal anti-calnexin (Stressgen). Secondary antibodies for immunofluorescence (Alexa Fluor-488 and Alexa Fluor-594) were from Molecular Probes. HRP-tagged secondary antibodies for immunoblotting were from Jackson Immunochemicals.
Cell-lines and the induction of apoptosis and autophagy
Cells were maintained in DMEM supplemented with 10% foetal bovine serum, at 37°C and 5% CO2. HeLa cells stably expressing GFP-GABARAP-L1 and A431 cells stably expressing YFP-LC3 were obtained following transient transfection by selecting positive clones after 500 μg/ml G418 treatment (Lane et al., 2002; Moss et al., 2006). HEK293 cells stably expressing GFP-LC3 were provided by Sharon Tooze (Cancer Research UK, London, UK). Cells were induced into apoptosis by treatment with 5 μg/ml anisomycin or 1 μM staurosporine, in the absence or presence of zVAD.fmk (50 μM). Autophagy was induced by incubation in nutrient-free and serum-free medium [140 mM NaCl; 1 mM CaCl2; 1 mM MgCl2; 5 mM glucose; 20 mM Hepes, pH 7.4; 1% BSA (Axe et al., 2008)]. Cells were then incubated for the indicated times at 37°C with 5% CO2.
Fluorescence microscopy, live-cell imaging and autophagosome quantitation
Wide-field fluorescence images were obtained using an Olympus IX-71 inverted microscope (60× Uplan Fluorite objective 0.65-1.25 NA, at maximum aperture) fitted with a CoolSNAP HQ CCD camera (Photometrics, Tucson, AZ) driven by MetaMorph software (Molecular Devices). Confocal images were obtained using a Leica AOBS SP2 microscope (63× PLAPO objective 1.4 NA) at 0.2 μm z-steps. For immunofluorescence, cells were fixed in 2% formaldehyde (methanol-free, EM grade; TAAB, Aldermaston, UK) followed by permeabilisation with 0.1% Triton X-100, or in –20°C methanol. Cells were routinely stained with DAPI and mounted in Mowiol containing 25 mg/ml DABCO anti-fade. Live-cell imaging was carried out using the Olympus IX-71 system. Halogen lamp illumination was used for both transmitted light and for epifluorescence to extend cell viability (Moss et al., 2006). Cells were maintained in CO2-independent DMEM (Invitrogen, Paisley, UK), at 37°C in 3-cm cell imaging dishes (MatTek, Ashland, MA). Automated autophagosome scoring was carried out using the Top Hat morphology filter in MetaMorph software, selecting for round objects of diameter 5 pixels (=1 μm) or less.
siRNA
HeLa cells were grown in 12-well plates to ∼30% confluency, and were then transfected with silencing oligonucleotides (MWG Biotech or Dharmacon, as indicated) using Oligofectamine (Invitrogen) according to the manufacturer's instructions. The following oligonucleotides were used: for Lamin, 5′-CUGGACUUCCAGAAGAACA-3′; for Atg4D, either (i) 5′-AGAUGACUUCCUGCUGUACTT-3′ or (ii) 5′-GUGGAAGUCUGUGGUCAUCTT-3′; for Atg4B, either (i) 5′-CUGAAGAUGACUUCAAUGATT-3′, (ii) 5′-AGGACGAGAUCUUGUCUGATT-3′ or (iii) 5′-GGAAGGACAGUUACUACUCTT-3′ (all MWG); for Atg5, either (i) 5′-GCAACUCUGGAUGGGAUUGTT-3′, (ii) 5′-AGAAUCAAGUGAUGAUUCATT-3′ (MWG) or (iii) 5′-GGAAUAUCCUGCAGAAGAA-3′ (Dharmacon). Oligonucleotides were reconstituted at 100 μM (MWG) or at 40 μM (Dharmacon) and used at 40 nM (lamin; Atg5) or 200 nM (Atg4D; Atg4B). Total protein extracts were obtained 72 hours post-transfection, and subjected to SDS-PAGE and immunoblotting using the relevant antibodies and anti-tubulin as a loading control. Alternatively, siRNA-treated cells were processed for immunofluorescence microscopy following incubation in starvation media, and autophagosome numbers quantitated as described above. Cell death was determined in fixed, siRNA-treated cells by assessing chromatin morphology following DAPI staining.
