Abstract
Background:
BCL3 is a putative oncogene encoding for a protein belonging to the inhibitory κB-family. We experienced that this putative oncogene was a common target gene for growth-promoting cytokines in myeloma cell lines.
Methods:
Gene expression of BCL3 was studied in 351 newly diagnosed myeloma patients, 12 patients with smouldering myeloma, 44 patients with monoclonal gammopathy of undetermined significance and 22 healthy individuals. Smaller material of samples was included for mRNA detection by RT-PCR, protein detection by Western blot and immunohistochemistry, and for cytogenetic studies. A total of eight different myeloma cell lines were studied.
Results:
Bcl-3 was induced in myeloma cell lines by interleukin (IL)-6, IL-21, IL-15, tumor necrosis factor-α and IGF-1, and its upregulation was associated with increased proliferation of the cells. In a population of 351 patients, expression levels of BCL3 above 75th percentile were associated with shorter 5-yr survival. When this patient population was divided into subgroups based on molecular classification, BCL3 was significantly increased in a poor risk subgroup characterized by overexpression of cell cycle and proliferation related genes. Intracellular localization of Bcl-3 was dependent on type of stimulus given to the cell.
Conclusion:
BCL3 is a common target gene for several growth-promoting cytokines in myeloma cells and high expression of BCL3 at the time of diagnosis is associated with poor prognosis of patients with multiple myeloma (MM). These data may indicate a potential oncogenic role for Bcl-3 in MM.
Keywords: multiple myeloma, Bcl-3, nuclear factor-κB
Myeloma cells are dependent on signals from the bone marrow micro-environment for proliferation and survival (1). Myeloma cell growth factors mediate redundant effects since one growth factor can substitute for another in vitro (2, 3). This observation led us to hypothesize that intracellular signals generated by myeloma growth factors target common genes important for growth and survival of myeloma cells. These genes may represent potential drug targets. Microarray experiments done in our laboratory showed that the myeloma cell growth factors interleukin (IL)-6, IL-21 and tumor necrosis factor (TNF)-α all induced expression of the putative oncogene BCL3 in the myeloma cell lines IH-1 and OH-2 (paper in preparation). These data supported previous studies done by Brocke-Heidrich et al. and Tsuyama et al., who showed that BCL3 is upregulated by IL-6 at mRNA level as well as at protein level in myeloma cell lines (4–6). However, the expression of BCL3 in myeloma patient samples has not been studied. Based on this we decided to study the role of BCL3 in primary myeloma cells as well as in myeloma cell lines in more detail.
BCL3 was first identified through its involvement in the t (14;19) (q32;q13) translocation in B-cell chronic lymphocytic leukemia (CLL) (7). Leukemic cells from these patients had increased levels of BCL3 mRNA, leading to the hypothesis that BCL3 is a proto-oncogene contributing to leukemogenesis (8). BCL3 is overexpressed in breast cancers, subtypes of lymphomas and nasopharyngeal carcinomas (9–12). Supporting the notion that BCL3 is an oncogene, Viatour et al. demonstrated that overexpression of BCL3 is sufficient to transform the mouse fibroblast cell line NIH3T3 and induce tumor growth subcutaneously in nude mice (13). Physiologically, BCL3 has been implicated to play a role during B-cell development and as a negative regulator of immune responses (14–16).
BCL3 encodes a protein denoted Bcl-3 that is a member of the inhibitory κB (IκB)-family (17, 18). The IκB proteins modulate the DNA-binding activity of nuclear factor-κB (NFκB), a family of transcription factors involved in apoptosis and cell growth (19). Activation of NFκB is implicated as an important mechanism for the development of antiapoptosis and drug resistance in multiple myeloma (20). Depending on context, Bcl-3 either activates or inhibits NFκB-dependent gene transcription through interactions with homodimers of NFκB p50 or p52 (14, 21–26). We here present evidence that BCL3 is overexpressed in myeloma cells from a subset of myeloma patients, and that the high expression of BCL3 at the time of diagnosis is associated with inferior prognosis. Furthermore, high expression of Bcl-3 in myeloma cell lines induced by growth-promoting cytokines is associated with increased proliferation of the cells. We propose that Bcl-3 contributes to regulation of NFκB-dependent gene transcription in myeloma cells, with potential oncogenic consequences.
Materials and methods
Cell lines and culture conditions
The human myeloma cell line ANBL-6 (gift from Dr D. Jelinek, Mayo Clinic, Rochester, MN, USA), IH-1 (3), INA-6 (gift from Dr M. Gramatzki, Erlangen, Germany), JJN-3 (gift from Dr J. Ball, Department of Immunology, University of Birmingham, UK), OH-2 (2), RPMI-8226 and U-266 (both from American Type Culture Collection, Rockville, MD, USA) were cultured as previously described (27). The CAG cell line (gift from Dr J Epstein, Little Rock, AK, USA) was grown in RPMI 1640 (Gibco, Paisley, UK) supplemented with l-glutamine 100 μg/mL, gentamicin 20 μg/mL (referred to as RPMI) and 10% heat inactivated fetal calf serum (FCS; HyClone, Logan, UT, USA). The cells were kept at 37°C in a humidified atmosphere containing 5% CO2, and were washed four times in Hanks` balanced salt solution (HBSS; Gibco, Paisley, UK) to deplete them of serum and cytokines before performing experiments.
