Abstract
Many details of structure, function and substrate specificity of eukaryotic proteasomal systems have been elucidated. This information far-exceeds that available for the archaeal and bacterial counterparts. While structural and functional studies have provided some insight into the workings of prokaryotic proteasomes, the question of substrate targeting and global cellular influence remain largely unaddressed. In this communication, we report an over 720-fold increase in the half-life of the DNA-sliding clamp protein proliferating cell nuclear antigen after knockout of the panA gene, encoding a proteasome-activating nucleotidase A, on the chromosome of the halophilic archaeon Haloferax volcanii. This discovery marks the first identification of a protein stabilized by an archaeal proteasome mutation and provides a starting point for investigations into substrate recognition mechanisms. The findings also begin to address the functional role of proteasomal systems within the scope of the archaeal cell.
Keywords: archaea, proteasome, halophile, DNA polymerase, sliding clamp, proteolysis
Introduction
Proteasomes are energy-dependent proteases distributed in all three domains of life: Bacteria, Archaea and Eucarya (Volker & Lupas, 2002). Although proteasomes from these three domains share a common architecture and core molecular mechanism, little is known regarding the relatedness of their biological roles within the cell. In eukaryotes, proteasomes often require substrates to be covalently modified with ubiquitin before their destruction and are essential for catalyzing the precisely timed and rapid turnover of key regulatory and metabolic proteins as well as maintaining general protein quality control (Raasi & Wolf, 2007). In contrast, the biological roles of the proteasomal systems of archaea and bacteria are poorly understood.
Proteasomes are composed of a 20S core particle of α- and β-type subunits that associate with ATPase regulatory particles to facilitate protein unfolding and degradation (reviewed in Maupin-Furlow et al., 2006). In archaea, proteasome-activating nucleotidase (PAN) proteins, closely related to the proteasomal regulatory particle triple-A type I proteins (Rpt) of eukaryotes, serve as ATPase components (Rabl et al., 2008). Recently, we described the disruption of a gene on the Haloferax volcanii chromosome (panA) encoding a PAN protein present at high levels throughout exponential and stationary phase (Kirkland et al., 2008). Disruption of this PAN paralog, PanA, has a pleiotropic effect on the (phospho)proteome of the cell and is anticipated to impede proteasome-mediated degradation of at least a subset of proteins identified by electrospray ionization tandem-mass spectrometry (MS/MS) to accumulate in the panA mutant compared with its parent strain (Kirkland et al., 2008). This list includes a number of proteins with orthologs known to be targets of the ubiquitin–proteasome system in eukaryotes. In particular, our efforts for this study focused on the DNA-sliding clamp protein proliferating cell nuclear antigen (PCNA) of H. volcanii, which was identified as (phospho)proteome fractions enriched from lysate of panA mutant and wild-type cells using immobilized metal affinity chromatography (IMAC) (Kirkland et al., 2008).
PCNA is a trimeric ring, which serves as a sliding DNA clamp to recruit leading and lagging strand DNA polymerases to the replication fork, aid in excision repair tasks and a myriad of other cellular functions (Moldovan et al., 2007; Watson et al., 2008). Previous reports in rice (Oryza sativa) (Yamamoto et al., 2004) and humans (Wang et al., 2006; Nikolaishvili-Feinberg et al., 2008) suggest a role for the proteasome in PCNA turnover and, in a broader sense, reveal a more important global function for the proteasome in regulating DNA replication, recombination and/or repair. Indeed, phosphorylation of the tyrosine residue 211 of PCNA modulates its modification by polyubiquitination and ultimately its stability in human cancer cell lines (Wang et al., 2006).
To assess whether PCNA is a target of the archaeal proteasome system, pulse-chase labeling with 35S-methionine and 35S-cysteine was coupled with immunoprecipitation and used to establish the rate of turnover of this protein in H. volcanii parent and panA mutant strains (wild type and ΔpanA, respectively). Elongation initiation factor 2, α subunit (eIF2α) was included for comparison, because this protein also accumulated in proteasome-deficient cells of H. volcanii.