Recombinant protein production and in vitro enzyme assays
In vitro synthesis of 35S-labeled recombinant proteins was performed using a coupled in vitro transcription-translation kit (Promega) according to the manufacturer's instructions. Translate (1 μl) was added to 9 μl of Hepes buffer (0.1% CHAPS, 10% (wt/vol) sucrose, 5 mM DTT, 2 mM EDTA, 50 mM Hepes, pH 7.4) containing recombinant caspases 2, 3, or 7 (Alexis Biochemicals) and incubated for 90 minutess at 30°C, or for the times indicated. Samples were diluted into SDS sample buffer and analysed by SDS-PAGE and autoradiography. Recombinant His-Atg8-myc and His-Atg4 paralogues were produced in BL21-(DE3)-RILP-Codon Plus Escherichia coli (Stratagene) and purified using ProBond nickel-chelating resin (Invitrogen). For Atg8 priming experiments, purified recombinant proteins were incubated for the indicated times at 30°C in HEPES buffer, and reactions were stopped by addition of SDS-PAGE sample buffer. For delipidation experiments, postnuclear supernatants (800 g) from GFP-GABARAP-L1 HeLa and YFP-LC3 A431 cells were prepared and membranes were harvested by centrifugation at 40,000 g for 30 minutes at 4°C. Membranes were resupended in Hepes buffer pH 7.4, and incubated for 5 hours at 30°C in the presence of recombinant Atg4 proteins.
Supplementary Material
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/122/14/2554/DC1
We acknowledge the support of the Medical Research Council in providing an Infrastructure Award to establish the School of Medical Sciences Cell Imaging Facility at Bristol University, and the Wolfson Foundation for recent funds to modernise and expand the Facility. We thank the anonymous referees for their comments, which significantly improved this manuscript, colleagues in the autophagy field for reagents, David Stephens for useful discussions, and Jade Cheng for critical reading of the manuscript. This work was supported by a Wellcome Trust Research Career Development Fellowship to J.D.L. (No. 067358). J.D.L. is a Research Councils UK Academic Fellow. Deposited in PMC for release after 6 months.
References
- Axe, E. L., Walker, S. A., Manifava, M., Chandra, P., Roderick, H. L., Habermann, A., Griffiths, G. and Ktistakis, N. T. (2008). Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182, 685-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baehrecke, E. H. (2005). Autophagy: dual roles in life and death? Nat. Rev. Mol. Cell Biol. 6, 505-510. [DOI] [PubMed] [Google Scholar]
- Botti, J., Djavaheri-Mergny, M., Pilatte, Y. and Codogno, P. (2006). Autophagy signaling and the cogwheels of cancer. Autophagy 2, 67-73. [DOI] [PubMed] [Google Scholar]
- Broustas, C. G., Gokhale, P. C., Rahman, A., Dritschilo, A., Ahmad, I. and Kasid, U. (2004). BRCC2, a novel BH3-like domain-containing protein, induces apoptosis in a caspase-dependent manner. J. Biol. Chem. 279, 26780-26788. [DOI] [PubMed] [Google Scholar]
- Chen, G., Cizeau, J., Vande Velde, C., Park, J. H., Bozek, G., Bolton, J., Shi, L., Dubik, D. and Greenberg, A. (1999). Nix and Nip3 form a subfamily of pro-apoptotic mitochondrial proteins. J. Biol. Chem. 274, 7-10. [DOI] [PubMed] [Google Scholar]
- Cuervo, A. M. (2004). Autophagy: in sickness and in health. Trends Cell Biol. 14, 70-77. [DOI] [PubMed] [Google Scholar]
- Fujita, N., Hayashi-Nishino, M., Fukumoto, H., Omori, H., Yamamoto, A., Noda, T. and Yoshimori, T. (2008). An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol. Biol. Cell 19, 4651-4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemelaar, J., Lelyveld, V. S., Kessler, B. M. and Ploegh, H. L. (2003). A single protease, Apg4B, is specific for the autophagy-related ubiquitin-like proteins GATE-16, MAP1-LC3, GABARAP, and Apg8L. J. Biol. Chem. 278, 51841-51850. [DOI] [PubMed] [Google Scholar]
- Holler, N., Zaru, R., Micheau, O., Thome, M., Attinger, A., Valitutti, S., Bodmer, J. L., Schneider, P., Seed, B. and Tschopp, J. (2000). Fas triggers an alternative, caspase-8-independent cell death pathway using the kinase RIP as effector molecule. Nat. Immunol. 1, 489-495. [DOI] [PubMed] [Google Scholar]