Patients and healthy individuals; separation of CD138+ plasma cells
Gene expression profiles (GEP) were studied in plasma cells from 351 patients with newly diagnosed multiple myeloma (MM), 12 patients with smouldering multiple myeloma (SMM), 44 patients with monoclonal gammopathy of undetermined significance (MGUS) and 22 healthy individuals. The patients were recruited from University of Arkansas for Medical Sciences, Little Rock, USA, and the use of patient samples in research studies was approved by the University of Arkansas for Medical Sciences (Little Rock, AR) Institutional Review Board. Details about classification of these patients have been previously described (28, 29).
We used patient samples from the Norwegian Research Biobank for multiple myeloma for studies of Bcl-3 expression by RT-PCR (10 patients) immunohistochemistry (18 patients with MM and three patients with plasmocytoma), Western blot (eight patients) and cIg-FISH (19 patients). The Norwegian Research Biobank for multiple myleoma has been approved by the Regional Ethics Committee of Middle Norway and Norwegian Health Authorities. All patients have signed informed consent to store samples in the Biobank and to use samples for research purposes. Separation of plasma cells before GEP was done as previously described (28).
Cytokines and antibodies
IL-6 was purchased from Biosource (Camarillo, CA, USA). IL-10, IL-15 and insulin-like growth factor (IGF)-1 were from R&D systems (Abingdon, UK). TNF-α was from Genetech (South San Francisco, CA, USA), and IL-21 was a gift from R.Holly, ZymoGenetics (Seattle, WA, USA). Hepatocyte growth factor (HGF) was purified from a medium conditioned by JJN-3 as described previously (30). All cytokines except HGF were of recombinant human type. Polyclonal anti-Bcl-3 (sc-185), polyclonal anti-p65 (sc-109) and monoclonal anti-p50 (sc-8414) for Western blotting were purchased from Santa Cruz Biotechnologies (Santa Cruz, CA, USA). Monoclonal anti-GAPDH and anti-Lamin B used as loading controls on Western blots were purchased from Abcam (ab8245; Cambridge, UK) and Calbiochem (Cat#NA12; Darmstadt, Germany), respectively.
Preparation of cRNA and microarray hybridization
RNA, cRNA preparation and hybridization to U133 Plus2 GeneChip microarrays (Affymetrix, Santa Clara, CA, USA) were performed as previously described (31).
Statistical analysis
Gene expression was analyzed as previously described (28, 31). Differences in expression of BCL3 between plasma cells from MM patients and normal plasma cells (NPC) were analyzed using One-Way anova test. Survival distributions were presented with the use of the Kaplan–Meier method and compared with the log-rank test. Statistical tests were performed with the software package spss 12.0 (SPSS, Chicago, IL, USA).
Analysis of BCL3 expression by quantitative real-time PCR (qPCR)
Plasma cells from 10 patients were isolated with CD138 antibodies conjugated to magnetic beads using a RoboSep (StemCell Technologies, Vancouver, BC, Canada) cell separation device. Total RNA isolated using the MirVana™ miRNA Isolation Kit (Applied Biosystems, Foster City, CA, USA). cDNA was synthesized using Taqman reverse transcriptase reagents from Roche-Applied Biosystems. qPCR of BCL3 was performed using StepOne Real-Time PCR System, (Applied Biosystems). BCL3 TaqMan primer (Hs00180403_m1, Taqman, Gene Expression Assays, Applied Biosystems) was used to detect BCL3 expression. The comparative Ct-method was used for quantization with GAPDH (HS99999905_m1) as housekeeping gene.
Western blotting
Cells (1.2 × 106) were seeded in 3 mL RPMI supplemented with 0.1% bovine serum albumin (BSA, Sigma-Aldrich, St. Louis, MO, USA) before Western blotting of whole cell lysates. The ANBL-6, IH-1, INA-6 and OH-2 cell lines were starved overnight before they were stimulated with cytokines as indicated at time point 0 (T0). At this time point, cells that were not stimulated with cytokines were harvested. The viability of these cells as measured by propidium iodide-staining was 60–80%, similar to the viability of cells grown in IL-6 enriched medium. CAG, JJN-3, RPMI-8226 and U-266 were stimulated with IL-6 immediately after seeding or left without cytokines in RPMI with 0.1% BSA for 24 h. After harvesting, dry pellets were kept at −80°C until further processing. Pellets were lysed in lysis buffer (10% sodium dodecyl sulfate, 10 mm Tris-HCL, pH 6.8) at 50°C using a Hamilton injector, mixed with NuPage LDS sample buffer (Invitrogen, Carlsbad, CA, USA) containing 0.1 m dithiothreitol, and heated for 2 min at 90°C. The lysates (40 μg of proteins) were then separated on 10% NuPage Bis-Tris gels (Invitrogen), followed by electrophoretic transfer to nitrocellulose membranes (Bio-Rad, Hercules, CA, USA). Membranes were blocked in 50 mm Tris buffered saline pH 7.5 containing 0.05% Tween 20 and 5% non-fat dried milk (Nestle, Vevey, Switzerland) and incubated with antibodies as indicated. After incubation with horseradish peroxidase-conjugated secondary antibodies (DakoCytomation, Glostrup, Denmark), the proteins were visualized by ECL Western blotting detection reagents (Amersham Biosciences, Buckinghamshire, UK), scanned (Canoscan D1250 U25, Canon Europa NV, Amstelveen, The Netherlands) and imported into adobe photoshop elements 4.0 software (Adobe Systems Inc, San Jose, CA, USA) for image acquisition. Cytosol-and nuclear extracts of IH-1 were prepared using Active Motifs nuclear extract kit (Active Motif, CA, USA) according to the manufacturer’s protocol. These cells were seeded in RPMI containing 10% FCS and cytokines as indicated for 24 h without previous starvation of cells.