Materials and methods
Antibody preparation
To overcome interdomain differences in protein structure, custom antibodies were raised against the PCNA and eIF2α polypeptides of H. volcanii. In brief, the coding regions for PCNA (HVO_0175) and eIF2α (HVO_0699) were amplified from the H. volcanii genome (April 2007 version, http://archaea.ucsc.edu/) by PCR using Accuprime GC-Rich DNA polymerase (Invitrogen, Carlsbad, CA) and the following primer pairs: PCNA, 5′-TCCTCTTCATATGTTCAAGGCCATCGTGAG-3′ and 5′-AAACTCGAGGTCACTCTGGATGCGCGGG-3′; eIF2α, 5′- TTGAGTTCATATGAAGTACAGCGGATGGCCTG-3′ and 5′-TTTCTCGAGCTCTTCGTCGCCGCTG-3′ (NdeI and XhoI sites in italics). The PCR fragments were cloned into the NdeI and XhoI sites of plasmid pET24b (Novagen, Madison, WI) to generate expression plasmids pJAM510 and pJAM511, respectively. The cloned DNA was sequenced to verify fidelity (UF ICBR DNA sequencing core, Gainesville, FL). Proteins were synthesized with C-terminal hexa-histidine tags (-His6) in recombinant Escherichia coli Rosetta (DE3) strains carrying these plasmids (pJAM510 and pJAM511). Proteins were purified from cell lysate by nickel column chromatography (Ni Sepharose 6 Fast Flow, Pharmacia) and reducing SDS-PAGE and used as antigens for the generation of polyclonal antibodies in rabbits, as previously described for other haloarchaeal proteins (Reuter et al., 2004).
Pulse-chase assay coupled with immunoprecipitation
Haloferax volcanii wild-type and panA deletion strains (DS70 and GG102, respectively) (see Wendoloski et al., 2001; Kirkland et al., 2008 for detailed genotypes) were grown in 200 mL minimal (Hv-Min) medium (Dyall-Smith, 2008) to an OD600 nm of 0.4–0.6 (200 r.p.m., 42 °C). Cells were collected by centrifugation at 10 000 g for 15 min at 40 °C and resuspended in 80 mL of prewarmed (42 °C) Hv-Min medium. Cells were incubated at 42 °C for 1 h with shaking (200 r.p.m.). Radioactively labeled methionine and cysteine (500 μCi of 35S-methionine and 35S-cysteine mixture) (EXPRE35S35S Protein Labeling Mix, Perkin Elmer) were added to the culture and incubated for an additional 20 min (42 °C, 200 r.p.m.). Samples were divided into 8–10-mL fractions in 15-mL conical tubes. Translation was arrested immediately in one tube (0 min) by addition of 1 mL of ‘stop’ solution [22% (w/v) sodium azide, 100 μg mL−1 puromycin] and incubation on ice. The remaining seven samples were incubated with 1 mL of ‘chase’ solution (8 mg mL−1‘cold’ methionine and cysteine, 100 μg mL−1 puromycin, 1 mg mL−1 protease inhibitor cocktail) at 42 °C and 200 r.p.m. Each sample was arrested at appropriate time intervals (1, 3, 5, 10, 30 and 60 min and 12 h) by the addition of 1 mL of ‘stop’ solution and chilling on ice. Each 10-mL sample was collected by centrifugation at 8000 g at room temperature (RT) for 10 min. Pellets with c. 1 mL of medium were resuspended and repelleted in 2 mL microcentrifuge tubes at 14 000 g at RT for 5 min. Cell pellets were frozen at −80 °C for a maximum of 48 h before immunoprecipitation.