- Hou, Y. C., Chittaranjan, S., Barbosa, S. G., McCall, K. and Gorski, S. M. (2008). Effector caspase Dcp-1 and IAP protein Bruce regulate starvation-induced autophagy during Drosophila melanogaster oogenesis. J. Cell Biol. 182, 1127-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura, Y., Kirisako, T., Takao, T., Satomi, Y., Shimonishi, Y., Ishihara, N., Mizushima, N., Tanida, I., Kominami, E., Ohsumi, M. et al. (2000). A ubiquitin-like system mediates protein lipidation. Nature 408, 488-492. [DOI] [PubMed] [Google Scholar]
- Jin, S. and White, E. (2007). Role of autophagy in cancer: management of metabolic stress. Autophagy 3, 28-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Dalton, V. M., Eggerton, K. P., Scott, S. V. and Klionsky, D. J. (1999). Apg7p/Cvt2p is required for the cytoplasm-to-vacuole targeting, macroautophagy, and peroxisome degradation pathways. Mol. Biol. Cell 10, 1337-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo, Y. and Kondo, S. (2006). Autophagy and cancer therapy. Autophagy 2, 85-90. [DOI] [PubMed] [Google Scholar]
- Lane, J. D., Lucocq, J., Pryde, J., Barr, F. A., Woodman, P. G., Allan, V. J. and Lowe, M. (2002). Caspase-mediated cleavage of the stacking protein GRASP65 is required for Golgi fragmentation during apoptosis. J. Cell Biol. 156, 495-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. B., Kim, S., Lee, J., Park, J., Lee, G., Kim, Y., Kim, J. M. and Chung, J. (2007). ATG1, an autophagy regulator, inhibits cell growth by negatively regulating S6 kinase. EMBO Rep. 8, 360-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leist, M. and Jaattela, M. (2001). Four deaths and a funeral: from caspases to alternative mechanisms. Nat. Rev. Mol. Cell Biol. 2, 589-598. [DOI] [PubMed] [Google Scholar]
- Levine, B. and Klionsky, D. J. (2004). Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell 6, 463-477. [DOI] [PubMed] [Google Scholar]
- Liang, X. H., Kleeman, L. K., Jiang, H. H., Gordon, G., Goldman, J. E., Berry, G., Herman, B. and Levine, B. (1998). Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J. Virol. 72, 8586-8596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockshin, R. A. and Zakeri, Z. (2004). Apoptosis, autophagy, and more. Int. J. Biochem. Cell Biol. 36, 2405-2419. [DOI] [PubMed] [Google Scholar]
- Lum, J. J., DeBerardinis, R. J. and Thompson, C. B. (2005). Autophagy in metazoans: cell survival in the land of plenty. Nat. Rev. Mol. Cell Biol. 6, 439-448. [DOI] [PubMed] [Google Scholar]
- Maiuri, M. C., Le Toumelin, G., Criollo, A., Rain, J. C., Gautier, F., Juin, P., Tasdemir, E., Pierron, G., Troulinaki, K., Tavernarakis, N. et al. (2007). Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J. 26, 2527-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino, G., Uria, J. A., Puente, X. S., Quesada, V., Bordallo, J. and Lopez-Otin, C. (2003). Human autophagins, a family of cysteine proteinases potentially implicated in cell degradation by autophagy. J. Biol. Chem. 278, 3671-3678. [DOI] [PubMed] [Google Scholar]
- Marino, G., Salvador-Montoliu, N., Fueyo, A., Knecht, E., Mizushima, N. and Lopez-Otin, C. (2007). Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in ATG4C/autophagin-3. J. Biol. Chem. 282, 18573-18583. [DOI] [PubMed] [Google Scholar]
- Martin, D. N., Balgley, B., Dutta, S., Chen, J., Rudnick, P., Cranford, J., Kantartzis, S., DeVoe, D. L., Lee, C. and Baehrecke, E. H. (2007). Proteomic analysis of steroid-triggered autophagic programmed cell death during Drosophila development. Cell Death Differ. 14, 916-923. [DOI] [PubMed] [Google Scholar]
- Mizushima, N., Yamamoto, A., Hatano, M., Kobayashi, Y., Kabeya, Y., Suzuki, K., Tokuhisa, T., Ohsumi, Y. and Yoshimori, T. (2001). Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 152, 657-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss, D. K., Betin, V. M., Malesinski, S. D. and Lane, J. D. (2006). A novel role for microtubules in apoptotic chromatin dynamics and cellular fragmentation. J. Cell Sci. 119, 2362-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra, D., Tanaka, A., Suen, D. F. and Youle, R. J. (2008). Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183, 795-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberstein, A., Jeffrey, P. D. and Shi, Y. (2007). Crystal structure of the Bcl-XL-beclin 1 peptide complex: beclin 1 is a novel BH3-only protein. J. Biol. Chem. 282, 13123-13132. [DOI] [PubMed] [Google Scholar]