Thymidine incorporation assays
To measure DNA synthesis, cells were seeded in triplets in 96-well cell culture plates (Corning Inc, Corning, NY, USA) at a density of 2 × 104 cells per well in RPMI supplemented with 0.1% BSA and cytokines as indicated. ANBL-6, IH-1,INA-6 and OH-2 were pulsed with 0.75 μCi methyl-[3H]-thymidine (NEN Life Science products, Boston, MA, USA) after 30 h of stimulation with cytokines and harvested 18 h later using a Micromate 96-well harvester (Packard, Meriden, CT, USA). CAG, JJN-3, RPMI-8226 and U-266 were pulsed with 0.75 μCi methyl-[3H]-thymidine for 4 h after 24 h of stimulation. Beta-radiation was measured with Matrix 96 counter (Packard).
Immunohistochemistry
Sections (4 μm) of formalin-fixed bone marrow biopsies were prepared and visualized as previously described (32). The sections were incubated with a monoclonal antibody specific for Bcl-3 (clone 1E8, Novocastra, Newcastle upon Tyne, UK) at a dilution of 1 : 25. Parallel sections were stained with anti CD138 (Dako, code no. M7228, clone MI 15, dilution 1 : 100). Only CD138+ cells were considered and the biopsies were defined as Bcl3+ when ≥20% of the CD138+ cells was stained.
Cytoplasmic immunoglobulin fluorescence in situ hybridization (cIg-FISH)
Cytospin of Lymphoprep-separated (Dynal, Oslo, Norway) bone marrow mononuclear cells from MM patients were fixed in acetic acid/methanol (1:3 v/v, −20°C, 40 min) and air-dried. 5 μL of BCL3 split probe (No.Y5411; DakoCytomation) was applied and the slide was sealed with coverslip and rubber cement. Hybridization was done in a Dako Hybridizer according to the manufacturer’s protocol. The cells were then incubated with 15% goat serum in phosphate buffered saline. Plasma cells were labeled with AMCA (7-amino-4-methylcoumarin-3-acetic acid)-conjugated anti-human IgG against cytoplasmic κ/λ-chains (Vector Laboratories, Burlingame, CA, USA). The slides were air dried in the dark before anti-fade (Vectashield had-set mounting medium without DAPI, Vector Laboratories) was added. The cells were scored as previously described (32). Normal locus was detected as a colocalization of red and green signal, and a split was detected as separate red and green signal.
Assessment of NFκB activity
Activation of the NFκB family members can be measured by their capacity to bind to consensus DNA binding sites (33). We used the TransAm NFκB p65 and p50 transcription Factor Assay Kit (Active Motif, CA, USA) to quantify the DNA binding activity of p50 and p65. The assay was performed according to the manufacturer’s protocol. Briefly, IH-1 cells were cultured for 24 h without previous starvation in RPMI containing 10% FCS and cytokines as indicated. Nuclear extracts were prepared and incubated in 96-well plates coated with an immobilized oligonucleotide containing the 5`-GGACTTTCC-3` consensus binding site for NFκB. Hybridization to the target oligonucleotide was detected by incubation with primary antibodies specific for p65 of p50, visualized by anti-IgG1-HRP and quantified at 450 nm. Specificity was confirmed by incubating with a wild type consensus oligonucleotide that competed with the substrate for binding to the immobilized oligonucleotide.
Results
Growth promoting cytokines induce Bcl-3 in myeloma cell lines
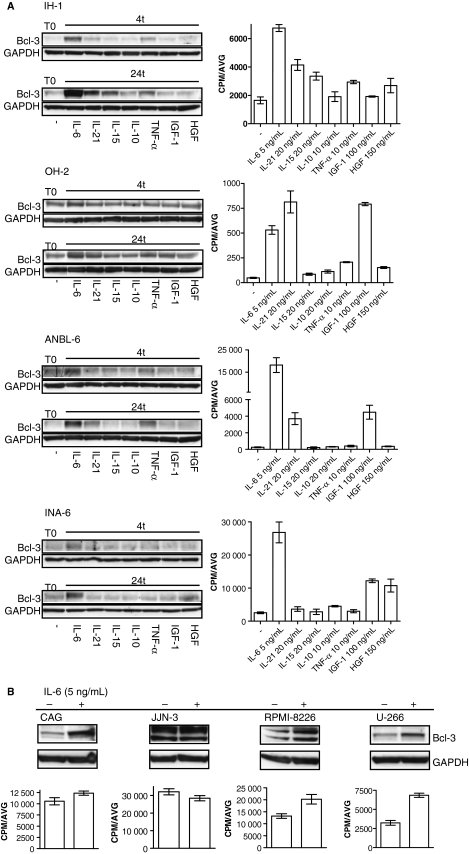
We stimulated the IL-6-dependent myeloma cell lines ANBL-6, IH-1, INA-6 and OH-2 with cytokines known to promote growth of myeloma cells (2, 3, 34–37). The cytokines were given in optimal concentrations for growth. Protein expression of Bcl-3 increased in these cell lines after 4 h of cytokine stimulation compared with baseline level, and increased even more after 24 h of stimulation (Fig. 1A). IL-6, IL-21 and TNF-α were the most potent inducers of Bcl-3. Cytokines that induced Bcl-3 also induced proliferation of the corresponding myeloma cell line (Fig. 1A). However, there were a few exceptions from this association. In ANBL-6, TNF-α increased the level of Bcl-3 without stimulating proliferation, whereas IGF-1 induced proliferation without increasing Bcl-3 expression. We also tested the effect of IL-6 on Bcl-3 expression in the IL-6 independent cell lines CAG, RPMI-8226, U-266 and JJN-3. Bcl-3 was induced in the cell lines where IL-6 gave a proliferative response. JJN-3 had constitutive expression of Bcl-3, and in this cell line IL-6 had no effect on Bcl-3 expression (Fig. 1B).