For immunoprecipitation, protein A–Sepharose beads (Pharmacia) [10 mg mL−1 phosphate-buffered saline (PBS) with 0.01% (w/v) sodium azide] were aliquoted into 1.8-mL Eppendorf tubes (75 μL per tube), washed once with cold PBS and resuspended in 1 mL of cold PBS. The beads were charged by addition of 10–12 μL of polyclonal antiserum for 4–12 h at 4 °C with continuous agitation and washed five times with cold PBS to remove unbound antibody. Cell pellets from pulse-chase labeling (described above) were prepared for immunoprecipitation by resuspension in 150 μL of denaturing lysis buffer [1% (w/v) sodium dodecyl sulfate (SDS), 50 mM Tris-Cl, pH 7.4, 5 mM EDTA, 10 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride (PMSF) and 300 mM NaCl], boiling for 10 min and addition of 1.35 mL nondenaturing lysis buffer [1% (v/v) Triton X-100, 50 mM Tris-Cl, pH 7.4, 5 mM EDTA, 0.02% (w/v) sodium azide, 10 mM iodoacetamide, 1 mM PMSF, 300 mM NaCl]. The lysate was incubated with 500 U of benzonase (2 μL) (Sigma-Aldrich) at RT for 30 min with occasional mixing. Samples were clarified by centrifugation at 10 000 g for 5 min, and the clarified lysate was added to the charged beads. The lysate/bead mixture was incubated at 4 °C with rocking for 3 h. Beads were washed five times with immunoprecipitation wash buffer [0.1% (v/v) Triton X-100, 50 mM Tris-Cl, pH 7.4, 300 mM NaCl, 5 mM EDTA, 0.02% (w/v) sodium azide, 0.1% (w/v) SDS and 0.1% (w/v) deoxycholine] and one final time with cold PBS. SDS-reducing dye [20 μL of 100 mM Tris-Cl, pH 6.8, 10% (v/v) β-mercaptoethanol, 2% (w/v) SDS, 10% (v/v) glycerol and 0.6 mg mL−1 bromophenol blue] was added. Samples were boiled for 10 min and centrifuged at 14 000 g for 5 min to remove the beads. Proteins in the supernatant were separated by 12% SDS-polyacrylamide gel electrophoresis (PAGE) (200 V for 45 min), and their migration was compared with Precision Plus Protein dual color standards (BioRad). Gels were incubated in fixing solution (10% methanol, 7% acetic acid) for 30 min and an intensifier solution (1 M salicylic acid) for 1 h. Gels were washed with dH2O, dried under vacuum for 1.5–2 h, and exposed to radiographic film for a minimum of 10 days at −80 °C. Band intensity was quantified using a Versa Doc 1000 with quantity one v. 4 software (BioRad).
Results and discussion
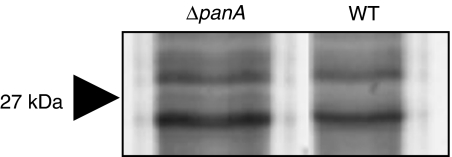
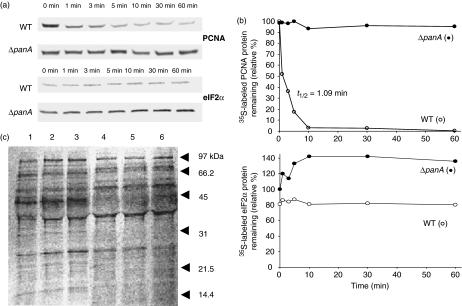
The DNA-sliding clamp protein PCNA was identified by MS/MS analysis of IMAC fractions purified from cell lysate of an H. volcanii panA mutant as described previously (Kirkland et al., 2008). IMAC fractions of the panA mutant (GG102) were compared with its parent strain (DS70) by 1-D gel electrophoresis, and regions of differential protein bands were excised and analyzed by MS/MS. The PCNA protein of 27 kDa was identified in the panA mutant IMAC fractions of gel slices in the 25–30-kDa region (Fig. 1), thus implicating PCNA as a potential substrate of the proteasomal system. To further investigate this finding, antibodies were raised against PCNA. Pulse-chase labeling with 35S-methionine and 35S-cysteine was coupled with immunoprecipitation to determine the rate of turnover of PCNA in H. volcanii parent (wild-type, WT) and panA mutant (ΔpanA) strains (Fig. 2). Similarly, the half-life of eIF2α was analyzed for comparison. The PCNA- and eIF2α-specific protein bands detected by immunoprecipitation after pulse-chase were estimated at 27 and 30 kDa, respectively, based on migration in SDS-PAGE gels. The sizes of these bands were consistent with the molecular masses calculated for the respective polypeptides deduced from the H. volcanii genome sequence. Protein bands of similar molecular mass were not detected by immunoprecipitation using PCNA- or eIF2α-preimmune serum (data not shown).
Fig. 1.
Comparison of IMAC-enriched proteins from cell lysate of Haloferax volcanii wild-type (WT) and panA deletion (ΔpanA) strains. Proteins enriched from cell lysate by IMAC were separated by reducing 12% SDS-PAGE and stained with SYPRO Ruby as described previously (Kirkland et al., 2008). Regions of reproducible and differential protein banding between the two strains such as the 25–30 kDa region shown were digested with trypsin and analyzed by MS/MS. PCNA DNA-sliding clamp protein of 27 kDa was identified by this semi-quantitative 1-D-gel approach.