- Pattingre, S. and Levine, B. (2006). Bcl-2 inhibition of autophagy: a new route to cancer? Cancer Res. 66, 2885-2888. [DOI] [PubMed] [Google Scholar]
- Pattingre, S., Tassa, A., Qu, X., Garuti, R., Liang, X. H., Mizushima, N., Packer, M., Schneider, M. D. and Levine, B. (2005). Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 122, 927-939. [DOI] [PubMed] [Google Scholar]
- Pyo, J. O., Jang, M. H., Kwon, Y. K., Lee, H. J., Jun, J. I., Woo, H. N., Cho, D. H., Choi, B., Lee, H., Kim, J. H. et al. (2005). Essential roles of Atg5 and FADD in autophagic cell death: dissection of autophagic cell death into vacuole formation and cell death. J. Biol. Chem. 280, 20722-20729. [DOI] [PubMed] [Google Scholar]
- Qu, X., Zou, Z., Sun, Q., Luby-Phelps, K., Cheng, P., Hogan, R. N., Gilpin, C. and Levine, B. (2007). Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell 128, 931-946. [DOI] [PubMed] [Google Scholar]
- Satoo, K., Noda, N. N., Kumeta, H., Fujioka, Y., Mizushima, N., Ohsumi, Y. and Inagaki, F. (2009). The structure of Atg4B-LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J. 28, 1341-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherz-Shouval, R., Sagiv, Y., Shorer, H. and Elazar, Z. (2003). The COOH terminus of GATE-16, an intra-Golgi transport modulator, is cleaved by the human cysteine protease HsApg4A. J. Biol. Chem. 278, 14053-14058. [DOI] [PubMed] [Google Scholar]
- Scherz-Shouval, R., Shvets, E., Fass, E., Shorer, H., Gil, L. and Elazar, Z. (2007). Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 26, 1749-1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, R. C., Juhasz, G. and Neufeld, T. P. (2007). Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr. Biol. 17, 1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu, S., Kanaseki, T., Mizushima, N., Mizuta, T., Arakawa-Kobayashi, S., Thompson, C. B. and Tsujimoto, Y. (2004). Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat. Cell Biol. 6, 1221-1228. [DOI] [PubMed] [Google Scholar]
- Shintani, T. and Klionsky, D. J. (2004). Autophagy in health and disease: a double-edged sword. Science 306, 990-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani, T., Mizushima, N., Ogawa, Y., Matsuura, A., Noda, T. and Ohsumi, Y. (1999). Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast. EMBO J. 18, 5234-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohecker, A. M., Yehiely, F., Chen, F. and Cryns, V. L. (2008). Caspase cleavage of HER-2 releases a Bad-like cell death effector. J. Biol. Chem. 283, 18269-18282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, K., Kirisako, T., Kamada, Y., Mizushima, N., Noda, T. and Ohsumi, Y. (2001). The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 20, 5971-5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida, I., Sou, Y. S., Ezaki, J., Minematsu-Ikeguchi, N., Ueno, T. and Kominami, E. (2004). HsAtg4B/HsApg4B/autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3- and GABAA receptor-associated protein-phospholipid conjugates. J. Biol. Chem. 279, 36268-36276. [DOI] [PubMed] [Google Scholar]
- Yorimitsu, T. and Klionsky, D. J. (2005). Autophagy: molecular machinery for self-eating. Cell Death Differ. 12 Suppl. 2, 1542-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura, K., Shibata, M., Koike, M., Gotoh, K., Fukaya, M., Watanabe, M. and Uchiyama, Y. (2006). Effects of RNA interference of Atg4B on the limited proteolysis of LC3 in PC12 cells and expression of Atg4B in various rat tissues. Autophagy 2, 200-208. [DOI] [PubMed] [Google Scholar]
- Yousefi, S., Perozzo, R., Schmid, I., Ziemiecki, A., Schaffner, T., Scapozza, L., Brunner, T. and Simon, H. U. (2006). Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat. Cell Biol. 8, 1124-1132. [DOI] [PubMed] [Google Scholar]
- Yu, L., Alva, A., Su, H., Dutt, P., Freundt, E., Welsh, S., Baehrecke, E. H. and Lenardo, M. J. (2004). Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science 304, 1500-1502. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.