Figure 1.
Protein expression of Bcl-3 in myeloma cell lines and corresponding cell proliferation after cytokine stimulation. Bcl-3 expression in whole cell lysates and corresponding myeloma cell proliferation. The same cytokine concentrations were used for Western blot as for [3H]-thymidine incorporation assay. Average counts per minute (CPM/AVG) are plotted along the y-axis on graph. Bars represent mean + SD of triplicate wells. (A) IL-6 dependent myeloma cell lines (IH-1, OH-2, ANBL-6, INA-6) were harvested after overnight starvation (T0) or stimulated with cytokines as indicated for 4 and 24 h. (B) IL-6-independent myeloma cell lines (CAG, JJN-3, RPMI-8226, U-266) were stimulated with IL-6 or left unstimulated for 24 h. GAPDH was used as loading control.
Expression of BCL3 in CD 138+ bone marrow plasma cells from MM patients is associated with poor outcome
We studied GEP of CD138+ bone marrow plasma cells from 351 newly diagnosed MM patients, 22 healthy individuals, 44 patients with MGUS and 12 patients with SMM. Based on ‘present’ detection call, BCL3 was present in 229 (65%) of MM plasma cell samples and in 11 (46%) of normal plasma cell samples (NPC; data not shown). By further comparison of BCL3 expression between NPC and primary MMs based on absolute intensity signal, there was no significant difference in BCL3 expression (median 445 (range: 23–2555) in MM plasma cells vs. median 534 (range: 41–970) in NPC, P = 0.7). However, when the expression of BCL3 among NPC, MGUS, SMM, and MM subgroups was compared, the level of BCL3 was significantly increased in one of the gene expression-defined high-risk subgroups, namely the PR (proliferation) subgroup (Fig. 2A) (28). When looking at outcome, Kaplan–Meier analysis revealed 5-yr event-free survival estimates of 37% (EFS, P = 0.0125) and 5-yr overall survival estimates of 42% (OS, P = 0.0030) among 88 patients with BCL3 expression above 75th percentile, compared with 52% and 72% among the other 263 patients respectively (Figs 2B and 2C). Our study population of myeloma patients was uniformly treated with tandem transplants (28).
Figure 2.
Expression of BCL3 and its protein in CD138+ MM plasma cells. (A) Box plot of BCL3 expression in NPC and in plasma cells from patients with MGUS, SMM and overt MM. The expression level of BCL3 is in addition shown for subgroups of MM patients based on molecular classification. Sample groups are along the x-axis and the natural log transformed Affymetrix-derived signal is plotted on the y-axis. The top, bottom and middle lines of each box correspond to the 75th percentile (top quartile), 25th percentile (bottom quartile) and 50th percentile (median), respectively. The whiskers extend from the 10th percentile (bottom decile) and 90th percentile (top decile). Open circles denote outliers within each group. The P-value states that BCL3 was significantly increased in the proliferation subgroup (PR) compared with the other subgroups as well as to the NPC, MGUS and SMM groups. MY, all MM patients; LB, low bone disease subgroup; MS, MMSET subgroup; HY, hyperdiploid subgroup; CD-1, CCND1 subgroup: CD-2, CCND3 subgroup: MF, MAF/MAFB subgroup. (B) and (C) Kaplan–Meier 5 yr estimates of EFS and OS showed inferior EFS and OS of MM patients with BCL3 expression levels above 75th percentile(top quartile). (D) Bcl-3 was detected in bone marrow biopsies by immunohistochemistry. The bone marrow biopsy illustrated here had an overall 20% Bcl-3+ plasma cells. (E) MM patient with an unbalanced BCL3 translocation detected by cIg-FISH. The red signal is upstream and the green signal is downstream of the BCL3 gene. Colocalization of the signals represents normal genes, while a single green signal represents an unbalanced translocation.
Bcl-3 is detected at mRNA and protein level in plasma cells from MM patients
Expression of BCL3 mRNA was studied by qPCR in samples from 10 randomly selected MM patients (Figure S1). All patients expressed BCL3, with six of the patients expressing four times or more the amount compared to the patient with the lowest expression level, confirming the Affymetrix data showing that a subset of MM patients expresses high amounts of BCL3. Expression of Bcl-3 at protein level was examined by two independent methods, immunohistochemistry in bone marrow biopsies and Western blot of purified myeloma cells. Of 18 newly diagnosed MM patients and in biopsies from three plasmocytoma patients, two biopsies (one bone marrow biopsy from a MM patient and one plasmocytoma biopsy) were positive for Bcl-3, with 20% and 40% Bcl-3+plasma cells, respectively (Fig. 2D). In addition, one MM bone marrow biopsy had 5–10% of Bcl3+ plasma cells. When looking at the Bcl-3 protein expression in CD138+ plasma cells from eight myeloma patients, we detected Bcl-3 in seven of eight samples with Western blot (data not shown).
The BCL3 gene is altered in malignant plasma cells from MM patients
To reveal whether alterations in the BCL3 gene locus could explain the upregulation of Bcl-3 in malignant plasma cells from MM patients, we examined bone marrow aspirates from 19 newly diagnosed MM patients using cIg-FISH and a commercial BCL3 split probe. We found that four patients had an extra copy of the BCL3 gene (data not shown). In addition, one t (4;14)+ patient had an unbalanced translocation involving an extra copy of the BCL3 locus (Fig. 2E).