Fig. 2.
Comparison of the half-lives of PCNA DNA-sliding clamp protein and translation initiation factor eIF2α in Haloferax volcanii wild-type (WT, ○) and panA deletion (ΔpanA, •) strains. (a) 35S-pulse-chase labeling was coupled with immunoprecipitation using anti-PCNA and anti-eIF2α polyclonal antibodies to monitor the respective half-lives of PCNA and eIF2α in wild-type and ΔpanA mutant strains over a 12-h time course (with 12 h results similar to 1 h, data not shown). These data are representative of at least three independent experiments. (b) A graphical representation of the PCNA- and eIF2α-specific protein band intensities in percentages relative to 0 min for each protein, were used to calculate protein half-lives. (c) Similar overall levels of 35S-label were incorporated into the bulk-protein of wild-type (lanes 4–6) and ΔpanA (lanes 1–3) strains. Total cell protein was radiolabeled with 35S-methionine and 35S-cysteine, separated by 12% SDS-PAGE and analyzed by autoradiography as described in Materials and methods.
Based on pulse-chase immunoprecipitation analysis, the PCNA protein had a surprisingly short half-life in wild-type cells of only c. 1 min with little protein remaining after 10 min (Fig. 2a and b). In contrast, no reduction in PCNA levels was detected over the course of 12 h for the panA mutant, which no longer synthesizes a functional PanA protein (Fig. 2a and b). Thus, the 35S-labeled PCNA was relatively stable in these panA mutant cells with a half-life >12 h. Interestingly, minute levels of PCNA persisted throughout the 12-h time course in the wild-type cells. Thus, although PCNA undergoes a high rate of protein turnover, low levels of this protein were retained within the cell for relatively long periods of time. Maintaining low levels of PCNA may reflect its critical role as a processivity-promoting factor in DNA replication and other functions that are essential for cell growth. Despite this observed, retention of PCNA protein in wild-type cells, it is clear that the levels of PCNA protein were reduced significantly (>90%) through a mechanism that requires synthesis of the PanA protein.
As a comparison, pulse-chase immunoprecipitation was also used to assess the half-life of eIF2α in wild-type and panA mutant strains (Fig. 2a and b). In contrast to PCNA, no appreciable reduction in the levels of eIF2α protein was observed for either strain over the 12-h time course. Although eIF2α was highly stable, the overall level of labeled eIF2α protein in cells possessing a fully functional proteasome was notably lower than that of the panA mutant. This difference is most likely due to a panA-dependent increase in the level of eIF2α-specific transcript and/or translation of the eIF2α-specific transcript, because similar levels of 35S-label were incorporated into the bulk protein of both strains as demonstrated by autoradiograph analysis of total cell protein separated by SDS-PAGE (Fig. 2c). These results are consistent with our previous MS/MS findings that the steady state levels of eIF2α protein are higher in the panA mutant compared with its parent.
In conclusion, the comparison of PCNA and eIF2α half-lives in the presence and absence of a functional proteasomal system (ΔpanA) corroborates and builds upon our results, which identified, by MS/MS, enhanced levels of both of these proteins after perturbation of PanA function. It also enabled us for the first time to attribute these proteomic results to panA-dependent differences in PCNA protein half-life vs. the translational/transcriptional effects seen for eIF2α. These results suggest that, much like eukaryotes, archaeal proteasomes and/or their associated ATPase regulatory proteins (i.e. PanA) are intimately linked to regulating the levels and ultimately function of PCNA protein. Because PCNA orchestrates several functions associated with DNA replication, recombination and repair, the finding that PCNA is stabilized in a panA mutant may explain our previous results that this same mutation reduces the growth rate of H. volcanii (Kirkland et al., 2008). Whether the H. volcanii PCNA protein is phosphorylated and this influences its stability, similar to the eukaryotic counterpart, remains to be determined. Our previous work demonstrated that PCNA was enriched from H. volcanii cell lysate by IMAC, a technique commonly used to purify phosphopeptides. Likewise, the Tyr211 residue shown to be phoshorylated in human and mouse PCNA orthologs (Wang et al., 2006) is conserved in a wide group of eukaryotic and archaeal species, including H. volcanii. However, a specific site(s) of phosphorylation is yet to be identified for this or other archaeal PCNA proteins.