Intracellular localization of Bcl-3 is dependent on type of stimulus given to the cell
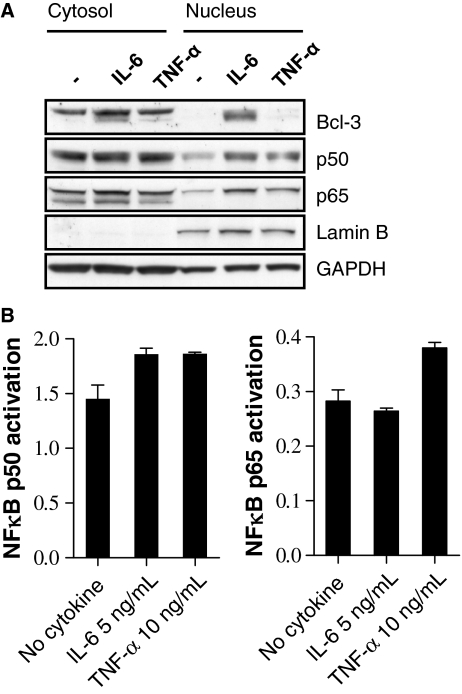
To be activated, Bcl-3 is translocated to the nucleus of the cells (26). We wanted to study if Bcl-3 is a nuclear protein in myeloma cells. We used IH-1 as a model cell line since Bcl-3 was highly inducible by cytokines in this cell line (Fig. 1A). After stimulation with IL-6, Bcl-3 was detected in the nuclear fraction of IH-1 (Fig. 3A). By contrast, Bcl-3 was detected only in the cytosolic fraction of TNF-α-stimulated cells. Both IL-6 and TNF-α have a proliferative effect on myeloma cells (Fig. 1A).
Figure 3.
Intracellular localization of Bcl-3 and activation of NFκB p50 and p65. (A) Bcl-3 was detected on Western blot in the nuclear fraction of IL-6-stimulated IH-1 cells, but was absent in the nuclear fraction of TNF-α- and unstimulated cells. The NFκB proteins p50 and p65 were present in all conditions. GAPDH was used as loading control. Presence of GAPDH in the nucleus is earlier reported (43). Lamin B was used to determine the purity of the extracts. (B) Activation of NFκB p50 in nuclear extracts of IH-1 after IL-6 stimulation, and p50 and p65 after TNF-α-stimulation. Bars represent mean + SD of duplicate wells. The y-axis denotes optical density (OD) at 450 nm. The experiments were repeated three times.
Bcl-3 has been shown to associate with homodimers of NFκB p50 or p52 in the nucleus and repress DNA-binding of p50/p65, the classical NFκB dimer activated by TNF-α (14, 21–26, 38). We therefore examined the intracellular localization of NFκB p50 and p65 in IH-1 after IL-6 and TNF-α stimulation, as well as their DNA-binding capacity under these conditions. We found that both these proteins were present in the nucleus after TNF-α and IL-6 stimulation (Fig. 3A). However, while TNF-α activated both p65 and p50, IL-6 activated p50 only (Fig. 3B). Hence, after IL-6 stimulation, p50 was activated, and this activation coincided with nuclear presence of Bcl-3. Simultaneously, there was a lack of p65 activation.
Discussion
This work shows for the first time that myeloma cells from MM patients express BCL3 and that high expression of BCL3 at the time of diagnosis is associated with reduced survival. Furthermore, in a comparison of molecularly defined subgroups, BCL3 expression was significantly increased in a high risk subgroup characterized by overexpression of cell cycle-and proliferation-related genes (28). Our data are consistent with the concept that high BCL3 expression is an event associated with poor outcome.
We have not shown that BCL3 is an independent adverse prognostic factor and the exact relevance of elevated expression of BCL3 in myeloma high-risk disease is currently unclear. Interestingly, Annunziata et al. has shown that the high risk subgroup with high expression of BCL3 has a very low average expression of NFκB signature genes (39). NFκB signature genes are genes that are activated by the classical and/or alternative NFκB pathway (39). It is therefore tempting to speculate that the nuclear presence of Bcl-3 as after IL-6 stimulation activates a different set of genes than activation of the classical and/or alternative NFκB pathway in myeloma cells.
The array data was confirmed with qPCR in a material with 10 patients. In another randomly selected material with eight patients with MM, Bcl-3 protein was detected in seven of eight samples by Western blot. We also examined the expression of Bcl-3 at protein level by immunohistochemical staining of biopsies from MM patients and patients with plasmacytoma, and found that 11% (two of 18) of the biopsies stained positive for Bcl-3. This proportion of Bcl-3+ plasma cell malignancies is similar to the proportion of B-cell lymphomas expressing Bcl-3 (6%) (9). The discrepancy between the proportion of patient samples positive for BCL3 at mRNA level and biopsies positive for Bcl-3 at protein level may be caused by lower sensitivity of immunostaining of biopsies, a well-known problem when using immunohistochemistry as detection method. However, the number of patient samples tested was small, and we cannot rule out that there is reduced translation or post-translational modifications.
Our data showed that stimulation of myeloma cells with various cytokines well-known to promote proliferation increased Bcl-3 expression. These data indicate that the gene encoding Bcl-3 is indeed a common target gene for growth promoting cytokines in myeloma cells. As growth-promoting cytokines are frequently present in the bone marrow of MM patients, cytokine signaling is a highly possible mechanism for upregulation of Bcl-3 in myeloma cells in vivo. Importantly, and in concordance with the patient data, we found that the induction of Bcl-3 by cytokines was associated with increased proliferation of myeloma cells. However, in line with the results from Brocke-Heidrich et al. (5), we could not show any effect on thymidine incorporation or apoptosis when down regulating Bcl-3 with siRNA in the INA-6 cell line (data not shown). These results are from only one cell line and further studies are needed to clarify the exact role of Bcl-3.
Additional potential mechanisms for upregulation of Bcl-3 in MM patients are gains/amplification of the BCL3 gene, as described in patients with anaplastic large cell lymphoma, or translocations involving chromosome 19, as found in patients with CLL (40, 41). Our studies substantiate that both these mechanism may be active in freshly isolated MM patients cells. The BCL3 gene is located at chromosome 19, a chromosome with frequent trisomy in MM patients having a hyperdiploid tumor (42).
In our study, intracellular localization of Bcl-3 was dependent of type of stimulus given to the cell. Nuclear accumulation of Bcl-3, as after IL-6 stimulation, has been shown to be associated with p50/Bcl-3- or p52/Bcl-3 dependent gene activation and subsequent proliferation in CYLD-lacking keratinocytes (26). It is possible that similar mechanisms are active in myeloma cells, and may explain why we observed activation of p50 in IH-1 after IL-6 stimulation.
The starting point of this study was our observation that IL-6, IL-21 and TNF-α induced expression of BCL3 in the myeloma cell lines OH-2 and IH-1. Our study demonstrates that also the protein, Bcl-3, is produced in cell lines by cytokine stimulation and is associated with increased proliferation of the cells. We show for the first time that Bcl-3 is present in a subset of MM patients, and that the high gene expression at the time of diagnosis is associated with inferior prognosis as demonstrated in a large cohort of newly diagnosed patients. Our results indicate a potential role for Bcl-3 in the development of multiple myeloma, and further studies are needed to clarify this.
Acknowledgments
This work was supported by grants from the Norwegian Cancer Society (A.T.B., U.M.F., T.B.R.), the Cancer Fund of St. Olavs University Hospital (A.T.B., A.S. M-B., A.W.), Odd Fellow`s Research Fund (A.T.B.), NIH grant CA55819 (B.B., J.D.S., F.Z.,) and CA97513 (J.D.S.), and the Lebow Fund to Cure Myeloma. We thank Toril Holien and Randi Røsbak for technical assistance with Western blot; Borgny Ytterhus, Department of Laboratory Medicine, Children`s and Women `s Health, Norwegian University of Science and Technology, for technical assistance with immunohistochemistry; and Harald Aarset, Department of Pathology and Molecular Genetic, St. Olavs University Hospital, Trondheim, Norway, for assistance with interpretation of immunohistochemistry.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Figure S1. BCL3 mRNA expression in samples from 10 MM patients.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- 1.Mitsiades CS, Mitsiades NS, Munshi NC, Richardson PG, Anderson KC. The role of the bone microenvironment in the pathophysiology and therapeutic management of multiple myeloma: interplay of growth factors, their receptors and stromal interactions. Eur J Cancer. 2006;42:1564–73. doi: 10.1016/j.ejca.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 2.Borset M, Waage A, Brekke OL, Helseth E. TNF and IL-6 are potent growth factors for OH-2, a novel human myeloma cell line. Eur J Haematol. 1994;53:31–7. doi: 10.1111/j.1600-0609.1994.tb00176.x. [DOI] [PubMed] [Google Scholar]
- 3.Brenne AT, Baade RT, Waage A, Sundan A, Borset M, Hjorth-Hansen H. Interleukin-21 is a growth and survival factor for human myeloma cells. Blood. 2002;99:3756–62. doi: 10.1182/blood.v99.10.3756. [DOI] [PubMed] [Google Scholar]
- 4.Brocke-Heidrich K, Kretzschmar AK, Pfeifer G, Henze C, Loffler D, Koczan D, Thiesen HJ, Burger R, Gramatzki M, Horn F. Interleukin-6-dependent gene expression profiles in multiple myeloma INA-6 cells reveal a Bcl-2 family-independent survival pathway closely associated with Stat3 activation. Blood. 2004;103:242–51. doi: 10.1182/blood-2003-04-1048. [DOI] [PubMed] [Google Scholar]
- 5.Brocke-Heidrich K, Ge B, Cvijic H, Pfeifer G, Loffler D, Henze C, McKeithan TW, Horn F. BCL3 is induced by IL-6 via Stat3 binding to intronic enhancer HS4 and represses its own transcription. Oncogene. 2006;25:7297–304. doi: 10.1038/sj.onc.1209711. [DOI] [PubMed] [Google Scholar]
- 6.Tsuyama N, Danjoh I, Otsuyama K, Obata M, Tahara H, Ohta T, Ishikawa H. IL-6-induced Bcl6 variant 2 supports IL-6-dependent myeloma cell proliferation and survival through STAT3. Biochem Biophys Res Commun. 2005;337:201–8. doi: 10.1016/j.bbrc.2005.09.036. [DOI] [PubMed] [Google Scholar]
- 7.McKeithan TW, Ohno H, Diaz MO. Identification of a transcriptional unit adjacent to the breakpoint in the 14; 19 translocation of chronic lymphocytic leukemia. Genes Chromosomes Cancer. 1990;1:247–55. doi: 10.1002/gcc.2870010310. [DOI] [PubMed] [Google Scholar]