Together, these findings address the functional role of proteasomal systems within the scope of the archaeal cell and provide a starting point for investigations into mechanisms of substrate recognition. The apparent link of archaeal proteasomes with the DNA-sliding clamp protein, PCNA, points to future comparison of proteasome function in mixed populations to that of synchronized cells. Recent advances in the study of the haloarchaeal cell cycle (Baumann et al., 2007) will facilitate this latter objective.
Acknowledgments
Thanks are due to Eric Ziegelbauer for technical assistance. This research was funded in part by grants from the National Institutes of Health (R01 GM057498) and Department of Energy (DE-FG02-05ER15650).
Statement
Re-use of this article is permitted in accordance with the Creative Commons Deed, Attribution 2.5, which does not permit commercial exploitation.
References
- Baumann A, Lange C, Soppa J. Transcriptome changes and cAMP oscillations in an archaeal cell cycle. BMC Cell Biol. 2007;8:21. doi: 10.1186/1471-2121-8-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyall-Smith M. The Halohandbook: Protocols for Halobacterial Genetics. 2008. p. 144. http://www.haloarchaea.com. [Google Scholar]
- Kirkland PA, Gil MA, Karadzic IM, Maupin-Furlow JA. Genetic and proteomic analyses of a proteasome-activating nucleotidase A mutant of the haloarchaeon Haloferax volcanii. J Bacteriol. 2008;190:193–205. doi: 10.1128/JB.01196-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maupin-Furlow JA, Humbard MA, Kirkland PA, Li W, Reuter CJ, Wright AJ, Zhou G. Proteasomes from structure to function: perspectives from archaea. Curr Top Dev Biol. 2006;75:125–169. doi: 10.1016/S0070-2153(06)75005-0. [DOI] [PubMed] [Google Scholar]
- Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Nikolaishvili-Feinberg N, Jenkins GS, Nevis KR, Staus DP, Scarlett CO, Unsal-Kacmaz K, Kaufmann WK, Cordeiro-Stone M. Ubiquitylation of proliferating cell nuclear antigen and recruitment of human DNA polymerase η. Biochemistry. 2008;47:4141–4150. doi: 10.1021/bi702329h. [DOI] [PubMed] [Google Scholar]
- Raasi S, Wolf DH. Ubiquitin receptors and ERAD: a network of pathways to the proteasome. Semin Cell Dev Biol. 2007;18:780–791. doi: 10.1016/j.semcdb.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Rabl J, Smith DM, Yu Y, Chang SC, Goldberg AL, Cheng Y. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol Cell. 2008;30:360–368. doi: 10.1016/j.molcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter CJ, Kaczowka SJ, Maupin-Furlow JA. Differential regulation of the PanA and PanB proteasome-activating nucleotidase and 20S proteasomal proteins of the haloarchaeon Haloferax volcanii. J Bacteriol. 2004;186:7763–7772. doi: 10.1128/JB.186.22.7763-7772.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volker C, Lupas AN. Molecular evolution of proteasomes. Curr Top Microbiol. 2002;268:1–22. doi: 10.1007/978-3-642-59414-4_1. [DOI] [PubMed] [Google Scholar]
- Wang SC, Nakajima Y, Yu YL, et al. Tyrosine phosphorylation controls PCNA function through protein stability. Nat Cell Biol. 2006;8:1359–1368. doi: 10.1038/ncb1501. [DOI] [PubMed] [Google Scholar]
- Watson JD, Baker TA, Bell SP, Gann A, Levine M, Losick R. Molecular Biology of the Gene. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2008. p. 841. [Google Scholar]
- Wendoloski D, Ferrer C, Dyall-Smith ML. A new simvastatin (mevinolin)-resistance marker from Haloarcula hispanica and a new Haloferax volcanii strain cured of plasmid pHV2. Microbiology. 2001;147:959–964. doi: 10.1099/00221287-147-4-959. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Kimura S, Mori Y, Oka M, Ishibashi T, Yanagawa Y, Nara T, Nakagawa H, Hashimoto J, Sakaguchi K. Degradation of proliferating cell nuclear antigen by 26S proteasome in rice (Oryza sativa L.) Planta. 2004;218:640–646. doi: 10.1007/s00425-003-1140-2. [DOI] [PubMed] [Google Scholar]