- 8.Ohno H, Takimoto G, McKeithan TW. The candidate proto-oncogene bcl-3 is related to genes implicated in cell lineage determination and cell cycle control. Cell. 1990;23:60. doi: 10.1016/0092-8674(90)90347-h. [DOI] [PubMed] [Google Scholar]
- 9.Canoz O, Rassidakis GZ, Admirand JH, Medeiros LJ. Immunohistochemical detection of BCL-3 in lymphoid neoplasms: a survey of 353 cases. Mod Pathol. 2004;17:911–7. doi: 10.1038/modpathol.3800140. [DOI] [PubMed] [Google Scholar]
- 10.Cogswell PC, Guttridge DC, Funkhouser WK, Baldwin AS., Jr Selective activation of NF-kappa B subunits in human breast cancer: potential roles for NF-kappa B2/p52 and for Bcl-3. Oncogene. 2000;19:1123–31. doi: 10.1038/sj.onc.1203412. [DOI] [PubMed] [Google Scholar]
- 11.Mathas S, Johrens K, Joos S, et al. Elevated NF-kappaB p50 complex formation and Bcl-3 expression in classical Hodgkin, anaplastic large-cell, and other peripheral T-cell lymphomas. Blood. 2005;106:4287–93. doi: 10.1182/blood-2004-09-3620. [DOI] [PubMed] [Google Scholar]
- 12.Thornburg NJ, Pathmanathan R, Raab-Traub N. Activation of nuclear factor-kappaB p50 homodimer/Bcl-3 complexes in nasopharyngeal carcinoma. Cancer Res. 2003;63:8293–301. [PubMed] [Google Scholar]
- 13.Viatour P, Dejardin E, Warnier M, et al. GSK3-mediated BCL-3 phosphorylation modulates its degradation and its oncogenicity. Mol Cell. 2004;16:35–45. doi: 10.1016/j.molcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 14.Carmody RJ, Ruan Q, Palmer S, Hilliard B, Chen YH. Negative regulation of toll-like receptor signaling by NF-kappaB p50 ubiquitination blockade. Science. 2007;317:675–8. doi: 10.1126/science.1142953. [DOI] [PubMed] [Google Scholar]
- 15.Franzoso G, Carlson L, Scharton-Kersten T, et al. Critical roles for the Bcl-3 oncoprotein in T cell-mediated immunity, splenic microarchitecture, and germinal center reactions. Immunity. 1997;6:479–90. doi: 10.1016/s1074-7613(00)80291-5. [DOI] [PubMed] [Google Scholar]
- 16.Schwarz EM, Krimpenfort P, Berns A, Verma IM. Immunological defects in mice with a targeted disruption in Bcl-3. Genes Dev. 1997;11:187–97. doi: 10.1101/gad.11.2.187. [DOI] [PubMed] [Google Scholar]
- 17.Kerr LD, Duckett CS, Wamsley P, Zhang Q, Chiao P, Nabel G, McKeithan TW, Baeuerle PA, Verma IM. The proto-oncogene bcl-3 encodes an I kappa B protein. Genes Dev. 1992;6:2352–63. doi: 10.1101/gad.6.12a.2352. [DOI] [PubMed] [Google Scholar]
- 18.Wulczyn FG, Naumann M, Scheidereit C. Candidate proto-oncogene bcl-3 encodes a subunit-specific inhibitor of transcription factor NF-kappa B. Nature. 1992;358:597–9. doi: 10.1038/358597a0. [DOI] [PubMed] [Google Scholar]
- 19.Karin M, Cao Y, Greten FR, Li ZW. NF-kappaB in cancer: from innocent bystander to major culprit. Nat Rev Cancer. 2002;2:301–10. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- 20.Mitsiades N, Mitsiades CS, Poulaki V, Chauhan D, Richardson PG, Hideshima T, Munshi N, Treon SP, Anderson KC. Biologic sequelae of nuclear factor-kappaB blockade in multiple myeloma: therapeutic applications. Blood. 2002;99:4079–86. doi: 10.1182/blood.v99.11.4079. [DOI] [PubMed] [Google Scholar]
- 21.Bours V, Franzoso G, Azarenko V, Park S, Kanno T, Brown K, Siebenlist U. The oncoprotein Bcl-3 directly transactivates through kappa B motifs via association with DNA-binding p50B homodimers. Cell. 1993;72:729–39. doi: 10.1016/0092-8674(93)90401-b. [DOI] [PubMed] [Google Scholar]
- 22.Caamano JH, Perez P, Lira SA, Bravo R. Constitutive expression of Bc1-3 in thymocytes increases the DNA binding of NF-kappaB1 (p50) homodimers in vivo. Mol Cell Biol. 1996;16:1342–8. doi: 10.1128/mcb.16.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Franzoso G, Bours V, Azarenko V, Park S, Tomita-Yamaguchi M, Kanno T, Brown K, Siebenlist U. The oncoprotein Bcl-3 can facilitate NF-kappa B-mediated transactivation by removing inhibiting p50 homodimers from select kappa B sites. EMBO J. 1993;12:3893–901. doi: 10.1002/j.1460-2075.1993.tb06067.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujita T, Nolan GP, Liou HC, Scott ML, Baltimore D. The candidate proto-oncogene bcl-3 encodes a transcriptional coactivator that activates through NF-kappa B p50 homodimers. Genes Dev. 1993;7:1354–63. doi: 10.1101/gad.7.7b.1354. [DOI] [PubMed] [Google Scholar]
- 25.Grundstrom S, Anderson P, Scheipers P, Sundstedt A. Bcl-3 and NFkappaB p50-p50 homodimers act as transcriptional repressors in tolerant CD4 + T cells. J Biol Chem. 2004;279:8460–8. doi: 10.1074/jbc.M312398200. [DOI] [PubMed] [Google Scholar]
- 26.Massoumi R, Chmielarska K, Hennecke K, Pfeifer A, Fassler R. Cyld inhibits tumor cell proliferation by blocking Bcl-3-dependent NF-kappaB signaling. Cell. 2006;125:665–77. doi: 10.1016/j.cell.2006.03.041. [DOI] [PubMed] [Google Scholar]
- 27.Ro TB, Holt RU, Brenne AT, Hjorth-Hansen H, Waage A, Hjertner O, Sundan A, Borset M. Bone morphogenetic protein-5, -6 and -7 inhibit growth and induce apoptosis in human myeloma cells. Oncogene. 2004;23:3024–32. doi: 10.1038/sj.onc.1207386. [DOI] [PubMed] [Google Scholar]
- 28.Zhan F, Huang Y, Colla S, et al. The molecular classification of multiple myeloma. Blood. 2006;108:2020–8. doi: 10.1182/blood-2005-11-013458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhan F, Barlogie B, Arzoumanian V, et al. Gene-expression signature of benign monoclonal gammopathy evident in multiple myeloma is linked to good prognosis. Blood. 2007;109:1692–700. doi: 10.1182/blood-2006-07-037077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borset M, Lien E, Espevik T, Helseth E, Waage A, Sundan A. Concomitant expression of hepatocyte growth factor/scatter factor and the receptor c-MET in human myeloma cell lines. J Biol Chem. 1996;271:24655–61. doi: 10.1074/jbc.271.40.24655. [DOI] [PubMed] [Google Scholar]
- 31.Zhan F, Hardin J, Kordsmeier B, et al. Global gene expression profiling of multiple myeloma, monoclonal gammopathy of undetermined significance, and normal bone marrow plasma cells. Blood. 2002;99:1745–57. doi: 10.1182/blood.v99.5.1745. [DOI] [PubMed] [Google Scholar]
- 32.Fagerli UM, Holt RU, Holien T, et al. Overexpression and involvement in migration by the metastasis-associated phosphatase PRL-3 in human myeloma cells. Blood. 2008;111:806–15. doi: 10.1182/blood-2007-07-101139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Renard P, Ernest I, Houbion A, Art M, Le CH, Raes M, Remacle J. Development of a sensitive multi-well colorimetric assay for active NFkappaB. Nucleic Acids Res. 2001;29:E21. doi: 10.1093/nar/29.4.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Derksen PW, de Gorter DJ, Meijer HP, Bende RJ, van DM, Lokhorst HM, Bloem AC, Spaargaren M, Pals ST. The hepatocyte growth factor/Met pathway controls proliferation and apoptosis in multiple myeloma. Leukemia. 2003;17:764–74. doi: 10.1038/sj.leu.2402875. [DOI] [PubMed] [Google Scholar]
- 35.Hjorth-Hansen H, Waage A, Borset M. Interleukin-15 blocks apoptosis and induces proliferation of the human myeloma cell line OH-2 and freshly isolated myeloma cells. Br J Haematol. 1999;106:28–34. doi: 10.1046/j.1365-2141.1999.01510.x. [DOI] [PubMed] [Google Scholar]
- 36.Jelinek DF, Witzig TE, Arendt BK. A role for insulin-like growth factor in the regulation of IL-6-responsive human myeloma cell line growth. J Immunol. 1997;159:487–96. [PubMed] [Google Scholar]
- 37.Lu ZY, Zhang XG, Rodriguez C, Wijdenes J, Gu ZJ, Morel-Fournier B, Harousseau JL, Bataille R, Rossi JF, Klein B. Interleukin-10 is a proliferation factor but not a differentiation factor for human myeloma cells. Blood. 1995;85:2521–7. [PubMed] [Google Scholar]
- 38.Borset M, Medvedev AE, Sundan A, Espevik T. The role of the two TNF receptors in proliferation, NF-kappa B activation and discrimination between TNF and LT alpha signalling in the human myeloma cell line OH-2. Cytokine. 1996;8:430–8. doi: 10.1006/cyto.1996.0059. [DOI] [PubMed] [Google Scholar]
- 39.Annunziata CM, Davis RE, Demchenko Y, et al. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 2007;12:115–30. doi: 10.1016/j.ccr.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McKeithan TW, Takimoto GS, Ohno H, Bjorling VS, Morgan R, Hecht BK, Dube I, Sandberg AA, Rowley JD. BCL3 rearrangements and t(14;19) in chronic lymphocytic leukemia and other B-cell malignancies: a molecular and cytogenetic study. Genes Chromosomes Cancer. 1997;20:64–72. [PubMed] [Google Scholar]
- 41.Nishikori M, Maesako Y, Ueda C, Kurata M, Uchiyama T, Ohno H. High-level expression of BCL3 differentiates t(2;5)(p23;q35)-positive anaplastic large cell lymphoma from Hodgkin disease. Blood. 2003;101:2789–96. doi: 10.1182/blood-2002-08-2464. [DOI] [PubMed] [Google Scholar]
- 42.Chng WJ, Ketterling RP, Fonseca R. Analysis of genetic abnormalities provides insights into genetic evolution of hyperdiploid myeloma. Genes Chromosomes Cancer. 2006;45:1111–20. doi: 10.1002/gcc.20375. [DOI] [PubMed] [Google Scholar]
- 43.Mazzola JL, Sirover MA. Subcellular localization of human glyceraldehyde-3-phosphate dehydrogenase is independent of its glycolytic function. Biochim Biophys Acta. 2003;1622:50–6. doi: 10.1016/s0304-4165(03)00117-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.